Abstract
l-Arginine and l-arginine-metabolizing enzymes play important roles in the biology of some types of myeloid cells, including macrophage and myeloid-derived suppressor cells. In this study, we found evidence that arginase 1 (Arg1) is required for the differentiation of mouse dendritic cells (DCs). Expression of Arg1 was robustly induced during monocyte-derived DC differentiation. Ectopic expression of Arg1 significantly promoted monocytic DC differentiation in a granulocyte-macrophage colony-stimulating factor culture system and also facilitated the differentiation of CD8α+ conventional DCs in the presence of Flt3 ligand. Knockdown of Arg1 reversed these effects. Mechanistic studies showed that the induced expression of Arg1 in differentiating DCs was caused by enhanced recruitment of histone deacetylase 4 (HDAC4) to the Arg1 promoter region, which led to a reduction in the acetylation of both the histone 3 and STAT6 proteins and subsequent transcriptional activation of Arg1. Further investigation identified a novel STAT6 binding site within the Arg1 promoter that mediated its regulation by STAT6 and HDAC4. These observations suggest that the cross talk between HDAC4 and STAT6 is an important regulatory mechanism of Arg1 transcription in DCs. Moreover, overexpression of Arg1 clearly abrogated the ability of HDAC inhibitors to suppress DC differentiation. In conclusion, we show that Arg1 is a novel regulator of myeloid DC differentiation.
INTRODUCTION
Dendritic cells (DCs) are specialized antigen-presenting cells that capture, process, and present antigens to T cells and thereby play important roles in both innate and adaptive immunity (1, 2). Differentiation of DCs from hematopoietic progenitor cells (HPCs) is regulated by complex signaling pathways that involve soluble cytokines and transcription factors (TFs) in bone marrow (BM) and peripheral lymphoid tissues (3, 4). Impaired DC differentiation facilitates the escape of tumor cells and invading pathogens from the host's immune surveillance (5–7). Although many molecular mechanisms linked to DC differentiation have been proposed, a complete picture has yet to be obtained (8–10).
l-Arginine is a conditionally essential amino acid in adult mammals because it is required under some special circumstances such as trauma, pregnancy, and infections. l-Arginine is metabolized by arginase (Arg) to produce urea and l-ornithine and by nitric oxide synthase to produce nitric oxide and l-citrulline. There are two known Arg isoforms, Arg1 and Arg2. Arg1 is constitutively expressed in the liver, where it participates in the urea cycle, while Arg2 is located in mitochondria in various cell types (11). Recently, it was reported that Arg1 expression is induced in myeloid cells exposed to Th2 cytokines such as interleukin-4 (IL-4) and IL-13. Induction of Arg1 or Nos2 has been used to distinguish macrophages activated through either the classical or the alternative pathway (12). Upregulation of Arg1 and Nos2 is closely associated with the suppressive activity of myeloid-derived suppressor cells (MDSCs), which causes l-arginine depletion and suppression of host T cell responses to tumor cells (13). These reports support the potential biological significance of Arg1 and l-arginine metabolism in myeloid cells. Their roles in DC biology, however, remain poorly understood.
Histone (de)acetylation is one of the major epigenetic mechanisms in eukaryotic cells, as it regulates gene transcription by condensing or relaxing chromatin structures, thus facilitating the recruitment of coactivators or corepressors to the transcriptional machinery (14–17). Histone acetylation-mediated gene transcription plays crucial roles in the function and differentiation of immune cells (18–21). Histone deacetylase (HDAC) inhibitors (HDACi), which are potential anticancer agents, have various effects on the immune system, including the suppression of DC differentiation and functions (22–25). HDACi treatment also causes defects in myeloid cell differentiation that lead to the accumulation of myeloid-derived suppressor cells (26). However, the mechanisms underlying HDACi-mediated DC suppression remain elusive. Elucidation of the relevant processes will benefit the understanding of DC differentiation, as well as shed light on the immunomodulatory role of HDACi in cancer therapy.
In our pilot study, the mechanism of HDACi-mediated inhibition of DC differentiation was studied by analyzing the mRNA profile of mouse HPCs cultured with granulocyte-macrophage colony-stimulating factor (GM-CSF), following treatment with the HDACi trichostatin A (TSA). We found that Arg1 expression was significantly downregulated by TSA, indicating a potential role for this enzyme in HDACi-mediated DC suppression. In this study, we demonstrate that Arg1 is a novel regulator of DC differentiation and that HDAC4 and STAT6 cooperate in the transcriptional regulation of Arg1 in differentiating DCs. Finally, we report that Arg1 mediates the suppressive effect of HDACi on DCs.
MATERIALS AND METHODS
Ethics statement.
This study was approved by the Ethics Review Board of Sun Yat-Sen University. Written informed consent was provided by the enrolled healthy donors. All mouse experiments were approved by the Sun Yat-Sen University Institutional Animal Care and Use Committee. All mice were maintained under specific-pathogen-free conditions and used at 6 to 8 weeks of age.
Mice and cell lines.
OT-II transgenic mice were kindly provided by Hui Zhang (Sun Yat-Sen University). All of the cell lines used, including HEK 293T, 32D, and WEHI-3B, were purchased from the American Type Culture Collection (ATCC) and cultured under the conditions recommended by the ATCC.
Reagents.
For the reagents used in this study, see the supplemental material.
Generation of mouse DCs in vitro.
For monocytic DCs, BM cells were collected from mouse tibiae and femurs and cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 50 μM 2-mercaptoethanol, 20 ng/ml GM-CSF, and 10 ng/ml IL-4 for 6 days. For some experiments, mouse HPCs were enriched from mouse BM with a Lineage depletion kit (Miltenyi Biotec, Teterow, Germany) and cultured under the same conditions described for BM cells. For steady-state DC differentiation, mouse BM cells were cultured in medium containing 100 ng/ml Flt3 ligand (Flt3L) for 9 days; the medium was refreshed every 2 to 3 days.
Flow cytometric analyses and cell sorting.
The procedures used for flow cytometric analyses and cell sorting were described previously (27, 28). Single-cell suspensions were prepared from cultured cells or mouse tissues and stained with fluorescein-conjugated antibodies. The proportions of the corresponding cell populations were evaluated with a FACSCalibur flow cytometer (Beckman Coulter, Fullerton, CA). Data were acquired as the fraction of labeled cells within a live-cell gate set for 50,000 events and analyzed by the CellQuest program (Becton Dickinson, Mountain View, CA). For flow cytometric sorting, the stained cells were isolated with a FACSAria cell sorter (BD LSR II; BD Biosciences, San Jose, CA).
qRT-PCR.
The experimental procedures used for quantitative reverse transcription (qRT)-PCR were described earlier (27); the sequences of the primers used are listed in Table S1 in the supplemental material.
l-Arginine measurement.
Amounts of l-arginine were determined by high-performance liquid chromatography (HPLC) as described previously (29). Briefly, l-arginine was extracted from culture supernatants by solid-phase extraction (SPE), derivatized with ortho-phthaldialdehyde (OPA), and separated by isocratic reverse-phase chromatography. The derivatized samples were injected into the column with an autosampler, and the flow rate was set at 1.0 ml/min. The excitation and emission wavelengths of the fluorescence detector were set at 420 and 483 nm, respectively.
Lentivirus transduction.
Lentiviral stock preparation and viral transduction were performed as described previously (30, 31). HEK 293T cells were transfected with lentiviral vectors and packaging plasmids (pCMV-ΔR8.2, pMD.G) by using Lipofectamine 2000. The culture supernatants were collected, concentrated, and stored at −80°C. BM cells or enriched HPCs were infected with a 30% volume of concentrated lentiviral stock solution with 8 μg/ml Polybrene. The medium was replaced with fresh medium at 3 h postinfection.
Adoptive cell transfer.
The procedure used for adoptive cell transfer was described earlier (32). Briefly, C57BL/6 mice were irradiated with 9.5 Gy administered (three times) with a 137Cs irridiator (MDS Nordion, Ottawa, Ontario, Canada) and used as recipients. Enriched HPCs were transduced with different lentiviral plasmids (with a green fluorescent protein [GFP] tag) and cultured with a cytokine cocktail to allow cell proliferation. Cells were then injected intravenously (i.v.) into recipient mice, and the proportions of myeloid cells among the GFP+ cells in the spleen were analyzed at 8 weeks posttransfer.
Antigen-specific T cell proliferation assay.
