Abstract
A new model of kinase regulation based on the assembly of hydrophobic spines has been proposed. Changes in their positions can explain the mechanism of kinase activation. Here, we examined mutations in human cancer for clues about the regulation of the hydrophobic spines by focusing initially on mutations to Phe. We identified a selected number of Phe mutations in a small group of kinases that included BRAF, ABL1, and the epidermal growth factor receptor. Testing some of these mutations in BRAF, we found that one of the mutations impaired ATP binding and catalytic activity but promoted noncatalytic allosteric functions. Other Phe mutations functioned to promote constitutive catalytic activity. One of these mutations revealed a previously underappreciated hydrophobic surface that functions to position the dynamic regulatory αC-helix. This supports the key role of the C-helix as a signal integration motif for coordinating multiple elements of the kinase to create an active conformation. The importance of the hydrophobic space around the αC-helix was further tested by studying a V600F mutant, which was constitutively active in the absence of the negative charge that is associated with the common V600E mutation. Many hydrophobic mutations strategically localized along the C-helix can thus drive kinase activation.
INTRODUCTION
Protein kinases, whose genes represent one of the largest gene families (1), have evolved to be dynamic molecular switches that regulate most biological processes (2). Typically in a basal inactive state, they are dynamically assembled into an active conformation by a complex set of regulatory events that can include recruitment to membranes, dimerization, phosphorylation, and/or translocation to the nucleus. Because protein kinases are associated with so many diseases, especially cancers, we have a large collection of structures, which allows us to explore their dynamic properties at the molecular level.
We previously identified two highly conserved structural entities known as the hydrophobic spines that are common to all kinases (2, 3). The first spine to be identified is referred to as the regulatory spine, or R-spine, and the assembled R-spine is a hallmark signature of every active kinase (3). The R-spine consists of four residues, RS1 to RS4, two from the C-lobe and two from the N-lobe. Each R-spine residue comes from a critical part of the kinase. RS1 is the histidine residue from the HRD motif in the catalytic loop. RS2 is the phenylalanine from the DFG motif in the activation segment. RS3 is a conserved aliphatic residue from the αC-helix, and RS4 is an aliphatic residue from the β4-strand. Assembly of the R-spine, which is typically mediated by a highly regulated set of events, results in the formation of the active conformation of the kinase.
The second spine, known as the catalytic spine, or C-spine, consists of a series of hydrophobic residues and is completed after ATP binds (2). Two of the conserved residues in the C-spine are from the N-lobe, and six are from the C-lobe. The binding of the adenine ring of ATP in its binding pocket brings the two parts of the C-spine together, allowing the two lobes of the kinase to close. Assembly of the R-spine defines activation, whereas assembly of the C-spine poises the kinase for catalysis.
Recently, we began to validate the functional importance of the spines as driving forces for the assembly of active kinases by using the RAF kinases (4). The RAFs are a family of kinases that include ARAF, BRAF, and CRAF (5, 6). Activation of RAF is initiated by activation of RAS, which recruits RAFs to the plasma membrane. Through a complex series of steps that involve both phosphorylation and dimerization, the RAF kinases are activated (7–9). We recently validated the role of assembly of the C-spine in helping to stabilize the active conformation by using a hydrophobic substitution of a C-spine residue (A70F) that we predicted would mimic ATP binding (4, 10). This change generated a conformation of the RAF C-spine that could constitutively dimerize and activate wild-type RAFs. This demonstrated that a catalytically inactive mutant can serve as an allosteric activator.
To mimic assembly of the R-spine, we used an aromatic substitution for the third residue of the R-spine (RS3F) in BRAF (4). This mutation resulted in a constitutively active kinase that did not require RAS, dimerization, or activation loop phosphorylation for its activity. Together, these data indicated that mutations that stabilize the closed and active or active-like conformation of BRAF by assembling the C- and/or R-spine can stimulate activity in cells via two different mechanisms. Mutations that lock the C-spine in the closed conformation are catalytically dead but can activate the kinase through allosteric mechanisms, while mutations that lock the R-spine in the active conformation result in a kinase with constitutive activity.
Since kinases are a common target for oncogenic mutations, we surveyed the sequenced cancer kinomes (http://www.sanger.ac.uk/genetics/CGP/cosmic/) for hydrophobic mutations that might function to influence the C-spine and R-spine in ways that might be similar to A70F and RS3F. To correlate the mutated sequence with structural motifs and secondary structure elements, we used the Protein Kinase Ontology (ProKinO) tool, which provides a conceptual representation of data, relating cancer mutations to conserved sequence and structural motifs associated with protein kinase regulation (11, 12). We identified a C-spine residue, valine 57 (V57), which is mutated to a phenylalanine (V57F). V57 is the second C-spine residue from the N-lobe and is in close proximity to A70, suggesting that the mutant with Phe will function like A70F. Replacing V57 with Phe in BRAF and CRAF abolished ATP binding but allowed the mutated protein to function as an allosteric activator for wild-type RAFs. Replacing these C-spine residues with Phe was critical; Met and Leu substitutions were not sufficient. We also identified Phe mutations in the RS3 position in BRAF and showed that Met and His can also function to stabilize the R-spine. Lastly, we identified another Phe mutation at L74. Analysis of L74F showed that it influences the R-spine even though it is not itself an R-spine residue. Like Ala70, it is located in β-strand 3, but it faces toward the C-helix. Replacing this residue with Phe leads to the assembly of the active kinase and activation of MEK and extracellular signal-regulated kinase (ERK) that is independent of Ras, dimerization, and activation loop phosphorylation. Based on these two mutations, L95F and L74F, we propose that hydrophobic packing along the entire C-helix is a driving force for the regulation and assembly of all active kinases. Because these three mutations were found not only in BRAF but also in other protein kinases, like ABL1 and the epidermal growth factor receptor (EGFR), we believe that these mechanisms for creating an oncogenic mutation that can drive tumorigenesis and hijack the normal regulatory pathway are likely to be common.
The importance of the hydrophobic space around the αC-helix was further tested by using a V600F mutant, which was constitutively active in the absence of the negative charge that is associated with the common V600E mutation. This mutation demonstrates that hydrophobicity alone at this key site can be sufficient to drive the assembly of the active kinase; the negative charge is not essential. We thus report here an array of strategic single hydrophobic mutations that can drive the assembly of active RAF and also a unifying theme for their common phenotype, “assembly of the R-spine and positioning of the αC-helix”.
MATERIALS AND METHODS.
Biochemicals.
The antibodies utilized in this study included the following: anti-phospho-ERK1/2, anti-phospho-MEK1/2, and anti-MEK1/2 (Cell Signaling Technology); anti-FLAG and antihemagglutinin (anti-HA; both from Sigma); anti-ERK2 (Santa Cruz Biotechnology). Horseradish peroxidase-labeled secondary antibodies were from Jackson Laboratories. All chemical reagents were obtained from Sigma.
Plasmids.
All mutations were generated by PCR and were tagged with either HA or FLAG, as described before (4). The pCDNA3.1(+) vector (Invitrogen) was used for transient expression.
Cell culture and transfection.
293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS). Cells were transfected using Lipofectamine 2000 (Invitrogen), harvested at 48 h, and lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with protease and phosphatase inhibitors. Whole-cell lysates were prepared through depletion of cell debris by a hard spin at 4°C.
Immunoprecipitation, in vitro kinase assay, and Western blotting.
Immunoprecipitations were performed as described before (10). Briefly, whole-cell lysates were mixed with either anti-HA or anti-FLAG beads (Sigma), rotated at 4°C for 60 min, and washed three times with RIPA buffer. For in vitro kinase assays, the immunoprecipitates were washed once with kinase reaction buffer (25 mM HEPES, 10 mM MgCl2, 0.5 mM Na3VO4, 0.5 mM dithiothreitol; pH 7.4) and then incubated with 20 μl of kinase reaction mixture (2 μg inactive MEK1 and 100 μM ATP in 20 μl of kinase reaction buffer) per sample at room temperature for 10 min. Kinase reactions were stopped by adding 5 μl per sample of 5× Laemmli sample buffer. Immunoblotting was carried out as described before (10).
Focus formation assay.
