Abstract
Although the majority of genomic binding sites for the insulator protein CCCTC-binding factor (CTCF) are constitutively occupied, a subset show variable occupancy. Such variable sites provide an opportunity to assess context-specific CTCF functions in gene regulation. Here, we have identified a variably occupied CTCF site in the Drosophila Ultrabithorax (Ubx) gene. This site is occupied in tissues where Ubx is active (third thoracic leg imaginal disc) but is not bound in tissues where the Ubx gene is repressed (first thoracic leg imaginal disc). Using chromatin conformation capture, we show that this site preferentially interacts with the Ubx promoter region in the active state. The site lies close to Ubx enhancer elements and is also close to the locations of several gypsy transposon insertions that disrupt Ubx expression, leading to the bx mutant phenotype. gypsy insertions carry the Su(Hw)-dependent gypsy insulator and were found to affect both CTCF binding at the variable site and the chromatin topology. This suggests that insertion of the gypsy insulator in this region interferes with CTCF function and supports a model for the normal function of the variable CTCF site as a chromatin loop facilitator, promoting interaction between Ubx enhancers and the Ubx transcription start site.
INTRODUCTION
There is considerable evidence indicating a major role for the CCCTC-binding factor (CTCF) in genome organization (reviewed in references 1 and 2). CTCF binds to insulator elements and is required for their function in blocking interactions between enhancers and promoters (3). It has been shown to be involved in the formation of chromatin loops (4), and CTCF binding is enriched at the boundaries of topological chromatin domains (5–8). However, it is remains to be determined how much of CTCF function is linked to a specifically architectural role in genome organization and how much is more directly involved in the control of gene expression.
CTCF was originally identified as a transcription factor (9). Subsequent genome-wide mapping of CTCF binding revealed that 20% of binding sites are within 2.5 kb upstream from transcription start sites (10) and that CTCF sites are enriched at gene promoters (11, 12). A current unifying hypothesis is that the molecular function of CTCF is to mediate chromosomal loop formation and that this may give rise to a variety of context-dependent roles; in some contexts, loop formation may serve an architectural purpose, and in others, it may be more intimately associated with gene regulation. One way to partition CTCF binding sites into possible functional classes is to differentiate between sites that are constantly occupied and sites that show variable occupancy. The first comparisons between whole-genome maps of CTCF binding in different cell lines indicated that the majority of sites are constitutively bound (10, 13, 14). However, more recent studies have revealed higher proportions of variable sites (15, 16) and, interestingly, the variable sites are preferentially associated with enhancers (12). However, very few individual variable CTCF sites have yet been analyzed, and more examples are required to build an understanding of their association with gene regulation.
The classical example of a variable CTCF site is at the imprinted control region (ICR) of the mammalian insulin-like growth factor 2 gene (Igf2)/H19 locus, where CTCF binding is regulated by DNA methylation of the binding sites. On the maternal chromosome, CTCF binds the unmethylated ICR and the enhancer-blocking action of CTCF prevents Igf2 expression. However, on the paternal chromosome, methylation of the ICR prevents CTCF binding and the lack of insulator function enables Igf2 expression (17–20). A second example involves a CTCF site in the chicken lysozyme locus, where CTCF binding is regulated by the chromatin structure. Activation of the lysozyme gene is linked to eviction of CTCF, and this is mediated through the transcription of a noncoding RNA, chromosome remodeling, and repositioning of a nucleosome over the CTCF binding site (21). Recently, in Drosophila, Wood et al. provided evidence for two classes of regulated insulator (22). In one class, the occupancy of DNA-binding insulator proteins [e.g., BEAF, CTCF, and Su(Hw)] at insulator sites is regulated. In a second class, the DNA-binding insulator proteins are constitutively bound, but the insulators are regulated by the variable recruitment of other components (e.g., CP190) required to build a functional insulator complex.
Here, we present an analysis of a variably occupied CTCF site in the Drosophila Bithorax complex (BX-C). The BX-C contains three Hox genes, Ultrabithorax (Ubx), abdominal A (abd-A), and Abdominal B (Abd-B) and has a clear regulatory domain structure with independent regulatory elements controlling gene expression in the parasegmental (PS) units along the anteroposterior axis of the developing embryo (reviewed in reference 23). The regulatory domains are separated by boundaries that constrain the activation of PS-specific enhancers. Genetic deletion of boundaries leads to inappropriate enhancer activation and ectopic expression of Hox genes. CTCF binding is associated with BX-C boundaries, and CTCF mutations cause misexpression of Abd-B (24–26). The CTCF binding at boundary elements appears to be constitutive, and this may fit with an architectural role for these sites. Here, we report the identification of a variable CTCF site within the Ubx gene that preferentially binds CTCF when the Ubx gene is active and is associated with different chromatin topologies in active and inactive states. We present a model where CTCF has a role in facilitating the interaction between Ubx enhancers and the Ubx promoter.
MATERIALS AND METHODS
Fly lines.
The wild-type Drosophila melanogaster strain Oregon R was used in the chromatin immunoprecipitation (ChIP)-array, ChIP-quantitative PCR (qPCR), and chromosome conformation capture (3C) experiments. In addition, homozygous bx83Ka mutants (27) from the bx83Ka/TM6B strain were used in ChIP-PCR and 3C experiments.
Antibodies.
The following antibodies were used in the ChIP experiments: anti-CTCF-C antiserum (24), anti-CP190 antiserum (28), anti-RNA polymerase II (RNA Pol II) (0.9 mg/ml affinity-purified IgG, ab5131; Abcam), and anti-GAGA factor antibody (0.2 mg/ml IgG, SC-98263; Santa Cruz Biotechnology).
Chromatin preparation.
Dissected head segments of late 3rd instar larvae were inverted and fixed with 2% formaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature. These were washed with twice with PBS–125 mM glycine–0.01% Triton X-100, followed by a single wash with PBS and then with PBS containing 1% protease inhibitor cocktail (catalog number P8340; Sigma). The T1 and T3 leg imaginal discs were then dissected, snap-frozen in liquid nitrogen, and stored at −80°C prior to use. Approximately 150 leg discs were combined in PBS–0.01% Triton X-100 and centrifuged in a microcentrifuge at 1,200 rpm for 1 min. The discs were resuspended in 20 μl cell lysis buffer {5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 8, 85 mM KCl, 0.5% NP-40} containing 1% protease inhibitor cocktail and homogenized using a motorized pestle at 2-min intervals for 8 min. After a brief microcentrifuge centrifugation (13,200 rpm for 10 s), the pellet was resuspended in 300 μl nuclear lysis buffer (50 mM Tris-HCl, pH 8.1, 10 mM EDTA-Na2, 1% SDS) with protease inhibitors and incubated for 20 min at room temperature. The extracts were sonicated in a Bioruptor standard device (Diagenode) at the high setting for 4 min 15 s (cycles of 30 s on, 30 s off), producing 0.5- to 3.0-kb fragments. One hundred-milliliter aliquots of chromatin extracts were flash-frozen in liquid nitrogen and then stored at −80°C prior to use.
ChIP.
ChIP was performed as described by Birch-Machin et al. (29). One hundred-milliliter aliquots of chromatin were precleared with 13 μl blocked Staphylococcus aureus cells (SAC) and mixed with 200 μl of IP dilution buffer (16.7 mM Tris-HCl, pH 8, 167 mM NaCl, 1% EDTA, 1.1% Triton X-100, 0.01% SDS) with protease inhibitors. Two microliters of antibody was added, and the mixture was incubated on a roller overnight at 4°C. Then, 13 μl of SAC was added to each IP reaction mixture and the samples were incubated for 35 min at 4°C on a roller. The mixture was centrifuged in a microcentrifuge at 13,200 rpm at room temperature, and the pellets were washed successively with 1 ml each of low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA-Na2, pH 8.0, 20 mM Tris-HCl, pH 8, 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA-Na2, pH 8, 20 mM Tris-HCl, pH 8, 500 mM NaCl), and LiCl buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA-Na2, pH 8.0, 10 mM Tris-HCl, pH 8.0) and twice with Tris-EDTA (TE) buffer, pH 8.0, for 5 min at 4°C on a roller for each solution. The immune-precipitated chromatin was then eluted twice from the SAC pellet with 300 μl of IP elution buffer (50 mM NaHCO3, 1% SDS) by vigorously vortexing for 15 min at room temperature. One microliter of RNase A (catalog number R4642; Sigma) and 24.3 μl of 4 M NaCl (0.3 M final concentration) were then added to the eluate, and the mixture was incubated for 4 h at 65°C to reverse the cross-linking. The DNA was then precipitated by adding 812 μl of 100% ethanol and incubating overnight at −20°C. The samples were centrifuged in a microcentrifuge at 4°C for 20 min, and the pellets were air dried for 1 h at room temperature. The pellets were resuspended in 100 μl TE buffer, followed by the addition of 25 μl of 5× PK buffer (50 mM Tris-HCl, pH 7.5, 25 mM EDTA-Na2, pH 8, 1.25% SDS) and 1.5 μl of 20 mg/ml proteinase K, incubated at 45°C for 2 h, and purified using the QIAquick PCR purification kit (catalog number 28104; Qiagen). The DNA was eluted in 30 μl of buffer EB and stored at −20°C until use.