Antigen-specific T cell proliferation was measured as described previously (33). DCs from cultured BM cells were treated with OVA323-339 peptide at 37°C for 3 h and then stimulated with lipopolysaccharide (100 ng/ml) for 2 h. CD4+ T cells from OT-II transgenic mice were stained with 2 μM carboxyfluorescein succinimidyl ester (CFSE) and cocultured with DCs at a 1:10 DC/T cell ratio for 72 h. The proliferation of T cells were analyzed by flow cytometry.
Coimmunoprecipitation and Western blotting.
For the coimmunoprecipitation and Western blotting procedures used in this study, see the supplemental material.
Arg activity assay.
Arg activity was measured as described previously (27).
Plasmid constructs and transfection assays.
For the plasmid constructs and transfection assays used in this study, see the supplemental material.
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed by following the instructions from Millipore (Billerica, MA). In brief, cultured BM cells were fixed with a 1% formaldehyde solution, lysed, and sheared by sonication. Cell lysates were precleared with protein G-agarose and immunoprecipitated with specific antibodies or an anti-IgG control. Antibody-chromatin complexes were collected with protein G-agarose. The DNA in the complex was recovered and quantitated by quantitative PCR (qPCR). Ten percent of the lysate before immunoprecipitation was used as the input control. Amplification of cyclophilin from the input was used as a loading control. For sequential ChIP assays, samples were initially immunoprecipitated with anti-STAT6 antibody; the complex was then reimmunoprecipitated with anti-HDAC4 antibody. DNA from both rounds of ChIP assays was purified and subjected to qPCR.
Methylated BSA-induced peritonitis.
For the methylated bovine serum albumin (BSA)-induced peritonitis procedure used in this study, see the supplemental material.
Statistical analyses.
Statistical analyses of differences between groups were done with unpaired t tests. The Prism version 5.0a (GraphPad, San Diego, CA) and SPSS Statistics 17.0 software packages were used. A P value of <0.05 was considered significant.
RESULTS
Induction of Arg1 expression during DC differentiation.
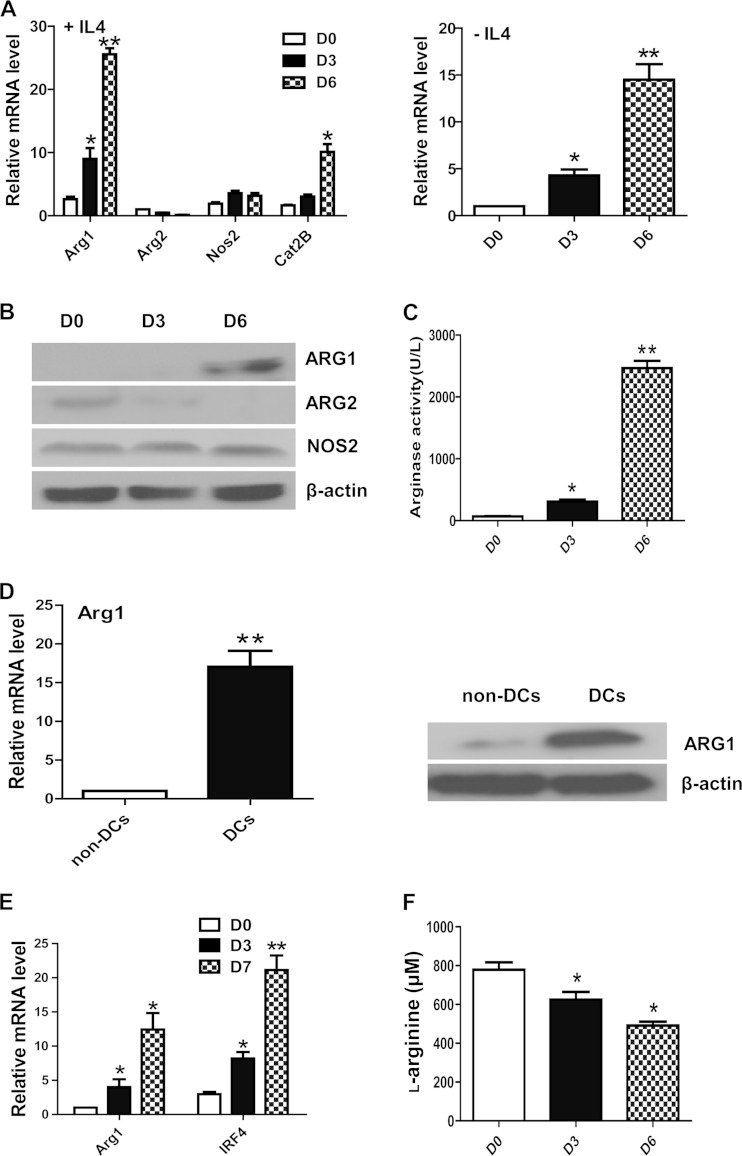
To determine the role of Arg1 in the regulation of DC differentiation, mouse HPCs were enriched from BM and cultured in medium containing GM-CSF with or without the cytokine IL-4 to induce monocytic DC differentiation. Cells were harvested on days 0, 3, and 6, and the expression of Arg1 was analyzed by qRT-PCR. DC differentiation was coupled with a drastic induction of Arg1 expression in the presence or absence of IL-4 (Fig. 1A). In contrast to the induction of Arg1, the expression of Arg2 was moderately downregulated during DC differentiation, while no noticeable change in the expression of the Nos2-encoding gene, another gene involved in l-arginine metabolism, was observed. Interestingly, the expression of the cationic amino acid transporter 2B (Cat2B), which is responsible for cellular uptake of l-arginine, was also significantly induced (Fig. 1A). The changes in the expression of the mRNAs for the respective proteins were further confirmed by Western blotting (Fig. 1B). Consistent with the induction of Arg1, the activity of Arg also strongly increased during the course of DC differentiation (Fig. 1C), indicating that Arg1 may be the dominant Arg isoform in this process. CD11c+ major histocompatibility complex class II-positive (MHCII+) DCs purified by flow cytometric sorting from BM cells cultured for 6 days displayed significantly higher levels of Arg1 expression and Arg activity than CD11c− MHCII− control cells from the same culture (Fig. 1D; see Fig. S1 in the supplemental material). The induction of Arg1 expression was also observed in the course of human monocyte-derived DC differentiation (Fig. 1E). These observations suggest that Arg1 is a potential regulator of DC differentiation.
FIG 1.
Induction of Arg1 during DC differentiation. (A to C) Enriched HPCs from mouse BM were cultured in medium containing GM-CSF and/or IL-4 to induce monocytic DC differentiation; cells were harvested at 0, 3, and 6 days; the expression of l-arginine-metabolizing enzyme-encoding genes was measured by qRT-PCR (A) and Western blotting (B); and Arg activity was determined in cell lysates (C). (D) DCs (CD11c+ MHCII+) and non-DCs (CD11c− MHCII−) were purified by flow cytometric sorting from cells cultured for 6 days, and Arg1 expression was evaluated by qRT-PCR (left) and Western blotting (right). (E) Arg1 expression in the course of human monocyte differentiation into DCs was determined by qRT-PCR. IRF4 was used as a positive control. (F) The l-arginine contents of supernatants from cultured BM cells were measured by HPLC. (A, C to F) The data represent the mean results ± the standard deviations from three independent experiments. The Western blot assays in panels B and D are representative of three independent experiments. *, P < 0.05; **, P < 0.01 (compared with the corresponding controls).
Considering the crucial role of Arg1 in l-arginine metabolism, we further examined whether l-arginine metabolism was changed during the course of DC differentiation. The contents of l-arginine in the supernatants from cultured BM cells were measured by HPLC. The results showed that the l-arginine concentration was significantly lower on days 6 and 3 than that on day 0 (Fig. 1F), indicating enhanced uptake and/or metabolism of l-arginine by differentiating DCs. These results support a potential role for Arg1 and l-arginine metabolism in DC differentiation.
Modulation of Arg1 expression affects monocyte-derived DC differentiation.