Immortalized RAF knockout mouse embryonic fibroblasts (MEFs) reconstituted with different spine mutants were plated at 5 × 103 cells per 60-mm dish and fed every other day. Twelve days later, cells were fixed with 2% formaldehyde and stained with Giemsa solution (Sigma).
Molecular modeling.
Molecular modeling of different BRAF mutants was based on the active structure (13) (PDBID 4E26) chain A with the six missing residues in the activation segment (amino acids [aa] 604 to 609) which was reconstructed using the ModLoop server (14). Mutant structures were optimized by the “minimize” procedure within the TINKER software package (http://dasher.wustl.edu/tinker/) with default convergence parameters.
RESULTS
Ontology searches for hydrophobic spine mutations in cancer.
Previously, we showed that mutations that stabilize the assembled form of the spines drive the active conformation of the RAF kinase, as evidenced by constitutive kinase or allosteric activity (4). Replacement of Ala70 with Phe blocked ATP binding and modeling, suggesting that it stabilizes a closed conformation of the C-spine (10). We also showed that replacement of the RS3 residue, Leu95, with Phe resulted in a constitutively active kinase; modeling suggested that the Phe could stabilize the active conformation of the R-spine (4). To determine whether such changes are important in disease, we used an ontology-based search tool, ProKinO (11, 12), to query for mutations that introduce a Phe residue in the hydrophobic core (Fig. 1). We did not identify phenylalanine substitutions at position A70, but we did identify a Phe substitution in BRAF and ERBB4 at V57, a C-spine residue that is spatially adjacent to A70. We thus decided to interrogate this position in greater detail.
FIG 1.
Phenylalanine substitutions in cancer genes. The ProKinO ontology tool was used to search the cancer kinome databases for all phenylalanine substitutions. The numbers refer to the residue position, based on the sequence of the catalytic subunit of PKA as the reference point. Different colors represent individual kinases.
Replacement of V57 with a phenylalanine generates a kinase-dead BRAF that is an allosteric activator.
A70 and V57 are both highly conserved residues that are contained in the N-terminal lobe of protein kinases and constitute the two top residues in the C-spine (2, 15). When ATP is bound, the two lobes of the kinase close over the adenine ring, with A70 and V57 interacting on the top of the adenine ring and Leu173 interacting on the bottom (Fig. 2A). Leu173 is replaced with Phe in BRAF and CRAF but is retained as a Leu in most other kinases. Previously, we showed that replacement of A70 with phenylalanine, but not that with Ile or Met, abolished ATP binding but stabilized KSR1 in an active-like conformation, allowing it to dimerize and allosterically activate wild-type RAF (10).
FIG 2.
A mutation that fuses the C-spine and blocks ATP binding generates a pseudokinase, allosteric activator. (A) Model showing the relationship between the C-spine residues and ATP. Using PKA numbering, in the N-lobe, V57 and A70 interact with the upper surface of the adenine ring, while L173 interacts with the bottom surface. In BRAF, the C-spine residue below is a Phe. (B) Phe replacement of V471 of BRAF is an activating mutation that requires dimerization for its effect. BRAF V471F, A481F, or V471F plus R509H were overexpressed in 293 cells and immunoblotted with antibodies to pERK, ERK2, or an epitope tag, HA, that was fused to BRAF. (C) Phe replacement of V363 of CRAF is an activating mutation that requires dimerization for its effect. Because the allosteric function of CRAF requires N-terminal acidic residues, acidic residues (DDEE) were substituted for residues 338 to 341. CRAF V363F, A373F, or V363F plus R401H were overexpressed in 293 cells and immunoblotted with antibodies to pERK, ERK2, or an epitope tag, HA, that was fused to CRAF. (D) In vitro kinase reactions of BRAF C-spine mutants. The wild type (WT) or V471F or A481F mutants of BRAF were overexpressed in cells, and immunoprecipitates were tested for kinase activity against MEK by using purified MEK. Kinase reactions were analyzed by immunoblotting with antibodies against pMEK1/2, MEK1/2, and the HA epitope tag. (E) In vitro kinase reactions of CRAF C-spine mutants. To generate constitutively active CRAF kinases, we used a construct encoding acidic residues (TESD) for the two activation loop phosphorylation sites (T491E and S494D). The V363F and A373F mutants were compared to the wild-type CRAF construct with the two activation loop substitutions (TESD). (F) Activator/receiver assays demonstrated that the BRAF C-spine mutants function as allosteric activators. BRAF and BRAF C-spine mutants (A481F and V471F) were coexpressed with a CRAF construct that lacks N-terminal acidic residues (AAFF). This prevents the CRAF construct from being able to function as an activator, but its kinase activity was still stimulated when it dimerized with a RAF protein that has an acidic N-terminal acidic domain. Various constructs were expressed alone or together, as indicated, as cell lysates were immunoblotted with antibodies to pERK, ERK2, and epitope tags (HA and FLAG) that were present on the receiver or the activator constructs, respectively. The ability to activate ERK required dimerization, as mutation of R401H blocked this effect. (G) Activator/receiver assays demonstrated that the CRAF C-spine mutants function as allosteric activators. This experiment was performed similar to that shown in panel F, except that the C-spine CRAF mutants with DDEE substituted for the N-terminal acidic domain residues 338 to 341 was used. (H) Only Phe substitutions abrogate kinase activity. The V363F substitution in CRAF was compared with Met or Leu substitutions. CRAF constructs with the designated substitution for V363 that also contained acidic substitutions at positions 338 to 341 and an acidic substitution in the activation loop (DDEE/TESD) were overexpressed in cells, and kinase activity measured in vitro toward purified MEK was measured as described for panel E.
To test whether Phe substitution of V57 behaved like the A70F mutant, we generated the V57F mutation in BRAF and CRAF and tested whether it retained kinase activity and whether it could allosterically activate wild-type RAFs (Fig. 2B and C). Like the A70F (A481F) mutant of BRAF, the V57F (V471F) mutant of BRAF was unable to phosphorylate MEK in vitro, confirming that it was kinase dead (Fig. 2B). To test CRAF activity, we used a mutated CRAF with acidic residues substituted for the phosphorylation sites in the activation loop (16) (T491E/S494D [TESD]) and overexpressed it in cells. While the CRAF TESD construct exhibited kinase activity for MEK in vitro, substitutions of Phe for A70 or V57 both abrogated kinase activity (Fig. 2C). This is consistent with our hypothesis that the V57F mutation impairs kinase activity by impairing ATP binding. When we overexpressed the V57F mutant of BRAF or CRAF in cells, however, it stimulated the activation of ERK (Fig. 2D and E). Impairing the ability of the BRAF or CRAF V57F mutant to dimerize by mutating a conserved Arg required for dimerization (+RH) (17) blocked its ability to activate ERK, suggesting that it functions by allosterically activating wild-type RAF molecules, similar to A70F.
We directly tested the ability of these mutants to allosterically activate RAF by using an activator/receiver assay that we developed previously (4). In this assay, the kinase-defective V57F mutant is coexpressed with a kinase-active construct that lacks the N-terminal acidic motif. Because it lacks the N-terminal acidic motif (AAFF), activation of the receiver requires that it dimerize with an RAF construct that possesses phosphorylation or acidic residues at the N-terminal acidic motif (DDEE) and is in an ATP-bound conformation. As expected, the A70F and V57F forms of BRAF (Fig. 2F) and CRAF (Fig. 2G) both could function as activators, and this required the conserved Arg residue that mediates dimerization (17).
Modeling confirmed, in principle, that the V57F mutant functions by completing the C-spine through the phenylalanine ring, which is very similar to the A70F mutant. We had previously tested various hydrophobic residue mutations at the A70 position and found that only a Phe substitution could abrogate ATP binding (10). Similarly, we tested whether large aliphatic amino acids, such as Met or Leu, could function in a similar way as Phe at the Val57 site. Our results for BRAF (Fig. 2H) showed that kinase activity was still present with replacements with Met or Leu, indicating that ATP binding was not blocked by substitution with Leu or Met. Table 1 shows the C-spine residues that pack against the adenine ring in several kinases. Notably, the RAFs already have a Phe at position 173.
TABLE 1.