CTCF ChIP-array.
Five microliters of CTCF ChIP and 5 μl of control ChIP DNA from T1 and T3 leg discs obtained from Oregon R larvae were amplified using the GenomePlex single-cell whole-genome amplification kit (product number WGA4; Sigma-Aldrich) according to the manufacturer's instructions. The samples were amplified for 21 cycles, and the amplified DNA purified using the QIAquick PCR purification kit. One microgram each of amplified ChIP and control DNA were labeled with Cy5 and Cy3 in the presence of Cy3- or Cy5-dCTP (GE Healthcare) using the BioPrime DNA labeling kit (Invitrogen) and hybridized onto Nimblegen ChIP-chip (ChIP with microarray technology) 2.1 M probe whole-genome tiling arrays according to the manufacturer's instructions.
Microarray data processing.
Two biological replicates were prepared for each sample with a Cy3/Cy5 dye swap for one biological replicate of each sample. ChIP DNA prepared with preimmune serum was used as the reference control to assay ChIP enrichment in the array experiments. Arrays were scanned and processed as previously described (30). The enrichment profiles were visualized using the Integrated Genome Browser (http://bioviz.org/igb/index.html). Patser position-specific weight matrix analysis was as described previously (24). Analysis of conservation used the PhastCons multiple alignment data available from http://genome.ucsc.edu.
Quantitative PCR.
Quantitative real-time PCR experiments were performed with a LightCycler 480 II (Roche Diagnostics) in 10-μl reaction mixtures using SYBR green PCR master mix (catalog number 04707516001; Roche). Each reaction mixture consisted of 5 μl SYBR green PCR master mix, 3 μl water, 1 μl 10 μM primer mix, and 1 μl DNA. Amplification was carried using the following conditions: 1 cycle at 95°C for 15 min and 45 cycles of 95°C for 10 s, 58°C for 10 s, and 72°C for 10 s. The primer pairs used for the amplification are listed in Table 1. Serial dilutions of Drosophila genomic DNA (100 to 0.01 ng/μl) were used as standards for quantification.
TABLE 1.
ChIP-qPCR primers
| IDa | Primer | Chr | Directionb | Position |
Sequence | |
|---|---|---|---|---|---|---|
| Start | End | |||||
| 0 | Neg | 3R | F | 12526683 | 12526702 | CCTAAATGGCAGAGGATTGG |
| R | 12526792 | 12526773 | AAATTCAGGATGCAGGATGC | |||
| 1 | R1 | 3R | F | 12528866 | 12528885 | ATCAGCAGCCGTTGAGTAGG |
| R | 12528971 | 12528952 | ATTCCTCAGCGACAAAGAGC | |||
| 2 | R2 | 3R | F | 12529660 | 12529679 | GAGTTGCCATAAAGCACTCG |
| R | 12529764 | 12529745 | TTCTCTTCGCAGCCTATTCC | |||
| 3 | R3 | 3R | F | 12529861 | 12529880 | TTACAGCCGACACCTCATCA |
| R | 12529987 | 12529968 | CTGGCTTGACACTGGGCTAC | |||
| 4 | R4 | 3R | F | 12530745 | 12530769 | CTCGCTGGTTCCTAATATGATATAC |
| R | 12530863 | 12530846 | GTGCCTTTCGGTGACTTC | |||
| 5 | R5 | 3R | F | 12531112 | 12531129 | GCACAGATTCCGTTGAGC |
| R | 12531253 | 12531234 | CCTTCTATGCTCTGCTCTCG | |||
| +ve | BXC-49 | 3R | F | 12760726 | 12760707 | ATCGATAAAAAGCGCCAACA |
| R | 12760565 | 12760584 | GCTCTTACTGCCCGATTCTG | |||
| −ve | SuVar 3-9 | 3R | F | 11087377 | 11087396 | AGCCGCTACTATTGCTTGGA |
| R | 11087573 | 11087554 | GCAGCGACAGCAGTATGAAA | |||
| Ubx-P | F-675 | 3R | F | 12559800 | 12559819 | AATACTTGGATTGCGCTTGC |
| R | 12560001 | 12559982 | TTTCCACTAGATTGGCGTCC | |||
Preparation of 3C DNA from T1 and T3 leg discs.
Approximately 450 each of the T1 and T3 leg discs from 3rd instar larvae were dissected and frozen as described above. The discs were thawed on ice and transferred to a 1.5-ml microcentrifuge tube. The pooled discs were briefly centrifuged at 13,200 rpm for 10 s. The excess liquid was discarded and the discs were resuspended in 20 μl lysis buffer (31) containing 10 mM Tris-Cl, pH 8.0, 10 mM NaCl, 0.2% Igepal CA360 (catalog number I8896; Sigma), and 10 μl/ml of protease inhibitor (Sigma). The discs were homogenized with a plastic motorized pestle at 2-min intervals for a total of 8 min. After a brief centrifugation, 500 μl of lysis buffer with 50 μl of protease inhibitor was added to the homogenate, and the suspension was centrifuged at 5,000 rpm for 5 min at room temperature.
The 3C DNA was prepared based on the protocol described by Hagège et al. (32). The leg disc lysate pellet was washed twice with ice-cold 1.2× NEBuffer 3 (catalog number B7003S; New England BioLabs) at 5,000 rpm for 5 min at room temperature. The pellet was then resuspended in 500 μl 1.2× NEBuffer 3 and 7.5 μl 20% SDS. The mixture was incubated at 37°C, 900 rpm for 1 h in a Thermomixer (catalog number 5355000038; Eppendorf). Then, 50 μl 20% Triton X-100 was added and the mixture further incubated at 37°C, 900 rpm for 1 h. The lysate was then digested with 400 U of DpnII at 37°C, 900 rpm overnight. The enzyme was inactivated by heat treatment at 65°C for 20 min, and the mixture was ligated at 16°C for 16 h in a 10-ml reaction mixture with 10,000 U of T4 DNA ligase (New England BioLabs). The ligated chromatin digest was then de-cross-linked and purified as described by Hagège et al. (32). The purified 3C DNA was resuspended in 50 μl TLE buffer (10 mM Tris-Cl, pH 8.0, 0.1 mM EDTA), and the DNA concentration was measured by using the Qubit dsDNA HS assay kit (catalog number Q32854; Invitrogen). 3C DNA samples were stored at −20° until use.
PCR amplification of 3C DNA.
3C interactions were determined according to the protocol of Dekker et al. (33). To investigate the chromatin conformation and interactions in the Ubx region in T1 and T3 leg discs, 29 primers spanning chromosome (Chr) 3R:12400341.0.12695484 were designed based on the expected fragments generated by DpnII digestion (Table 2). In addition, primer pairs located in DpnII fragments containing the CTCF differential peak in Ubx, the Ubx promoter, and the Mcp region were also designed to serve as anchor fragment internal primers (Table 3).
TABLE 2.