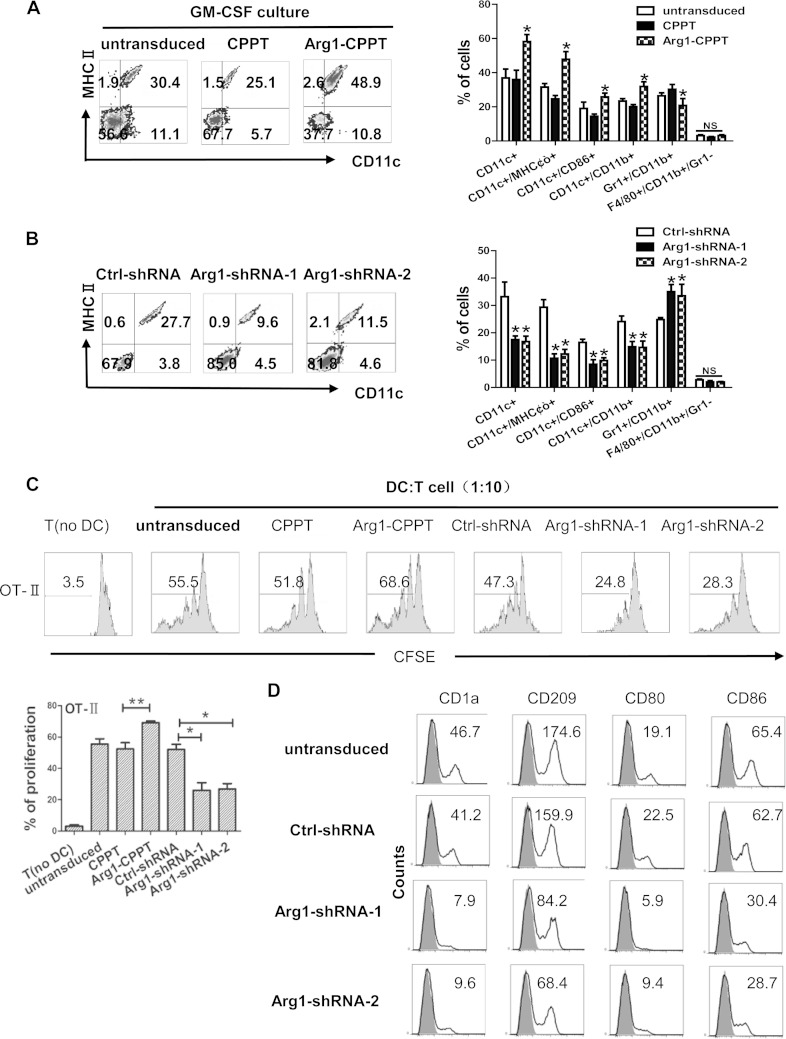
To study the role of Arg1 in the differentiation of monocyte-derived DCs, mouse HPCs were infected with lentiviral plasmids expressing Arg1 cDNA or the empty vector cPPT (human immunodeficiency virus type 1 central polypurine tract with a GFP tag) and cultured under conditions that induce DC differentiation. The lentiviral infection efficiency was approximately 20% in mouse HPCs (data not shown), and Arg1 overexpression was confirmed by Western blotting (see Fig. S2 in the supplemental material). The percentages of distinct cell populations among the GFP+ cells were evaluated by flow cytometric analysis. This revealed that overexpression of Arg1 significantly increased the proportion of CD11c+ MHCII+ and CD11c+ CD86+ DCs, while it caused a reduction in the percentage of immature myeloid cells (IMCs; Gr1+ CD11b+); no effect on macrophage (F4/80+ CD11b+ Gr1−) differentiation was observed (Fig. 2A). Consistently, knockdown of Arg1 by two independent short hairpin RNAs (shRNAs) suppressed DC differentiation and facilitated the accumulation of IMCs (Fig. 2B). The total cell numbers remained largely unchanged among the different groups (see Fig. S3). As further support, administration of l-arginine enhanced DC differentiation, while the Arg inhibitor Nω-hydroxy-l-arginine (NOHA) clearly suppressed DC differentiation (see Fig. S4). These observations suggest that Arg1 is essential for GM-CSF-derived DC differentiation. We further studied whether Arg1-modulated DCs displayed any difference in the stimulation of T cells responses, which is a hallmark of DC activity. Indeed, DCs with Arg1 overexpression were more potent stimulators of T cell proliferation, while DCs with Arg1 knockdown showed the reverse changes, with respect to both antigen-specific (Fig. 2C) and non-antigen-specific (see Fig. S5) responses.
FIG 2.
Modulation of Arg1 expression affects monocyte-derived DC differentiation. (A and B) Enriched mouse HPCs were infected with lentiviral plasmids expressing Arg1 cDNA (A), two independent Arg1 shRNAs (B), or the empty vectors (with a GFP tag). Untransduced HPCs were used as controls. Cells were cultured for 6 days, and the proportions of the indicated subpopulations among the GFP+ cells were evaluated by flow cytometric analysis. The left half of each panel is from a single experiment, and the right half of each panel represents the mean results ± the standard deviations from three independent experiments. (C) Antigen-specific T cell proliferation. DCs from panels A and B were stimulated with OVA peptide and then cocultured with T cells isolated from OT-II transgenic mice for 3 days at a 1:10 ratio. T cell proliferation was measured by CFSE dilution. T cell culture alone was used as a negative control. The data in the top part of the panel are from a single experiment, and the graph at the bottom represents the mean results ± the standard deviations from three independent experiments. (D) Human CD14+ monocytes were transduced with Arg1 shRNAs or the empty vector and cultured for 7 days to induce DC differentiation. The expression of DC surface markers was evaluated by flow cytometric analysis of the GFP+ cells. The data are representative of three independent experiments. *, P < 0.05; **, P < 0.01 (compared with the corresponding controls).
Consistent with the observed Arg1 induction during the differentiation of human monocytes into DCs, inhibition of Arg1 by shRNAs significantly downregulated the expression of surface markers related to human DCs (Fig. 2D), suggesting an impairment of DC generation.
Modulation of Arg1 expression affects steady-state DC differentiation.
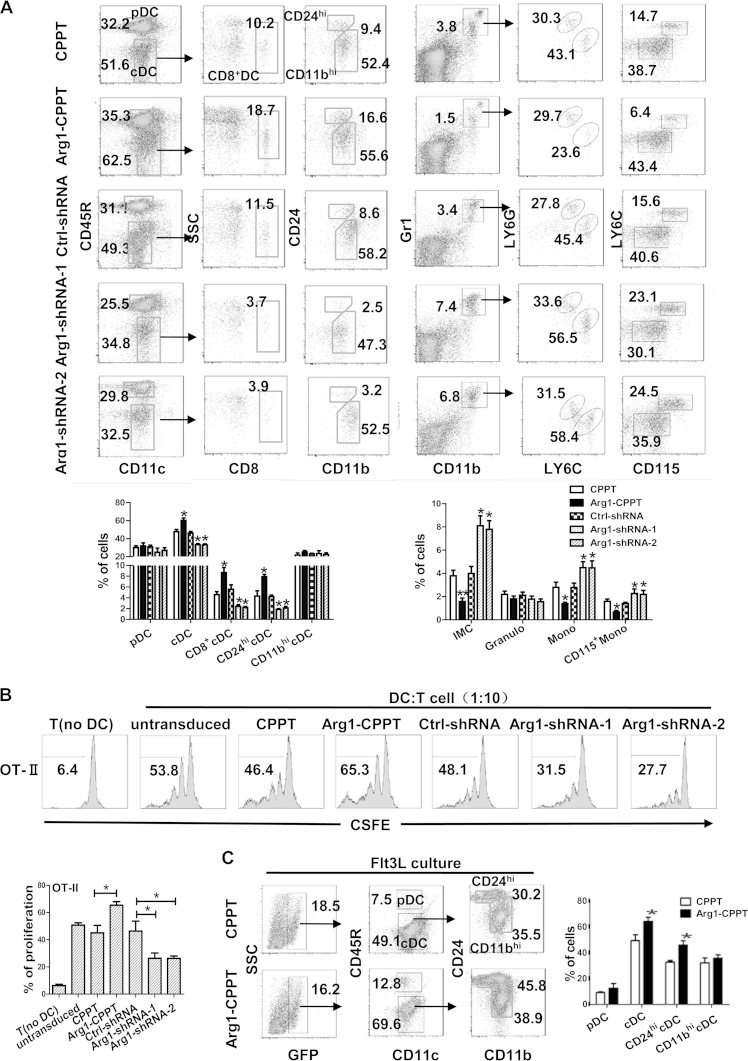
We next investigated whether Arg1 affects the differentiation of plasmacytoid DCs (pDCs) and conventional DCs (cDCs) under steady-state conditions. We first performed adoptive-transfer experiments; for these, enriched HPCs were transduced with lentiviruses (with a GFP tag) that expressed Arg1 cDNA or Arg1 shRNA, as well as the corresponding empty vectors, and then cultured in a cytokine cocktail to allow cell proliferation but not differentiation (34). The cultured HPCs were then transferred into lethally irradiated recipient mice by i.v. injection. The levels of splenic DC subsets and other types of myeloid cells among the donor cells (GFP+) were determined by flow cytometric analysis at 8 weeks posttransfer. This showed that ectopic expression of Arg1 significantly increased the proportions of splenic CD8α+ cDCs (CD11c+ CD45R− CD8α+) and CD24hi cDCs (CD11c+ CD45R− CD24hi), while the levels of splenic CD11bhi cDCs (CD11c+ CD45R− CD11bhi) and pDCs (CD11c+ CD45R+) remained largely unaffected; knockdown of Arg1 by shRNA caused the reverse phenotypes in DC subsets (Fig. 3A). In line with the changes in myeloid DC differentiation, we observed a clear reduction in the frequencies of IMCs (CD11b+ Gr1+) upon Arg1 overexpression. Further analysis showed that CD115+ monocytic cells (CD115+ CD11b+ LY6C+), not granulocytes (CD11b+ LY6G+), were affected by Arg1 expression. These observations were further confirmed in Arg1 shRNA-transduced cells (Fig. 3A). Additional functional assays showed that cDCs generated by Arg1 overexpression displayed a significantly greater ability to stimulate antigen-specific T cell responses, while knockdown of Arg1 had the opposite effects, compared to the corresponding controls (Fig. 3B).
