Abstract
The LIM-only protein FHL2 is expressed in smooth muscle cells (SMCs) and inhibits SMC-rich-lesion formation. To further elucidate the role of FHL2 in SMCs, we compared the transcriptomes of SMCs derived from wild-type (WT) and FHL2 knockout (KO) mice. This revealed that in addition to the previously recognized involvement of FHL2 in SMC proliferation, the cholesterol synthesis and liver X receptor (LXR) pathways are altered in the absence of FHL2. Using coimmunoprecipitation experiments, we found that FHL2 interacts with the two LXR isoforms, LXRα and LXRβ. Furthermore, FHL2 strongly enhances transcriptional activity of LXR element (LXRE)-containing reporter constructs. Chromatin immunoprecipitation (ChIP) experiments on the ABCG1 promoter revealed that FHL2 enhances the association of LXRβ with DNA. In line with these observations, we observed reduced basal transcriptional LXR activity in FHL2-KO SMCs compared to WT SMCs. This was also reflected in reduced expression of LXR target genes in intact aorta and aortic SMCs of FHL2-KO mice. Functionally, the absence of FHL2 resulted in attenuated cholesterol efflux to both ApoA-1 and high-density lipoprotein (HDL), in agreement with reduced LXR signaling. Collectively, our findings demonstrate that FHL2 is a transcriptional coactivator of LXRs and points toward FHL2 being an important determinant of cholesterol metabolism in SMCs.
INTRODUCTION
Vascular smooth muscle cells (SMCs) are crucial for proper vascular function, and phenotypic modulation of SMCs contributes to postangioplasty restenosis and atherosclerosis (1, 2). Cholesterol is an essential component of the cell membrane and vital for normal cellular function (3), but excessive lipid accumulation in arterial wall cells, including SMCs, is an early event during atherosclerosis (4). Lipid-filled SMCs are the predominant foam cell type in early human atherosclerotic lesions and have been shown to promote lesion development (5, 6). At a later stage, the stability of the atherosclerotic plaque depends largely on the abundance and reparative capacity of SMCs, owing to their ability to produce a lesion-stabilizing fibrous cap (2, 7). Understanding how dysregulated cholesterol metabolism in SMCs affects their function is therefore essential in order to identify novel targets to regulate their responses.
FHL2, also known as DRAL or SLIM3, is the second member of the four-and-a-half-LIM-domain protein family (8, 9), which possesses an amino-terminal half LIM domain followed by four complete LIM domains. LIM proteins mediate protein-protein interactions and sometimes bind DNA or mediate nuclear localization (10, 11). Multiple functions have been ascribed to FHL2, likely a result of its interaction with over 50 different proteins that are involved in most major signaling pathways (8). FHL2 lacks any obvious enzymatic and direct DNA binding activities; nevertheless, it has been shown to act as a transcriptional coactivator or corepressor of multiple transcription factors in a cell type- and cellular-context-dependent manner (8, 9). As such, FHL2 has been reported to regulate the transcriptional activity of NF-κB, cyclic AMP-responsive element binding protein (CREB), androgen receptor (AR), and Nur77, among others (8, 9, 11–13). The extensive interactome of FHL2 and its intricate regulatory role in multiple cellular processes illustrate the multifaceted function of this protein.
Previous studies have shown that FHL2 is expressed in the heart and in the vessel wall in endothelial cells and SMCs, among other tissues (9, 14). In SMCs, FHL2 is transcriptionally regulated by serum response factor (SRF) and participates in SRF-dependent transcription by stabilizing myocardin factors (14). Additionally, FHL2 antagonizes RhoA-mediated induction of smooth muscle (SM) actin and SM22α by competing with the coactivator myocardin-related transcription factor (MRTF) A for SRF binding (15). Significantly, FHL2 inhibits SMC proliferation and has a protective function in SMC-rich-lesion formation (16, 17). The latter activity of FHL2 involves enhanced activation of the extracellular signal-regulated kinase 1/2 (ERK1/2)—cyclin D1 signaling pathway (17). Microarray profiling in mouse livers shows that FHL2 regulates genes involved in signal transduction, cytoskeleton formation, and cardiovascular function (18). Very recently, FHL2 was shown to be associated with lipid metabolism in pigs (19).
The liver X receptors (LXRs) α (NR1H3) and β (NR1H2) are members of the nuclear hormone receptor superfamily and regulate genes involved in cholesterol metabolism and inflammation (20). LXRs are transcription factors that heterodimerize with the retinoid X receptor (RXR). LXRα is highly expressed in several tissues that are important for cholesterol and lipid metabolism, such as liver, adipose tissue, intestine, and macrophages, whereas LXRβ is expressed ubiquitously (21). Oxysterols are the endogenous ligands for LXRs, and several synthetic ligands have been identified (e.g., T0901317 and GW3965) (22, 23). Both LXRα and LXRβ directly regulate several important genes in reverse cholesterol transport and lipid metabolism, including members of the ATP-binding cassette (ABC) superfamily of membrane transporters such as ABCA1, ABCG1, lipoprotein lipase (LPL), inducible degrader of low-density-lipoprotein receptor (IDOL), and sterol regulatory element-binding protein 1c (SREBP1c) (24). Although ligands induce LXR activity, transcriptional coregulators play a crucial role in specificity of the downstream signaling events. A number of transcriptional coregulators have been found to play an important role in lipid metabolism, including SRC1, PGC-1α, and TIF2 (24, 25); however, identification of LXR coregulators and the underlying molecular mechanisms requires further research.
Given the established role of FHL2 in SMCs, the present study aimed to characterize the molecular mechanism by which FHL2 exerts its atheroprotective function in SMCs in greater detail. Using transcriptional profiling, we identified alteration in the cholesterol synthesis and LXR pathways in the absence of FHL2. We provide evidence that in SMCs, FHL2 acts as a transcriptional coactivator of LXRs and that this may underlie, in part, its atheroprotective function.
MATERIALS AND METHODS
Microarray profiling.
Aortic SMCs were isolated from wild-type (WT) and FLH2 knockout (FHL2-KO) mice as described previously (17). Aortic SMCs isolated from 3 WT and 3 FHL2-KO mice were grown to confluence and made quiescent in serum-free medium for 48 h. Subsequently, the cells were lysed and used for microarray profiling. RNA was isolated using the Aurum total-RNA isolation kit (Bio-Rad), and samples were sent to ServiceXS (Leiden, The Netherlands) for further microarray processing. In brief, to assess the quality of the samples, the concentration of the RNA was determined using the Nanodrop ND1000 spectrophotometer. The Agilent Bioanalyzer was used to analyze the quality and integrity of the RNA samples, and the Illumina TotalPrep-96 RNA amplification kit was used to generate biotin-labeled (biotin-16-UTP) amplified cRNA. The biotinylated cRNA samples were hybridized onto Illumina MouseWG-6 v2 arrays, which were scanned using the Illumina iScan array scanner, and the data were retrieved by using Illumina's Genomestudio v. 2011.1 software.
Data normalization and analyses.