Spine and spine-related residues
| Location | Residues in protein |
||||||
|---|---|---|---|---|---|---|---|
| PKA | BRAF | CRAF | RSK | MEK1 | ERK2 | KSR | |
| C-spine | Val 57, Ala 70, Met 128, Leu 172, Leu 173, Ile 174, Leu 227, Met 231 | Val 471, Ala 481, Leu 537, Ile 582, Phe 583, Leu 584, Val 645, Leu 649 | Val 363, Ala 373, Leu 429, Ile 474, Phe 475, Leu 476, Tyr 539, Leu 541 | Val 82, Ala 98, Leu 155, Ile 199, Leu 200, Leu 201, Leu 266, Met 261 | Val 82, Ala 95, Leu 151, Ile 196, Leu 197, Leu 198, Ser 252, Met 256 | Val 39, Ala 52, Leu 112, Leu 155, Leu 156, Leu 157, Ile 217, Met 221 | Val 680, Ala 690, Leu 747, Val 792, Phe 793, Tyr 794, Ile 863, Leu 867 |
| R-spine | Leu 95, Leu 106, Tyr 164, Phe 185 | Leu 505, Phe 516, His 574, Phe 595 | Leu 397, Met 409, His 466, Phe 487 | Leu 122, Tyr 135, Tyr 191, Leu 214 | Leu 118, Phe 129, His 188, Phe 209 | Leu 75, Ile 86, His 147, Phe 168 | Tyr 714, Phe 721, His 784, Phe 804 |
| Shield residues | Val 104, Met 118, Met 120 | Leu 514, Ile 526, Met 528 | Leu 404, Ile 419, Thr 421 | Val 131, Leu 145, Leu 147 | Val 127, Ile 141, Met 143 | Ile 84, Ile 103, Gln 105 | Val 723, Ile 737, Thr 739 |
| Other residues | Leu 74, Thr 88, Arg 190 | Leu 485, Phe 498, Val 600 | Leu 377, Phe 390, Val 492 | Leu 102, Thr 110, Glu 217 | Ile 99, Ile 111, Gln 214 | Ile 56, Thr 68, Val 173 | Ile 694, Phe 707, Glu 809 |
Methionine and histidine can substitute for Leu95 to generate a constitutively active RAF kinase.
The R-spine consists of four residues, and all four must be aligned in an active kinase (Fig. 3A). This is the hallmark signature of an active kinase (15). RS1 is usually the histidine from the conserved HRD motif (Y164 in protein kinase A [PKA]; H574 in BRAF). RS2 is the Phe from the DFG motif (Phe185 in PKA; Phe595 in BRAF). RS3 (Leu95 in PKA; Leu505 in BRAF) is from the αC-helix, and RS4 (Leu106 in PKA; Phe516 in BRAF) comes from the β4-strand. Figure 3A shows the residue numbers in BRAF, CRAF, and PKA that correspond to the R-spine residues. The corresponding residues in every kinase were designated in our earlier analysis of the R-spine residues (18) and in the ProKinO database (12).
FIG 3.
Generation of constitutively active kinases through R-spine mutations. (A) The nomenclature of the R-spine residues in PKA, BRAF, and CRAF. (B) Testing different hydrophobic residue substitutions at the RS3 residue in BRAF. Replacement of L505 with Phe, Met, or His was tested by overexpression in 293 cells and compared to wild-type BRAF. Cell lysates were immunoblotted with antibodies to pERK1/2, ERK2, and HA, the epitope tag fused to BRAF. (C) Testing different hydrophobic residue substitutions at the RS3 residue in CRAF. Replacements of L397 with Phe, Met, or His were tested by overexpression in 293 cells and compared to wild-type CRAF. Cell lysates were immunoblotted with antibodies to pERK1/2, ERK2, and HA, the epitope tag, fused to BRAF. (D and E) The RS3 substitution generated a constitutively active kinase that was independent of dimerization (BRAF R509H and CRAF R401H) and activation loop phosphorylation (BRAF T599A/S602A, CRAF T491A and S494A). Results for experiments with the indicated mutants were analyzed as described above.
Since RS4 is already a Phe in BRAF, we searched the cancer kinome for RS3 mutations in BRAF (Fig. 1). Although we did not find any Phe mutations in BRAF, we did identify a His substitution in BRAF for RS3 (L505) and found that many kinases, especially the tyrosine kinases, have a Met at the RS3 position in their native state (Table 2). To test whether other larger hydrophobic residues in the RS3 position would lead to RAF activation, we replaced the RS3 Leu with either a His or Met for BRAF and CRAF and compared these mutants to the mutant with the Phe substitution. Overexpression of each of these mutants in cells resulted in ERK activation, consistent with the generation of constitutively active kinases (Fig. 3B and C). Impairing dimerization (RH) or mutating the activation loop phosphorylation sites (T491A/S494A [TASA]) did not impair ERK activation, confirming that these are truly constitutively active kinases that act independently of dimerization and activation loop phosphorylation (Fig. 3D and E). Consistent with these observations, we found that the Met substitution was even better than Phe in generating the constitutively active kinase.
TABLE 2.
Natural Met or His RS3 residues in the human kinome
| Family and protein | Sequencea |
|---|---|
| TK kinase family | |
| ACK | PEAMDDFIR-EVNAMHSLD-HRNLIRLYGVVLTP-- |
| TNK1 | GTELGDFLR-EVSVMMNLE-HPHVLRLHGLVLGQ-- |
| EGFR | -KANKEILD-EAYVMASVD-NPHVCRLLGICLTS-- |
| HER2/ErbB2 | -KANKEILD-EAYVMAGVG-SPYVSRLLGICLTS-- |
| HER4/ErbB4 | -KANVEFMD-EALIMASMD-HPHLVRLLGVCLSP-- |
| SYK | -ALKDELLA-EANVMQQLD-NPYIVRMIGICEAE-- |
| ZAP70 | -ADTEEMMR-EAQIMHQLD-NPYIVRLIGVCQAE-- |
| FAK | --VREKFLQ-EALTMRQFD-HPHIVKLIGVITEN-- |
| PYK2 | --NKEKFMS-EAVIMKNLD-HPHIVKLIGIIEEE-- |
| EphA3 | -KQRRDFLG-EASIMGQFD-HPNIIRLEGVVTK--- |
| EphA5 | -KQRRDFLG-EASIMGQFD-HPNIIHLEGVVTK--- |
| EphA4 | -KQRRDFLS-EASIMGQFD-HPNIIHLEGVVTK--- |
| EphA6 | -RQRRDFLR-EASIMGQFD-HPNIIRLEGVVTKRSF |
| EphA7 | -KQRRDFLC-EASIMGQFD-HPNVVHLEGVVTR--- |
| EphB1 | -KQRRDFLS-EASIMGQFD-HPNIIRLEGVVTK--- |
| EphB2 | -KQRRDFLS-EASIMGQFD-HPNVIHLEGVVTK--- |
| EphB3 | -RQRRDFLS-EASIMGQFD-HPNIIRLEGVVTK--- |
| EphB4 | -RQRREFLS-EASIMGQFE-HPNIIRLEGVVTN--- |