FIG 3.
Modulation of Arg1 expression affects steady-state DC differentiation. (A to C) Enriched mouse HPCs were infected with lentiviral plasmids (with a GFP tag) expressing Arg1 cDNA and Arg1 shRNA, and empty vectors were used as controls. Cells were cultured with a cytokine cocktail for 5 days and then adoptively transferred to lethally irradiated mice (n = 6). The percentages of splenic DC subsets among the GFP+ CD11c+ cells (left) and other types of myeloid cells among the GFP+ cells (right) were determined by flow cytometric analysis. The data in the upper part of each panel are from a single experiment, and the graph at the bottom represents the mean results ± the standard deviations from the six mice analyzed. (B) cDCs purified from spleen of recipient mice in panel A were stimulated with OVA peptide and then cocultured with CD4+ T cells from OT-II mice at a 1:10 ratio for 72 h, and T cell proliferation was evaluated by CFSE dilution. (C) BM cells were transduced with lentivirus expressing Arg1 or a vector control and then cultured with Flt3L for 9 days. The percentages of DC subsets were evaluated by flow cytometric analysis. The data in the left part of the panel are from a single experiment, and the graph on the right represents the mean results ± the standard deviations from four independent experiments. *, P < 0.05; **, P < 0.01 (compared with the corresponding controls).
The Flt3L culture system represents a cellular model of the generation of steady-state DCs in vitro (35). We therefore utilized this system to confirm the effect of Arg1 on the differentiation of steady-state DCs. Ectopic expression of Arg1 led to significantly higher levels of CD24hi cDCs, the splenic CD8α+ cDC equivalent, while no changes were observed in the levels of pDCs and CD11bhi cDCs upon Arg1 overexpression (Fig. 3C). As further support, the expression of Arg1 in human cDC2 (mouse CD8α+ cDC equivalent) was much higher than that in other DC subsets (see Fig. S6 in the supplemental material). These observations collectively demonstrate that Arg1 plays an essential role in DC differentiation at steady state and that CD8α+ cDCs are particularly responsive to Arg1 activity.
Expression of Arg1 is regulated by HDAC4-mediated histone acetylation.
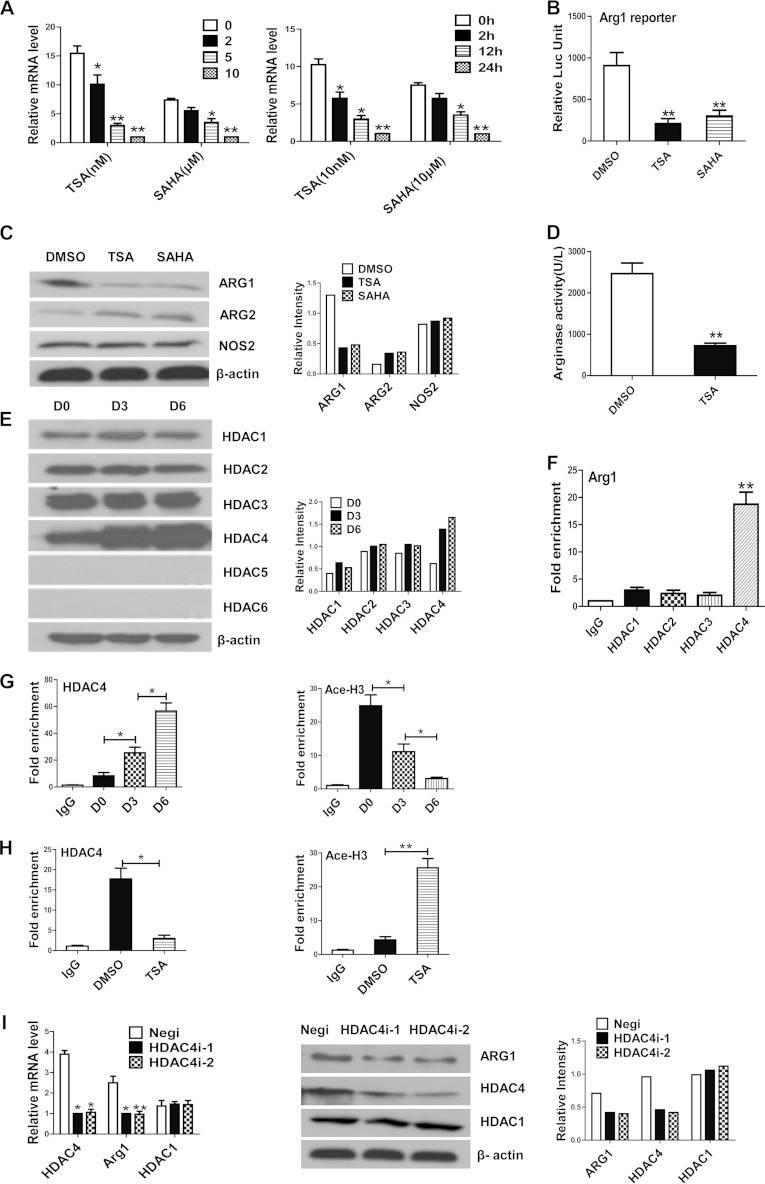
We next performed mechanistic studies to examine the transcriptional regulation of Arg1 in differentiating DCs. First, we investigated whether epigenetic mechanisms participate in this process. To do this, we administered HDACi, including TSA and suberoylanilide hydroxamic acid (SAHA), or the DNA methylation inhibitor 5-aza-2′-deoxycytidine (AZA) to cultured mouse HPCs and determined the expression of Arg1 by qRT-PCR. This revealed that both TSA and SAHA dramatically decreased Arg1 expression in a time- and dose-dependent manner (Fig. 4A), while no effect of AZA was observed (data not shown). The suppressive effect of TSA on Arg1 expression was further confirmed by a reporter activity assay (Fig. 4B) and Western blotting (Fig. 4C). The expression of Arg2, however, was induced by TSA to some extent (Fig. 4C), indicating that distinct Arg isoforms may be differentially regulated by histone acetylation in DCs. The total cell numbers did not show noticeable changes upon HDACi treatment (see Fig. S3 in the supplemental material). Consistent with the downregulation of Arg1 expression, the Arg activity in DCs was reduced by TSA treatment (Fig. 4D). These data indicate that Arg1 is subject to regulation by histone acetylation in DCs.
FIG 4.
Expression of Arg1 is subject to regulation by histone acetylation. (A) Enriched HPCs were cultured in medium containing GM-CSF and IL-4 in the presence of HDACi or dimethyl sulfoxide (DMSO), and Arg1 expression was determined by qRT-PCR in cells treated with different concentrations for 12 h (left) or for different times (right). (B) 32D cells transfected with an Arg1 reporter were treated with HDACi or DMSO, and luciferase activity was measured at 48 h posttransfection. (C) The expression of l-arginine-metabolizing enzymes in BM cells treated with HDACi or DMSO for 72 h was determined by Western blotting. (D) Arg activity in BM cells treated with TSA or DMSO. (E) The expression of distinct HDAC members in cultured BM cells was determined by Western blotting. (F) ChIP assays were performed with BM cells cultured for 6 days with antibodies against HDACs or with an IgG control. Recruitment of HDACs to the Arg1 promoter was determined by qPCR in eluted DNA. Data were normalized to the input DNA and are presented as fold increases over the values obtained with IgG. (G and H) ChIP assays with antibodies against HDAC4 or Ace-H3 were performed with BM cells cultured for different numbers of days (G) or treated with TSA for 3 days (H). The data represent fold increases over the values obtained with the IgG control after normalization to the input DNA. (I) BM cells were transfected with two independent siRNAs for HDAC4 or the negative control (Negi), and the expression of HDAC4 and Arg1 was evaluated by qRT-PCR (left) and Western blotting (right). HDAC1 was used as a negative control. (C, E, and I) The Western blot assay data shown are representative of three independent experiments; the relative intensities of bands normalized to that of β-actin are shown on the right. (A, B, D, and F to I) The data represent the mean results ± the standard deviations from three independent experiments. *, P < 0.05; **, P < 0.01 (compared with the corresponding controls).