Analyses were carried out with packages from Bioconductor in the statistical software package R (version 2.14.0). Normexp-by-control background correction, quantile normalization, and log2 transformation (26) were performed on the Illumina sample and control probe profiles using the limma package (version 3.10.2). The arrayQualityMetrics package (version 3.10.0) was used to ensure that the microarray data were of good quality. Gene-wise linear models were fitted using the limma package. Differential gene expression between the FHL2-KO and WT SMC samples was assessed via a moderated t test. Only probes detected on at least one array were included in the differential expression analysis. Resulting P values were corrected for multiple testing using the Benjamini-Hochberg false-discovery rate. The illuminaMousev2.db package (version 1.12.2) was used to update the probe annotation provided by GEO. The data set with differentially expressed genes with a P value of <0.05 was analyzed using Ingenuity pathway analysis (IPA; Ingenuity Systems) to test for enriched canonical pathways and networks and to identify upstream transcriptional regulators. The background set for the gene enrichment analyses consisted of all genes detected on at least one array. Significant expression on the Illumina MouseWG-6 v2 array and statistical association for mapping of genes to functions and pathways were assessed using the right-tailed Fisher's exact test.
Plasmids and chemicals.
pCMX-hLXRα, pCMX-hLXRβ-Flag, pCMX-hRXRα, an LXR response element (LXRE)-luciferase reporter (pTK.LXRE3-luc), and several deletion mutants of FHL2 have been described before (9, 27). The pCMV-LIM2-4-Myc construct was made using the primers described before (9). pGL3-hABCA1-luc was kindly provided by Johan Auwerx (EPFL, Lausanne, Switzerland) and was described previously (28). The LXR agonists T0901317 and GW3965 were purchased from Sigma-Aldrich. [3H]cholesterol was purchased from Amersham; high-density lipoprotein (HDL) and ApoA1 were purchased from Calbiochem.
Cell culture and transfection.
HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 20 mM glucose, supplemented with 10% serum and penicillin-streptomycin (Invitrogen). Aortic SMCs were cultured in DMEM—F-12 medium (Invitrogen) supplemented with 20% fetal calf serum (FCS) and penicillin-streptomycin. SMCs were used at passages 6 to 8 and were characterized by SM α-actin expression (1A4; Dako) showing uniform fibrillar staining. Culturing of human SMCs has been described before (9). HeLa cells were transfected using Lipofectamine 2000 (Invitrogen), and SMCs were transfected using Fugene6 transfection reagent (Roche).
Coimmunoprecipitation assays and Western blot analysis.
Coimmunoprecipitation assays and Western blot analysis were described previously (9). Briefly, HeLa cells were cotransfected with appropriate plasmids and incubated for 48 h. The cells were lysed in NP-40 lysis buffer supplemented with complete protease inhibitor mixture (Roche Applied Science). For endogenous coimmunoprecipitation assays, SMC lysates from WT and FHL2-KO mice (1.5 mg total protein each) were used. Cell lysates were precleared for 1 h at 4°C with protein A-Sepharose (GE Healthcare) and then incubated overnight with the pulldown antibody and protein A-Sepharose. Immunoprecipitates were washed three times in lysis buffer, and bound protein was eluted by boiling in SDS loading buffer before electrophoresis on 12% SDS-polyacrylamide gels. SMC lysates were prepared with NP-40 lysis buffer (with protease inhibitors) and samples were boiled in SDS loading buffer before electrophoresis on 12% SDS-polyacrylamide gels. Proteins were transferred using a transfer system (Bio-Rad) to polyvinylidene difluoride (PVDF) membranes (Bio-Rad) that were incubated with appropriate blocking agents, primary antibodies, and fluorescently conjugated secondary antibodies, followed by scanning using an Odyssey infrared imaging system (Licor Biosciences). Antibodies applied in this study were antihemagglutinin (anti-HA) (12CA5; Roche Applied Science), anti-Flag (Invitrogen), anti-LXRα (Abcam), anti-LXRβ (Abcam), anti-FHL2 (Thermo Scientific), antitubulin (Cedarlane Labs), and anti-β-actin (Cell Signaling).
Luciferase assays.
HeLa cells were transiently transfected with LXRE luciferase reporter plasmids and ABCA1 promoter constructs (WT and DR4 mutant; kindly provided by Herbert Stangl, Vienna, Austria) (29) together with pCMX-LXRα, pCMX-Flag-LXRβ, and pCMV-HA-FHL2 or pCMV-mock. The Renilla reporter plasmid pRL-TK (Promega) was cotransfected as an internal control for transfection efficiency. SMCs were cotransfected with pTK.LXRE3-luc or pGL3-hABCA1-luc and Renilla reporter plasmids by Fugene6 (Roche Applied Science). Luciferase activity measurements were performed using the dual-luciferase reporter assay system (Promega) and Glomax Multi detection system (Promega) according to the manufacturer's protocol. Each experiment (in duplicate) was repeated at least three times.
Chromatin immunoprecipitation (ChIP) assays.
ChIP assays in HeLa cells were performed using the Magnify ChIP system (Invitrogen) as described previously (9). The following primers were used to amplify the ABCG1 promoter by quantitative real-time PCR (qRT-PCR): 5′-TTCTGTGGACAGGTACTAGGT-3′ (sense) and 5′-CCACAAACATAGGTAGTCCAG-3′ (antisense).
qRT-PCR.
HeLa cells and SMCs were treated with the LXR agonists T0901317 and GW3965 for 8 h before lysis. In some experiments, SMCs were incubated in serum-free medium (SFM), lipoprotein-deficient serum (LPDS), or LPDS supplemented with simvastatin and mevalonate and then treated with 25-OH-cholesterol or desmosterol. Total RNA from mouse aorta was prepared with TRIzol (Invitrogen). Total RNA from cells was isolated with a total-RNA minikit (Bio-Rad). cDNA synthesis was performed with iScript (Bio-Rad), followed by real-time PCR using the MyIQ system (Bio-Rad). Primers used for real-time PCR are detailed in Table S3 in the supplemental material. The acidic ribosomal phosphoprotein P0 was measured as an internal control for cDNA content of the samples.
FHL2 knockdown in SMCs.
Recombinant lentiviral particles of short hairpin RNAs (shRNAs) targeting FHL2 were produced, concentrated, and titrated as described previously (9, 17). Two different mouse shRNAs that target different regions in the FHL2 mRNA (shFHL2#1 target sequence, GATGGGAAGATGGTTTGGAAT; shFHL2#2 target sequence, CTGTGACTTGTACGCTAAGAA) were used for generation of lentiviruses. shRNAs targeting human FHL2 and the scrambled control shRNA have been described before (9). Lentiviral infection in cultured aortic SMCs was performed as described previously (17). Transduction efficiency was determined by immunofluorescence and qRT-PCR.
Cell proliferation assays.
Bromodeoxyuridine (BrdU) incorporation assays were performed as described previously (17). For both BrdU and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assays, cells were seeded in a 96-well plate at a density of 3 × 103 cells/well and incubated overnight. Cells were made quiescent by incubation in medium without FCS for 48 h and then treated with GW3965 overnight followed by incubation with FCS (10% [vol/vol]) for 24 h. After the incubation, cells were incubated with BrdU reagent for 16 h or 10 μl of MTT reagent (5 mg/ml) for 3 h at 37°C. In MTT assays, 100 μl of isopropanol was added to each well and incubated for 15 min. Colorimetric analysis was performed with an enzyme-linked immunosorbent assay (ELISA) plate reader. Each experiment (in quadruplicate) was repeated at least three times.
