Abstract
The retinoblastoma protein (pRb/p105) tumor suppressor plays a pivotal role in cell cycle regulation by blockage of the G1-to-S-phase transition. pRb tumor suppressor activity is governed by a variety of posttranslational modifications, most notably phosphorylation by cyclin-dependent kinase (Cdk) complexes. Here we report a novel regulation of pRb through protein arginine methyltransferase 4 (PRMT4)-mediated arginine methylation, which parallels phosphorylation. PRMT4 specifically methylates pRb at the pRb C-terminal domain (pRb Cterm) on arginine (R) residues R775, R787, and R798 in vitro and R787 in vivo. Arginine methylation is important for efficient pRb Cterm phosphorylation, as manifested by the reduced phosphorylation of a methylation-impaired mutant, pRb (R3K). A methylmimetic form of pRb, pRb (R3F), disrupts the formation of the E2F-1/DP1-pRb complex in cells as well as in an isolated system. Finally, studies using a Gal4–E2F-1 reporter system show that pRb (R3F) expression reduces the ability of pRb to repress E2F-1 transcriptional activation, while pRb (R3K) expression further represses E2F-1 transcriptional activation relative to that for cells expressing wild-type pRb. Together, our results suggest that arginine methylation negatively regulates the tumor suppressor function of pRb during cell cycle control, in part by creating a better substrate for Cdk complex phosphorylation and disrupting the interaction of pRb with E2F-1.
INTRODUCTION
Members of the retinoblastoma protein (pRb) family, also known as “pocket proteins,” are key regulators of cell cycle progression by serving as gatekeepers of the G1-to-S-phase transition (1, 2). Indeed, inactivating mutations of pRb function have been observed in a wide array of cancers. Of the three family members (pRb/p105, pRB2/p130, and pRBL1/p107), pRb/p105 has been most extensively studied for its definitive role in tumor suppression and its ability to bind and inhibit the neoplastic transforming transcriptional activator E2F-1 (3–6). In normal quiescent cells and early-G1 cells, most pRb is in its active hypophosphorylated form and suppresses the activity of E2F-1 through protein-protein interactions via its pocket domain and C-terminal domain (pRb Cterm) (3, 7–10). Upon mitogen stimulation, pRb is spatially and temporally phosphorylated throughout the G1 phase, initially by the cyclin D/cyclin-dependent kinase 4 (Cdk4) or cyclin D/Cdk6 complex and later by the cyclin E/Cdk2 or cyclin A/Cdk2 complex (11–14). Phosphorylation of conserved serine (S) and threonine (T) residues located in the pRb pocket domain and pRb Cterm results in dissociation of the E2F-1 transactivation domain and E2F-1/DP1 coiled-coil/marked box (CM) domain, respectively (9, 13, 15, 16). Release of E2F-1/DP1 permits E2F-1-mediated transactivation of S-phase-specific genes and induction of DNA replication for cell cycle progression (17). Accordingly, others have reported that although the pRb pocket domain is necessary for growth suppression, pRb Cterm is required for both maximum E2F-1 repression and growth suppression (8, 18, 19). Seven of the 16 Cdk serine and threonine phosphorylation consensus sites are located at the pRb Cterm (S780, S788, S795, S807/811, T821, and T826), in which phosphorylation at S788 and S795 is required for complete pRb–E2F-1/DP1 complex disruption (9, 20, 21).
In addition to phosphorylation, various posttranslation modifications (PTMs), including acetylation (22–24), sumoylation (25), ubiquitylation (26, 27), and, more recently, lysine methylation (28, 29), have been reported to regulate pRb activity during cell proliferation, differentiation, and the DNA damage response (30). For example, pRb monomethylation on K810 by lysine methyltransferases Set7/9 and Smyd2 inhibits Cdk-directed phosphorylation and is required for cell cycle arrest in response to DNA damage, while monomethylation on K873 by the same methylase results in enhanced pRb interaction with the heterochromatin protein (HP-1), which is important for pRb-induced cell cycle arrest and augmentation of pRb-dependent differentiation (28, 29). Studies by Saddic et al. have demonstrated that monomethylation of pRb by Smyd2 at K860 increases the interaction with the malignant brain tumor (MBT) methyl-binding domain of the L3MBTL1 transcriptional repressor and that K860 methylation is dynamically regulated throughout cell proliferation and differentiation and in the presence of a DNA-damaging agent (31). In light of these discoveries and the established cross talk among PTMs on histone and nonhistone proteins, we hypothesized that pRb activity may also be regulated by arginine methylation.
In contrast to the role of pRb in cell cycle inhibition, overexpression of protein arginine methyltransferase 4 (PRMT4) (also known as coactivator-associated arginine methylase 1 [CARM1]) and its methylase activity have been implicated in many types of cancers, including colorectal, prostate, and breast cancers, and most prominently in metastatic breast cancer (32). PRMT4 is a type I arginine methyltransferase, which asymmetrically dimethylates specific arginine residues on proteins (33). It was originally characterized as a coactivator of the p160 steroid receptor coactivator (SRC) protein family, facilitating the transcriptional activation of nuclear hormone target genes (34). PRMT4-directed methylation of histone H3 at R17 and R26 (H3R17 and H3R26) at promoter regions has been strictly associated with gene activation and thus serves as a coactivator for many cancer-related transcription factors, such as c-Fos, p53, and NF-κB (35–38). As an example, in breast cancer cells, recruitment of PRMT4 by the estrogen receptor α (ER-α)/ACTR complex to promoter regions and subsequent histone methylation on H3R17 and H3R26 residues leads to upregulation of E2F-1 and cyclin E gene expression, contributing to breast cancer cell proliferation (39, 40). Recently, Wang et al. revealed another factor contributing to PRMT4-induced pathogenicity during breast cancer progression and metastasis: protein arginine methylation on R1064 of BAF155, a core subunit of the SWI/SNF chromatin-remodeling complex. Methylation of BAF155 positively controls expression of the c-Myc pathway (41). With these pieces of evidence, it is important to understand the complete range of molecular mechanisms utilized by PRMT4 in promoting cancer cell proliferation.
Here we investigate the functional importance of arginine methylation for pRb regulation by mutating methylated arginine (R) residues into lysine (K) residues, thus rendering pRb methylation defective while retaining the positive charge. In addition, we have examined the function of methylation by replacing PRMT4 methylation target site R residues with phenylalanine (F) residues to mimic the bulky hydrophobic properties of methylation, a strategy that has been used successfully in other studies (42). From our analysis, we have determined that methylation of R775, R787, and R798 residues on pRb Cterm are important for pRb phosphorylation, E2F-1 dissociation, and E2F-1 transcriptional activation.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney 293T (HEK 293T) epithelial cells and the human breast adenocarcinoma cell line MCF-7 were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) in the presence of 5% CO2. Isogenic U2OS osteosarcoma cells stably expressing wild-type pRb [pRb (WT)], a methylmimetic form of pRb in which R residues are replaced with F residues [pRb (R3F)], or a methylation-impaired pRb mutant in which R residues are mutated to K residues [pRb (R3K)] were generated with a Flp-In recombination system (Invitrogen). The pcDNA5FRT/TO transfer vector encoding Flag-pRb (WT), Flag-pRb (R3K), or Flag-pRb (R3F) was cotransfected with plasmid pOG44 into FRT/TO U2OS cells. Hygromycin (250 μg/ml) was then added to select transfectants. The resultant cells were maintained in DMEM containing 10% FBS and 250 μg/ml hygromycin B (Invitrogen).
Plasmids.