3C PCR primers
| ID | Primer | Chr | Position |
Sequence | |
|---|---|---|---|---|---|
| Start | End | ||||
| 1 | 223 | 3R | 12400341 | 12400360 | GCGAGACGATAAACGACGAC |
| 2 | 237 | 3R | 12412997 | 12413016 | AAGAAGTGGTAAAGTGGCGG |
| 3 | 372 | 3R | 12444906 | 12444925 | CTGTGCATCTCCACCACATC |
| 4 | 396 | 3R | 12449306 | 12449325 | CAGAAGCTGCCTCTCGTAGG |
| 5 | 444 | 3R | 12465581 | 12465600 | CAAAGCCACCTTCCTGAAAC |
| 6 | 478 | 3R | 12474725 | 12474744 | ATCTCGCCCAGCACTATTTG |
| 7 | 504 | 3R | 12480871 | 12480890 | TTTGAGTGGGTTAAGCTGCC |
| 8 | 559 | 3R | 12508313 | 12508332 | TAAATACGAAGTGCATGCGG |
| 9 | 589 | 3R | 12529861 | 12529880 | TTACAGCCGACACCTCATCA |
| 10 | 590 | 3R | 12530474 | 12530494 | GGAACACGCATATAGCATTGG |
| 11 | 636 | 3R | 12549178 | 12549196 | TTTGAAATGCAAACACGGC |
| 12 | 674 | 3R | 12559159 | 12559178 | GGAGGCCTGTTCAAAGTACG |
| 13 | 675 | 3R | 12559351 | 12559332 | CAAAGGAGGCAAAGGAACAG |
| 14 | 677 | 3R | 12561570 | 12561589 | CGAGAAGACCCAGAGCAAAG |
| 15 | 698 | 3R | 12574489 | 12574509 | AAGAAATATGCGTTTCCCACC |
| 16 | 699 | 3R | 12575770 | 12575788 | CGCCAGACAATGGAAACTG |
| 17 | 745 | 3R | 12592412 | 12592433 | GTGCTATCAACTCGCTTTCTTG |
| 18 | 751 | 3R | 12593896 | 12593915 | CTCTTTGTTAGCGGAGGCAG |
| 19 | 789 | 3R | 12608923 | 12608942 | TAAGCGAGTGCGTGTCATTC |
| 20 | 842 | 3R | 12625282 | 12625303 | TCATCTGGAACTGGTTCTATCG |
| 21 | 858 | 3R | 12633588 | 12633607 | AATCCGGTTGTGAAACAAGG |
| 22 | 875 | 3R | 12640691 | 12640710 | TCAGTCTCACAGCCATTTCG |
| 23 | 899 | 3R | 12649777 | 12649797 | GCATGTGCATTTAAGGAGTGG |
| 24 | 918 | 3R | 12657009 | 12657031 | CCAGTTAATGTGCTTCCTACCTG |
| 25 | 918 | 3R | 12657020 | 12657043 | GCTTCCTACCTGTCTATTTGTTGG |
| 26 | 919 | 3R | 12658026 | 12658046 | GTGTCGAGTTTCGGTTGAGTC |
| 27 | 923 | 3R | 12660715 | 12660734 | AAATGTTTGGACGGGAAATG |
| 30 | 961 | 3R | 12683796 | 12683817 | GCTTTAACTTTAACCTCTGGCG |
| 31 | 983 | 3R | 12695484 | 12695507 | CTGCTCTGCTTATCAGTTTATTGG |
TABLE 3.
Anchor fragment internal primers for 3C PCR
| Anchor position | Fragment ID | Chr | Directiona | Position |
Sequence | |
|---|---|---|---|---|---|---|
| Start | End | |||||
| Ubx promoter | 675 | 3R | F | 12559800 | 12559819 | AATACTTGGATTGCGCTTGC |
| R | 12560001 | 12559982 | TTTCCACTAGATTGGCGTCC | |||
| Variable CTCF site_1 | 589 | 3R | F | 12529861 | 12529880 | TTACAGCCGACACCTCATCA |
| R | 12529987 | 12529968 | CTGGCTTGACACTGGGCTAC | |||
| Variable CTCF site_2 | 590 | 3R | F | 12530221 | 12530240 | AGGGTTAATTCGTTCATCGC |
| R | 12530362 | 12530343 | CTGATGATGACGCTGTTGTG | |||
| Mcp | 983 | 3R | F | 12694755 | 12694774 | ATTGTATGTATCCGCTCCGC |
| R | 12694917 | 12694898 | AAGCCCTTATTTGCAGACCC | |||
F, forward; R, reverse.
For each anchor fragment investigated, individual 10 μM primer mixes composed of the anchor fragment internal primers and individual anchor primer/target primer pairs were prepared. The 3C PCRs were carried out in a 25-μl mixture using the Thermo-Start Taq DNA polymerase kit (product number AB-1057; Thermo Scientific). Each reaction mixture contained 18.3 μl water, 2.5 μl 10× PCR buffer, 1.5 μl 25 mM MgCl2, 0.5 μl 10 mM deoxynucleoside triphosphate (dNTP) mix, 0.2 μl Taq DNA polymerase, 1 μl 10 μM primer mix, and 1 μl (1 ng/μl) of 3C DNA sample. Amplification was carried out in an iCycler 582BR thermal cycler (Bio-Rad) using a touchdown protocol with 1 cycle at 95°C for 15 min and then 10 cycles at 95°C for 30 s, annealing from 69 to 59°C for 30 s, and 72°C for 30 s. This was followed by 30 cycles at 95°C for 30 s, 59°C for 30 s, and 72°C for 30 s, followed by a final extension at 72°C for 10 min. PCR products were then subjected to electrophoresis on a 2% agarose gel in 0.5× Tris-borate-EDTA (TBE).
Quantification of 3C PCR products.
Gel images were digitized, and the bands were quantified using ImageJ software (http://imagej.nih.gov/ij). The relative interaction between the different primer pairs was then expressed as the ratio of the signal strength between the anchor/target 3C PCR product and the anchor fragment PCR product. The relative interactions between the 3C primer pairs and each specific anchor fragment were plotted to visualize interactions.
Microarray data accession number.
The ChIP-array data have been submitted to GEO under accession number GSE62234.
RESULTS
Identification of a variably occupied CTCF site in the Ubx gene.
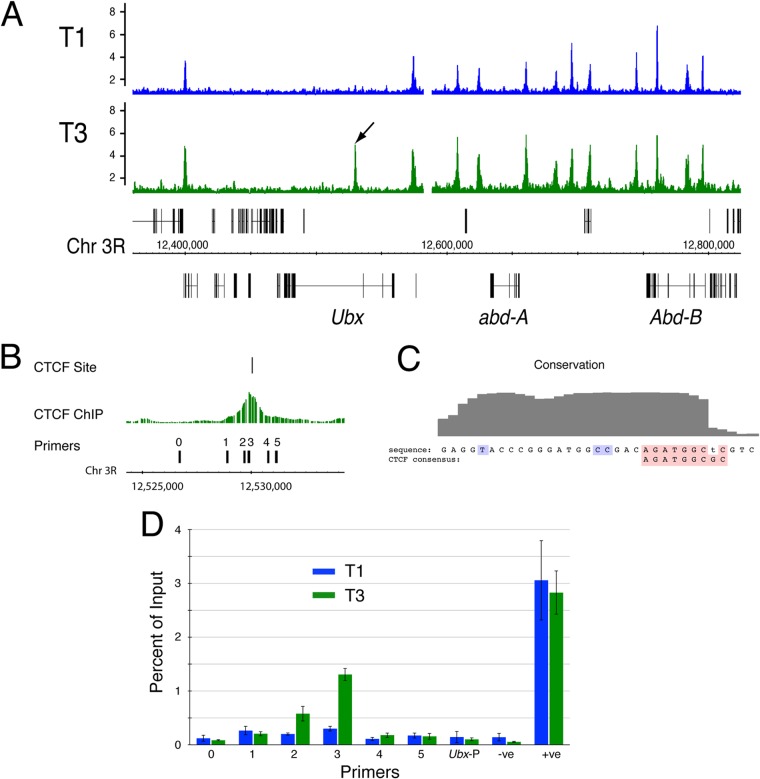
The individual Hox genes of the BX-C are expressed in different segments along the anteroposterior axis (23), presenting a useful experimental system for the isolation of in vivo tissues with different states of gene expression in sufficient quantities for genomic analysis. Here, we have used the imaginal discs from Drosophila larvae to compare the genome-wide CTCF binding profile in leg imaginal discs from the 1st thoracic segment (T1) to that of leg discs from the 3rd thoracic segment (T3). The Hox gene Ubx is not expressed in T1 but is active in T3. The other two genes of the BX-C, abd-A and Abd-B, are inactive in both T1 and T3. The activity state of these BX-C genes is regulated by Polycomb (Pc) silencing, which imposes a repressive chromatin state on inactive genes. Comparing the CTCF ChIP-array profiles of the T1 leg disc and the T3 leg disc, we find that the profiles are generally extremely similar, with very few clear differential peaks. However, we identify a clear differential CTCF binding peak in the Ubx gene (Fig. 1A). There is strong CTCF binding at this position in the T3 leg disc, where Ubx is expressed, but we find little binding at this site in the T1 leg disc, where the Ubx gene is repressed. In contrast, the binding of CTCF in the repressed abd-A and Abd-B regions is very similar in both discs.