Cholesterol efflux assay.
Cholesterol efflux assays were performed as previously described with minor modifications (30). Briefly, SMCs from WT and FHL2-KO mice were seeded into 24-well-plate wells at a concentration of 3 × 105 cells per ml and allowed to adhere overnight, followed by incubation for 24 h in DMEM—F-12 plus 20% FCS plus [3H]cholesterol. Cells were then washed and incubated for an additional 18 h with equilibration medium (DMEM—F-12 plus 0.2% bovine serum albumin [BSA]) supplemented with T0901317 (5 μM) or GW3965 (5 μM) where indicated. Efflux was initiated by the addition of efflux medium (DMEM—F-12 plus 0.2% BSA) plus either vehicle, ApoAI (10 μg/ml), or HDL (100 μg/ml). Radioactivity in the medium plus cells was measured by liquid scintillation counting. This assay was performed in quadruplicate and repeated three times. Values are expressed as means and standard deviations (SD).
Statistical analysis.
Data are reported as means and SD and were analyzed with the unpaired Student t test. P values of <0.05 were considered significant.
Microarray data accession number.
The microarray data have been deposited in NCBI Gene Expression Omnibus in a MIAME compliant format and are accessible under GEO series accession number GSE56860.
RESULTS
Expression profiling reveals attenuated LXR-RXR signaling in FHL2-KO SMCs.
To understand the function of FHL2 in murine SMCs, RNA was isolated from three independent SMC isolates after serum stimulation, derived from different FHL2-KO and WT mice. Transcriptional profiling revealed that >2,500 genes were differentially expressed in FHL2-KO compared with WT SMCs (P < 0.05; change ≥ 1.4-fold), of which 843 genes remained differentially expressed after correction for multiple testing (adjusted P value < 0.05). Of these genes, 373 were upregulated and 470 were downregulated in FHL2-KO compared with WT SMCs. The top 30 up- and downregulated genes are shown in Fig. 1 and in Table S1 in the supplemental material. Ingenuity pathway analysis (IPA) was performed to identify canonical pathways with a statistically significant enrichment of differentially expressed genes, and the top 25 canonical pathways indicate involvement of FHL2 in lipid and cholesterol metabolism (see Fig. S1A in the supplemental material). The top-ranking canonical pathway is the “super-pathway of cholesterol biosynthesis,” which comprises 87 genes, 13 of which show changed expression in FHL2-KO SMCs compared with WT cells. The second pathway, “LPS/IL-1 mediated inhibition of RXR function,” concerns retinoid X receptor (RXR), which is a nuclear receptor that is activated by 9-cis retinoic acid and is an obligate heterodimerization partner for a subfamily of nuclear receptors, including the liver X receptors (LXR). LXRs are important regulators of cholesterol and fatty acid homeostasis (24), and interestingly, our transcriptional analysis also identified the LXR/RXR pathway as being sensitive to FHL2 in SMCs. Subsequent generation of networks of the top canonical pathways with at least 4 genes in common divided these pathways into two networks centered around cholesterol synthesis and metabolism and signaling (see Fig. S1B in the supplemental material).
FIG 1.

Changes in mRNA abundance in FHL2-KO compared to WT SMCs, as determined by microarray analysis. This heat map shows the top 30 up- and downregulated genes in FHL2-KO compared with WT SMCs. Green indicates downregulation, and red indicates upregulation.
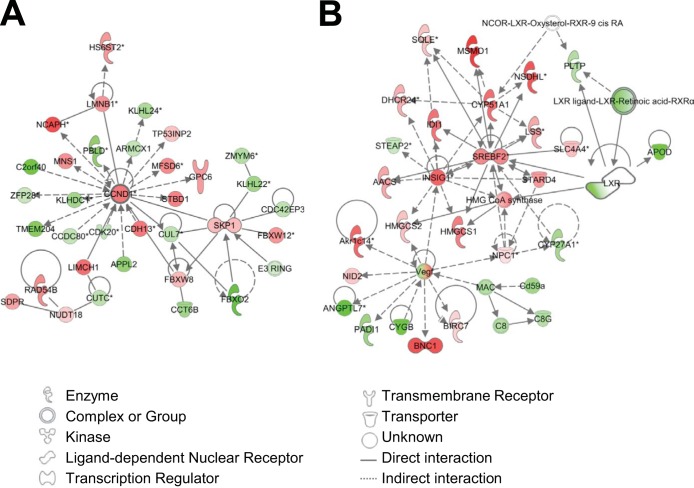
IPA gene network analysis, which examines the intermolecular connections among interacting genes based on functional knowledge input, was also performed, and the top 25 networks are shown in Table 1. The highest-scoring network concerns cell cycle regulation and is centered around cyclin D1, which is known to be regulated by FHL2 in SMCs (Table 1; Fig. 2A) (17). In line with the canonical pathway analysis, four of the networks were found to involve lipid metabolism (Table 1, networks 2, 15, 24, and 25), and one of these (network 15) indicates a role for FHL2 in LXR-RXR signaling and cholesterol metabolism (Fig. 2B; also, see Fig. S2 in the supplemental material).
TABLE 1.
Functional clusters associated with the highest-score networks identified by Ingenuity pathway analysis and their scores for gene categories in FHL2-KO versus WT SMCsa
| Network | Top diseases and functions | Scoreb |
|---|---|---|
| 1 | Cell cycle, connective tissue disorders, developmental disorder | 40 |
| 2 | Hereditary disorder, neurological disease, lipid metabolism | 36 |
| 3 | Drug metabolism, molecular transport, nucleic acid metabolism | 35 |
| 4 | RNA posttranscriptional modification, protein synthesis, connective tissue disorders | 33 |
| 5 | Cellular response to therapeutics, cancer, developmental disorder | 31 |
| 6 | Cell cycle, cellular assembly and organization, amino acid metabolism | 31 |
| 7 | Cellular development, cardiovascular system development and function, hematological system development and function | 31 |
| 8 | Posttranslational modification, embryonic development, organ development | 31 |
| 9 | Skeletal and muscular system development and function, tissue development, cellular movement | 30 |
| 10 | Cellular movement, cellular growth and proliferation, embryonic development | 30 |
| 11 | Developmental disorder, hereditary disorder, neurological disease | 29 |
| 12 | Cellular assembly and organization, DNA replication, recombination, and repair, cell cycle | 29 |
| 13 | Cancer, developmental disorder, hereditary disorder | 29 |
| 14 | Cancer, cardiovascular disease, cellular assembly and organization | 29 |
| 15 | Lipid metabolism, small molecule biochemistry, vitamin and mineral metabolism | 28 |
| 16 | Amino acid metabolism, posttranslational modification, small molecule biochemistry | 28 |
| 17 | Cellular development, cellular growth and proliferation, hematological system development and function | 27 |
| 18 | Cell-to-cell signaling and interaction, nervous system development and function, cellular assembly and organization | 26 |
| 19 | Cell-to-cell signaling and interaction, cellular assembly and organization, cellular function and maintenance | 26 |
| 20 | Cellular function and maintenance, small molecule biochemistry, molecular transport | 26 |
| 21 | Embryonic development, organismal development, organ development | 26 |
| 22 | Cellular assembly and organization, cellular compromise, cell-to-cell signaling and interaction | 26 |
| 23 | Skeletal and muscular system development and function, behavior, cellular movement | 26 |
| 24 | Lipid metabolism, molecular transport, small molecule biochemistry | 25 |
| 25 | Protein synthesis, lipid metabolism, small molecule biochemistry | 25 |
Pathways involving cell cycle regulation are underlined, and pathways involving lipid and cholesterol metabolism are in bold.