The cloning strategy employed throughout these studies involved the cloning of fragments into several modified vectors containing a CpoI restriction enzyme site generated within each polylinker. This strategy has been described previously (43, 44). Fragments were digested with CpoI, desalted (Qiagen, Valencia, CA), and cloned into CpoI-digested vectors for expression in mammalian cells (pcDNA3-CpoI), baculoviral expression (pFastBac-CpoI), and glutathione S-transferase (GST) fusion expression (pGEX-2T-CpoI). The clones resulting from all procedures possess an N-terminal Flag, hemagglutinin (HA), or GST tag. Wild-type pRb and wild-type PRMT4 expression vectors were constructed as described above, using cDNA prepared from U2OS cells. These plasmids served as templates for site-directed mutagenesis yielding the enzymatically inactive mutant PRMT4 (EQ), pRb (R3K), and pRb (R3F) mutants and for the construction of truncated pRb fragments by PCR. The oligonucleotide sequences used throughout this study are available upon request. To generate recombinant proteins of Flag-pRb and Flag-PRMT4 in the baculovirus protein expression system, pRb and PRMT4 were subcloned into pFastBac1. Successive overlapping N-terminal pRb deletion mutants encoding pRb Del-1 (residues 40 to 150), pRb Del-2 (residues 100 to 250), pRb Del-3 (residues 200 to 350), pRb Del-4 (residues 300 to 450), pRb Del-5 (residues 400 to 550), pRb Del-6 (residues 500 to 650), pRb Del-7 (residues 600 to 750), pRb Del-8 (residues 700 to 850), or pRb Del-9 (residues 800 to 928) were cloned into a GST-tagged pGEX-2T vector for expression of recombinant protein in Escherichia coli.
Methylation assay.
In vitro methylation assays were carried out by incubation of 500 nM (final concentration) purified Flag-PRMT4 (WT) or Flag-PRMT4 (EQ) and full-length Flag-Rb protein family members (0.5 μg to 2 μg) or GST-pRb fusion deletion proteins (1 μg) with 55 μCi S-adenosyl-l-[methyl-3H]methionine as a cosubstrate. Methylation reactions were carried out in volumes of 20 μl for each reaction in 1× methylation buffer (150 mM NaCl, 20 mM Tris [pH 7.5], 1 mM EDTA, 0.02% Triton) at 37°C for periods ranging from 3 h to overnight. In vitro methylation assays for mass spectrometry (MS) analyses were conducted similarly except for the use of 40 μM cold S-adenosyl-l-methionine as a cosubstrate. The peptides used in these studies had the following sequences: biotin-ILQYASTRPPTLSPI, biotin-SPIPHIPRSPYKFPS, and biotin-KFPSSPLRIPGGNIY.
Immunoprecipitation (IP) and Western blot analysis.
For the detection of endogenous pRb methylation, MCF-7 cells were transfected with short hairpin RNA (shRNA) targeting either green fluorescent protein (GFP) as a control or PRMT4. Forty-eight hours posttransfection, the cells were harvested and were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 6.7], 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1× protease inhibitor cocktail (Roche). Five hundred micrograms of total-cell lysates (TCLs) was incubated overnight at 4°C with an antibody raised against pRb peptide dimethylated on R787 (pRb R787-Me2). Immune complexes were captured with protein A+G–agarose beads. Precipitated beads were washed, and pRb methylation was detected by probing for pRb. Immunoblot analysis was performed by using an anti-pRb R787-Me2 antibody for the detection of methylation of recombinant proteins. For pRb phosphorylation levels, U2OS cells stably expressing pRb (WT) or pRb (R3K) were collected in phospholysis buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 0.1% SDS, 1% Triton X-100, 0.5% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM NaF, 2 mM Na3VO4, 10% glycerol) supplemented with 1 mM PMSF, 1× protease inhibitor cocktail (Roche), and 1× phosphatase inhibitor cocktail (Roche). Immunoblot analysis for pRb Cterm phosphorylation was performed using an antibody against pRb phosphorylated on S788 [phospho-pRb (S788)] (Pierce), phospho-pRb (S795) (Cell Signaling), phospho-pRb (S807/811) (Cell Signaling), phospho-pRb (T821) (Cell Signaling), or phospho-pRb (T826) (Cell Signaling) and an affinity-purified rabbit polyclonal antibody against pRb (AnaSpec Inc.). For pRb and E2F-1 coimmunoprecipitation experiments, the isogenic U2OS cell lines stably expressing either pRb (WT) or pRb (R3F) were compared. Similarly, isogenic U2OS cell lines stably expressing pRb (WT) transfected with either a control shRNA (shControl) plasmid or a PRMT4 shRNA (shPRMT4) plasmid were used to examine the interaction between E2F-1 and pRb. The cells were collected in phospholysis buffer, and 750 μg of TCLs was incubated with anti-Flag–M2 agarose beads (Sigma) overnight at 4°C. The beads were washed three times in phospholysis buffer, and an anti-E2F-1 antibody (Cell Signaling) was used to probe for the interaction.
Purification of recombinant protein.
Spodoptera frugiperda Sf9 cells were maintained in Ex-Cell 420 medium (JRH Biosciences), and recombinant baculoviruses were generated as described previously (44). Recombinant baculovirus bacmid DNA was transfected into Sf9 cells by using FuGene 6 (Roche), and recombinant viruses were subsequently amplified twice. Expression of recombinant proteins was confirmed by immunoblotting with an anti-Flag monoclonal antibody (Sigma). Large-scale cultures of Sf9 cells (100 ml) were infected with recombinant baculovirus at a multiplicity of infection (MOI) of 0.1 to 1.0, and cells were harvested 48 h after infection. Recombinant proteins were purified as described previously (43). The purity and amount of protein were measured by SDS-PAGE and Coomassie blue staining, using bovine serum albumin (BSA) as a standard.
Preparation and purification of GST fusion proteins.
GST-pRb deletion mutants were transformed and expressed in Escherichia coli strain BL21. Briefly, bacterial cells (250 ml) were cultured in Luria broth for each construct. Protein expression was induced with 0.5 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were harvested, washed once in phosphate-buffered saline (PBS), and then lysed by sonication in PBS containing 1% Triton X-100 and 0.3× BugBuster lysis buffer (Novagen). After clearing by centrifugation at 7,000 × g for 15 min at 4°C, glutathione-Sepharose beads (200 μl of a 1:1 slurry in PBS) were added to the lysates for affinity purification. After overnight incubation at 4°C with rotation, the beads were washed four times in PBS containing 1% Triton X-100 and 0.3× BugBuster lysis buffer. The proteins immobilized on the glutathione-agarose beads were quantified by Coomassie blue staining, using BSA as a protein standard.
Mass spectrometry analysis.
For matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis, in vitro methylation reactions were carried out on the peptides listed above. The resulting mixtures were desalted through a C18 ZipTip (Millipore) according to the manufacturer's instructions. The bound peptides were eluted with 2 μl buffer containing 70% acetonitrile with 0.1% trifluoroacetic acid (TFA). Eluates were spotted directly onto MALDI sample plates and were analyzed with a MALDI-TOF micro MX mass spectrometer (ABI 4700) at the Campus Mass Spectrometry Facilities at UC Davis.
For liquid chromatography (LC)-tandem mass spectrometry (MS-MS) analysis, the GST-pRb (700–850) fusion protein (GST fused to pRb amino acids 700 to 850) was subjected to an in vitro methylation assay and was resolved by 10% SDS-PAGE. After Coomassie brilliant blue (CBB) staining, the band corresponding to GST-pRb (700–850) was cut out and was submitted to the UC Davis Proteomics Core Facility.
Database searching.
All MS-MS samples were analyzed using Mascot (version 2.4.01; Matrix Science, London, United Kingdom). The search was conducted with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 20 ppm. The iodoacetamide derivative of cysteine was specified in Mascot as a fixed modification. Methylation of arginine, oxidation of methionine, dimethylation of arginine, and phosphorylation of serine, threonine, and tyrosine were specified in Mascot as variable modifications.
Criteria for protein identification.