FIG 1.
A variably occupied CTCF site in the Ubx gene. (A) CTCF binding profiles from T1 (Ubx inactive) and T3 (Ubx active) leg imaginal discs. The arrow indicates the variably occupied CTCF site. Ubx, abd-A, and Abd-B are transcribed from right to left. (B) The CTCF ChIP peak aligns with a match to the CTCF position-specific weight matrix. The positions of the PCR primers used in the PCR whose results are reported in panel D are shown. (C) PhastCons conservation plot across 15 insect species (http://genome.ucsc.edu). The sequence at the variable CTCF site is compared with the Drosophila consensus (red) (36). The conserved CC motif (34) and conserved T in module number 1 of Rhee and Pugh (35) are indicated in blue. (D) ChIP-PCR confirming the differential binding of CTCF at the variable site. Ubx-P is at the Ubx promoter; for the −ve and +ve primers, see Table 1. Error bars show standard errors of the means.
The variably occupied CTCF site lies in an intron within the Ubx transcription unit. Motif analysis with the CTCF position-weight matrix revealed a strong sequence match at this position (Fig. 1B). It has been proposed that CTCF sites serving different functions may be identifiable at the sequence level, and subfamilies of CTCF binding sites have been identified. We examined the variable site for sequence features that might place it in a defined subfamily. In general, the variable site has features associated with high occupancy, having, in addition to a strong match to the core motif (Patser score = 12.3), the conserved T of module number 1 described by Rhee and Pugh and the CC motif (Fig. 1C) that are both associated with higher levels of CTCF binding (34, 35). The variable site is on the edge of a sequence block that is highly conserved across 15 insect genomes (Fig. 1C), and CTCF binding at this site is clearly identified in pupal-stage chromatin from four Drosophila species (D. melanogaster, D. simulans, D. yakuba, and D. pseudoobscura) covering a range of evolutionary divergence of up to 25 million years (36).
We validated the differential CTCF binding at this site using quantitative PCR with a set of primer pairs spanning the CTCF peak (Fig. 1B and D). We see clearly enriched CTCF binding in T3 versus T1 leg disc chromatin specifically at this CTCF site.
Protein complex formation at the variable CTCF site.
To investigate whether the DNA binding protein CTCF is involved in building a protein complex together with other insulator proteins or transcription factors at this site, we analyzed the binding of other protein components (Fig. 2). Centrosomal protein 190 (CP190) does not bind DNA directly but associates with CTCF [and other DNA-binding insulator components, such as Su(Hw)] through a BTB domain interaction and is required for the enhancer-blocking function of insulator complexes (25, 37, 38) and for looping interactions of CTCF insulators (22). We find no evidence for CP190 association with the variable CTCF site in T1 leg disc chromatin, but CP190 is significantly associated with this site in T3 leg disc chromatin. This suggests that differential binding of CTCF in T3 enables the formation of a protein complex involving proteins associated with insulator function.
FIG 2.
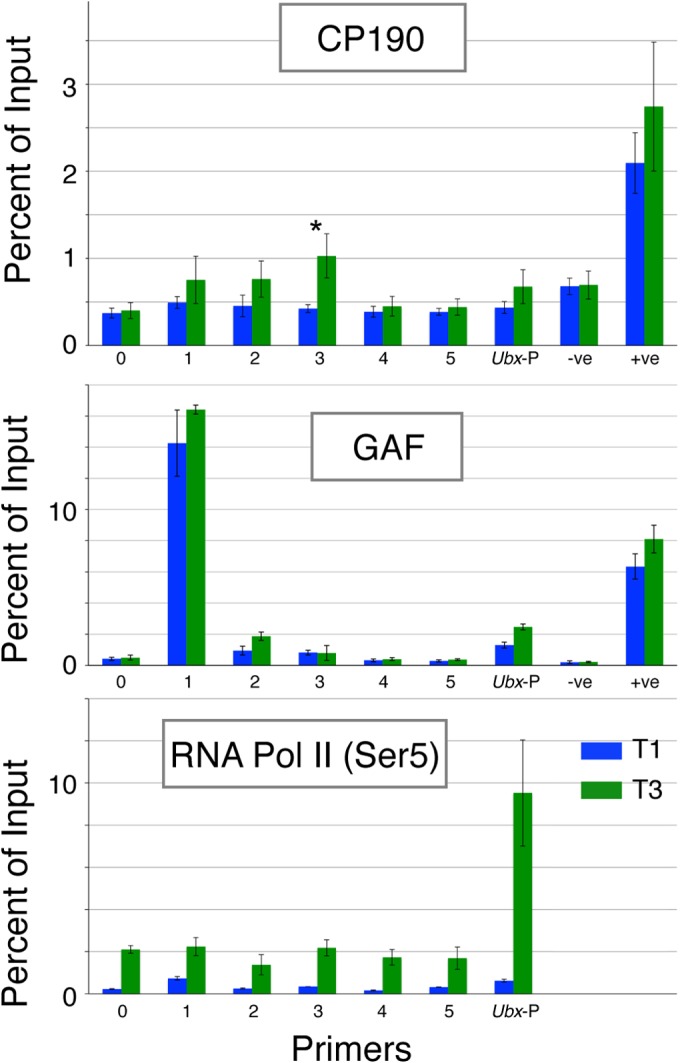
ChIP-PCR analysis of binding of CP190, GAF, and RNA Pol II (Ser5) in the region of the variably occupied CTCF site. RNA Pol II (Ser5) refers to the Ser5-phosphorylated form of RNA Pol II. The results for T1 and T3 chromatin are color coded as shown in the key. Primers are as described in the legend to Fig. 1 and shown in that figure. *, P = 0.02 (t test). Error bars show standard errors of the means.
GAGA factor (GAF) appears to participate in a diverse range of transcriptional processes and is required for the activity of some insulators (39–41). GAF does not bind at the variable CTCF site, but there is substantial binding in the region of primer pair 1 that lies about 1 kb away from the CTCF site (Fig. 2). This strong GAF binding is similar in both T1 and T3 leg imaginal disc chromatin. We also examined the binding of the insulator components Su(Hw), mod(mdg4 isoform N), and BEAF32 but found no evidence for binding in the region of the variable CTCF site in leg discs (data not shown).
Intronic CTCF sites have been implicated in splicing regulation and Pol II pausing (42). We examined the binding profile of Pol II across the region spanning the variable CTCF site and at the Ubx promoter using an antibody that recognizes the Ser5-phosphorylated (Ser5P) Pol II (Fig. 2). Pol II-Ser5P is found preferentially bound across the region in T3 versus T1 discs, which fits with the specific Ubx expression in T3; however, there is no pronounced peak at the CTCF site, and thus, we see no evidence of Pol II pausing at this site. At the promoter, Pol II-Ser5P shows strong binding in T3 and no binding in T1, indicating the engagement of Pol II with the active promoter and a lack of paused Pol II when the Ubx promoter is inactive.
Chromatin topology in the active and inactive states.