−Log10(P value).
FIG 2.
Network analysis of the expression profiles of WT and FHL2-KO SMCs. Data analyses revealed that networks related to cell cycle regulation (A) and lipid metabolism (B) are affected by FHL2 deficiency. Interactions between genes are shown as explained in the legend. Red symbols indicate that genes are upregulated in FHL2-KO SMCs compared to WT cells, green symbols indicate downregulation of genes, and white indicates proteins with unchanged expression.
Finally, IPA upstream regulator analysis was used to identify upstream transcriptional regulators and revealed that LXR is among the 14 identified transcription factors that are predicted to be inhibited in FHL2-KO SMCs (see Table S2 in the supplemental material). Taken together, the results of our transcriptional analysis of WT and FHL2-KO SMCs suggest that FHL2 may influence lipid handling in these cells, in part through LXR-regulated pathways.
FHL2 interacts with LXRα and LXRβ.
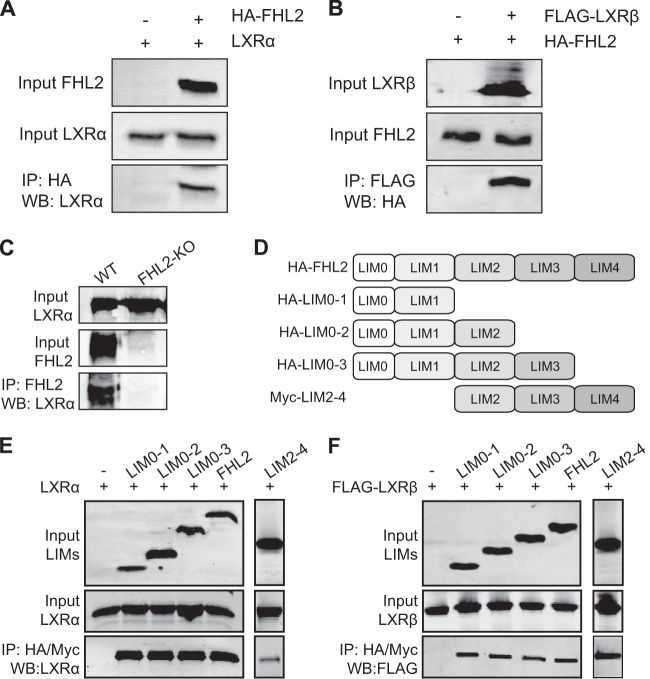
Recent studies have established that FHL2 can interact with several nuclear receptors, including the androgen receptor and Nur77 (9, 12). In view of our transcriptional analysis, we therefore hypothesized that FHL2 may interact with LXRs to regulate its transcriptional activity. We tested this by performing coimmunoprecipitation experiments with FHL2 and the two LXR isoforms in HeLa cells. As shown in Fig. 3A and B, we observed that FHL2 interacted with both LXRα and LXRβ. Furthermore, the interaction between FHL2 and both LXRs was not altered by the addition of two LXR synthetic ligands (T0901317 and GW3965; data not shown). To further substantiate these findings, we performed coimmunoprecipitation experiments in cultured SMCs from WT and FHL2-KO mice. We found that FHL2 interacts with LXRα in WT SMCs. As a control, we also included FHL2-KO SMCs (Fig. 3C). To delineate which LIM domain of FHL2 is involved in the interaction with LXRs, coimmunoprecipitation assays were performed with truncated FHL2 constructs. A schematic representation of various FHL2 mutants is shown in Fig. 3D. Our data show that each LIM domain can already bind both LXRα and LXRβ (Fig. 3E and F).
FIG 3.
FHL2 interacts with LXRs. (A and B) HA-tagged FHL2 and LXRα (A) and FLAG-tagged LXRβ (B) were expressed in HeLa cells, as indicated. Whole-cell extracts were immunoprecipitated, and the bound protein was analyzed by Western blotting with the indicated antibodies. Data are representative of at least three independent experiments. (C) Whole-cell extracts from WT and FHL2-KO SMCs were prepared and immunoprecipitated with anti-FHL2 antibody and analyzed by Western blotting with anti-LXRα antibody. (D) Schematic representation of various truncated mutants of FHL2. (E and F) Cells were cotransfected with expression vectors encoding LXRα (C) or LXRβ (D) and FHL2 mutants, as indicated. Whole-cell extracts were prepared and immunoprecipitated (IP) with anti-HA or Myc antibody. Input samples and immunoprecipitated samples were resolved by 12% SDS-PAGE and analyzed by Western blotting with the indicated antibodies. Data are representative of at least three independent experiments.
FHL2 enhances the transcriptional activity of LXRs.
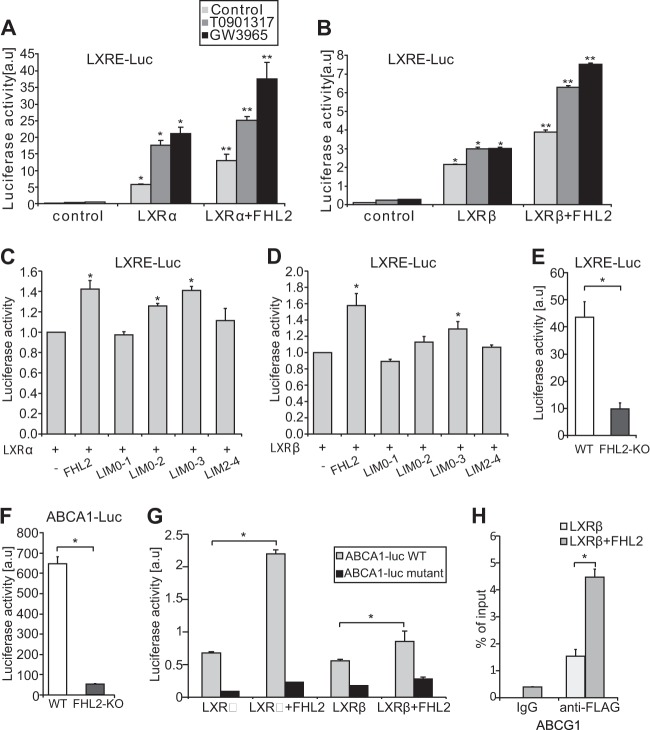
In view of its interaction with LXRs, and in order to determine the effect on activity, we tested the impact of FHL2 on LXR-dependent transcriptional activity, using a surrogate reporter construct containing an LXRE coupled to luciferase. The transcriptional activity of both LXRα and LXRβ was significantly increased in cells coexpressing full-length FHL2 protein (Fig. 4A and B). We also observed that FHL2 further enhanced LXR transcriptional activity induced by the synthetic ligands T0901317 and GW3965 (Fig. 4A and B). As all truncated FHL2 mutants interact with both LXRα and LXRβ, we then investigated their effect on LXR transcriptional activity. Interestingly, we found that in addition to full-length FHL2, only the LIM0-2 and LIM0-3 mutants of FHL2 enhanced LXRα-mediated transcriptional activity and that only the LIM0-3 mutant of FHL2 is able to transactivate LXRβ-mediated transcriptional activity, suggesting the differential regulation of LXRs by FHL2 (Fig. 4C and D). To functionally assess the consequence of FHL2 loss on LXR signaling in SMCs, we transfected aortic SMCs derived from either WT or FHL2-KO mice with the LXR reporter construct and human ABCA1 promoter constructs. In line with the overexpression studies, FHL2-KO SMCs displayed reduced transcriptional activity of an LXRE-containing reporter construct as well as of an ABCA1 promoter construct (Fig. 4E and F). Furthermore, the transcriptional activity of both LXRα and LXRβ was enhanced by FHL2 on the ABCA1 promoter reporter (Fig. 4G). Consistent with this, FHL2 failed to increase the LXRα and LXRβ-mediated transcriptional activity on a DR4-mutated ABCA1 promoter construct (Fig. 4G), suggesting functional association of FHL2 with LXRs. Collectively, these findings suggest that FHL2 enhances the transcriptional activity of LXRs both in HeLa cells and in SMCs.