| EphA8 | -RQRRDFLS-EASIMGQFD-HPNIIRLEGVVTR--- |
| EphA2 | -KQRVDFLG-EAGIMGQFS-HHNIIRLEGVISK--- |
| EphA1 | -GQWWNFLR-EATIMGQFS-HPHILHLEGVVTK--- |
| DDR1 | -NARNDFLK-EVKIMSRLK-DPNIIRLLGVCVQD-- |
| DDR2 | -NARNDFLK-EIKIMSRLK-DPNIIHLLSVCITD-- |
| MUSK | -DMQADFQR-EAALMAEFD-NPNIVKLLGVCAVG-- |
| ROR1 | -QQWMEFQQ-EASLMAELH-HPNIVCLLGAVTQE-- |
| ABL | ---VEEFLK-EAAVMKEIK-HPNLVQLLGVCTRE-P |
| ARG | ---VEEFLK-EAAVMKEIK-HPNLVQLLGVCTLE-P |
| HCK | ---VEAFLA-EANVMKTLQ-HDKLVKLHAVVTK--E |
| LYN | ---VQAFLE-EANLMKTLQ-HDKLVRLYAVVTRE-E |
| LCK | ---PDAFLA-EANLMKQLQ-HQRLVRLYAVVTQ--E |
| BLK | ---PEAFLG-EANVMKALQ-HERLVRLYAVVTK--E |
| SRC | ---PEAFLQ-EAQVMKKLR-HEKLVQLYAVVSE--E |
| YES | ---PEAFLQ-EAQIMKKLR-HDKLVPLYAVVSE--E |
| FYN | ---PESFLE-EAQIMKKLK-HDKLVQLYAVVSE--E |
| FGR | ---PKAFLE-EAQVMKLLR-HDKLVQLYAVVSE--E |
| FRK | ---PNDFLR-EAQIMKNLR-HPKLIQLYAVCTLE-D |
| BRK | ---HQQMLQSEIQAMKKLR-HKHILALYAVVSVG-D |
| BTK | ---EDEFIE-EAKVMMNLS-HEKLVQLYGVCTKQ-R |
| BMX | ---EDEFFQ-EAQTMMKLS-HPKLVKFYGVCSKE-Y |
| TEC | ---EEDFIE-EAKVMMKLT-HPKLVQLYGVCTQQ-K |
| TXK | ---EEDFIE-EAKVMMKLS-HSKLVQLYGVCIQR-K |
| ITK | ---EEDFIE-EAEVMMKLS-HPKLVQLYGVCLEQ-A |
| CSK | ---AQAFLA-EASVMTQLR-HSNLVQLLGVIVEEKG |
| CTK | ---AQAFLD-ETAVMTKMQ-HENLVRLLGVILHQ-- |
| FGFR2 | -KDLSDLVS-EMEMMKMIGKHKNIINLLGACTQ-DG |
| FGFR3 | -KDLSDLVS-EMEMMKMIGKHKNIINLLGACTQ-GG |
| FGFR1 | -KDLSDLIS-EMEMMKMIGKHKNIINLLGACTQ-DG |
| FGFR4 | -KDLADLVS-EMEVMKLIGRHKNIINLLGVCTQ-EG |
| FMS | -DEKEALMS-ELKIMSHLGQHENIVNLLGACTH-GG |
| PDGFRa | -SEKQALMS-ELKIMTHLGPHLNIVNLLGACTK-SG |
| PDGFRb | -SEKQALMS-ELKIMSHLGPHLNVVNLLGACTK-GG |
| FLT3 | -SEREALMS-ELKMMTQLGSHENIVNLLGACTL-SG |
| AXL | -SELEDFLS-EAVCMKEFD-HPNVMRLIGVCFQGSE |
| MER | -REIEEFLS-EAACMKDFS-HPNVIRLLGVCIEMSS |
| TYRO3 | -SDIEEFLR-EAACMKEFD-HPHVAKLVGVSLRSRA |
| MET | -GEVSQFLT-EGIIMKDFS-HPNVLSLLGICLRSEG |
| RON | -QQVEAFLR-EGLLMRGLN-HPNVLALIGIMLPPEG |
| ROS | -QEKIEFLK-EAHLMSKFN-HPNILKQLGVCLLNEP |
| INSR | -RERIEFLN-EASVMKGFT-CHHVVRLLGVVSKGQP |
| IGF1R | -RERIEFLN-EASVMKEFN-CHHVVRLLGVVSQGQP |
| IRR | -RECIEFLK-EASVMKAFK-CHHVVRLLGVVSQGQP |
| JAK1 | --ISLAFFE-AASMMRQVS-HKHIVYLYGVCVRD-- |
| TYK2 | --IALAFYE-TASLMSQVS-HTHLAFVHGVCVRG-- |
| JAK2 | --YSESFFE-AASMMSKLS-HKHLVLNYGVCVCG-- |
| JAK3 | --CMESFLE-AASLMSQVS-YRHLVLLHGVCMAG-- |
| SuRTK106 | --EVQDFLG-RIQFHQYLGKHKNLVQLEGCCTEK-- |
| TKL kinase family | |
| ANKRD3 | --ERMELLE-EAKKMEMAKFRYILPVYGICREP-- |
| SgK288 | S-DVNYLIE-EAAKMKKIKFQHIVSIYGVCKQP-- |
| RIPK3 | ----KAISR-EVKAMASLDNEFVLRLEGVIEKVN- |
| RIPK1 | ----EALLE-EAKMMNRLRHSRVVKLLGVIIEEG- |
| LIMK2 | ----KTFLT-EVKVMRSLDHPNVLKFIGVLYKDK- |
| LIMK1 | ----RTFLK-EVKVMRCLEHPNVLKFIGVLYKDK- |
| TESK1 | ----GNTLR-EVQLMNRLRHPNILRFMGVCVHQG- |
| TESK2 | ----ANMLK-EVQLMNRLSHPNILR---------- |
| IRAK4 | -EELKQQFDQEIKVMAKCQHENLVELLGFSSDGD- |
| MLKL | AIVRQTFNK-EIKTMKKFESPNILRIFGICIDETV |
| STK kinase family | |
| MAP3K1 | QEEVVEALREEIRMMSHLN-HPNIIRMLGATC--- |
| MAP3K5 | DSRYSQPLHEEIALHKHLK-HKNIVQYLGSFS--- |
| MAP3K7 | DSRYSQPLHEEIALHKYLK-HRNIVQYLGSVS--- |
| MAP3K6 | DSRFSQPLHEEIALHRRLR-HKNIVRYLGSAS--- |
| MST1 | ---DLQEIIKEISIMQQCD-SPHVVKYYGSYFKNT |
| MST2 | ---DLQEIIKEISIMQQCD-SPYVVKYYGSYFKNT |
| KHS2 | -GEDFAVVQQEIIMMKDCK-HPNIVAYFGSYLRRD |
| PAK1 | -QPKKELIINEILVMRENK-NPNIVNYLDSYLVGD |
| PAK3 | -QPKKELIINEILVMRENK-NPNIVNYLDSYLVGD |
| PAK2 | -QPKKELIINEILVMKELK-NPNIVNFLDSYLVGD |
| PAK4 | -QQRRELLFNEVVIMRDYQ-HENVVEMYNSYLVGD |
| PAK5 | -QQRRELLFNEVVIMRDYH-HDNVVDMYSSYLVGD |
| PAK6 | -QQRRELLFNEVVIMRDYQ-HFNVVEMYKSYLVGE |
| OSR1 | -TS-MDELLKEIQAMSQCH-HPNIVSYYTSFVVKD |
| STLK3 | -TS-MDELLKEIQAMSQCS-HPNVVTYYTSFVVKD |
| CMGC kinase family | |
| JNK1 | ----RAYRELVLMKCV---NHKN-----IIGLLNVFTPQ |
| JNK3 | ----RAYRELVLMKCV---NHKN-----IISLLNVFTPQ |
| GSK3A | ----FKNRELQIMRKLD---HCN-----IVRLRYFFYSS |
| GSK3B | ----FKNRELQIMRKLD---HCN-----IVRLRYFFYSS |
| CAMK kinase family | |
| MARK2 | LQKLFREVRIMKVLN----HPNIVKLFEVIETEK----- |
| MARK1 | LQKLFREVRIMKILN----HPNIVKLFEVIETEK----- |
| MARK3 | LQKLFREVRIMKILN----HPNIVKLFEVIETQK----- |
| MARK4 | LQKLFREVRIMKGLN----HPNIVKLFEVIETEK----- |
| QIK | LEKIYREVQIMKMLD----HPHIIKLYQVMETKS----- |
| SIK | LEKIYREVQLMKLLN----HPHIIKLYQVMETKD----- |
| QSK | LKKIFREVQIMKMLC----HPHIIRLYQVMETER----- |
| NuaK1 | MVHIRREIEIMSSLN----HPHIISIYEVFENKD----- |
| NuaK2 | LMHIRREIEIMSSLN----HPHIIAIHEVFENSS----- |
| NIM1 | QRLLSREISSMEKLH----HPNIIRLYEVVETLS----- |
| SNRK | TGHLFQEVRCMKLVQ----HPNIVRLYEVIDTQT----- |
| TSSK4 | -KFLPREIQVMKVLR----HKYLINFYRAIESTS----- |
| caMLCK | REDVKNEINIMNQLS----HVNLIQLYDAFESKH----- |
| SgK085 | KEEVKNEISVMNQLD----HANLIQLYDAFESKN----- |
| skMLCK | KEMVLLEIEVMNQLN----HRNLIQLYAAIETPH----- |
| smMLCK | KENIRQEISIMNCLH----HPKLVQCVDAFEEKA----- |
| MAPKAPK2 | CPKARREVELHWRASQ---CPHIVRIVDVYEN------L |
| MAPKAPK3 | SPKARQEVDHHWQASG---GPHIVCILDVYEN------M |
| MAPKAPK5 | RPKARNEVRLHMMCAT---HPNIVQIIEVFANSVQFPHE |
| AGC kinase family | |
| ROCK2 | RSDSAFFWEERD------IMAFANS------PWVVQLFYAF |
| ROCK1 | RSDSAFFWEERD------IMAFANS------PWVVQLFYAF |
| PKG1 | TRQQEHIRSEKQ------IMQGAHS------DFIVRLYRTF |
| PDK1 | ENKVPYVTRERD------VMSRLDH------PFFVKLYFTF |
| YANK2 | RDEVRNVFRELQ------IMQGLEH------PFLVNLWYSF |
| YANK1 | RNEVRNVFKELQ------IMQGLEH------PFLVNLWYSF |
The Met (M) or His (H) residue at the RS3 position is underlined; in addition, the His residues are indicated in boldface. The sequences proceed from the αC to the β4 regions (the rightmost sequence portion is the β4 region).