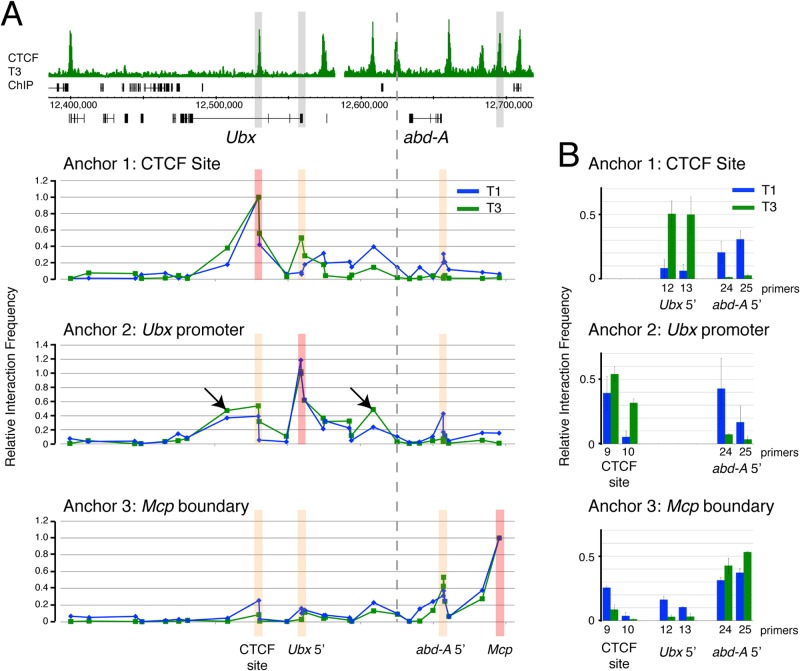
We next investigated whether the variable CTCF-dependent protein complex that assembles on the active Ubx gene is associated with alteration in chromosomal topology between the inactive and active states of Ubx transcription. We used chromosome conformation capture (3C) (33) to analyze interactions from the viewpoint of the variable CTCF site as an anchor fragment and 28 nearby target sites, including the Ubx promoter, the abd-A promoter, and CTCF sites across the Ubx and abd-A regions. The overall interaction profiles are shown in Fig. 3A, and the interaction scores for selected primers closest to particular features, e.g., the Ubx promoter and the abd-A promoter, are detailed in Fig. 3B. We find that the variable CTCF site shows a marked preferential interaction with the Ubx promoter in the Ubx active (T3) state (Fig. 3B, anchor 1, Ubx 5′ primers). In contrast, the interaction of the variable CTCF site with the repressed abd-A promoter shows the reverse preference; in T3, there is no interaction, but in the Ubx inactive state (T1), the variable CTCF site is associated with the repressed abd-A promoter (Fig. 3B, anchor 1, abd-A 5′ primers).
FIG 3.
Chromatin interactions in the BX-C in T1 and T3. (A) 3C interactions at 29 sites in the BX-C. Top, overview of the BX-C, showing the T3 CTCF ChIP profile with 3C anchor positions highlighted in gray. The graphs below show the 3C profiles. The genomic sites of anchors 1 (primer 589), 2 (primer 675), and 3 (primer 983) are indicated. Anchor positions are indicated by red bars, and orange bars indicate positions whose results are detailed in panel B. Arrows in anchor 2 data indicate interactions of the Ubx promoter with sites in the abx (left) and pbx (right) regulatory regions. The dashed line indicates the boundary between the Ubx and abd-A regulatory domains (61). Primers are listed in Tables 2 and 3. (B) T1 versus T3 comparisons focusing on selected primers that are closest to key genomic features; for the interactions between anchors and the variable CTCF site, we show data for primers 9 and 10; for the Ubx promoter, primers 12 and 13, and for the abd-A promoter primers 24 and 25. Error bars show standard errors of the means.
Since using the variable CTCF site as the 3C anchor indicated a specific preferential interaction with the Ubx promoter in the active state, we next examined interaction from the viewpoint of a 3C anchor at the Ubx promoter. This confirmed the preferential interaction between the variable CTCF site and the Ubx promoter in the active (T3) state (Fig. 3B, anchor 2, CTCF site primers). In contrast, in T1, the repressed Ubx promoter shows evidence of a preferential interaction with the repressed abd-A promoter.
We also examined a third viewpoint using a 3C anchor at the Mcp boundary element, which contains a CTCF binding site and is in the repressed abd-A domain in both T1 and T3. The Mcp anchor shows a peak of interaction with the abd-A promoter in both T1 and T3 but shows a preferential interaction with the Ubx promoter and the variable CTCF site in the inactive (T1) state (Fig. 3B, anchor 3). Since there is little CTCF associated with the variable site in the inactive state, these interactions may involve the nearby Polycomb response element (bx-PRE) (Fig. 4).
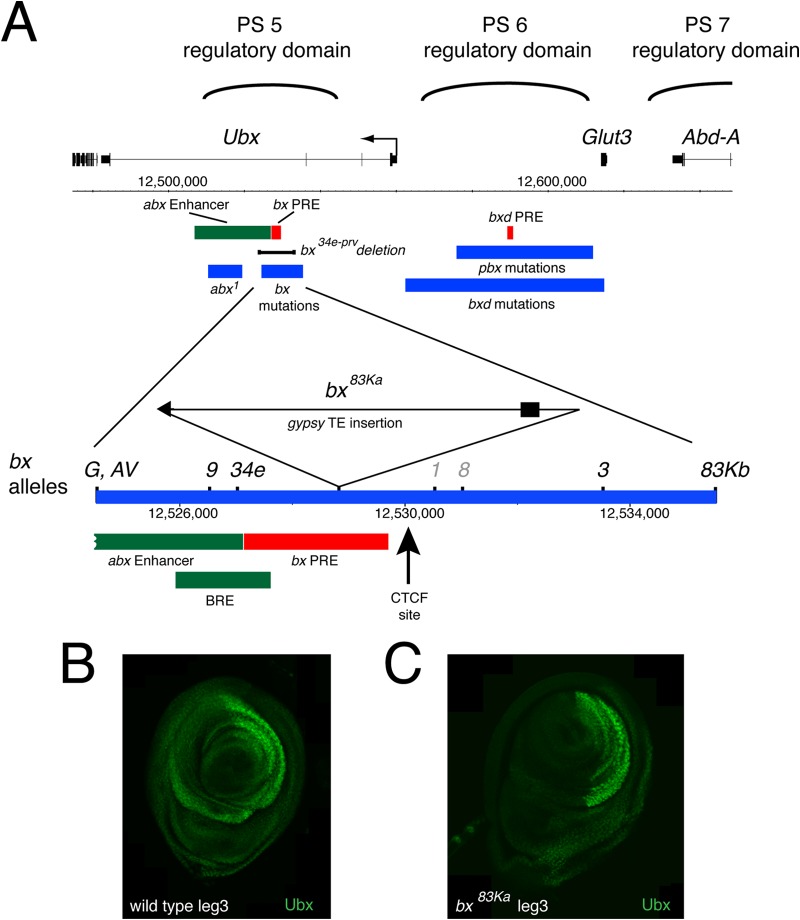
FIG 4.
Ubx regulation and bx mutations. (A) Map of the Ubx regulatory region. Regulatory regions defined by mutation are in blue. The rectangle on the gypsy transposable element indicates Su(Hw) binding sites. Coordinates are indicated for abx enhancer (abx20 [45]), bx and bxd PREs (62), pbx and bxd mutations (61), abx1 and bx alleles (27), and BRE (44). The gypsy insertion in bx83Ka was mapped by sequencing: the insertion is at Chr 3R:12528835, with a 6-bp duplication of the target site at position 12528830 to 12528835. In addition to the indicated cluster of bx alleles (gypsy-associated alleles in black and non-gypsy alleles in gray), there is also an outlier, bxF31, associated with an I element insertion at approximately position 12516500 (27). (B and C) Immunofluorescence labeling of Ubx expression in wild-type (B) and bx83Ka (C) T3 leg imaginal discs. The discs are oriented with the anterior to the left; the in bx83Ka T3 leg imaginal disc, Ubx expression is strongly reduced in the anterior compartment.
Overall, the 3C analysis indicates that the Ubx region adopts a different chromatin topology in the active versus the inactive state. The active (T3) state is characterized by increased interaction between the variable CTCF site and the Ubx promoter and decreased association of both the variable CTCF site and the Ubx promoter with repressed regions, specifically, the abd-A promoter and the Mcp boundary element.
Chromatin topology in the bx83Ka mutation.
The variable CTCF site lies close to the bx-PRE (43), the BRE embryonic enhancers (44), and the abx enhancers (45), which are active both in the embryo and in imaginal discs (Fig. 4). This arrangement, together with the interaction between the variable CTCF site and the Ubx promoter, suggests a model where the variable CTCF site may play a role in facilitating interaction between the abx/bx enhancers and the Ubx promoter. Deletion of a 9.5-kb region that includes the variable CTCF site gives a bx phenotype (bx34e-prv) (27) caused by decreased Ubx expression in T3 discs, and it is intriguing that the variable CTCF site lies in the heart of the region defined by the cluster of bx mutations. There is a strong connection between bx mutations and insulator function since, of the 10 bx mutations, seven are caused by the insertion of gypsy transposable elements (27, 46), which carry a cluster of binding sites for the Su(Hw) insulator protein, the most studied insulator in Drosophila (47). These gypsy-induced bx alleles are all suppressed in a su(Hw) mutant background (27, 46), indicating that it is not simply the presence of the 7.5-kb gypsy element but, rather, the binding of the Su(Hw) insulator protein that causes the bx mutant phenotype. This suggests that this region is topologically sensitive and that the gypsy insertions may interfere with interactions between the abx/bx enhancers and the Ubx promoter. Specifically, in terms of the above-described model for the function of the variable CTCF site, the insertion of a second topological regulator, Su(Hw), in this region may interfere with the interaction between the variable CTCF site and the Ubx promoter.