To address how histone acetylation may regulate Arg1 transcription in DCs, we first examined the expression of multiple HDAC family members, since these enzymes are responsible for removing acetyl groups from histones and other proteins. The expression of the HDAC1 to -4 proteins was relatively abundant in BM cells at all of the time points examined. The expression of HDAC1 to -3 did not change noticeably over the course of DC differentiation, while HDAC4 was gradually induced in this process. The expression of HDAC5 and -6 was undetectable in Western blot assays at all of the time points examined (Fig. 4E). We further investigated which HDACs could bind to the Arg1 promoter by performing ChIP assays. In comparison with HDAC1 to -3, significantly higher recruitment of HDAC4 to the Arg1 promoter (approximately from bp −453 to −572 upstream of the transcription start site [TSS]) was observed in cultured BM cells (Fig. 4F). Importantly, HDAC4 recruitment was clearly enhanced in the course of DC differentiation, which was coupled with a clear reduction in the acetylated histone 3 (Ac-H3) level within the same Arg1 locus in ChIP assays (Fig. 4G). Meanwhile, administration of TSA suppressed HDAC4 recruitment but enhanced the level of Ac-H3 present on the Arg1 promoter (Fig. 4H). As further confirmation, knockdown of HDAC4 with two independent small interfering RNAs (siRNAs) suppressed Arg1 expression at both the mRNA and protein levels (Fig. 4I). These observations collectively indicate that HDAC4-mediated histone acetylation plays a critical role in the regulation of Arg1 in DCs.
STAT6 cooperates with HDAC4 in the transcriptional regulation of Arg1.
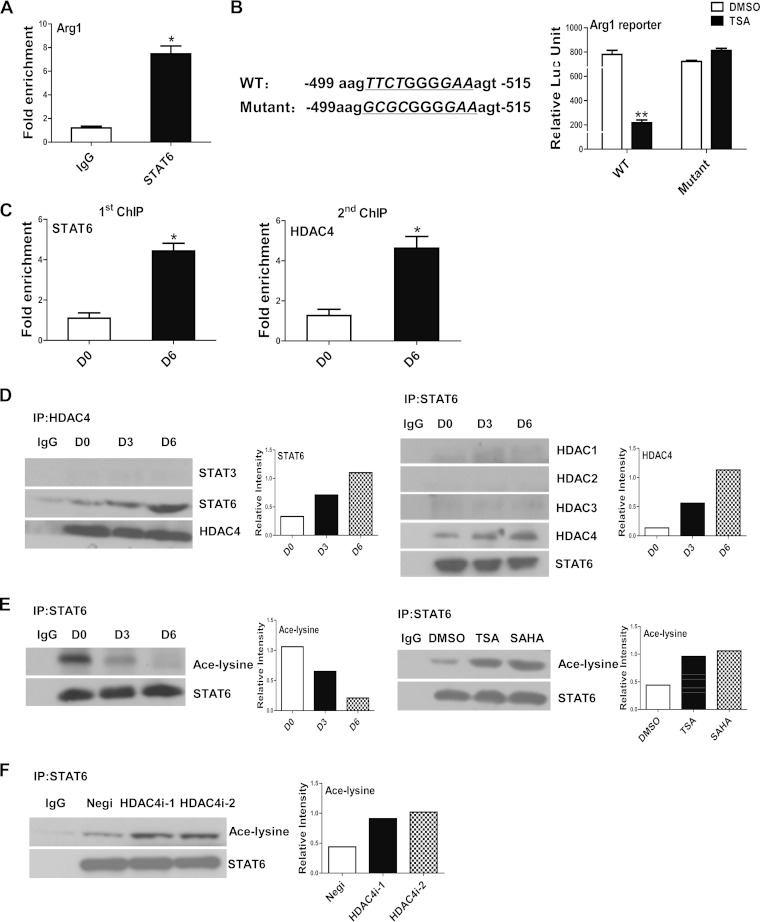
As one of the key epigenetic modifications that occur during transcriptional regulation, histone acetylation usually facilitates the activity or DNA-binding function of TFs (36). To investigate how HDAC4 modulates Arg1 expression, we first studied which TFs might exhibit cross talk with HDAC4 in this process. To this end, we first examined the Arg1 regulatory region surrounding the HDAC4 binding site that was revealed by ChIP assays. This revealed a potential STAT6 binding site (approximately from bp −502 to −512 upstream of the TSS). A ChIP assay further confirmed the binding of STAT6 protein to this locus (Fig. 5A). this STAT6 binding site was distinct from a previously identified STAT6 binding site (from bp −2938 to −2948) (37). We further constructed Arg1 reporter plasmids containing wild-type (WT) or mutant STAT6 binding sites. Transfection assays with these constructs showed that TSA administration suppressed WT Arg1 reporter activity in 32D myeloid cells as expected, while mutation of the STAT6 binding site completely abrogated this effect (Fig. 5B), suggesting that the STAT6 binding site was necessary and sufficient for histone acetylation-mediated Arg1 regulation. A further sequential two-step ChIP assay confirmed the corecruitment of the HDAC4 and STAT6 proteins to the Arg1 promoter in cultured BM cells, with greater enrichment at day 6 than at day 0 (Fig. 5C). Results from immunoprecipitation assays showed that HDAC4 specifically interacted with STAT6 but not STAT3 and that this interaction gradually increased during the course of DC differentiation (Fig. 5D). These data demonstrated that HDAC4 and STAT6 cooperate in order to regulate Arg1 transcription in DCs.
FIG 5.
Cross talk between HDAC4 and STAT6 in the regulation of Arg1. (A) ChIP assay performed with BM cells cultured for 6 days with anti-STAT6 or anti-IgG antibody. The presence of the Arg1 promoter containing the potential STAT6 binding site was measured by qPCR. The data represent fold increases over the values obtained with the IgG control. (B) The sequences of the WT and mutant STAT6-binding sites within the Arg1 promoter are indicated (left). 32D cells transfected with reporter plasmids were treated with TSA or DMSO, and luciferase activity was measured at 48 h posttransfection. (C) Two-step ChIP assays with BM cells harvested on day 0 or 6 were performed sequentially with antibodies against STAT6 and HDAC4 as described in Materials and Methods. The data represent fold increases over the values obtained with the IgG control. (D) Lysates from cultured BM cells were immunoprecipitated with anti-HDAC4 (left) or anti-STAT6 (right) antibody, and the presence of STAT or HDAC protein was determined by Western blotting. (E) Acetylation of STAT6 as determined by immunoprecipitation (IP) in BM cells cultured for various numbers of days (left) or treated with TSA for 3 days (right). (F) 32D cells were transfected with two independent siRNAs for HDAC4 or a negative-control siRNA, and the acetylation level of STAT6 was determined by immunoprecipitation. The data in panels A to C represent the mean results ± the standard deviations of three independent experiments. The data in panels D to F represent three independent experiments, and the relative intensities of the bands are shown in the bar graphs to the right of the blots. *, P < 0.05; **, P < 0.01 (compared with the corresponding controls).
In addition to being phosphorylated, some STAT proteins are also acetylated (23, 38). We therefore examined whether the interaction with HDAC4 would change the acetylation status of the STAT6 protein. Consistent with the enhanced interaction between HDAC4 and STAT6 in differentiating DCs, the level of STAT6 acetylation was significantly lower on day 6 than on day 0 in cultured BM cells. Conversely, HDACi treatment enhanced STAT6 acetylation (Fig. 5E). In contrast to the changes in STAT6 acetylation, the phosphorylation of STAT6 remained similar under all of the experimental conditions tested (see Fig. S7 in the supplemental material). Immunoprecipitation assays showed that knockdown of HDAC4 with specific siRNAs clearly enhanced the level of STAT6 acetylation in the 32D cell line (Fig. 5F). These results indicate that HDAC4 modulates STAT6 acetylation in DCs.
These observations collectively demonstrate that the recruitment of HDAC4 may cause deacetylation of both the histone H3 and STAT6 proteins in the Arg1 promoter, which then leads to transactivation of Arg1 during the course of DC differentiation.