Replacement of L74 with phenylalanine generates a constitutively active kinase.
We hypothesized that other hydrophobic mutations might function to enhance kinase activity by stabilizing the kinase hydrophobic core and, in particular, the R-spine. We noted that Phe substitutions were present at other positions in BRAF, EGFR, ABL1, KIT, ERBB4, and AURA (Fig. 1). We noted one substitution in particular, Leu74Phe, that was found in BRAF but that did not have an obvious relationship with either the C-spine or the R-spine. A Phe replacment of the homologous residue in BRAF is known to cause cranio-facial syndrome (19). L74 is located in the loop between the β3-strand and the N terminus of the C-helix (Fig. 4A).
FIG 4.
L74 functions to couple the C-spine to the R-spine. (A) Model showing the position of L74 in PKA (left), how ATP interacts with both the C- and R-spines (center), and how L485 in BRAF potentially interacts with F498 in the C-helix. (B) L74 replacement by Phe in BRAF and CRAF results in a constitutively active kinase that is independent of dimerization and activation loop phosphorylation. The indicated mutants were overexpressed in 293 cells, and cell lysates were analyzed as described above. (C) Replacement of L74 with other hydrophobic residues also generates constitutively active forms of CRAF. (D) Mutation of F498 in the C-helix abrogates the ability of L485F to induce BRAF activation. (E) The F498A mutant of BRAF can still be activated. An activator/receiver assay was performed using BRAF V471F as the activator and BRAF F498A as the receiver. Similar amounts of active ERK were induced with either BRAF or F498A BRAF as the receiver. (F) Models showing the series of hydrophobic interactions along the surface of the C-helix facing the β3-strand in PKA (left) and BRAF (right). In PKA, R-spine residues (RS2 to -4) interact with the gatekeeper residue (M120), which interacts with C-spine residues V57 and A70. Note how F350 from the C-tail interacts with L74, filling the space between L74 and the C-spine. On the right is the same surface of BRAF. In this case, F485 interacts with F498, which is facing it from the C-helix. Also note the smaller gatekeeper residue (T529), which does not completely fill the space between the R-spine and the C-spine.
The L74F substitution in BRAF (L485F) and CRAF (L397F) was tested by overexpressing it in 293 cells and then comparing the level of ERK activation with that of the RS3F mutation (Fig. 4B). The L74F mutants of CRAF and BRAF were comparable in activity to the previously characterized RS3F mutants and were constitutively active, as evidenced by measurement of in vitro kinase activity toward MEK (Fig. 4C and data not shown). We also tested other hydrophobic amino acids. Methionine but not isoleucine was as effective as Phe in inducing the constitutively active phenotype (Fig. 4D).
Modeling of the L74F mutation allowed us to appreciate its unique location relative to the C-spine and the R-spine and, in particular, to the αC-helix. L74 is located just after β3 in the loop between the β3-strand and the αC-helix (Fig. 4A). It directly interacts with the N terminus of the αC-helix, reinforcing its importance for positioning the αC-helix for kinase regulation. Leu74 faces in the same direction as the salt bridge between Glu91 in the αC-helix and Lys72 in the β3-strand. Formation of the salt bridge positions L74 to interact with the N-terminal portion of the C-helix, specifically, with Val79 and Leu116 in PKA (Fig. 4A). At the C terminus of the αC-helix is another cluster of hydrophobic residues that flank the conserved Glu91 (Fig. 4F). This cluster of hydrophobic residues includes two R-spine residues (highlighted in red in the figure): the RS3 residue in the C-helix (PKA F185; BRAF F516) and the RS4 residue in the β4-strand (PKA L106; BRAF F516), as well as three residues from β5 (L116, M118, and the gatekeeper residue M120 in PKA; L525, I527, and T529 in BRAF). We recently showed that Met118, Met120, and Val104 are critical “shell” residues that influence the assembly of the R-spine (18). There is thus an extended surface on the αC-helix that allows for multiple contacts between the αC-helix, the hydrophobic spines, and the glycine loop.
Modeling Phe at position 74 (L485) in various conformations in BRAF suggested that it interacts with Phe498 in the αC-helix (Fig. 4F). This residue is Thr88 in PKA but is typically a hydrophobic residue, often Phe, Ile, or Leu, in non-AGC kinases. In AGC kinases, however, the same pocket is filled by a hydrophobic motif from the C terminus of the kinase (F347/F350) (see Fig. 6A, below). This hydrophobic motif is conserved in all AGC kinases. Modeling suggested that the interaction of L485F of BRAF (L74F) with Phe498 would function to facilitate movement of the αC-helix inwards, helping to position RS3 in the R-spine and the formation of the Lys-Glu salt bridge. Since the catalytic Lys makes contacts with the α- and β-phosphate groups of ATP (Fig. 4A, middle panel), this series of hydrophobic interactions helps to bridge the R-spine with ATP and the C-spine, as well as position the γ-phosphate for transfer.
FIG 6.
Modeling the effect of V600E and V600F mutations on the conformation of the activation loop in BRAF. In an effort to explore the space that might be visited by the mutations at V600, we replaced V600 in the wild-type BRAF structure (PDBID 4E26) with Glu and Phe and then minimized the structure by using ModLoop server (14). Mutant structures were optimized by minimize procedure from the TINKER software package (http://dasher.wustl.edu/tinker/) with default convergence parameters. Since the activation loop was disordered in these structures, we added in the missing residues prior to minimization. Wild-type BRAF with the built-in activation loop was also minimized (C). In both V600E (A) and V600F (B), the hydrophobic nodule was strengthened, although more so with the Phe. In both cases, the mutated residue interacted with K507. With the Glu, there was an ion pair, while the Phe made an amino aromatic interaction with Lys507. The ion pair with V600E and K507 has been validated in crystal structures, while the Phe mutation is just a prediction. Following minimization, the activation loop became ordered with Arg603 flipping down to bridge multiple sites in the activation loop. (C) The initial model for minimization was based on a wild-type structure where R603 in the activation loop was ordered.
We tested this hypothesis by mutating BRAF Phe498 to Ala (Fig. 4E). Substitution with Ala abrogated the constitutive kinase activity of L74F, suggesting that the interaction between F498 and L485F is critical. To prove that the mutation did not impair kinase activity, we tested its ability to be activated by dimerization by using a receiver and activator assay (Fig. 5E), which we have previously described. Dimerization of the F498A mutant with a C-spine mutation of BRAF (kinase inactive) resulted in MEK and ERK activation. This suggested that the L485F/F498A mutant is still active, as it can be activated by allosteric mechanisms, and supports the idea that the L74F mutant functions by helping to position and stabilize the αC-helix.
FIG 5.