Scaffold (version 4.0.7; Proteome Software Inc., Portland, OR) was used to validate MS-MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at a >80.0% probability by the PeptideProphet algorithm (45). Protein identifications were accepted if they could be established at a >99.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the ProteinProphet algorithm (46). Proteins that contained similar peptides and could not be differentiated on the basis of MS-MS analysis alone were grouped to satisfy the principles of parsimony.
Cell proliferation assay.
U2OS cells stably expressing pRb (WT) or pRb (R3F) and U2OS control cells were seeded at 2.5 × 104 per well onto 12-well plates. Cell growth was monitored daily for 4 days by addition of the MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] reagent followed by solubilization buffer according to the manufacturer's protocol (Roche Cell Proliferation kit). The plates were analyzed by measuring the optical density of cells at 570 nm with the background absorbance at 630 nm subtracted.
Luciferase assay.
HEK293T cells were plated onto 12-well plates at 1.5 × 105/well. The cells were transfected with an expression plasmid encoding the GAL4 DNA binding domain fused to the E2F-1 transactivation domain (residue 190 to the C terminus of the protein) and with a pRb (WT), pRb (R3F), or pRb (R3K) expression vector, along with a GAL4 DNA binding site promoter luciferase reporter plasmid (pFR [Promega]) in triplicate. In this reporter system, the E2F1 transactivation domain is recruited to the promoter through GAL4 DNA binding and activates luciferase expression. pRb (WT)-, pRb (R3F)-, and pRb (R3K)-mediated transcriptional repression was then monitored. Cell lysates were prepared with 1× passive lysis buffer (Promega) 48 h after transfection. Luciferase assays were performed according to the manufacturer's protocol by using a Lumat LB 9501 luminometer (Wallac Inc., CA). At least three independent determinations were performed at each setting.
RESULTS
PRMT4 methylates the pRb C terminus in vitro.
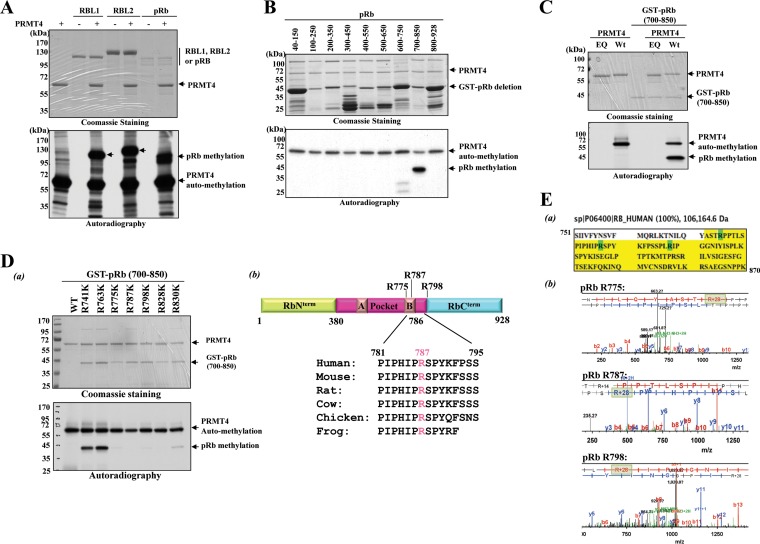
After the discovery of p53 lysine methylation (47), a myriad of nonhistone proteins were identified as substrates for lysine and arginine methyltransferases. In searching for other target proteins, our lab has generated purified baculovirus-derived methyltransferases consisting of 28 lysine methyltransferases (see Fig. S1 in the supplemental material) (data not shown) and 8 out of 9 putative human arginine methyltransferases (48). With the recombinant proteins in hand, we screened our protein collections of cellular transcription regulators, including Stat1, Stat3, RBP-Jκ, NF-κB family proteins, nuclear hormone receptors, and the pRb protein family, to see whether any one of them could serve as a substrate (data not shown). In our search, we observed that PRMT4 could methylate all three retinoblastoma protein (pRb) family members, as shown by the methylated higher-molecular-weight bands corresponding to pRb's in the autoradiogram in Fig. 1A. Of note, PRMT4 automethylation was detected at ∼60 kDa. In order to deduce the region of methylation, a series of overlapping GST-pRb deletion mutants, each mutant consisting of only those 150 amino acids, was generated in E. coli and was subjected to PRMT4 methylation in vitro by incubation with the baculovirus-purified PRMT4 enzyme along with radiolabeled S-adenosylmethionine (SAM) as the methyl donor. Methylation was then analyzed by autoradiography after SDS-PAGE analysis. The results revealed that PRMT4 methylated pRb within the C terminus, encompassing amino acid residues 700 to 850 (Fig. 1B). To confirm that PRMT4 was responsible for this methylation, the GST-pRb (700–850) fusion protein was incubated either alone, with PRMT4 (WT), or with PRMT4 (EQ), a catalytically inactive mutant (49–51). Methylation reactions with PRMT4 (EQ) or PRMT4 (WT) alone confirm that PRMT4 (EQ) is indeed inactive and that PRMT4 (WT) is highly active (Fig. 1C, 1st and 2nd lanes, respectively). Only in the presence of PRMT4 (WT) was the GST-pRb (700–850) fusion protein methylated; GST-pRb (700–850) alone or with PRMT4 (EQ) was not (Fig. 1C, 3rd and 4th lanes, respectively). These results demonstrate that PRMT4 indeed methylates pRb Cterm in vitro and that the arginine(s) responsible is located between amino acid residues 700 and 850.
FIG 1.
PRMT4 methylates pRb on the C-terminal domain in vitro. (A) In vitro methylation reactions of purified full-length pRb family proteins (0.5 μg to 2 μg) incubated with S-adenosyl-l-[methyl-3H]methionine as a cosubstrate in the absence or presence of 1 μg PRMT4 (WT). Methylation was detected by autoradiography (bottom), and protein loading was detected by CBB staining after resolution by 8% PAGE (top). (B) (Bottom) Autoradiogram of PRMT4 (WT)-dependent in vitro methylation assay on E. coli-derived overlapping recombinant GST-fused pRb deletion mutants. (Top) CBB staining was carried out as a loading control. (C) In vitro methylation assays with enzymatically inactive PRMT4 (EQ) alone (1st lane), PRMT4 (WT) alone (2nd lane), and the GST-pRb (700–850) fusion protein either alone (3rd lane) or in the presence of PRMT4 (EQ) (4th lane) or PRMT4 (WT) (5th lane). Reactions were stopped in SDS sample buffer, and proteins were separated by 10% PAGE, followed by autoradiography for the detection of methylation (bottom) and CBB staining for a loading control (top). (D) (a) Arginine (R)-to-lysine (K) point mutations were introduced for each R residue within the GST-pRb (700–850) fusion protein, and the resulting point mutants were subjected to in vitro methylation assays. Methylation was detected by autoradiography (bottom) after separation by 10% SDS-PAGE. CBB staining was applied for a protein loading control (top). (b) (Top) Schematic representation of full-length pRb and the relative positioning of the arginine methylation sites. (Bottom) Conservation of the R787 amino acid residue in various species as indicated. (E) (a) The GST-pRb (751–870) fusion protein was subjected to an in vitro methylation assay and was resolved by 10% SDS-PAGE. After CBB staining, the band corresponding to the GST-pRb fusion protein was cut out and was analyzed by liquid chromatography-mass spectrometry at the UC Davis Proteomics Core Facility. The protein sequence covered in the analysis is highlighted in yellow. Dimethyl-modified residues are highlighted in green. (b) In vitro methylation reaction of the pRb (700–850) fusion protein, PRMT4 (WT), and cold SAM followed by LC-MS-MS analysis. Only the spectra from digested peptides harboring the methylated R residues identified are included in this figure. Increases of 28 Da in the mass-to-charge ratio are highlighted in light green.
PRMT4 methylates pRb at R775, R787, and R795 in vitro.