FIG 4.
FHL2 acts as a coactivator of LXRs. (A and B) FHL2 enhances the activity of LXRα (A) and LXRβ (B) on the LXRE-luciferase reporter. LXR activity was further enhanced by two synthetic LXR ligands, T0901317 and GW3965. *, P ≤ 0.05, compared to control; **, P ≤ 0.05, compared to LXR α or LXRβ. (C and D) Effect of distinct FHL2 mutants on transcriptional activity of LXRα (C) or LXRβ (D) on the LXRE-luciferase reporter was assayed. (E and F) WT and FHL2-KO SMCs were transfected with indicated reporter plasmid. In FHL2-KO SMCs LXR activity is reduced on LXRE-reporter plasmids (E) and ABCA-1 promoter-reporter plasmids (F). (G) FHL2 increased LXR-mediated transcription of an ABCA1 promoter-reporter plasmid, unless the DR4 element was mutated in the ABCA1 promoter-reporter plasmid in HeLa cells. In all luciferase experiments, the transfection efficiency was normalized using Renilla luciferase, which was cotransfected with the reporter plasmids. (H) ChIP analyses were performed with ABCG1 promoter-specific PCR primers and with FLAG antibody following overexpression of LXRβ and FHL2 and pretreatment with GW3965 in HeLa cells. All experiments were performed in triplicate and repeated at least three times. Values are means and SD. *, P ≤ 0.05.
To further understand the enhanced effect of FHL2 on LXR transcriptional activity, we performed ChIP experiments on the ABCG1 promoter, which was analyzed by qRT-PCR (Fig. 4H). As shown in Fig. 4H, LXRβ binds to the ABCG1 promoter, and this association is significantly enhanced upon overexpression of FHL2. These results demonstrate that FHL2 enhances binding of LXRs to the LXREs in their target gene promoters, which reveals the mechanism of potentiation of LXR transcriptional activity by FHL2.
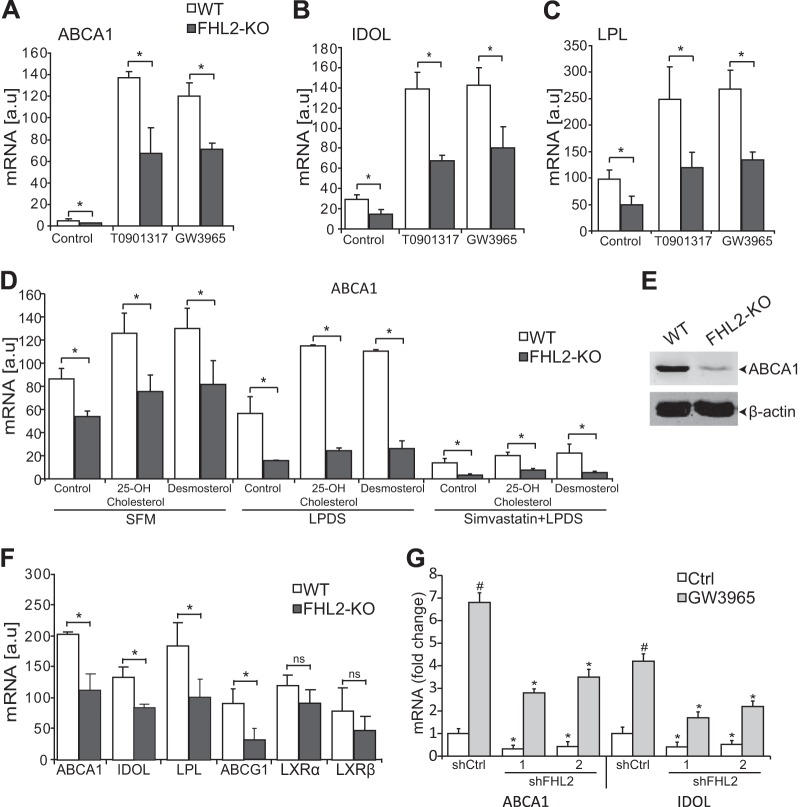
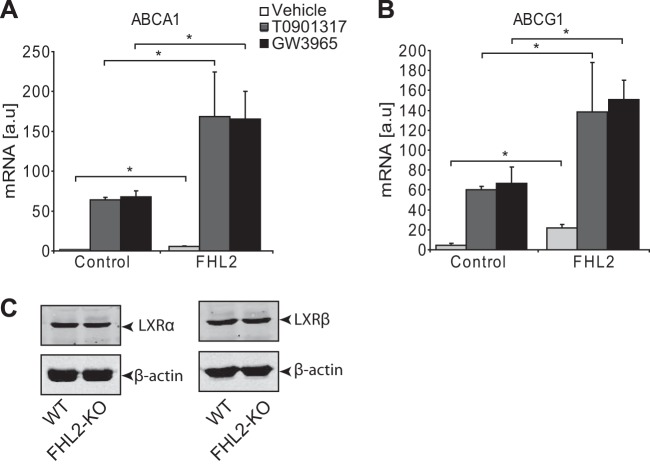
To test the significance of our findings, we turned to primary SMCs from WT and FHL2-KO mice. We treated cells with LXR agonists and determined the expression of established LXR target genes. As shown in Fig. 5, LXR agonists upregulated ABCA1, IDOL, and LPL in both WT and FHL2-KO SMCs (Fig. 5A to C). However, consistent with the hypothesis of FHL2 being required for maximal LXR activation, the expression level of the selected LXR regulated genes did not reach that observed in WT cells and was also lower at baseline (Fig. 5A to C). In addition, FHL2-KO SMCs displayed reduced protein expression of ABCA1 (Fig. 5E). To test whether FHL2 also modulates LXR target genes in the presence of endogenous ligands, we treated SMCs from WT and FHL2-KO mice with 25-OH-cholesterol and desmosterol in serum-free medium (SFM) and lipoprotein-deficient serum (LPDS). FHL2-KO SMCs showed reduced ABCA1 mRNA expression following treatment with natural ligands under both SFM and LPDS conditions (Fig. 5D). We then sought to investigate whether FHL2 regulates LXR target genes directly; we treated SMCs with simvastatin in LPDS. We found that simvastatin decreased endogenous mRNA expression of ABCA1. Interestingly, FHL2-KO SMCs showed a further decrease in mRNA expression of ABCA1 following treatment with natural ligands (Fig. 5D). Furthermore, we also confirmed decreased LXR target gene expression in FHL2-KO SMCs in RNA samples from freshly isolated mouse aortas to rule out confounding tissue culture-related effects (Fig. 5F). To assess whether deletion of FHL2 in adult SMCs also affects LXR target gene expression, we performed knockdown experiments. Knockdown of FHL2 in human SMCs using two independent shRNAs also showed a significant decrease in mRNA expression of ABCA1 and IDOL (Fig. 5G). Importantly, this is not limited to SMCs, as overexpression of FHL2 in HeLa cells increases the mRNA levels of ABCA1 and ABCG1 as well (Fig. 6A and B). A simple explanation for our observations would be that FHL2 controls the expression level of LXRs themselves. However, this can be ruled out, as both the transcript and protein levels of LXRα and LXRβ were unchanged in SMCs from KO mice (Fig. 6C). Collectively, our results point to the requirement of FHL2 for maximal LXR signaling in SMCs.