Arg1 mediates the inhibitory effect of HDACi on DC differentiation.
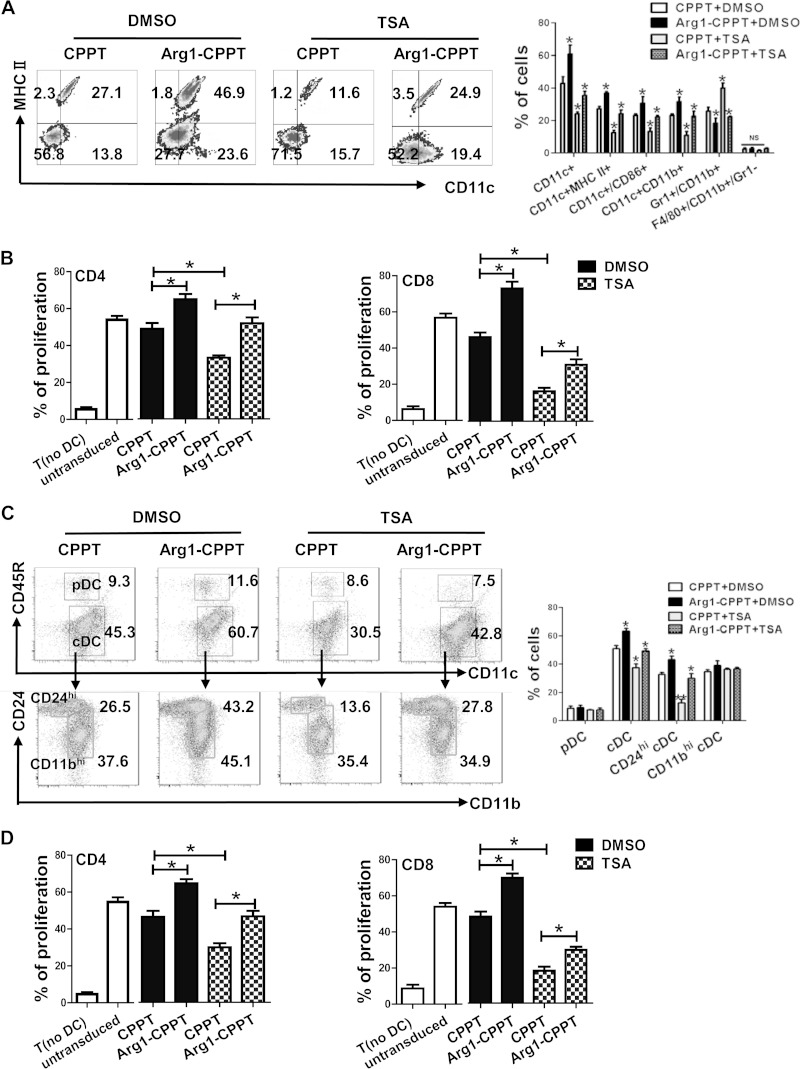
HDACi can suppress DC differentiation and function (17, 24). On the basis of our findings that HDACi treatment suppresses Arg1 expression and that ectopic expression of Arg1 enhances DC differentiation, we therefore hypothesized that Arg1 may mediate the suppressive effect of HDACi on DC differentiation. To test this possibility, we first confirmed the effects of HDACi on DC differentiation both in vitro and in vivo. For in vitro assays, TSA (10 nM) was administered at day 0 of a culture containing either GM-CSF or Flt3L in order to induce the differentiation of monocyte-derived or steady-state DCs, respectively; for in vivo experiments, an mouse inflammation model was used to generate monocyte-derived DCs (33, 39), or steady-state DCs were evaluated in the spleens of naive mice. TSA (5 mg/kg) was administered for 3 consecutive days. Flow cytometric analysis showed that TSA treatment significantly suppressed the differentiation of steady-state cDCs (see Fig. S8 in the supplemental material), as well as monocyte-derived DCs (see Fig. S9), both in vitro and in vivo. We next investigated whether ectopic expression of Arg1 could counteract the suppressive effects of TSA on DCs. In the context of GM-CSF culture conditions, mouse HPCs were infected with lentivirus expressing Arg1 or a vector control and then treated with TSA for 5 days. We observed that ectopic expression of Arg1 almost completely abrogated the suppressive effect of TSA on the differentiation of monocyte-derived DCs (Fig. 6A). The ability of TSA to block DC-dependent stimulation of T cell proliferation was also abrogated by the overexpression of Arg1 (Fig. 6B). In the Flt3L culture system, mouse BM cells were transduced with lentivirus expressing Arg1 or the vector and then treated with TSA for 10 days. This revealed that Arg1 abrogates the effect of TSA on cDC differentiation (Fig. 6C) and blocks their functional activity (Fig. 6D). Supplementation of l-arginine, however, had no effect on TSA-mediated DC suppression (data not shown). Taken together, these observations indicate that Arg1 mediates the inhibitory effect of HDACi on DCs.
FIG 6.
Arg1 mediates the inhibitory effect of TSA on DC differentiation. (A) BM were transduced with lentiviral plasmids expressing Arg1 or the empty vector cPPT and then cultured with TSA or DMSO, as indicated, in medium containing GM-CSF and IL-4 for 6 days. The proportions of distinct subpopulations among the GFP+ cells were analyzed by flow cytometry. (B) Allogeneic mixed-lymphocyte reaction (MLR) cells from panel A were cocultured with T cells from allogeneic BALB/c mice at a 1:10 ratio for 3 days, and T cell proliferation was measured by CFSE dilution; T cells cultured alone were used as controls. (C) BM cells were infected with lentiviral plasmids expressing Arg1 or the empty vector cPPT and then cultured with DMSO or TSA in medium containing Flt3L for 9 days. The proportions of DC subsets were analyzed by flow cytometry of the GFP+ cells. (D) Allogeneic MLR. cDCs isolated from panel C were cocultured with allogeneic T cells at a 1:10 ratio for 3 days, and T cell proliferation was measured by CFSE dilution. (A, C) The left half of each panel shows representative results from a single experiment, and the right half shows the mean results ± the standard deviations of three independent experiments. The data in panels B and D represent the mean results ± the standard deviations of three independent experiments. *, P < 0.05; **, P < 0.01 (compared with the corresponding controls).
DISCUSSION
Although the role of Arg1 and l-arginine metabolism in immune responses has been well recognized, little is known about their effects on DC biology (11). Our study demonstrated that Arg1, one of the key l-arginine-metabolizing enzymes, is a novel regulator of myeloid DC differentiation and shed new light on the immunoregulatory roles of Arg1 and l-arginine metabolism.
Our results support the idea that the level of Arg1 expression specifically impacts CD8α+ cDC differentiation. No effect on CD11b+ DCs or pDCs was observed upon the modulation of Arg1 expression. Arg1 expression is much higher in CD8α+ cDCs from Flt3L-cultured mouse BM and equivalent human cDC2, compared with other DC subsets. However, CD8α+ cDCs from fresh mouse spleen express a low level of Arg1 (see Fig. S10 in the supplemental material), which is consistent with the results from ImmGen (http://www.immgen.org/). The differential pattern of Arg1 expression in CD8α+ cDCs from in vivo and in vitro sources indicates that a complicated regulatory mechanism may dictate Arg1 expression in mice and deserves further investigation. Conditional knockout of Arg1 in myeloid cells would be ideal for fully understanding the role of Arg1 in DC differentiation.
Histone modification-based transcriptional regulation plays a fundamental role in cell fate determination, including lineage commitment of hematopoietic stem/progenitor cells (40, 41). However, little is known about the role of histone code-mediated gene regulation in the differentiation of DCs (24), the exploration of which will be crucial in delineating the transcriptional network that controls DC differentiation. Our present study identified Arg1 as a transcriptional target of histone acetylation in differentiating DCs. The fine tuning of Arg1 in this process was mediated by the dynamic recruitment of HDAC4 and STAT6 to a regulatory region within the Arg1 promoter. This highlights the importance of the cross talk between TFs and epigenetic pathways in DC development. The mechanism of dynamic HDAC4 recruitment to the Arg1 locus in differentiating DCs deserves further investigation.