In order to identify the methylation site(s) within pRb Cterm, each arginine (R) residue in the GST-pRb (700–850) fusion protein was mutated to a lysine (K) residue, and the mutants were subjected to PRMT4 methylation reactions in vitro. As shown from the autoradiogram in Fig. 1Da, the R775K, R798K, R828K, and R830K point mutants displayed very weak methylation, while R787K mutation completely abolished methylation by PRMT4, indicating that R787 was the major methylation site. Interestingly, R787 is conserved throughout pRb's from many species, suggesting that pRb methylation may have a conserved function in different species (Fig. 1Db). To verify these methylation sites, the GST-pRb (700–850) fusion protein was subjected to an in vitro methylation assay followed by liquid chromatography-mass spectrometry (LC-MS) analysis. While almost full coverage of the C-terminal protein sequence was obtained (highlighted in Fig. 1Ea), the spectra in Fig. 1Eb demonstrated a 28-Da increase in the mass-to-charge ratio on the R775, R787, and R798 residues but not on the R828 or R830 residue, confirming that dimethylation occurs only on residues R775, R787, and R798. We speculated that conformational changes induced by the mutations introduced at R828 and R830 may have contributed to the reduced methylation.
Verification of pRB methylation with peptides.
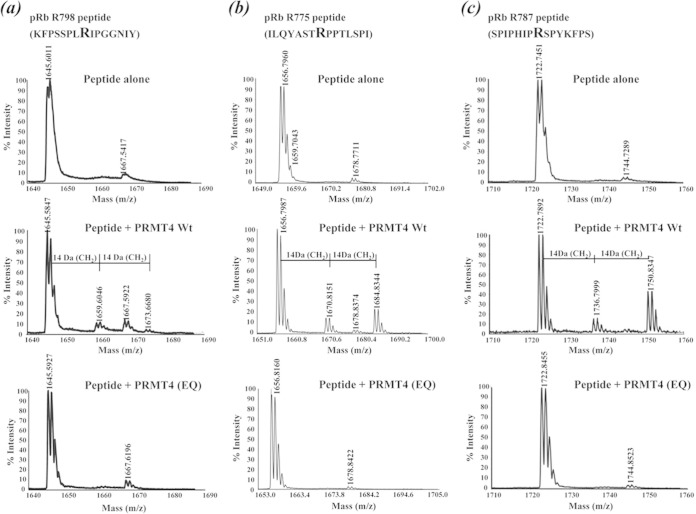
To further verify these methylation sites, a series of individual 15-mer synthetic peptides harboring the methylation sites identified were incubated either alone, with PRMT4 (WT), or with PRMT4 (EQ) for in vitro methylation reactions. The reactions were then analyzed by MALDI-TOF mass spectrometry. The spectra obtained from the peptide harboring the R798 residue showed peaks with 14-Da (monomethyl intermediate) and 28-Da (dimethylation) increases in the mass-to-charge ratio when the peptide was incubated with PRMT4 (WT) but not when it was incubated alone or in the presence of PRMT4 (EQ) (Fig. 2). Similar results were obtained for the peptides harboring the R775 or R787 residue (Fig. 2b and c). These results show the type I methyltransferase enzymatic activity of PRMT4 (33) and further confirm that monomethylation and dimethylation occur on R775, R787, and R798 in vitro.
FIG 2.
Verification of pRB methylation with peptide substrates. Spectra are derived from MALDI-TOF mass spectrometry analysis of 15-mer synthetic peptides harboring the R798 (a), R775 (b), and R787 (c) residues after in vitro methylation reactions with the peptide alone, the peptide plus PRMT4 (WT), or the peptide plus PRMT4 (EQ).
PRMT4 methylates pRb R787 in vivo.
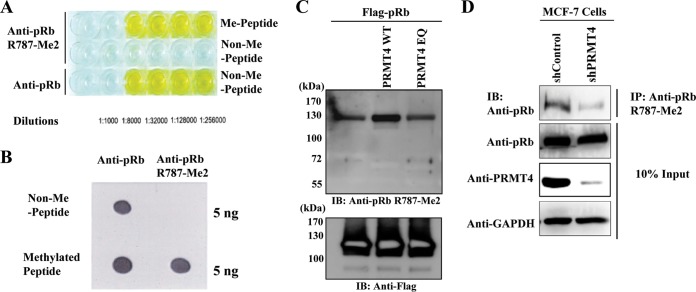
To assess whether pRb is methylated in vivo, we first raised an affinity-purified rabbit polyclonal antibody against a 15-mer asymmetric arginine 787-dimethyl (R787-Me2) peptide. As shown by the results of the enzyme-linked immunosorbent assay (ELISA) and the dot blot assay in Fig. 3A and B, respectively, the antibody specifically recognized the R787-Me2 peptide but not the nonmethylated peptide. To determine whether a purified antibody recognizes full-length methylated pRb, immunoblot analysis was performed with purified pRb after in vitro methylation reactions with either PRMT4 (WT) or PRMT4 (EQ). With an anti-pRb R787-Me2 polyclonal antibody, detection of pRb methylation was much more robust when pRb was incubated with PRMT4 (WT) than when it was incubated alone or with PRMT4 (EQ) (Fig. 3C). Finally, knockdown of PRMT4 by transient transfection of short hairpin RNA targeting endogenous PRMT4 in MCF-7 human breast adenocarcinoma cells reduced levels of methylated pRb by immunoprecipitation with an anti-pRb R787-Me2 antibody and immunoblotting with an anti-pRb antibody. Equal expression of pRb was also confirmed by immunoblotting (Fig. 3D). Together, these results demonstrate that endogenous pRb is dimethylated at residue R787 by PRMT4 in vivo.
FIG 3.
PRMT4 methylates pRb R787 in vivo. (A and B) ELISA analysis (A) and dot blot analysis (B) of an affinity-purified polyclonal antibody raised against a 15-mer pRb R787-Me2 peptide on a nonmethylated peptide (non-Me) versus a pRb arginine-dimethylated peptide (Me). (C) In vitro methylation reaction mixtures consisting of Flag-tagged full-length pRb, PRMT4 (WT) or PRMT4 (EQ), and cold SAM were assembled. pRb methylation was detected using an anti-pRb R787-Me2 polyclonal antibody. Input pRB is shown by anti-Flag immunoblotting (IB). (D) MCF-7 cells were transfected with shRNA vectors targeting either GFP, as a negative control, or PRMT4, to achieve endogenous PRMT4 knockdown. Cell lysates were immunoprecipitated with an anti-pRb R787-Me2 polyclonal antibody and were immunoblotted with an anti-pRb antibody. As a control, 10% input is included.
PRMT4 interacts with and methylates pRb in vivo.
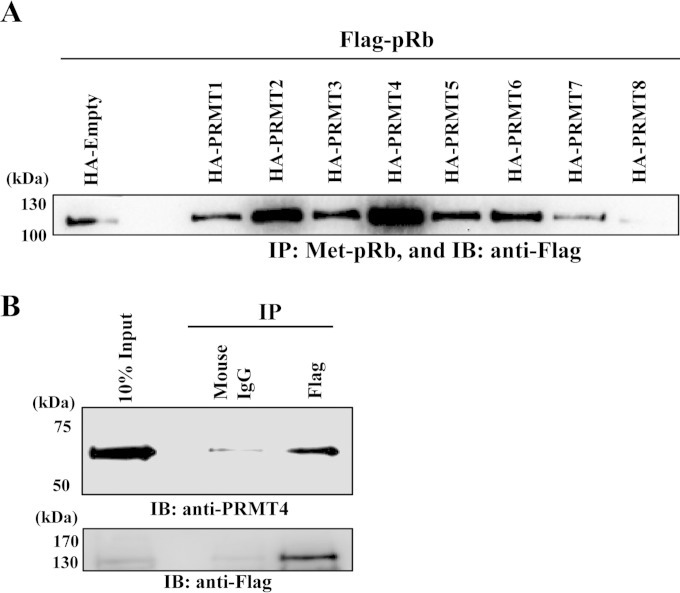
With a methyl-specific antibody in hand, we examined if another arginine methylase also targets pRb R787 for methylation in vivo. The pRb expression plasmid was cotransfected with PRMT expression vectors, and methyl pRb was enriched by co-IP and was probed with an anti-Rb antibody. The results showed that in addition to PRMT4, PRMT2 increased signal intensity, indicating that PRMT2 may also target the same residue of pRb for methylation (Fig. 4A), although in vitro methylation reactions with purified PRMT2 failed to demonstrate pRb methylation (data not shown). Importantly, both PRMT2 and PRMT4 have been demonstrated to interact with pRb (52) (Fig. 4B), attesting to the specificity of the methylation.