FIG 5.
FHL2 deficiency attenuates LXR target gene expression. (A to C) SMCs derived from WT and FHL2-KO mice were treated with LXR agonists and assayed by qRT-PCR for mRNA expression of ABCA1 (A), IDOL (B), and LPL (C). (D) qRT-PCR was performed to assess mRNA expression of ABCA1 in SMCs from WT and FHL2-KO mice after incubation in SFM, LPDS, or LPDS and simvastatin followed by treatment with the natural ligands 25-hydroxycholesterol and desmosterol. (E) Western blot analysis of ABCA1 expression in WT and FHL2-KO SMCs. β-Actin was used as a loading control. (F) qRT-PCR was performed to assess mRNA expression of LXRs and LXR target genes in the aortas from WT and FHL2-KO mice. (G) Expression of ABCA1 and IDOL in human SMCs was determined by qRT-PCR after knockdown of FHL2 using lentiviral particles encoding shFHL2#1 and shFHL2#2. shCtrl is a scramble control. All experiments were performed in triplicate and repeated at least three times. Values are means and SD. #, P < 0.05 for nontreated versus agonist-treated cells; *, P ≤ 0.05 for shCtrl versus shFHL2; ns, not significant.
FIG 6.
FHL2 regulates LXR target genes. (A and B) HeLa cells were transfected with an empty vector or an FHL2-encoding vector and treated with LXR agonists. qRT-PCR was performed to assess mRNA expression of ABCA1 (A) and ABCG1 (B). (C) Western blot analysis to show LXRα (left) and LXRβ (right) protein expression in WT and FHL2-KO SMCs. β-Actin was used as a loading control.
FHL2 is not involved in LXR-mediated cell proliferation but is crucial for LXR-mediated cholesterol efflux.
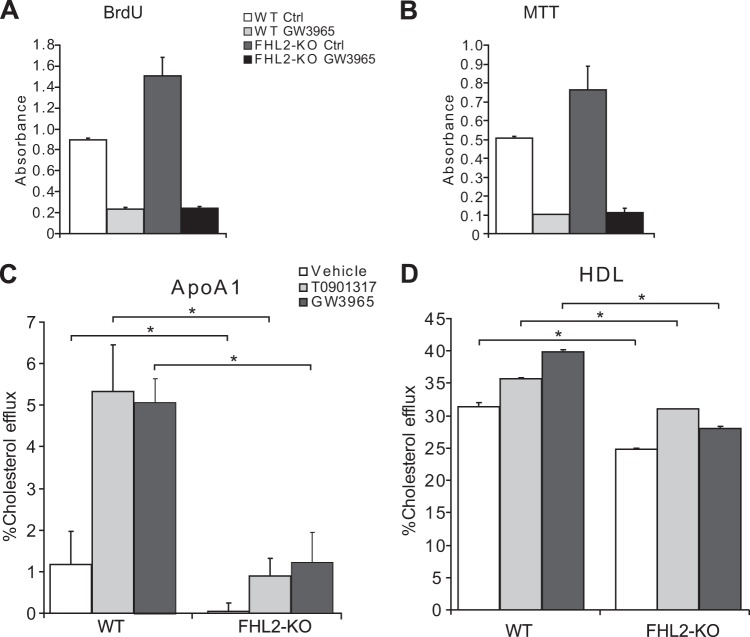
In our previous study, we demonstrated that FHL2-KO SMCs proliferate faster than their WT counterparts (17). As LXRs are also involved in regulation of cell proliferation, we sought to investigate this by performing cell proliferation assays. Consistent with our previous results, FHL2-KO SMCs proliferate faster, as determined by BrdU and MTT assays (Fig. 7A and B). However, pretreatment with GW3965 decreased proliferation in both WT and FHL2-KO SMCs, suggesting that FHL2 is not involved in LXR-mediated antiproliferative effects (Fig. 7A and B).
FIG 7.
FHL2 is not involved in LXR-mediated cell proliferation but is crucial for LXR-mediated cholesterol efflux. (A and B) Cell proliferation of WT and FHL2-KO SMCs was assessed by BrdU incorporation (A) or by MTT assay (B) following treatment with GW3965. (C and D) SMCs derived from WT and FHL2-KO mice were incubated with [3H]cholesterol and treated with LXR agonists. Cholesterol efflux was then measured in the presence of ApoAI (10 μg/ml) (C) or HDL (100 μg/ml) (D), without and with the LXR agonists T0901317 and GW3965. The assay was performed in quadruplicate and repeated three times. Values are means and SD. *, P ≤ 0.05.
The LXR-regulated gene ABCA1 is a crucial mediator of cellular cholesterol efflux in SMCs (30, 31). We therefore measured both ApoAI- and HDL-mediated cholesterol efflux in WT and FHL2-KO SMCs. To stimulate maximal cholesterol efflux, we also treated the cells with LXR ligands. As might have been anticipated, both basal cholesterol efflux and ligand-stimulated cholesterol efflux to ApoAI and HDL were diminished in the absence of FHL2 (Fig. 7C and D). These results further support the notion that FHL2 is an important determinant of LXR activity in SMCs.
DISCUSSION
The phenotypic switch of SMCs from contractile, quiescent cells to synthetic, highly proliferative cells plays a crucial role in the pathogenesis of vascular disease, including atherosclerosis and in-stent restenosis (1, 2). FHL2 is expressed in vascular SMCs and endothelial cells (9, 14) and has been shown to regulate SMC phenotype and inhibit SMC-rich-lesion formation in mice (16, 17). In the present study, we provide evidence that FHL2 is involved not only in SMC proliferation and differentiation but also in the regulation of cholesterol metabolism of SMCs through its interaction with LXRs. Our conclusions are based on the following principal findings: (i) transcriptome analyses revealed a modulation of cholesterol pathways in FHL2-KO SMCs compared with WT SMCs; (ii) FHL2 interacts with LXRs; (iii) the transcriptional activity of LXRs and the expression of endogenous LXR target genes are enhanced by FHL2; (iv) FHL2 enhances binding of LXRβ to LXR target genes; (v) FHL2 deficiency decreases cholesterol efflux in SMCs.