We observed robust induction of Arg1 during the course of DC differentiation, and modulation of Arg1 expression could significantly affect the differentiation of both monocyte-derived DCs and steady-state cDCs but not steady-state pDCs; these data indicate that Arg1 has a universal role in the development of myeloid DCs. In contrast to the induction of Arg1 expression, Arg2 was moderately downregulated in differentiating DCs. These results indicate that a potential balance may exist between different Arg isoforms in DC development, although the significance of Arg2 in DCs remains unknown. Interestingly, Cat2B, one of the members of the l-arginine transporter cationic amino acid transporter (CAT) family, was specifically induced in differentiating DCs. Induction of Arg1 is strictly linked with the upregulation of CAT proteins, which form functional units that are responsible for the majority of l-arginine consumption in certain cells types, including macrophages and MDSCs (11, 42–44). The induction of both Arg1 and Cat2B during the course of DC differentiation indicates that they may cooperate to facilitate the uptake and metabolism of l-arginine in this process. The clear reduction in the amounts of l-arginine in culture supernatants from differentiating DCs supports this possibility.
The molecular mechanism underlying Arg1 transcriptional regulation in DCs was delineated in this study. Th2 cytokines are known to induce the expression of Arg1 in macrophages through JAK-mediated STAT6 phosphorylation (37, 45–47). The regulation of Arg1 in DCs is clearly distinct from that in macrophages. First, the induction of Arg1 was not completely dependent on IL-4 in DCs. Addition of IL-4 increased the fold induction of Arg1 compared with that observed with GM-CSF alone, suggesting that both of the cytokines GM-CSF and IL-4 may participate in the regulation of Arg1 in DCs. Second, the phosphorylation of STAT6 was unchanged during the course of DC differentiation, although both GM-SCF and IL-4 could cause STAT6 phosphorylation in a BM culture (see Fig. S11 in the supplemental material). The level of STAT6 acetylation, however, was dynamically regulated. Third, an HDAC4-mediated epigenetic mechanism participated in the Arg1-dependent regulation of DCs. The recruitment of HDAC4 caused the acetylation of both the STAT6 and histone H3 proteins within the Arg1 regulatory locus, therefore leading to its transactivation.
Acetylation of STAT proteins plays important roles in the modulation of their transcriptional activity (48, 49). It was reported that the HDAC inhibitor TSA causes hyperacetylation of STAT3 and decreases the expression of cyclin D1 (50). Shankaranarayanan et al. demonstrated that acetylation of STAT6 by CBP/p300 is required for the transcriptional activation of the gene for 15-LOX-1 (51). These observations suggest that acetylation of STAT proteins regulates transcription machinery in a gene-specific manner. Our study demonstrates that STAT6 acetylation in DCs suppresses Arg1 transcription in differentiating DCs. Knockdown of STAT6 consistently led to a clear reduction of Arg1 expression (see Fig. S12 in the supplemental material). Taken together, our observations indicate that different types of myeloid cells may exploit Arg1 activation via distinct mechanisms.
Although our study demonstrates an essential role for Arg1 in the differentiation of myeloid DCs, the underlying mechanism remains unclear. We performed genomic mRNA profiling to screen DCs for potential targets of Arg1. The expression of several TFs important for CD8α+ DC development was induced by Arg1 overexpression, including BATF2, BATF3, and Nfil3 (52–54) (see Fig. S13 in the supplemental material). The exact role of these TFs in Arg1-mediated DC differentiation, however, deserves further investigation.
In addition to their direct effects on cancer cells (55), HDACi are known to have broad immunomodulatory effects. Although their role in the differentiation and function of myeloid cells is well documented (23, 24, 26), the detailed mechanisms are still poorly understood. In this study, we confirmed the effects of HDACi on the suppression of myeloid DC generation, as well as the enhanced accumulation of IMCs in mice. Further studies suggest that Arg1 mediates the suppressive effect of HDACi on DC differentiation. However, the possibility that HDACi and Arg1 function independently in regulating DC differentiation could not be completely excluded; mutation of the Arg1 promoter that does not contain the binding site for HDAC4 in vivo would be helpful in elucidating whether Arg1 mediates the effect of HDACi on DCs. Since HDACi have broad immunomodulatory properties, they could have significant potential impacts on pathological conditions such as cancer, inflammation, and autoimmune diseases. Our study provides a greater molecular insight into their mode of action that may be useful in the development of novel immunotherapeutic strategies for these disorders.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the following to J.Z.: the Natural Science Foundation of Guangdong (S2011020006072), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme grant GDUPS, 2014, National Key Basic Research Program of China (973 Program, 2012CB524900), the Guangdong Innovative Research Team Program (2009010058), the Key Research Projects of National 12th Five-year Plan for the Prevention and Treatment of Major Infectious Diseases (2012ZX10001003), the National Natural Science Foundation of China (81072397 and 31270921), and the Fundamental Research Funds for the Central Universities 111 Project (B12003). H.Y. is supported by grants from the National Natural Science Foundation of China (31170809) and the Hundred Talents Program of the Chinese Academy of Sciences (2012OHTP07).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00805-14.
REFERENCES
- 1.Banchereau J, Steinman RM. 1998. Dendritic cells and the control of immunity. Nature 392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 2.Mellman I, Steinman RM. 2001. Dendritic cells: specialized and regulated antigen processing machines. Cell 106:255–258. doi: 10.1016/S0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 3.Zenke M, Hieronymus T. 2006. Towards an understanding of the transcription factor network of dendritic cell development. Trends Immunol 27:140–145. doi: 10.1016/j.it.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Satpathy AT, Murphy KM, W KC. 2011. Transcription factor networks in dendritic cell development. Semin Immunol 23:388–397. doi: 10.1016/j.smim.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gabrilovich D. 2004. Mechanisms and functional significance of tumour-induced dendritic-cell defects. Nat Rev Immunol 4:941–952. doi: 10.1038/nri1498. [DOI] [PubMed] [Google Scholar]
- 6.Fitzgerald-Bocarsly P, Jacobs ES. 2010. Plasmacytoid dendritic cells in HIV infection: striking a delicate balance. J Leukoc Biol 87:609–620. doi: 10.1189/jlb.0909635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moretto MM, Lawlor EM, Khan IA. 2008. Aging mice exhibit a functional defect in mucosal dendritic cell response against an intracellular pathogen. J Immunol 181:7977–7984. doi: 10.4049/jimmunol.181.11.7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmid MA, Kingston D, Boddupalli S, Manz MG. 2010. Instructive cytokine signals in dendritic cell lineage commitment. Immunol Rev 234:32–44. doi: 10.1111/j.0105-2896.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- 9.Onai N, Manz MG. 2008. The STATs on dendritic cell development. Immunity 28:490–492. doi: 10.1016/j.immuni.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Zhou J, Cheng P, Youn J I, Cotter MJ, Gabrilovich DI. 2009. Notch and wingless signaling cooperate in regulation of dendritic cell differentiation. Immunity 30:845–859. doi: 10.1016/j.immuni.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bronte V, Zanovello P. 2005. Regulation of immune responses by l-arginine metabolism. Nat Rev Immunol 5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- 12.Gordon S. 2003. Alternative activation of macrophages. Nat Rev Immunol 3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 13.Gabrilovich DI, Nagaraj S. 2009. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 15.Struhl K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev 12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 16.Eberharter A, Becker PB. 2002. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep 3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Lint C, Emiliani S, Verdin E. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr 5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 18.Weishaupt H, Sigvardsson M, Attema JL. 2010. Epigenetic chromatin states uniquely define the developmental plasticity of murine hematopoietic stem cells. Blood 115:247–256. doi: 10.1182/blood-2009-07-235176. [DOI] [PubMed] [Google Scholar]