FIG 4.
PRMT4 interacts with pRb and is primarily responsible for pRb arginine methylation in vivo. (A) HEK293T cells were cotransfected with an HA-tagged empty vector or individual HA-tagged PRMTs with Flag-pRb expression plasmids. Five hundred micrograms of each cell lysate was immunoprecipitated (IP) with 3 μg of anti-pRb R787-Me2 overnight, and methylated Flag-pRb was detected using an anti-pRb monoclonal antibody. (B) Immunoprecipitation with either anti-mouse IgG or anti-Flag antibody M2 from cell lysates prepared from isogenic U2OS cells expressing Flag-pRb (WT). Interaction with PRMT4 was detected using an anti-PRMT4 antibody.
A pRb methylation-defective mutant decreases pRb phosphorylation.
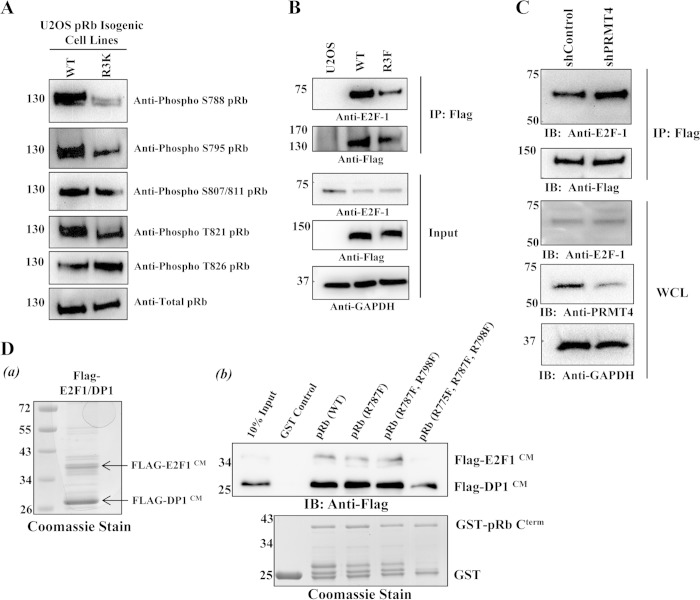
Because the growth-inhibitory function of pRb is governed largely by Cdk phosphorylation (5), and because the arginine methylation sites that we mapped are located adjacent to pRb Cterm Cdk phosphorylation sites, we sought to assess whether methylation can influence pRb Cterm phosphorylation, which has been the case for many arginine-methylated proteins (53–55). To test this, pRb Cterm phosphorylation was examined in U2OS osteosarcoma cell lines stably expressing pRb (WT) or the methylation-defective mutant pRb (R3K) by immunoblot analysis using the pRb phosphorylation-specific antibodies indicated in Fig. 5A. U2OS cells were used because this cell line has been widely used for Rb studies (56). The results showed that cells expressing pRb (R3K) had reduced phosphorylation on residues S788, S795, and T821 but not on S807/S811 or T826. These results indicate that arginine methylation may play a role in efficient pRb Cterm phosphorylation and thus in cell proliferation.
FIG 5.
pRb arginine methylation decreases E2F-1 binding in vivo and in vitro. (A) Immunoblot (IB) analysis was performed on total-cell lysates from isogenic U2OS cells expressing Flag-pRb (WT) or Flag-pRb (R3K) by using the indicated pRb phosphorylation-specific antibodies. (B) U2OS control cells or isogenic cell lines expressing either Flag-pRb (WT) or Flag-pRb (R3F) were lysed in phospholysis buffer; whole-cell lysates were immunoprecipitated (IP) with anti-Flag–M2 agarose beads overnight; and immunoprecipitates were probed for interactions with an anti-E2F-1 antibody. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (C) The U2OS cell line stably expressing pRb (WT) was transfected with an shControl or shPRMT4 vector. Forty-eight hours later, whole-cell lysates (WCL) were immunoprecipitated with anti-Flag–M2 agarose beads overnight, and immunoprecipitates were probed for interactions with an anti-E2F-1 antibody. (D) (a) CBB staining of baculovirus-derived Flag-tagged E2F-1/DP1 used for in vitro GST pulldown assays. (b) GST pulldowns were performed by incubation of baculovirus-derived Flag-tagged E2F-1/DP1 coiled-coil and marked box domains (labeled CM) with the immobilized wild-type GST-pRb (700–850) deletion protein or the GST-pRb (700–850) deletion protein harboring either single or combinatorial phenylalanine substitutions. Proteins were probed for interactions by immunoblot analysis using an anti-Flag antibody.
pRb arginine methylation decreases E2F-1 binding in vivo and in vitro.
There are at least three E2F-1/DP1 binding sites on pRb. One of them is reported as participating in a bipartite interaction between the pRb Cterm and E2F-1/DP1 coiled-coil/marked box (CM) domains. pRb phosphorylation at S788 and S795 essentially eliminates this bipartite interaction (9). Since S788 and S795 pRb Cterm phosphorylation was reduced on cells expressing the methylation-defective mutant pRb (R3K), we tested the idea that methylation may be important for E2F-1 dissociation by coimmunoprecipitation assays. Indeed, immunoprecipitation of Flag-pRb followed by immunoblot analysis using an anti-E2F-1 antibody revealed that precipitated Flag-pRb (R3F) bound to E2F-1 to a much lower extent than precipitated Flag-pRb (WT) (Fig. 5B). As a negative control, U2OS cells that did not express exogenous Flag-pRb were run in parallel and did not coprecipitate E2F-1, indicating the specificity of the immunoprecipitations. Furthermore, transient knockdown of endogenous PRMT4 in the isogenic U2OS cell line stably expressing pRb (WT) increased E2F-1 binding over that for cells transiently transfected with the shControl vector (Fig. 5C). To confirm this observation and to examine whether arginine methylation was directly or indirectly (through Cdk phosphorylation) responsible for the dissociation of E2F-1, an in vitro GST pulldown assay was employed to test the direct effects of methylation on the pRb–E2F-1/DP1 interaction. We generated recombinant baculoviruses expressing the Flag-tagged E2F-1 or DP1 coiled-coil and marked box (CM) domain [E2F-1 (CM) or DP1 (CM)], and recombinant proteins were copurified from Sf9 insect cells after coinfection. The resulting purified Flag-E2F-1/DP1 (CM) fusion proteins (Fig. 5Da) were incubated with the wild-type GST-pRb (700–850) fusion protein or with single or combinatorial R-to-F mutants derived from pRb (700–850) as indicated in Fig. 5Db. The results showed that GST-pRb (700–850) harboring R775F, R787F, and R795F substitutions bound to Flag-E2F-1/DP1 (CM) to a much lesser extent than wild-type GST-pRb (700–850) or other GST-pRb mutants. Collectively, these results demonstrate that methylation of all three arginine residues identified may decrease E2F-1 binding in vitro and in vivo and that the dissociation can be independent of pRb phosphorylation.
Arginine methylation inhibits the tumor suppressor function of pRb.