Hydrophobic interactions function to stabilize the BRAF activation loop. (A) Phe replacement of V600 generates a constitutively active kinase that activates ERK1/2 when overexpressed in cells. Wild-type (WT) and V600F BRAF were overexpressed in cells, and cell lysates were immunoblotted with the indicated antibodies. (B) The activity of the V600F mutant is independent of dimerization and activation loop phosphorylation. The role of dimerization was tested by replacing R401 with histidine. The role of activation loop phosphorylation was tested by replacing T599 and S602 with alanine to prevent their phosphorylation. Constructs were overexpressed in cells, and cell lysates were immunoblotted with the indicated antibodies. (C) In vitro kinase assays of wild-type and V600F BRAF constructs. The indicated constructs were overexpressed in cells, and BRAF immunoprecipitates were assessed for kinase activity against purified MEK, as described for Fig. 2E. The lanes shown are from the same gel.
V600F is an activating mutation.
In considering residues that contribute to RAF activation, we next turned our attention to V600, which lies in β9 at the beginning of the activation loop (aa 187 to 192) and interfaces with the αC-helix. Replacement of BRAF V600 with Glu is the most common activating BRAF mutation found in cancer, and the negative charge has been hypothesized to be important for bridging the activation loop and the αC-helix (20).We noted a Phe substitution in BRAF at this residue, although it is rare, presumably because the switch would require more than one base change. Replacement of V600 of BRAF with Phe generated a strong constitutively active kinase (Fig. 5A and C) that was not significantly affected when its ability to dimerize or for phosphorylation of the activation loop phosphorylation sites was blocked (Fig. 5B). Preliminary modeling studies suggested that a Phe at position 600 would not only strengthen a hydrophobic module that bridges the activation loop to the αC-helix (Fig. 6) but also would show an intriguing amino acid-aromatic interaction with Lys507. These findings further emphasize the importance of the hydrophobic space that surrounds the αC-helix and stabilizes the active kinase, and they demonstrate once again how single strategic hydrophobic mutations far from the active site can hijack a complex regulatory pathway to create an oncogenic mutant.
Transforming abilities of BRAF and CRAF mutants.
To validate and compare the strengths of various mutations, we performed transformation assays for BRAF and CRAF mutants (Fig. 7). We first tested the V57F C-spine mutation in BRAF (V471F) and found that it was only weakly transforming. We did not test the C-spine mutation of CRAF, because it would not be expected to be transforming without phosphorylation of the NtA (4). We then tested the L95M mutation of BRAF and CRAF. In the case of BRAF (L505M), this resulted in a transforming activity that was stronger than V471F but clearly weaker than that of V600E. For CRAF (L397M), this resulted in a level of transformation that was above the background but less than a known transforming version of RAF (N-terminally truncated CRAF). We next tested the L74F forms of BRAF and CRAF. The BRAF (L485F) mutant showed moderate transforming activity, while the CRAF (L377F) mutant showed weak transforming activity. This suggested that the presence of the N-terminal acidic sequence in BRAF may enhance the functional activity of the L74F mutant. Lastly, we compared the V600F mutant of BRAF. This mutant showed strong transforming activity, close to the activity seen with V600E. Thus, the BRAF mutants showed a range of transforming activities ranging from V600F (strong activity) to L505M to L485F, and finally V471F (weak activity).
FIG 7.
Measurement of transforming abilities of spine mutants of BRAF and CRAF in a focus-forming assay. Wild-type spine mutants or transforming mutants of BRAF and CRAF were cloned into a retroviral vector and used to reconstitute either BRAF- or CRAF-deficient immortalized MEFs. The expression of mutants was normalized by GFP cell sorting, and the focus formation assay was carried out as described in Materials and Methods.
DISCUSSION
Protein kinases have evolved to be dynamic and tightly regulated molecular switches where activation is achieved by the assembly of hydrophobic spines (15). While the C-spine is typically assembled for catalysis by the binding of ATP, assembly of the R-spine, which is tightly coupled with positioning of the αC-helix, is highly dynamic and regulated; most of the concepts for the kinase activation mechanism are embedded within the R-spine (3, 18). In most kinases, it is the disassembled and inactive R-spine conformation that is most stable, and the initial step in activation is typically release of an inhibitory domain. This release then allows the kinase to toggle between an active (assembled R-spine) and an inactive (disassembled R-spine) conformation. Shifting the equilibrium to favor the assembled R-spine often requires dimerization and phosphorylation of the activation loop. Because the kinase is often surrounded by phosphatases, the active conformation can be easily reversed, thereby creating an effective switch mechanism. We have shown here how easily this switch mechanism can be hijacked by a variety of single hydrophobic mutations to drive disease.
Having previously defined two different mechanisms whereby Phe replacement of C-spine and R-spine residues of RAF could generate active conformations that were capable of downstream activation (4, 10), we searched the kinase cancer genomes for Phe mutations to see if similar mechanisms might actually be used to drive disease. We found several examples that suggested that changes in the hydrophobic switch can potentially explain activation of genes in the cancer kinome and that mutational stabilization of the hydrophobic spine is a recurring theme in abnormal kinase activation in cancers. Val57Phe mutations, for example, were found in four kinases, and because Val57, like Ala70, is a C-spine residue, the phenylalanine substitution was predicted to mimic the adenine ring of ATP and fuse the C-spine into an “active-like” conformation (10). We confirmed that the V57F mutant impairs kinase activity but could function as an allosteric activator for wild-type BRAF or CRAF, likely by stabilizing the active, dimerization-competent conformation.
Our previous R-spine mutant, L95F (4), was not identified in the cancer kinome database (http://www.sanger.ac.uk/genetics/CGP/cosmic/), but we did find several Met and His substitutions at this site in the cancer kinome, and these mutations in BRAF and CRAF were activating mutations similar to L95F (4). In addition, we examined a nearby hydrophobic residue, L74, which in BRAF is associated with cranio-facial syndrome when the Leu is replaced with Phe (19). This mutation generated a constitutively active kinase in both BRAF and CRAF that was not dependent on dimerization or activation loop phosphorylation, suggesting that it stabilizes the R-spine. Hydrophobicity in the activation loop also appeared to stabilize the R-spine, as BRAF V600F was also found to be constitutively active and independent of dimerization. These findings not only establish that hydrophobic mutations can be used to create cancer-driving mutations but also provide insights into the overall mechanism of activation.
V600 is highly unusual. It is clearly a hot spot for disease mutations, with V600E being the most common substitution in melanoma and colon carcinoma (20). Surprisingly, although there are many structures of inhibitors bound to either wild-type or V600E BRAF, there is no structure of BRAF with ATP bound and in no structure is the activation loop ordered. To interrogate the effect of these V600 mutations, we used minimization of the wild-type BRAF structure following replacement of V600 with either Glu or Phe. We began with the wild-type type structure, which showed the activation loop ordered through Arg603 (PDBID 4E26). In most of the other structures, the activation loop ends with Val600. We then filled in the remaining activation loop residues by using the ModLoop program (14). In the starting wild-type structure (Fig. 6C), Arg603 is positioned facing up, close to K507, suggesting that the negative change introduced by V600E would further stabilize this interaction. Surprisingly, after minimization of both mutants, Arg603 flipped down to create a very stable activation loop (Fig. 6A and B). The minimized structure confirmed the interaction of V600E with K507 that has been seen in several crystal structures, but the activation loop has never been ordered in any published crystal structure to date. The modeled activation loop shown here is intriguing, not only because of the many interactions Arg603 is capable of making, but also because it would explain why phosphorylation of T599 and S602 is important. Both would not only stabilize the downward position of Arg603, but also, via introduction of a phosphate at T599, would very likely drag the HRD Arg into the center of the activation loop. This could also explain the unusual location of the activation loop phosphorylation sites at the beginning of the activation loop, which is distinct from most other kinases. Clearly, this model needs to be validated in a crystal structure, but it provides an intriguing hypothesis.