Our transcriptome analyses confirm that FHL2 regulates SMC proliferation, in line with our recent observations showing modulation of the ERK-cyclin D1 signaling pathway by FHL2 (17). Unexpectedly, the data also indicated that FHL2 modulates cholesterol pathways in SMCs. In the current study, we demonstrate that FHL2 influences cholesterol metabolism by interacting with both LXR isoforms and that this protein-protein interaction is independent of the presence of synthetic ligands. Although SMCs contain endogenous ligands for LXR, we assume that these may not influence the interaction between FHL2 and LXRs.
FHL2 has been shown to interact with more than 50 different proteins, including transcription factors and structural proteins in a cell type- and context-dependent manner (8, 9). The activity of the nuclear receptors estrogen receptor and Nur77 is reduced by FHL2, whereas FHL2 is a coactivator of androgen receptor, liver homolog 1 (LRH-1), steroidogenic factor 1 (SF-1), PPARα, and, as shown in this study, LXRs (8, 32). In contrast, the activity of glucocorticoid, mineralocorticoid, and progesterone receptors is not influenced by FHL2 (8). LIM domains contain a tandem cysteine-rich Zn2+ finger motif and are important for protein-protein interactions, may bind DNA, or are crucial in nuclear localization (8, 33, 34). FHL2 does not bind DNA directly and does not possess a characteristic LXXLL motif, a hallmark of many coregulators that are involved in the regulation of nuclear receptors (8, 12, 35). We reported previously that all four LIM domains of FHL2 can independently bind the nuclear receptor Nur77 (9). LXRα and LXRβ show high amino acid sequence homology in their DNA- and ligand-binding domains (78%), but their interactions with FHL2 are dissimilar. FHL2-LXRβ interaction is independent of the first two LIM domains of FHL2, whereas LXRα binds the first LIM domain of FHL2. Further analyses may reveal the exact involvement of the individual LIM domains in FHL2-LXR complex formation. The sharing of a coregulator between LXRs and other nuclear receptors may be a mechanism to coordinate and integrate signals derived from different signal transduction pathways in the cell.
Both LXR isoforms LXRα and LXRβ are present and functional in SMCs (30, 36). We show here that FHL2 enhances the transcriptional activity of LXRs in SMCs by activating transcription of several LXR target genes. Of note, FHL2 clearly enhances basal LXR activity in the absence of ligands, whereas treatment with LXR agonists, particularly efficacious synthetic compounds, appears to override the requirement for FHL2. We observed decreased cholesterol efflux in FHL2-deficient SMCs, and therefore, we investigated whether the expression of ABCA1, a major lipid transport protein in the plasma membrane that mediates cellular cholesterol export, might be altered in the absence of FHL2. As expected, deficiency of FHL2 results in greatly reduced ABCA1 expression, which is caused by decreased LXR-mediated gene transcription. This potent transcriptional repression of ABCA1 occurs in cultured cells as well as in the vascular wall in vivo in FHL2-deficient SMCs. Recently, FHL2 has been shown to regulate genes involved in signal transduction, cytoskeleton, and cardiovascular function in mouse livers (18). As LXRα plays a key role in liver, it will be of interest to further investigate the association of FHL2 and LXRα function in liver lipid metabolism.
Well-characterized coactivators of LXRs include SRC-1 and p300, which regulate LXR transcriptional activity, at least partly, by their histone acetyltransferase activity (37). FHL2 and p300 are also known to interact, and while this paper was in preparation, Ramayo-Caldas et al. demonstrated that genetic variation in both FHL2 and p300 genes is associated with lipid metabolism and control of energy homeostasis in pigs (19, 38). Hence, we propose that FHL2 is instrumental in formation of LXR multiprotein transcriptional complexes and may be considered a new key component of lipid homeostasis.
In summary, we have shown that FHL2 regulates cholesterol metabolism, at least partly, through interaction with and activation of LXRs and its specific LXR target genes in SMCs. FHL2 enhances ApoAI- and HDL-mediated efflux of cholesterol by regulating ABCA1 expression at the transcriptional level. These findings emphasize the importance of FHL2 in SMCs as an integrator of lipid transport and cellular signals that regulate cellular lipid homeostasis through modulation of LXRs.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the research program of the BioMedical Materials institute, cofunded by the Dutch Ministry of Economic Affairs as a part of Project P1.02 NEXTREAM.
We thank Johan Auwerx, EPFL, Switzerland, and Herbert Stangl, Vienna, Austria, for providing hABCA1-luc constructs.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00525-14.
REFERENCES
- 1.Orr AW, Hastings NE, Blackman BR, Wamhoff BR. 2010. Complex regulation and function of the inflammatory smooth muscle cell phenotype in atherosclerosis. J Vasc Res 47:168–180. doi: 10.1159/000250095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Owens GK, Kumar MS, Wamhoff BR. 2004. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev 84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 3.Maxfield FR, van Meer G. 2010. Cholesterol, the central lipid of mammalian cells. Curr Opin Cell Biol 22:422–429. doi: 10.1016/j.ceb.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstein JL, Brown MS. 1977. The low-density lipoprotein pathway and its relation to atherosclerosis. Annu Rev Biochem 46:897–930. doi: 10.1146/annurev.bi.46.070177.004341. [DOI] [PubMed] [Google Scholar]
- 5.Balis JU, Haust MD, More RH. 1964. Electron-microscopic studies in human atherosclerosis; cellular elements in aortic fatty streaks. Exp Mol Pathol 90:511–525. [DOI] [PubMed] [Google Scholar]
- 6.Katsuda S, Boyd HC, Fligner C, Ross R, Gown AM. 1992. Human atherosclerosis. III. Immunocytochemical analysis of the cell composition of lesions of young adults. Am J Pathol 140:907–914. [PMC free article] [PubMed] [Google Scholar]
- 7.Weissberg PL. 2000. Atherogenesis: current understanding of the causes of atheroma. Indian Heart J 52:467–472. [PubMed] [Google Scholar]
- 8.Johannessen M, Moller S, Hansen T, Moens U, Van GM. 2006. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell Mol Life Sci 63:268–284. doi: 10.1007/s00018-005-5438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurakula K, van der Wal E, Geerts D, van Tiel CM, de Vries CJ. 2011. FHL2 protein is a novel co-repressor of nuclear receptor Nur77. J Biol Chem 286:44336–44343. doi: 10.1074/jbc.M111.308999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Mourabit H, Muller S, Tunggal L, Paulsson M, Aumailley M. 2004. Analysis of the adaptor function of the LIM domain-containing protein FHL2 using an affinity chromatography approach. J Cell Biochem 92:612–625. doi: 10.1002/jcb.20096. [DOI] [PubMed] [Google Scholar]