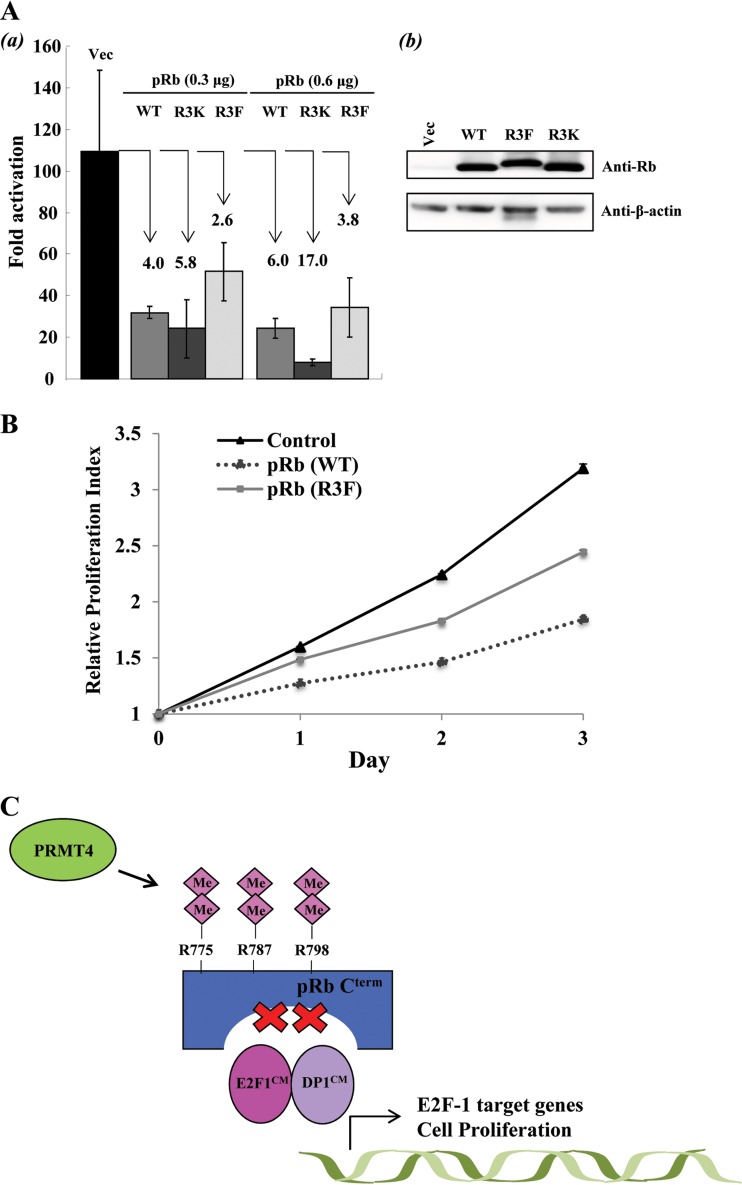
As mentioned above, phosphorylation and subsequent E2F-1 dissociation allow for the transcriptional activation of S-phase regulatory genes by E2F-1. This prompted us to investigate the effects of methylation on transcriptional activation by E2F-1. A luciferase reporter assay was conducted using a luciferase reporter plasmid containing three Gal4 DNA binding sites upstream of a minimal TATA box along with a Gal4-E2F-1 plasmid consisting of the E2F-1 transactivation domain and pRb binding sites. The plasmids were cotransfected into U2OS cells with either an empty vector or a pRb (WT), pRb (R3K), or pRb (R3F) mammalian expression plasmid (Fig. 6Ab). From the luciferase assay, cotransfection with increasing amounts (0.3 μg and 0.6 μg) of the pRb (WT) expression plasmid repressed E2F-1 transcription 4- and 6-fold, respectively, relative to that with an empty vector (Fig. 6Aa). With the pRb (R3K) expression plasmid, 5- and 17-fold repression was observed. However, the pRb (R3F) mutant significantly impaired the pRb transrepression function, repressing transcription 3- to 4-fold, in agreement with the results of GST pulldown analyses. These results strongly suggest that methylation decreases the transrepression of E2F-1 transactivation by pRb.
FIG 6.
Arginine methylation inhibits the tumor suppressor function of pRb. (A) (a) Luciferase reporter assays were employed to measure E2F-1 transcriptional activation by cotransfecting HEK293T cells with increasing amounts of DNA plasmids (0.3 μg and 0.6 μg) encoding Flag-pRb (WT), Flag-pRb (R3K), or 3×Flag-pRb (R3F) with expression plasmids encoding the GAL4 DNA binding domain fused to the E2F-1 transactivation domain (residue 190 to the C terminus) and a GAL4 DNA binding site promoter luciferase reporter plasmid. Luciferase activity was measured in arbitrary units by using a Lumat LB 9501 luminometer. The data are averages for triplicate wells. Error bars represent standard deviations (n = 3). The degree of repression is shown for each Rb construct. (b) Immunoblot analysis showing transient expression levels of pRb (WT) and mutants. (B) Cell proliferation studies were performed on U2OS control cells and isogenic U2OS cells expressing either pRb (WT) or pRb (R3F) by using the MTT colorimetric assay. Error bars represent standard deviations (n = 3). The relative proliferation index for each day was determined by measuring the optical density of the cells at 570 nm with a reference filter of 630 nm and using the absorbance from day zero as a standard. (C) Possible model for PRMT4-induced cell cycle progression. During the G1-to-S-phase transition, PRMT4-mediated arginine dimethylation of pRb may facilitate efficient Cdk-dependent phosphorylation of pRb Cterm. Arginine dimethylation also inactivates the tumor suppressor activity of pRb independently of phosphorylation, resulting in E2F-1 dissociation and activation of E2F-1 cell cycle target genes. Me, methylation.
Finally, we examined the effects of the pRb (R3F) mutation on cell growth in pRb-proficient cell lines. MTT assays were performed on U2OS cells and isogenic U2OS cell lines stably expressing either pRb (WT) or the methylmimetic pRb (R3F) mutant, in which pRb methylation sites R775, R787, and R798 were converted to phenylalanine (F). As shown in Fig. 6B, stable isogenic expression of pRb (WT) significantly reduced cell proliferation from that of control U2OS cells, an effect that was expected in view of the tumor suppressor activity of pRb. On the other hand, isogenic cells stably expressing pRb (R3F) displayed higher levels of proliferation than cells expressing pRb (WT), indicating that arginine methylation may impair the cell growth-inhibitory function of pRb (Fig. 6C).
DISCUSSION
Much like other PTMs, lysine methylation and arginine methylation are beginning to be recognized as signal transducers. As such, methylation regulates various cellular processes, including protein-protein interactions, RNA processing, transportation, protein turnover, and protein localization (33). Although recent advances in spectrometric and in silico analyses allow us to identify naturally methylated protein in cells, there are limited techniques for identifying the enzyme(s) responsible (57). One of the methods for identifying methylated substrates and the responsible methylase is to reconstitute in vitro methylation reactions (58). We have cloned and purified a total of 31 lysine and arginine methyltransferases and investigated whether these enzymes can methylate proteins of interest. We are interested in pRb family proteins, because they are critical tumor suppressors and multifunctional proteins much like histones.
In this study, we searched for transcription regulators that can serve as substrates for lysine and arginine methylation in order to expand our knowledge in the growing field of protein methylation. From our screen, we found that PRMT4 methylates pRb Cterm on R775, R787, and R795 in vitro and on R787 in vivo. These findings constitute the first evidence of methylation on pRb arginine residues and establish an inhibitory role for pRb arginine methylation, in part by permitting efficient Cdk-dependent phosphorylation of pRb Cterm on S788, S795, and T821. Independently of phosphorylation, pRb arginine methylation also leads to the dissociation of E2F-1 and promotes transcriptional activation by E2F-1 in dividing cells.
pRb lysine methylation on residues K810 and K873 by Set7/9, and on K860 by Smyd2, has been demonstrated to maintain the cell cycle-inhibitory function of pRb either by inhibiting Cdk binding and subsequent phosphorylation or by providing a docking site/increasing the interaction with transcription repressor-associated proteins such as HP-1 and L3MBTL1 (28, 29, 31). On the other hand, methylation of pRb residue K810 by Smyd2 enhances pRb S807/811 phosphorylation, increasing the transcriptional activity of E2F and promoting cell cycle progression (60). Markham et al. have demonstrated that acetylation on K873/K874 leads to the release of E2F-1 from pRb under DNA-damaging conditions (24). All these studies support our evidence that PTMs other than the conventional pRb phosphorylation can dictate pRb tumor suppressor function through mechanisms of cross talk between PTMs and modulation of protein-protein interactions with pRb-associated proteins.