While most attention has focused on the negative charge of V600E binding to K507 (Fig. 6A), Asp, Lys, Arg, Leu, Met, and even Phe mutations also occur at this position. When Marais and coworkers replaced V600 with Asp, Lys, and Arg, they found that all mutants were activating to various extents (21). They concluded that the hydrophobicity of the Val was important for stabilizing the inactive conformation and that all of these mutations worked by destabilizing the inactive conformation. Our results showing that V600F was constitutively active, not requiring dimerization or activation loop phosphorylation, argue against this hypothesis and suggest that hydrophobic interactions can be dominant and important for stabilizing the active conformation of BRAF. However, it should be emphasized that the inactive state seen in all of the BRAF structures available so far indicates a truncated kinase domain that has been crystallized in the presence of inhibitors. Furthermore, there are several conformations for the DFG motif in these different structures.
These results allow us to characterize the hydrophobic space of the N-lobe in new ways and allow us to predict how other cancer mutations might be activating, and why. Some general rules emerge from our analyses regarding packing of the hydrophobic residues in and around the two spines.
Fusing the C-spine.
To create a “fused” C-spine, one can scrutinize the residues that line the adenine-binding pocket and, in particular, those that sandwich the ring (A70, V57, and L173) (Fig. 2A and Table 1). Most kinases have a Leu at position 173, whereas the RAFs have a Phe (Table 1). Introducing another Phe in RAF is thus sufficient to seriously interfere with ATP binding. In the case of PKA, phenylalanines in both positions are required to produce a strong pseudokinase phenotype (unpublished results). Examination of sequences in this pocket in VRK3 allowed us to predict that it would be a pseudokinase unable to bind ATP. This has been validated experimentally, and the structure shows how bulky side chains fill the adenine pocket and “fuse” the spine (10, 22, 23). Other pseudokinases may require ATP binding to snap the kinase into the correct conformation, but they may not require the transfer of the phosphate (23). Fusing the C-spine residues would allow any kinase to be tested, whether the transfer of the phosphate was required for downstream signaling or whether noncatalytic functions that allow the kinase to function as an allosteric activator would be sufficient (4, 10, 23).
Stabilizing the R-spine.
Based on a crystal structure of inactive BRAF that showed that the RS2 Phe (in the DFG motif) was displaced, we previously postulated that a Phe replacement of the RS3 residue would stabilize the R-spine of BRAF and result in a constitutively active kinase (4). Substitution of Phe for RS3 did indeed generate a constitutively active kinase that was independent of dimerization and activation loop phosphorylation. Searching the cancer kinome, we did not identify any RS3 Phe substitutions in BRAF; however, we did find a His substitution in the RS3 position of BRAF, and Met is also found at this position in other cancer genes. We thus tested His, as well as Met, in this position and found that both substitutions resulted in constitutively active BRAF. Mining the cancer kinome showed that Met (21.26%) and His (3.13%) are found in the RS3 position in a large number of kinases (Table 2). Met, which is significantly longer and more hydrophobic than Leu, is also more flexible than Phe. Thus, and in contrast to the C-spine mutations where Phe is required, Met was as good or better than Phe at stabilizing the BRAF R-spine. The RAFs also have a Phe at the RS4 position, so this might explain why the Met is even more effective, since stacking of three phenylalanine rings is rare (24). Thus, scrutinizing the R-spine and shell residues carefully allows one to predict how to stabilize the R-spine (18) (Table 1).
Hydrophobic packing around the αC-helix drives the assembly of the R-spine.
Assembly of the R-spine has several functional consequences. It orients the Asp in the DFG motif so that it is poised to accept the second Mg2+ ion, which interacts with the γ-phosphate of ATP and also positions the entire αC-helix so that it is poised for catalysis. This includes forming the conserved salt bridge between K72 and E91 as well as creating a stable interface between the C-helix and the activation loop. Frequently, this requires phosphorylation of the activation loop. Our results forced us to consider the function of the αC-helix in its entirety, highlighting the critical importance of aligning this helix to mediate catalysis. This helix defines the switch mechanism that drives kinase activation. Our study of how hydrophobic mutations function as cancer drivers highlighted the importance of the hydrophobic space surrounding the αC-helix.
The αC-helix was earlier referred to as a signal integration motif (SIM), because the side chains that radiate out from the αC-helix are in contact with many of the conserved motifs and residues that are essential for the active kinase (Fig. 8) (25). The discovery of the R-spine (3), especially L95 (RS3), which is embedded near the end of the αC-helix, added further significance to the role of the αC-helix and in particular to the hydrophobic residues that surround it. Our present study shows the power of a single hydrophobic mutation in driving the assembly of the R-spine and the concomitant assembly the αC-helix, and it also emphasizes the allosteric range of the αC-helix.
FIG 8.
The C-helix functions as a signal integration motif that is distinct for PKA (left) and BRAF (right). The upper panels show the C-helix as a wheel and highlight the interactions of the various surfaces. For both kinases, the left surface is highly conserved and contains E91/E501, which forms the salt bridge with the catalytic lysine (middle panel). This surface also includes the RS3 residue (middle panel, PKA L95, BRAF L505). The lower surface shows how distinct residues in PKA and BRAF interact with the activation loop. The right and upper surfaces are unique for every kinase and interact with key regulatory domains (bottom panel). In the case of PKA, this surface interacts with residues from both the N and C termini of the kinase. In the case of BRAF, this surface forms the dimer interface and functions in the allosteric activation mechanism.
Comparison of the αC-helices of PKA and BRAF highlight not only the hydrophobic packing but also the unique surfaces, one side containing conserved core interactions and the other side associated with linkers and tails that are highly regulated and unique to each kinase (Fig. 8). In addition, key functions are distributed across the entire helix. In PKA (Fig. 8, left), the conserved surface includes three critical residues, L95 (RS3), E91, and H87, where each represents fundamental interactions with core regions that are conserved in all kinases. At the C terminus is L95 (RS3), which is part of the R-spine, and we found that strengthening the hydrophobic properties of this residue with Phe, His, or Met in BRAF was a dominant mutation that resulted in a constitutively active kinase. E91 in the middle of the αC-helix is the conserved partner to K72, and this electrostatic interaction links β3 and ATP binding to the αC-helix. L95, E91, and K72 are all essential for every kinase; mutation to Ala is inactivating (18, 26). At the N terminus is H87, which in PKA interacts with the activation loop. In all kinases the N terminus of the αC-helix interacts in some critical way with the activation loop and the DFG/β9 segment. In PKA, H87 at the N terminus of the αC-helix interacts directly with the phosphate on the activation loop and explains why activation loop phosphorylation is so important for assembling and/or stabilizing the active kinase (27, 28).
The opposite surface of the αC-helix, in contrast, interacts with the linkers and/or tails that flank the kinase core. In PKA, these are the N- and C-terminal tails. The hydrophobic surfaces and interaction for the αC-helix in BRAF (Fig. 8, right) highlight the BRAF-specific packing at the dimer interface. It is interesting how the DFG-β9 segment, where so many BRAF mutations overall are located, is positioned relative to the αC-helix. This motif serves as a “velcro” strip that links the DFG motif with the assembled activation loop and the αC-helix. V600 in BRAF is in this motif. The L74F mutation illustrates the extended hydrophobic space in the core and shows how these residues pack against the N-terminal part of the αC-helix. The V600F substitution bridges the activation loop with the αC-helix and the N-terminal acidic motif, which is part of the dimer interface. Understanding how hydrophobic residues control all parts of the C-helix makes us also appreciate that this helix is an extended allosteric sensor. Packing of the hydrophobic C-tail in PKA achieves the same thing as the L74F mutation in BRAF: -it extends the hydrophobic surface that is buttressed against the C-helix.
Conclusions.
Here, we have used the cancer kinome to help support the importance of the hydrophobic spine hypothesis for kinase activation. We searched for Phe replacements and tested some of them for their ability to stimulate kinase activity in cells. Only a limited number of kinases showed frequent Phe mutations. Two of these, BRAF and EGFR, are known to be able to be activated via an allosteric, catalytically independent mechanism (4, 21, 29). One of the residues that we tested in BRAF, V57F, did create a pseudokinase that could activate the ERK pathway in cells. The other two residues that we tested, L74 and L95F RS3, functioned to constitutively enhance the catalytic activity of BRAF. Modeling one of these residues, L74, revealed a newly appreciated network of hydrophobic residues that extends across the length of the αC-helix and involves the salt bridge, the gatekeeper residue, and two R-spine residues (Fig. 9). A final mutant, V600F, showed how another strategic hydrophobic mutation can be a cancer driver. Activating the kinase switch is clearly mediated by multiple hydrophobic interactions. Molecular dynamics simulations of these changes may provide new insights into the role of hydrophobic residues in the activation of protein kinases.