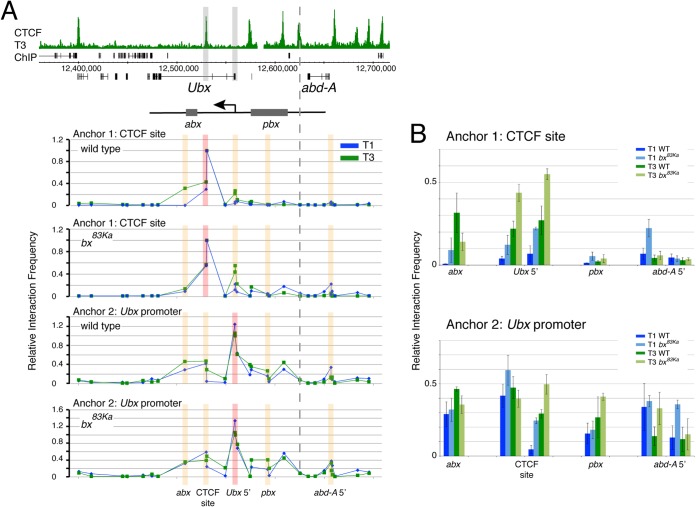
To test this hypothesis, we examined the effect of a bx mutation on chromatin topology by carrying out 3C analysis on homozygous bx83Ka T1 and T3 leg discs. The phenotype of bx mutations is a loss of Ubx expression in the anterior compartment of the T3 imaginal discs, haltere and T3 leg (Fig. 4B and C) (48). In the anterior compartment, Ubx expression may depend on interactions between the promoter and the downstream enhancers, abx and bx, whereas in the posterior compartment, the Ubx promoter may contact the upstream pbx region. This fits with the presence of both upstream and downstream preferential interactions with the Ubx promoter in the active state that we observed in the 3C analysis (Fig. 3A, arrows). The bx mutations might be expected to interfere specifically with the downstream interaction.
In the 3C analysis, we find that the mutation has several effects on chromatin topology in the Ubx region (Fig. 5). First, contrary to the expectations of the model, the gypsy insertion enhances interaction between the variable CTCF site and the Ubx promoter. This enhancement is seen in both T1 and T3, although the interaction remains stronger in T3 (Fig. 5B, anchor 1, Ubx5′ primers, and anchor 2, CTCF site primers). Second, fitting the predictions of the model, the preferential interaction seen in the active state (T3) between the downstream abx enhancer region and the variable CTCF site is lost in the mutant (Fig. 5B, anchor 1, abx primer). Similarly, for the interaction between the abx enhancer region and the Ubx promoter (Fig. 5B, anchor 2, abx primer), there is evidence for stronger interaction in T3 than in T1 in the wild type, and this differential is lost in the mutant. Also fitting the model, in contrast to the abx region, the pbx region preferentially interacts with the Ubx promoter in the active state (T3) in the bx83Ka mutant (Fig. 5B, anchor 2, pbx primer).
FIG 5.
Chromatin interactions in the BX-C in T1 and T3, comparing data for the wild type and the bx83Ka mutant. (A) 3C interactions at 29 sites in the BX-C. Top, overview of the BX-C showing the T3 CTCF ChIP profile with 3C anchor positions highlighted in gray. The positions of the abx and pbx regulatory regions are indicated, corresponding to abx1 deletion (27) and pbx deletions (61). The graphs below show the 3C profiles. The genomic sites of anchors 1 (primer 590) and 2 (primer 675) are indicated. Anchor positions are indicated by red bars, and orange bars indicate positions whose results are detailed in panel B. The dotted line indicates the boundary between the Ubx and abd-A regulatory domains (61). Primers are listed in Tables 2 and 3. (B) Comparisons of interactions at specific sites focusing on selected primers that are closest to key genomic features; for the interactions between anchors and the abx region, we show data for primer 8; for the variable CTCF site, primers 9 (left bars) and 10 (right bars); for the Ubx promoter, primers 12 (left bars) and 13 (right bars); for the pbx region, primer 17; and for the abd-A promoter, primers 24 (left bars) and 25 (right bars). Error bars show standard errors of the means.
Overall, although some predictions of the model are borne out, it appears that the effects of the gypsy insertion are more complex than simply blocking interactions between the variable CTCF site and the Ubx promoter.
The bx83Ka insertion affects protein binding in flanking regions.
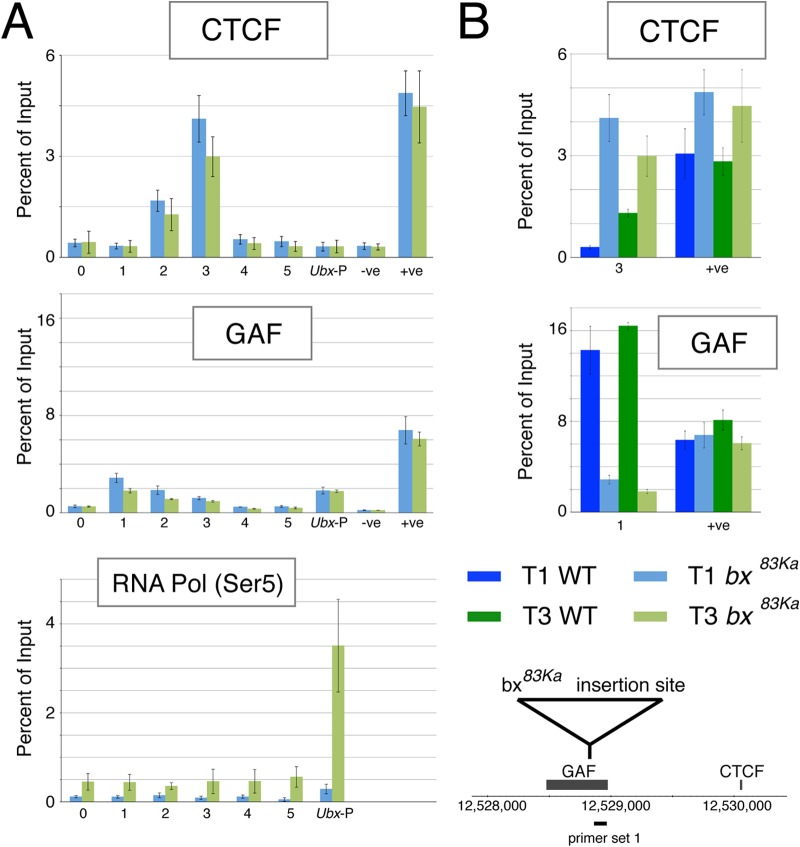
To investigate this further, we examined protein binding in the region of the variable CTCF site in homozygous bx83Ka T1 and T3 leg discs (Fig. 6). Strikingly, we find that, in the mutant, CTCF is strongly associated with the site not only in T3 but also in T1. In addition, we find that the gypsy insertion in the bx83Ka mutation also strongly affects GAF binding; compared to the wild type, it is markedly reduced in both T1 and T3. Pol II binding shows, as expected, clear occupancy in the T3 discs, where Ubx is expressed in posterior compartment cells.
FIG 6.
Binding of CTCF, GAF, and RNA Pol II (Ser 5) in the region of the variably occupied CTCF site in the bx83Ka mutant. (A) ChIP-PCR analysis of T1 and T3 in the bx83Ka mutant. RNA Pol (Ser5) refers to the Ser5-phosphorylated form of RNA Pol II. Data are in blue for T1 chromatin and in green for T3 chromatin. Primers are indicated on the x axes and are as described in the legend to Fig. 1 and shown in that figure. (B) Comparison of wild type versus bx83Ka at T1 and T3 for the CTCF peak (primer 3) and for the GAF peak (primer 1). Data are color coded as shown in the key. Error bars indicate standard errors of the means. The GAF binding interval is from reference 63.
Overall, perhaps the most striking effect of the bx83Ka insertion is the increase in CTCF binding at the variable CTCF site, particularly in T1. This indicates that the gypsy insulator can affect the loading of insulator proteins onto a nearby site, and this fits with an increased association between the variable insulator site and the Ubx promoter. It is possible that this interaction excludes the abx regulatory region, since the preferential contact between the abx regulatory region and the variable CTCF site seen in the active state in the wild type is lost in the mutant.