Our screening with a methyl-specific antibody indicated that PRMT2 might be able to target pRb for methylation, although methylation reactions with purified PRMT2 could not demonstrate pRb methylation. We speculated that a cofactor(s) may be required for PRMT2 to be activated. Importantly, we and others have demonstrated that both PRMT2 and PRMT4 interact physically with pRb (52; this study). In the study of PRMT2, Yoshimoto et al. found that PRMT2 interacts physically with pRb via its S-adenosylmethionine binding domain and inhibits E2F transcriptional activity and entry into S phase (52). Although the authors did not test the possibility that E2F repression is mediated through PRMT2-targeted pRb arginine methylation, it would be interesting to test if PRMT2-mediated pRb arginine methylation or the physical interaction counteracts PRMT4 function. This type of mechanism has certainly been observed in the work of Zheng et al. demonstrating that E2F-1 arginine methylation by PRMT1 antagonizes PRMT5-mediated E2F-1 methylation, and vice versa (61). The study showed that methylation on residue R109 by PRMT1 inhibits cell growth and promotes apoptosis, while methylation on residues R111 and R113 favors cell proliferation after DNA-damaging events (61).
Our series of biochemical and overexpression studies demonstrated that PRMT4 negatively regulates the tumor suppressor function of pRb. It will be important to identify which pRb target genes are subjected to PRMT4-utilized regulation.
PRMT4 overexpression and aberrant expression have been reported to be key regulators in promoting cancer cell proliferation (62). In breast cancer, the proposed mechanisms that implicate PRMT4 as a driving force for cell proliferation include the upregulation of cell cycle genes such as E2F1 and cyclin E genes through histone H3R17 dimethylation, arginine methylation of the BAF155 protein (core subunit of the SWI/SNF chromatin-remodeling complex), protein methylation of the oncogenic coactivator ACTR, and coactivation of breast cancer-related transcription factors (e.g., ER-α) (39–41, 62, 63). In addition, two independent studies have demonstrated that PRMT4 methyltransferase activity is governed by phosphorylation on conserved S228 and S217 residues in mitotic cells, further implicating PRMT4 as a critical regulator of cell cycle progression (64, 65). Thus, our studies on PRMT4-targeted pRb arginine methylation may provide another possible mechanism for the oncogenic effects of PRMT4 in cancer cell proliferation, in part through the negative regulation of the tumor suppressor protein pRb.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (CA14779, to Y.I.) and by an American Cancer Society Research Scholar grant (RSG-13-383-01-MPC) to Y.I. This work was also partially supported by a grant from the Department of Defense (W81XWH1110575, to Y.I.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00945-14.
REFERENCES
- 1.Weinberg RA. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 2.Massagué J. 2004. G1 cell-cycle control and cancer. Nature 432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 3.Lees JA, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K. 1993. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol 13:7813–7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shan B, Zhu X, Chen PL, Durfee T, Yang Y, Sharp D, Lee WH. 1992. Molecular cloning of cellular genes encoding retinoblastoma-associated proteins: identification of a gene with properties of the transcription factor E2F. Mol Cell Biol 12:5620–5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graña X, Garriga J, Mayol X. 1998. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene 17:3365–3383. [DOI] [PubMed] [Google Scholar]
- 6.DeCaprio JA, Ludlow JW, Lynch D, Furukawa Y, Griffin J, Piwnica-Worms H, Huang CM, Livingston DM. 1989. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell 58:1085–1095. doi: 10.1016/0092-8674(89)90507-2. [DOI] [PubMed] [Google Scholar]
- 7.Helin K, Harlow E, Fattaey A. 1993. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol 13:6501–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiebert SW. 1993. Regions of the retinoblastoma gene product required for its interaction with the E2F transcription factor are necessary for E2 promoter repression and pRb-mediated growth suppression. Mol Cell Biol 13:3384–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubin SM, Gall AL, Zheng N, Pavletich NP. 2005. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release. Cell 123:1093–1106. doi: 10.1016/j.cell.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 10.Lee C, Chang JH, Lee HS, Cho Y. 2002. Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor. Genes Dev 16:3199–3212. doi: 10.1101/gad.1046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchkovich K, Duffy LA, Harlow E. 1989. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell 58:1097–1105. doi: 10.1016/0092-8674(89)90508-4. [DOI] [PubMed] [Google Scholar]
- 12.Lundberg AS, Weinberg RA. 1998. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol 18:753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown VD, Phillips RA, Gallie BL. 1999. Cumulative effect of phosphorylation of pRB on regulation of E2F activity. Mol Cell Biol 19:3246–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittnacht S. 1998. Control of pRB phosphorylation. Curr Opin Genet Dev 8:21–27. doi: 10.1016/S0959-437X(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 15.Burke JR, Deshong AJ, Pelton JG, Rubin SM. 2010. Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J Biol Chem 285:16286–16293. doi: 10.1074/jbc.M110.108167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burke JR, Hura GL, Rubin SM. 2012. Structures of inactive retinoblastoma protein reveal multiple mechanisms for cell cycle control. Genes Dev 26:1156–1166. doi: 10.1101/gad.189837.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeGregori J, Kowalik T, Nevins JR. 1995. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol 15:4215–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin XQ, Chittenden T, Livingston DM, Kaelin WG Jr. 1992. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev 6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 19.Hiebert SW, Chellappan SP, Horowitz JM, Nevins JR. 1992. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev 6:177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- 20.Knudsen ES, Wang JY. 1996. Differential regulation of retinoblastoma protein function by specific Cdk phosphorylation sites. J Biol Chem 271:8313–8320. doi: 10.1074/jbc.271.14.8313. [DOI] [PubMed] [Google Scholar]
- 21.Burke JR, Liban TJ, Restrepo T, Lee HW, Rubin SM. 2014. Multiple mechanisms for E2F binding inhibition by phosphorylation of the retinoblastoma protein C-terminal domain. J Mol Biol 426:245–255. doi: 10.1016/j.jmb.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen DX, Baglia LA, Huang SM, Baker CM, McCance DJ. 2004. Acetylation regulates the differentiation-specific functions of the retinoblastoma protein. EMBO J 23:1609–1618. doi: 10.1038/sj.emboj.7600176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan HM, Krstic-Demonacos M, Smith L, Demonacos C, La Thangue NB. 2001. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat Cell Biol 3:667–674. doi: 10.1038/35083062. [DOI] [PubMed] [Google Scholar]
- 24.Markham D, Munro S, Soloway J, O'Connor DP, La Thangue NB. 2006. DNA-damage-responsive acetylation of pRb regulates binding to E2F-1. EMBO Rep 7:192–198. doi: 10.1038/sj.embor.7400591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ledl A, Schmidt D, Muller S. 2005. Viral oncoproteins E1A and E7 and cellular LxCxE proteins repress SUMO modification of the retinoblastoma tumor suppressor. Oncogene 24:3810–3818. doi: 10.1038/sj.onc.1208539. [DOI] [PubMed] [Google Scholar]
- 26.Kalejta RF, Shenk T. 2003. Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc Natl Acad Sci U S A 100:3263–3268. doi: 10.1073/pnas.0538058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ying H, Xiao ZX. 2006. Targeting retinoblastoma protein for degradation by proteasomes. Cell Cycle 5:506–508. doi: 10.4161/cc.5.5.2515. [DOI] [PubMed] [Google Scholar]