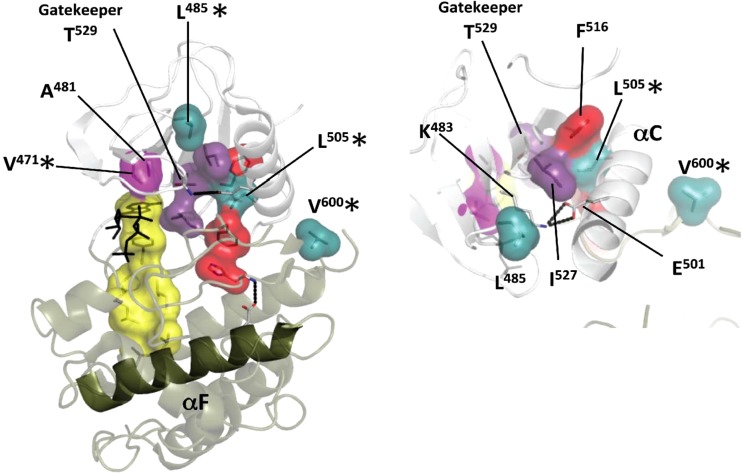
FIG 9.
Position of the spine mutants on the BRAF structure. On the left, the mutations that were introduced into BRAF C-spine residues (A481 and V471) are shown in pink. These mutations block ATP binding but generate pseudokinases that could serve as allosteric activators. Mutations of hydrophobic residues in and around the R-spine, including L485, V600, and L505, are shown in teal. These mutations produce a constitutively active phenotype, where activation is independent of Ras, dimerization, and activation loop phosphorylation. On the right, the “shell” residues that surround the R-spine (I527, T529, and V471) are shown in purple and include the “gatekeeper” T529. The assembled R-spine is shown in red, while the C-spine is shown in yellow with the exception of the mutated residues. The rotated view on the right shows a series of hydrophobic residues that surround the C-helix and can be manipulated to drive the assembly of the R-spine that leads to constitutive activation. The conserved K72-E91 ion pair is also shown. Mutations that are found in cancer genes are indicated with an asterisk.
ACKNOWLEDGMENTS
The work was supported by the Howard Hughes Medical Institute (A. S. Shaw and S. S. Taylor) and by grants from the NIH (AI57966 [A. S. Shaw], DK96688 [A. S. Shaw], and GM19301 [S. S. Taylor]). H. S. Meharena was supported by NSF (DGE1144086).
REFERENCES
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Taylor SS, Kornev AP. 2011. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci 36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornev AP, Taylor SS, Ten Eyck LF. 2008. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci U S A 105:14377–14382. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu J, Stites EC, Yu H, Germino EA, Meharena HS, Stork PJS, Kornev AP, Taylor SS, Shaw AS. 2013. Allosteric activation of functionally asymmetric RAF kinase dimers. Cell 154:1036–1046. doi: 10.1016/j.cell.2013.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baccarini M. 2005. Second nature: biological functions of the Raf-1 “kinase.” FEBS Lett 579:3271–3277. doi: 10.1016/j.febslet.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 6.Matallanas D, Birtwistle M, Romano D, Zebisch A, Rauch J, von Kriegsheim A, Kolch W. 2011. Raf family kinases: old dogs have learned new tricks. Genes Cancer 2:232–260. doi: 10.1177/1947601911407323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrison DK, Cutler RE. 1997. The complexity of Raf-1 regulation. Curr Opin Cell Biol 9:174–179. doi: 10.1016/S0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 8.Ory S, Morrison DK. 2004. Signal transduction: IMPlications for Ras-dependent ERK signaling. Curr Biol 14:R277–R278. doi: 10.1016/j.cub.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 9.Wimmer R, Baccarini M. 2010. Partner exchange: Protein-protein interactions in the Raf pathway. Trends Biochem Sci 35:660–668. doi: 10.1016/j.tibs.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Hu J, Yu H, Kornev AP, Zhao J, Filbert EL, Taylor SS, Shaw AS. 2011. Mutation that blocks ATP binding creates a pseudokinase stabilizing the scaffolding function of kinase suppressor of Ras, CRAF and BRAF. Proc Natl Acad Sci U S A 108:6067–6072. doi: 10.1073/pnas.1102554108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gosal G, Kochut KJ, Kannan N. 2011. Prokino: an ontology for integrative analysis of protein kinases in cancer. PLoS One 6:e28782. doi: 10.1371/journal.pone.0028782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ManChon U, Talevich E, Katiyar S, Rasheed K, Kannan N. 2014. Prediction and prioritization of rare oncogenic mutations in the cancer kinome using novel features and multiple classifiers. PLoS Comput Biol 10:e1003545. doi: 10.1371/journal.pcbi.1003545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin J, Xie P, Ventocilla C, Zhou G, Vultur A, Chen Q, Liu Q, Herlyn M, Winkler J, Marmorstein R. 2012. Identification of a novel family of BRAF(V600E) inhibitors. J Med Chem 55:5220–5230. doi: 10.1021/jm3004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiser A, Sali A. 2003. ModLoop: automated modeling of loops in protein structures. Bioinformatics 19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 15.Kornev AP, Taylor SS. 2009. Pseudokinases: functional insights gleaned from structure. Structure 17:5–7. doi: 10.1016/j.str.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang BH, Guan KL. 2000. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. EMBO J 19:5429–5439. doi: 10.1093/emboj/19.20.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajakulendran T, Sahmi M, Lefrançois M, Sicheri F, Therrien M. 2009. A dimerization-dependent mechanism drives RAF catalytic activation. Nature 461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 18.Meharena HS, Chang P, Keshwani MM, Oruganty K, Nene AK, Kannan N, Taylor SS, Kornev AP. 2013. Deciphering the structural basis of eukaryotic protein kinase regulation. PLoS Biol 11:e1001680. doi: 10.1371/journal.pbio.1001680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez-Viciana P, Tetsu O, Tidyman WE, Estep AL, Conger BA, Cruz MS, McCormick F, Rauen KA. 2006. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science 311:1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- 20.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JWC, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. 2002. Mutations of the BRAF gene in human cancer. Nature 417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 21.Wan PTC, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Project CG, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R. 2004. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116:855–867. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 22.Scheeff ED, Eswaran J, Bunkoczi G, Knapp S, Manning G. 2009. Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure 17:128–138. doi: 10.1016/j.str.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw AS, Kornev AP, Hu J, Ahuja LG, Taylor SS. 2014. Kinases and pseudokinases: lessons learned from RAF. Mol Cell Biol 34:1538–1546. doi: 10.1128/MCB.00057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanzarotti E, Biekofsky RR, Estrin DA, Marti MA, Turjanski AG. 2011. Aromatic-aromatic interactions in proteins: beyond the dimer. J Chem Infect Model 51:1623–1633. doi: 10.1021/ci200062e. [DOI] [PubMed] [Google Scholar]
- 25.Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan M, Taylor SS. 2001. Dynamics of cAMP-dependent protein kinase. Chem Rev 101:2243–2270. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 26.Iyer GH, Garrod S, Woods VL, Taylor SS. 2005. Catalytic independent functions of a protein kinase as revealed by a kinase-dead mutant: study of the Lys72His mutant of cAMP-dependent kinase. J Mol Biol 351:1110–1122. doi: 10.1016/j.jmb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Steichen JM, Iyer GH, Li S, Saldanha SA, Deal MS, Woods VL, Taylor SS. 2010. Global consequences of activation loop phosphorylation on protein kinase A. J Biol Chem 285:3825–3832. doi: 10.1074/jbc.M109.061820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steichen JM, Kuchinskas M, Keshwani MM, Yang J, Adams JA, Taylor SS. 2012. Structural basis for the regulation of protein kinase A by activation loop phosphorylation. J Biol Chem 287:14672–14680. doi: 10.1074/jbc.M111.335091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. 2006. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]