DISCUSSION
We have identified a variably occupied CTCF binding site in the Ubx gene in the Drosophila BX-C. This site lies close to characterized Ubx regulatory elements, and we find that CTCF occupancy is associated with a specific interaction between the variable site and the Ubx promoter in the transcriptionally active state. These observations suggest a model that CTCF binding at this site facilitates interaction between the regulatory elements and the Ubx promoter.
This model is supported by our studies on the bx83Ka mutation, where the insertion of a gypsy insulator close to the variable CTCF site disrupts the chromatin topology. One explanation for the effect of the gypsy insertion on Ubx expression is that the gypsy insulator acts as an enhancer blocker, preventing interactions between the Ubx promoter and regulatory elements (e.g., abx) lying beyond the insulator insertion site (49). However, a simple enhancer blocking model does not fit with the enhanced interaction we see between the variable CTCF site and the Ubx promoter in the bx83Ka mutant, nor does it explain the tight clustering of gypsy insertions with a bx phenotype within a specific 11-kb region centered on the variable CTCF site. Our analysis shows that the bx83Ka insertion does not simply introduce an insulator but also has effects on flanking regions. In particular, the bx83Ka insertion affects the binding of CTCF at the variable CTCF site, leading to clearly enhanced CTCF occupancy in both T1 and T3 discs. In the case of bx83Ka, the gypsy insertion also lies close to a GAF ChIP binding peak and results in loss of GAF binding in both T1 and T3 discs. This effect on GAF binding is difficult to interpret functionally; GAF has a role in Ubx expression, as the GAF gene Trl interacts with Ubx alleles (50). However, Trl mutant clones in imaginal discs do not appear to affect Ubx expression (51, 52). The topological changes associated with the bx83Ka insertion include enhanced interactions between the variable CTCF site and the Ubx promoter in both T1 and T3 and loss of the preferential interaction between the variable CTCF site and the distant abx regulatory region in T3. This suggests that the insertion of a gypsy insulator may stabilize CTCF binding and promote interactions with the Ubx promoter but in a manner that excludes interactions with distant regulatory elements. Hence, the gypsy Su(Hw) insulator element may indeed act as an enhancer blocker, but it may do so in collaboration with a CTCF complex. We speculate that the involvement of CTCF in the mechanism that generates the mutant phenotype explains the observed clustering of gypsy insertions with bx phenotypes around the variable CTCF site.
Although our observations indicate a likely role for CTCF in facilitating enhancer-promoter interaction in Ubx regulation, functional studies will be required to confirm the role of CTCF and its importance for Ubx expression. In this regard, we have looked for genetic interaction between CTCF and Ubx. As null CTCF mutants are lethal, we investigated whether the Ubx haplo-insufficient phenotype is enhanced by heterozygosity for CTCF. We have not seen clear enhancement in this situation, and further work will be required to test the proposed CTCF role.
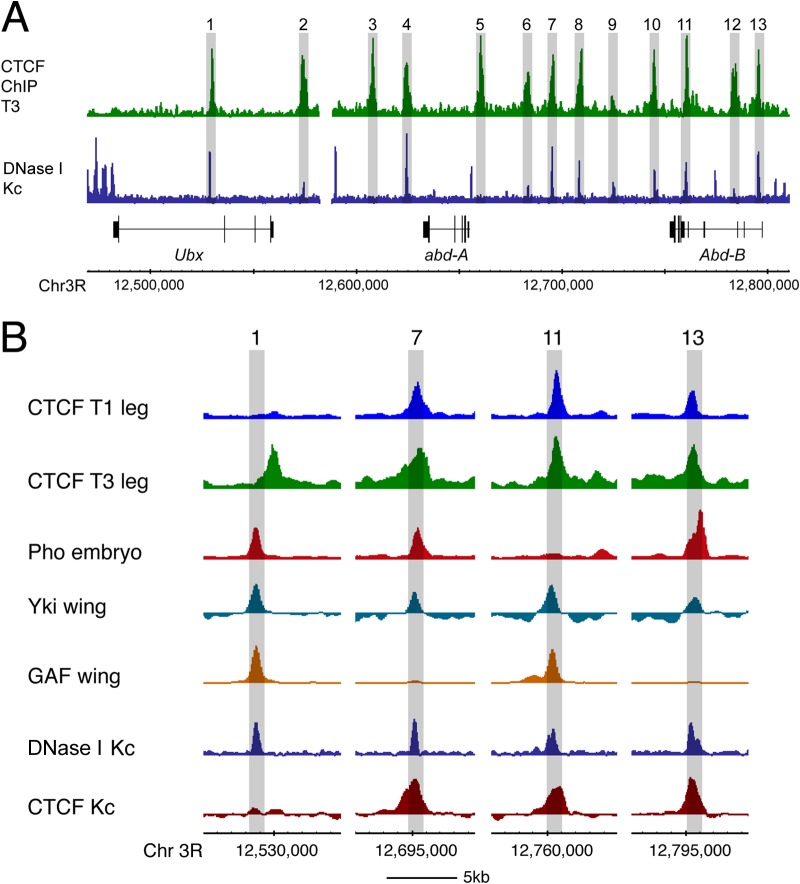
Why are some CTCF binding sites constitutive and others variably occupied? The occupancy of CTCF sites across the BX-C sheds light on this issue but initially presents a puzzle. CTCF sites within the abd-A and Abd-B domains are occupied even when these domains are silenced by Pc-mediated repression, whereas the variable CTCF site in the Ubx gene is only occupied when the Ubx domain is derepressed. This raises questions about the ability of CTCF to access its binding site in different chromatin states. There is evidence that CTCF binding is sensitive to the chromatin configuration. In particular, CTCF binding is affected by nucleosome positioning, and CTCF is unable to bind if its target site is covered by a nucleosome (21, 53). Examination of chromatin accessibility within the repressed abd-A and Abd-B domains by DNase I sensitivity reveals that CTCF sites generally correspond to small regions of DNase I accessibility within the repressed domains (Fig. 7A), indicating that CTCF is bound at sites of open, potentially nucleosome-free chromatin. Interestingly, these sites are bound by other factors, for example, Yki and GAF, so it is unclear which factor or factors are responsible for initiating and establishing open chromatin at these positions. Importantly, the presence of other factors indicates that CTCF is not necessarily responsible for pioneering binding at these sites in repressed chromatin. The variable CTCF site in Ubx supports the idea that CTCF on its own may not be able to bind to repressed chromatin, and it is intriguing that in this particular case, the adjacent DNase I site, occupied by Yki, GAF, and Pho, does not extend over the CTCF site (Fig. 7B). Occupancy of the variable site may be dependent on Pc derepression of the Ubx domain, enabling nucleosome remodeling to expose the CTCF site for binding. A different perspective is given by the finding that, although CTCF does not bind to the variable site in the repressed Ubx domain in T1 in the wild type, it does bind in the context of the bx83Ka mutant. The insertion of the gypsy transposon carrying the Su(Hw)-dependent gypsy insulator may stabilize CTCF binding at the variable binding site, perhaps through a general function of insulator complexes to facilitate loading of insulator components at nearby sites. Overall, our studies point to a view of CTCF binding where CTCF is in competition with nucleosomes for site occupancy. In the repressed state in T1, the nucleosome is dominant and there is very little CTCF binding to the variable site. CTCF binding may be enhanced either by decreasing nucleosome occupancy, associated with the opening of the Ubx domain in T3, or by local interactions between insulator complexes stabilizing CTCF binding.
FIG 7.
Chromatin accessibility and protein binding at CTCF sites in the BX-C. (A) In the repressed BX-C in the Drosophila Kc167 cell line (Kc), DNase I profiling reveals specific accessible sites in the repressed chromatin. Thirteen CTCF sites, bound in T3 chromatin, are numbered; 11 of the 13 are associated with DNase I sensitivity peaks. (B) Close-up of selected sites from the experiment whose results are shown in panel A; the binding peaks of several regulators align with the DNase I sites. The variable CTCF site (site 1) is offset from this alignment, whereas other, constitutive CTCF sites are more closely aligned with the DNase I sites. Data from the CTCF T1 and T3 leg are from this paper; data for Pho are from reference 64; data for Yki and GAF are from reference 63; data for DNase I Kc167 are from reference 65; and data for CTCF Kc167 are from ModENCODE (www.modencode.org) data set 908.