- 28.Carr SM, Munro S, Kessler B, Oppermann U, La Thangue NB. 2011. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. EMBO J 30:317–327. doi: 10.1038/emboj.2010.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munro S, Khaire N, Inche A, Carr S, La Thangue NB. 2010. Lysine methylation regulates the pRb tumour suppressor protein. Oncogene 29:2357–2367. doi: 10.1038/onc.2009.511. [DOI] [PubMed] [Google Scholar]
- 30.Macdonald JI, Dick FA. 2012. Posttranslational modifications of the retinoblastoma tumor suppressor protein as determinants of function. Genes Cancer 3:619–633. doi: 10.1177/1947601912473305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saddic LA, West LE, Aslanian A, Yates JR III, Rubin SM, Gozani O, Sage J. 2010. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem 285:37733–37740. doi: 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Roberts CW. 2014. CARMA: CARM1 methylation of SWI/SNF in breast cancer. Cancer Cell 25:3–4. doi: 10.1016/j.ccr.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bedford MT, Clarke SG. 2009. Protein arginine methylation in mammals: who, what, and why. Mol Cell 33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 35.Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. 2002. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep 3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Covic M, Hassa PO, Saccani S, Buerki C, Meier NI, Lombardi C, Imhof R, Bedford MT, Natoli G, Hottiger MO. 2005. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-κB-dependent gene expression. EMBO J 24:85–96. doi: 10.1038/sj.emboj.7600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fauquier L, Duboe C, Jore C, Trouche D, Vandel L. 2008. Dual role of the arginine methyltransferase CARM1 in the regulation of c-Fos target genes. FASEB J 22:3337–3347. doi: 10.1096/fj.07-104604. [DOI] [PubMed] [Google Scholar]
- 38.An W, Kim J, Roeder RG. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 39.El Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D, Theillet C, Vandel L, Bedford MT, Sardet C. 2006. Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the cyclin E1 gene. Proc Natl Acad Sci U S A 103:13351–13356. doi: 10.1073/pnas.0605692103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frietze S, Lupien M, Silver PA, Brown M. 2008. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res 68:301–306. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- 41.Wang L, Zhao Z, Meyer MB, Saha S, Yu M, Guo A, Wisinski KB, Huang W, Cai W, Pike JW, Yuan M, Ahlquist P, Xu W. 2014. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell 25:21–36. doi: 10.1016/j.ccr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dillon MB, Rust HL, Thompson PR, Mowen KA. 2013. Automethylation of protein arginine methyltransferase 8 (PRMT8) regulates activity by impeding S-adenosylmethionine sensitivity. J Biol Chem 288:27872–27880. doi: 10.1074/jbc.M113.491092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Izumiya Y, Ellison TJ, Yeh ET, Jung JU, Luciw PA, Kung HJ. 2005. Kaposi's sarcoma-associated herpesvirus K-bZIP represses gene transcription via SUMO modification. J Virol 79:9912–9925. doi: 10.1128/JVI.79.15.9912-9925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Izumiya Y, Izumiya C, Hsia D, Ellison TJ, Luciw PA, Kung HJ. 2009. NF-κB serves as a cellular sensor of Kaposi's sarcoma-associated herpesvirus latency and negatively regulates K-Rta by antagonizing the RBP-Jκ coactivator. J Virol 83:4435–4446. doi: 10.1128/JVI.01999-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 46.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 47.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D. 2004. Regulation of p53 activity through lysine methylation. Nature 432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 48.Campbell M, Chang PC, Huerta S, Izumiya C, Davis R, Tepper CG, Kim KY, Shevchenko B, Wang DH, Jung JU, Luciw PA, Kung HJ, Izumiya Y. 2012. Protein arginine methyltransferase 1-directed methylation of Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen. J Biol Chem 287:5806–5818. doi: 10.1074/jbc.M111.289496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR. 2002. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol Cell Biol 22:3621–3632. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parfitt DE, Zernicka-Goetz M. 2010. Epigenetic modification affecting expression of cell polarity and cell fate genes to regulate lineage specification in the early mouse embryo. Mol Biol Cell 21:2649–2660. doi: 10.1091/mbc.E10-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee YH, Bedford MT, Stallcup MR. 2011. Regulated recruitment of tumor suppressor BRCA1 to the p21 gene by coactivator methylation. Genes Dev 25:176–188. doi: 10.1101/gad.1975811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshimoto T, Boehm M, Olive M, Crook MF, San H, Langenickel T, Nabel EG. 2006. The arginine methyltransferase PRMT2 binds RB and regulates E2F function. Exp Cell Res 312:2040–2053. doi: 10.1016/j.yexcr.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 53.Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, Mukai H, Kasuya Y, Fukamizu A. 2008. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell 32:221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 54.Kowenz-Leutz E, Pless O, Dittmar G, Knoblich M, Leutz A. 2010. Crosstalk between C/EBPβ phosphorylation, arginine methylation, and SWI/SNF/Mediator implies an indexing transcription factor code. EMBO J 29:1105–1115. doi: 10.1038/emboj.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arita K, Isogai S, Oda T, Unoki M, Sugita K, Sekiyama N, Kuwata K, Hamamoto R, Tochio H, Sato M, Ariyoshi M, Shirakawa M. 2012. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc Natl Acad Sci U S A 109:12950–12955. doi: 10.1073/pnas.1203701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park YB, Park MJ, Kimura K, Shimizu K, Lee SH, Yokota J. 2002. Alterations in the INK4a/ARF locus and their effects on the growth of human osteosarcoma cell lines. Cancer Genet Cytogenet 133:105–111. doi: 10.1016/S0165-4608(01)00575-1. [DOI] [PubMed] [Google Scholar]
- 57.Bedford MT, Richard S. 2005. Arginine methylation an emerging regulator of protein function. Mol Cell 18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Levy D, Liu CL, Yang Z, Newman AM, Alizadeh AA, Utz PJ, Gozani O. 2011. A proteomic approach for the identification of novel lysine methyltransferase substrates. Epigenetics Chromatin 4:19. doi: 10.1186/1756-8935-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reference deleted.
- 60.Cho HS, Hayami S, Toyokawa G, Maejima K, Yamane Y, Suzuki T, Dohmae N, Kogure M, Kang D, Neal DE, Ponder BA, Yamaue H, Nakamura Y, Hamamoto R. 2012. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia 14:476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng S, Moehlenbrink J, Lu YC, Zalmas LP, Sagum CA, Carr S, McGouran JF, Alexander L, Fedorov O, Munro S, Kessler B, Bedford MT, Yu Q, La Thangue NB. 2013. Arginine methylation-dependent reader-writer interplay governs growth control by E2F-1. Mol Cell 52:37–51. doi: 10.1016/j.molcel.2013.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Y, Bedford MT. 2013. Protein arginine methyltransferases and cancer. Nat Rev Cancer 13:37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 63.Streubel G, Bouchard C, Berberich H, Zeller MS, Teichmann S, Adamkiewicz J, Muller R, Klempnauer KH, Bauer UM. 2013. PRMT4 is a novel coactivator of c-Myb-dependent transcription in haematopoietic cell lines. PLoS Genet 9:e1003343. doi: 10.1371/journal.pgen.1003343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng Q, He B, Jung SY, Song Y, Qin J, Tsai SY, Tsai MJ, O'Malley BW. 2009. Biochemical control of CARM1 enzymatic activity by phosphorylation. J Biol Chem 284:36167–36174. doi: 10.1074/jbc.M109.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Higashimoto K, Kuhn P, Desai D, Cheng X, Xu W. 2007. Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proc Natl Acad Sci U S A 104:12318–12323. doi: 10.1073/pnas.0610792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.