- 19.Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Attema JL, Papathanasiou P, Forsberg EC, Xu J, Smale ST, Weissman IL. 2007. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc Natl Acad Sci U S A 104:12371–12376. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang S, Aune TM. 2007. Dynamic changes in histone-methylation ‘marks’ across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat Immunol 8:723–731. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- 22.Reddy P, Sun Y, Toubai T, Duran-Struuck R, Clouthier SG, Weisiger E, Maeda Y, Tawara I, Krijanovski O, Gatza E, Liu C, Malter C, Mascagni P, Dinarello CA, Ferrara JL. 2008. Histone deacetylase inhibition modulates indoleamine 2,3-dioxygenase-dependent DC functions and regulates experimental graft-versus-host disease in mice. J Clin Invest 118:2562–2573. doi: 10.1172/JCI34712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y, Chin YE, Weisiger E, Malter C, Tawara I, Toubai T, Gatza E, Mascagni P, Dinarello CA, Reddy P. 2009. Cutting edge: negative regulation of dendritic cells through acetylation of the nonhistone protein STAT-3. J Immunol 182:5899–58903. doi: 10.4049/jimmunol.0804388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nencioni A, Beck J, Werth D, Grunebach F, Patrone F, Ballestrero A, Brossart P. 2007. Histone deacetylase inhibitors affect dendritic cell differentiation and immunogenicity. Clin Cancer Res 13:3933–3941. doi: 10.1158/1078-0432.CCR-06-2903. [DOI] [PubMed] [Google Scholar]
- 25.Licciardi PV, Karagiannis TC. 2012. Regulation of immune responses by histone deacetylase inhibitors. ISRN Hematol 2012:690–901. doi: 10.5402/2012/690901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosborough BR, Castellaneta A, Natarajan S, Thomson AW, Turnquist HR. 2012. Histone deacetylase inhibition facilitates GM-CSF-mediated expansion of myeloid-derived suppressor cells in vitro and in vivo. J Leukoc Biol 91:701–709. doi: 10.1189/jlb.0311119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin A, Cai W, Pan T, Wu K, Yang Q, Wang N, Liu Y, Yan D, Hu F, Guo P, Chen X, Chen L, Zhang H, Tang X, Zhou J. 2013. Expansion of monocytic myeloid-derived suppressor cells dampens T cell function in HIV-1-seropositive individuals. J Virol 87:1477–1490. doi: 10.1128/JVI.01759-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharabi AB, Aldrich M, Sosic D, Olson EN, Friedman AD, Lee SH, Chen SY. 2008. Twist-2 controls myeloid lineage development and function. PLoS Biol 6(12):e316. doi: 10.1371/journal.pbio.0060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teerlink T, Nijveldt RJ, de Jong S, van Leeuwen PA. 2002. Determination of arginine, asymmetric dimethylarginine, and symmetric dimethylarginine in human plasma and other biological samples by high-performance liquid chromatography. Anal Biochem 303:131–137. doi: 10.1006/abio.2001.5575. [DOI] [PubMed] [Google Scholar]
- 30.Kutner RH, Zhang XY, Reiser J. 2009. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc 4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- 31.Shen L, Evel-Kabler K, Strube R, Chen SY. 2004. Silencing of SOCS1 enhances antigen presentation by dendritic cells and antigen-specific anti-tumor immunity. Nat Biotechnol 22:1546–1553. doi: 10.1038/nbt1035. [DOI] [PubMed] [Google Scholar]
- 32.Haviernik P, Zhang Y, Bunting KD. 2008. Retroviral transduction of murine hematopoietic stem cells. Methods Mol Biol 430:229–241. doi: 10.1007/978-1-59745-182-6_16. [DOI] [PubMed] [Google Scholar]
- 33.Kamphorst AO, Guermonprez P, Dudziak D, Nussenzweig MC. 2010. Route of antigen uptake differentially impacts presentation by dendritic cells and activated monocytes. J Immunol 185:3426–3435. doi: 10.4049/jimmunol.1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luskey BD, Rosenblatt M, Zsebo K, Williams DA. 1992. Stem cell factor, interleukin-3, and interleukin-6 promote retroviral-mediated gene transfer into murine hematopoietic stem cells. Blood 80:396–402. [PubMed] [Google Scholar]
- 35.Naik SH, Proietto AI, Wilson NS, Dakic A, Schnorrer P, Fuchsberger M, Lahoud MH, O'Keeffe M, Shao Q X, Chen WF, Villadangos J A, Shortman K, Wu L. 2005. Cutting edge: generation of splenic CD8+ and CD8− dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J Immunol 174:6592–6597. doi: 10.4049/jimmunol.174.11.6592. [DOI] [PubMed] [Google Scholar]
- 36.Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. 2008. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell 29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pauleau AL, Rutschman R, Lang R, Pernis A, Watowich SS, Murray PJ. 2004. Enhancer-mediated control of macrophage-specific arginase I expression. J Immunol 172:7565–7573. doi: 10.4049/jimmunol.172.12.7565. [DOI] [PubMed] [Google Scholar]
- 38.Kramer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Guhrs KH, Stauber RH, Bohmer FD, Heinzel T. 2009. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev 23:223–235. doi: 10.1101/gad.479209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naik SH, Metcalf D, van Nieuwenhuijze A, Wicks I, Wu L, O'Keeffe M, Shortman K. 2006. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat Immunol 7:663–671. doi: 10.1038/ni1340. [DOI] [PubMed] [Google Scholar]
- 40.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Singh SK, Williams CA, Klarmann K, Burkett SS, Keller J R, Oberdoerffer P. 2013. Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. J Exp Med 210:987–1001. doi: 10.1084/jem.20121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mann GE, Yudilevich DL, Sobrevia L. 2003. Regulation of amino acid and glucose transporters in endothelial and smooth muscle cells. Physiol Rev 83:183–252. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez PC, Zea AH, DeSalvo J, Culotta KS, Zabaleta J, Quiceno DG, Ochoa J B, Ochoa AC. 2003. l-Arginine consumption by macrophages modulates the expression of CD3 zeta chain in T lymphocytes. J Immunol 171:1232–1239. doi: 10.4049/jimmunol.171.3.1232. [DOI] [PubMed] [Google Scholar]
- 44.Highfill SL, Rodriguez PC, Zhou Q, Goetz CA, Koehn BH, Veenstra R, Taylor PA, Panoskaltsis-Mortari A, Serody J S, Munn DH, Tolar J, Ochoa AC, Blazar BR. 2010. Bone marrow myeloid-derived suppressor cells (MDSCs) inhibit graft-versus-host disease (GVHD) via an arginase-1-dependent mechanism that is up-regulated by interleukin-13. Blood 116:5738–5747. doi: 10.1182/blood-2010-06-287839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray MJ, Poljakovic M, Kepka-Lenhart D, Morris SJ. 2005. Induction of arginase I transcription by IL-4 requires a composite DNA response element for STAT6 and C/EBPbeta. Gene 353:98–106. doi: 10.1016/j.gene.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Osorio EY, Travi BL, Da CA, Saldarriaga OA, Medina AA, Melby PC. 2014. Growth factor and Th2 cytokine signaling pathways converge at STAT6 to promote arginase expression in progressive experimental visceral leishmaniasis. PLoS Pathog 10(6):e1004165. doi: 10.1371/journal.ppat.1004165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheldon KE, Shandilya H, Kepka-Lenhart D, Poljakovic M, Ghosh A, Morris SJ. 2013. Shaping the murine macrophage phenotype: IL-4 and cyclic AMP synergistically activate the arginase I promoter. J Immunol 191:2290–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Icardi L, De Bosscher K, Tavernier J. 2012. The HAT/HDAC interplay: multilevel control of STAT signaling. Cytokine Growth Factor Rev 23:283–291. doi: 10.1016/j.cytogfr.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 49.Zhuang S. 2013. Regulation of STAT signaling by acetylation. Cell Signal 25:1924–1931. doi: 10.1016/j.cellsig.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pang M, Ma L, Liu N, Ponnusamy M, Zhao TC, Yan H, Zhuang S. 2011. Histone deacetylase 1/2 mediates proliferation of renal interstitial fibroblasts and expression of cell cycle proteins. J Cell Biochem 112:2138–2148. doi: 10.1002/jcb.23135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shankaranarayanan P, Chaitidis P, Kuhn H, Nigam S. 2001. Acetylation by histone acetyltransferase CREB-binding protein/p300 of STAT6 is required for transcriptional activation of the 15-lipoxygenase-1 gene. J Biol Chem 276:42753–42760. doi: 10.1074/jbc.M102626200. [DOI] [PubMed] [Google Scholar]
- 52.Tussiwand R, Lee WL, Murphy TL, Mashayekhi MWKC, Albring JC, Satpathy AT, Rotondo J A, Edelson BT, Kretzer NM, Wu X, Weiss LA, Glasmacher E, Li P, Liao W, Behnke M, Lam SS, Aurthur CT, Leonard WJ, Singh H, Stallings CL, Sibley LD, Schreiber RD, Murphy KM. 2012. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 490:502–507. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edelson BT, KC W, Juang R, Kohyama M, Benoit LA, Klekotka PA, Moon C, Albring J C, Ise W, Michael DG, Bhattacharya D, Stappenbeck TS, Holtzman MJ, Sung SS, Murphy TL, Hildner K, Murphy KM. 2010. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med 207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kashiwada M, Pham NL, Pewe LL, Harty J T, Rothman PB. 2011. NFIL3/E4BP4 is a key transcription factor for CD8alpha(+) dendritic cell development. Blood 117:6193–6197. doi: 10.1182/blood-2010-07-295873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prince HM, Bishton MJ, Harrison SJ. 2009. Clinical studies of histone deacetylase inhibitors. Clin Cancer Res 15:3958–3969. doi: 10.1158/1078-0432.CCR-08-2785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.