Our data also provide a view of the in vivo 3-dimensional organization of the BX-C, comparing the situation in T1, where all three BX-C genes are inactive, with the situation in T3, where Ubx is active and abd-A and Abd-B are inactive. In the active Ubx state, both the variable CTCF site and the Ubx promoter engage in long-range interactions over a range of about 100 kb, but the interactions we see are nevertheless confined to the Ubx domain. In the repressed state, the variable CTCF site and the Ubx promoter show more association with distant repressed regions outside the Ubx domain (Fig. 3). This fits with previous studies, both in Drosophila (54, 55) and in the mammalian Hox complexes (56–60), which support the idea of regulatory domains as dynamic topological structures where repressed domains cluster together and expressed domains are segregated into a separate compartment.
ACKNOWLEDGMENTS
The work was supported by the Wellcome Trust (grant 089834/Z/09/Z), and E.R. was supported by the Erasmus Programme.
We thank Bettina Fischer in the FlyChIP genomic facility for support and Sarah Bray for comments on the manuscript.
REFERENCES
- 1.Phillips-Cremins JE, Corces VG. 2013. Chromatin insulators: linking genome organization to cellular function. Mol Cell 50:461–474. doi: 10.1016/j.molcel.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phillips JE, Corces VG. 2009. CTCF: master weaver of the genome. Cell 137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell AC, West AG, Felsenfeld G. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387–396. doi: 10.1016/S0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 4.Splinter E, Heath H, Kooren J, Palstra R-J, Klous P, Grosveld F, Galjart N, De Laat W. 2006. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev 20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou C, Li L, Qin ZS, Corces VG. 2012. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell 48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J, Barillot E, Blüthgen N, Dekker J, Heard E. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 9.Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. 1990. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5:1743–1753. [PubMed] [Google Scholar]
- 10.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nègre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RAH, Stein L, Henikoff S, Kellis M, White KP. 2010. A Comprehensive map of insulator elements for the Drosophila genome. PLoS Genet 6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuddapah S, Jothi R, Schones DE, Roh T-Y, Cui K, Zhao K. 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Tian Y, Shu W, Bo X, Wang S. 2012. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS One 7:e41374. doi: 10.1371/journal.pone.0041374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, Thurman RE, Kaul R, Myers RM, Stamatoyannopoulos JA. 2012. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res 22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell AC, Felsenfeld G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 18.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 19.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi C-F, Wolffe A, Ohlsson R, Lobanenkov VV. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10:853–856. doi: 10.1016/S0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 20.Szabó PE, Tang S-HE, Rentsendorj A, Pfeifer GP, Mann JR. 2000. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol 10:607–610. doi: 10.1016/S0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- 21.Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C. 2008. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol Cell 32:129–139. doi: 10.1016/j.molcel.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, Jones KC, Corces VG. 2011. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol Cell 44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda RK, Karch F. 2006. The ABC of the BX-C: the bithorax complex explained. Development 133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- 24.Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. 2007. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet 3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RAH, Renkawitz-Pohl R, Saumweber H, Renkawitz R. 2007. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J 26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, Burke LJ, Renkawitz-Pohl R, Ohlsson R, Zhou J, Renkawitz R, Lobanenkov V. 2005. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep 6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peifer M, Bender W. 1986. The anterobithorax and bithorax mutations of the bithorax complex. EMBO J 5:2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitfield WG, Millar SE, Saumweber H, Frasch M, Glover DM. 1988. Cloning of a gene encoding an antigen associated with the centrosome in Drosophila. J Cell Sci 89(Pt 4):467–480. [DOI] [PubMed] [Google Scholar]
- 29.Birch-Machin I, Gao S, Huen D, McGirr R, White RAH, Russell S. 2005. Genomic analysis of heat-shock factor targets in Drosophila. Genome Biol 6:R63. doi: 10.1186/gb-2005-6-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Sharnouby S, Redhouse J, White RAH. 2013. Genome-wide and cell-specific epigenetic analysis challenges the role of polycomb in Drosophila spermatogenesis. PLoS Genet. 9:e1003842. doi: 10.1371/journal.pgen.1003842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forné T. 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 33.Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 34.Essien K, Vigneau S, Apreleva S, Singh LN, Bartolomei MS, Hannenhalli S. 2009. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol 10:R131. doi: 10.1186/gb-2009-10-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee HS, Pugh BF. 2011. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni X, Zhang YE, Nègre N, Chen S, Long M, White KP. 2012. Adaptive evolution and the birth of CTCF binding sites in the Drosophila genome. PLoS Biol 10:e1001420. doi: 10.1371/journal.pbio.1001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerasimova TI, Lei EP, Bushey AM, Corces VG. 2007. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell 28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pai C-Y, Lei EP, Ghosh D, Corces VG. 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell 16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Belozerov VE, Majumder P, Shen P, Cai HN. 2003. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J 22:3113–3121. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohtsuki S, Levine M. 1998. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev 12:3325–3330. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. 2004. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168:1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. 2011. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orlando V, Jane EP, Chinwalla V, Harte PJ, Paro R. 1998. Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J 17:5141–5150. doi: 10.1093/emboj/17.17.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qian S, Capovilla M, Pirrotta V. 1991. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J 10:1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon J, Peifer M, Bender W, O'Connor M. 1990. Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J 9:3945–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modolell J, Bender W, Meselson M. 1983. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A 80:1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spana C, Harrison DA, Corces VG. 1988. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev 2:1414–1423. doi: 10.1101/gad.2.11.1414. [DOI] [PubMed] [Google Scholar]
- 48.White RAH, Wilcox M. 1985. Regulation of the distribution of Ultrabithorax proteins in Drosophila. Nature 318:563–567. doi: 10.1038/318563a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pirrotta V, Chan CS, McCabe D, Qian S. 1995. Distinct parasegmental and imaginal enhancers and the establishment of the expression pattern of the Ubx gene. Genetics 141:1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. 1994. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature 371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 51.Bejarano F, Busturia A. 2004. Function of the Trithorax-like gene during Drosophila development. Dev Biol 268:327–341. doi: 10.1016/j.ydbio.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 52.Brown JL, Fritsch C, Mueller J, Kassis JA. 2003. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 130:285–294. doi: 10.1242/dev.00204. [DOI] [PubMed] [Google Scholar]
- 53.Kanduri M, Kanduri C, Mariano P, Vostrov AA, Quitschke W, Lobanenkov V, Ohlsson R. 2002. Multiple nucleosome positioning sites regulate the CTCF-mediated insulator function of the H19 imprinting control region. Mol Cell Biol 22:3339–3344. doi: 10.1128/MCB.22.10.3339-3344.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cléard F, Moshkin Y, Karch F, Maeda RK. 2006. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet 38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- 55.Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. 2007. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol 9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 56.Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. 2013. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340:1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- 57.Chambeyron S, Bickmore WA. 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev 18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA. 2010. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferraiuolo MA, Rousseau M, Miyamoto C, Shenker S, Wang XQD, Nadler M, Blanchette M, Dostie J. 2010. The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res 38:7472–7484. doi: 10.1093/nar/gkq644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. 2011. The dynamic architecture of Hox gene clusters. Science 334:222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 61.Bender W, Lucas M. 2013. The border between the Ultrabithorax and abdominal-A regulatory domains in the Drosophila bithorax complex. Genetics 193:1135–1147. doi: 10.1534/genetics.112.146340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ringrose L, Rehmsmeier M, Dura J-M, Paro R. 2003. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev Cell 5:759–771. [DOI] [PubMed] [Google Scholar]
- 63.Oh H, Slattery M, Ma L, Crofts A, White KP, Mann RS, Irvine KD. 2013. Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell Rep 3:309–318. doi: 10.1016/j.celrep.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kwong C, Adryan B, Bell I, Meadows L, Russell S, Manak JR, White R. 2008. Stability and dynamics of Polycomb target sites in Drosophila development. PLoS Genet 4:e1000178. doi: 10.1371/journal.pgen.1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas S, Li X-Y, Sabo PJ, Sandstrom R, Thurman RE, Canfield TK, Giste E, Fisher W, Hammonds A, Celniker SE, Biggin MD, Stamatoyannopoulos JA. 2011. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol 12:R43. doi: 10.1186/gb-2011-12-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]