- 11.Lange S, Auerbach D, McLoughlin P, Perriard E, Schafer BW, Perriard JC, Ehler E. 2002. Subcellular targeting of metabolic enzymes to titin in heart muscle may be mediated by DRAL/FHL-2. J Cell Sci 115:4925–4936. doi: 10.1242/jcs.00181. [DOI] [PubMed] [Google Scholar]
- 12.Muller JM, Isele U, Metzger E, Rempel A, Moser M, Pscherer A, Breyer T, Holubarsch C, Buettner R, Schule R. 2000. FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J 19:359–369. doi: 10.1093/emboj/19.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong Z, Ding L, Sun J, Cao J, Lin J, Lu Z, Liu Y, Huang C, Ye Q. 2010. Synergistic repression of estrogen receptor transcriptional activity by FHL2 and Smad4 in breast cancer cells. IUBMB Life 62:669–676. doi: 10.1002/iub.367. [DOI] [PubMed] [Google Scholar]
- 14.Hinson JS, Medlin MD, Taylor JM, Mack CP. 2008. Regulation of myocardin factor protein stability by the LIM-only protein FHL2. Am J Physiol Heart Circ Physiol 295:H1067–H1075. doi: 10.1152/ajpheart.91421.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philippar U, Schratt G, Dieterich C, Muller JM, Galgoczy P, Engel FB, Keating MT, Gertler F, Schule R, Vingron M, Nordheim A. 2004. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol Cell 16:867–880. doi: 10.1016/j.molcel.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 16.Neuman NA, Ma S, Schnitzler GR, Zhu Y, Lagna G, Hata A. 2009. The four-and-a-half LIM domain protein 2 regulates vascular smooth muscle phenotype and vascular tone. J Biol Chem 284:13202–13212. doi: 10.1074/jbc.M900282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurakula K, Vos M, Otermin Rubio I, Marinković G, Buettner R, Heukamp LC, Stap J, de Waard V, van Tiel CM, de Vries CJ. 2014. The LIM-only protein FHL2 reduces vascular lesion formation involving inhibition of proliferation and migration of smooth muscle cells. PLoS One 9:e94931. doi: 10.1371/journal.pone.0094931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng CF, Xu JY, Li MS, Tsui SK. 2014. Identification of FHL2-regulated genes in liver by microarray and bioinformatics analysis. J Cell Biochem 115:744–753. doi: 10.1002/jcb.24714. [DOI] [PubMed] [Google Scholar]
- 19.Ramayo-Caldas Y, Ballester M, Fortes MR, Esteve-Codina A, Castello A, Noguera JL, Fernandez AI, Perez-Enciso M, Reverter A, Folch JM. 2014. From SNP co-association to RNA co-expression: novel insights into gene networks for intramuscular fatty acid composition in porcine. BMC Genomics 15:232. doi: 10.1186/1471-2164-15-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kidani Y, Bensinger SJ. 2012. Liver X receptor and peroxisome proliferator-activated receptor as integrators of lipid homeostasis and immunity. Immunol Rev 249:72–83. doi: 10.1111/j.1600-065X.2012.01153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calkin AC, Tontonoz P. 2010. Liver X receptor signaling pathways and atherosclerosis. Arterioscler Thromb Vasc Biol 30:1513–1518. doi: 10.1161/ATVBAHA.109.191197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins JL, Fivush AM, Watson MA, Galardi CM, Lewis MC, Moore LB, Parks DJ, Wilson JG, Tippin TK, Binz JG, Plunket KD, Morgan DG, Beaudet EJ, Whitney KD, Kliewer SA, Willson TM. 2002. Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J Med Chem 45:1963–1966. doi: 10.1021/jm0255116. [DOI] [PubMed] [Google Scholar]
- 23.Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, Lustig KD, Shan B. 2000. Role of LXRs in control of lipogenesis. Genes Dev 14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao C, Dahlman-Wright K. 2010. Liver X receptor in cholesterol metabolism. J Endocrinol 204:233–240. doi: 10.1677/JOE-09-0271. [DOI] [PubMed] [Google Scholar]
- 25.Herzog B, Hallberg M, Seth A, Woods A, White R, Parker MG. 2007. The nuclear receptor cofactor, receptor-interacting protein 140, is required for the regulation of hepatic lipid and glucose metabolism by liver X receptor. Mol Endocrinol 21:2687–2697. doi: 10.1210/me.2007-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi W, Oshlack A, Smyth GK. 2010. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res 38:e204. doi: 10.1093/nar/gkq871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. 2005. Downregulation of liver X receptor-alpha in mouse kidney and HK-2 proximal tubular cells by LPS and cytokines. J Lipid Res 46:2377–2387. doi: 10.1194/jlr.M500134-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Brendel C, Schoonjans K, Botrugno OA, Treuter E, Auwerx J. 2002. The small heterodimer partner interacts with the liver X receptor alpha and represses its transcriptional activity. Mol Endocrinol 16:2065–2076. doi: 10.1210/me.2001-0194. [DOI] [PubMed] [Google Scholar]
- 29.Langmann T, Porsch-Ozcürümez M, Heimerl S, Probst M, Moehle C, Taher M, Borsukova H, Kielar D, Kaminski WE, Dittrich-Wengenroth E, Schmitz G. 2002. Identification of sterol-independent regulatory elements in the human ATP-binding cassette transporter A1 promoter: role of Sp1/3, E-box binding factors, and an oncostatin M-responsive element. J Biol Chem 277:14443–14450. doi: 10.1074/jbc.M110270200. [DOI] [PubMed] [Google Scholar]
- 30.Delvecchio CJ, Bilan P, Nair P, Capone JP. 2008. LXR-induced reverse cholesterol transport in human airway smooth muscle is mediated exclusively by ABCA1. Am J Physiol Lung Cell Mol Physiol 295:L949–L957. doi: 10.1152/ajplung.90394.2008. [DOI] [PubMed] [Google Scholar]
- 31.Wang N, Silver DL, Costet P, Tall AR. 2000. Specific binding of ApoA-I, enhanced cholesterol efflux, and altered plasma membrane morphology in cells expressing ABC1. J Biol Chem 275:33053–33058. doi: 10.1074/jbc.M005438200. [DOI] [PubMed] [Google Scholar]
- 32.Matulis CK, Mayo KE. 2012. The LIM domain protein FHL2 interacts with the NR5A family of nuclear receptors and CREB to activate the inhibin-alpha subunit gene in ovarian granulosa cells. Mol Endocrinol 26:1278–1290. doi: 10.1210/me.2011-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arber S, Caroni P. 1996. Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes Dev 10:289–300. doi: 10.1101/gad.10.3.289. [DOI] [PubMed] [Google Scholar]
- 34.Ng EK, Chan KK, Wong CH, Tsui SK, Ngai SM, Lee SM, Kotaka M, Lee CY, Waye MM, Fung KP. 2002. Interaction of the heart-specific LIM domain protein, FHL2, with DNA-binding nuclear protein, hNP220. J Cell Biochem 84:556–566. doi: 10.1002/jcb.10041. [DOI] [PubMed] [Google Scholar]
- 35.Xu L, Glass CK, Rosenfeld MG. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev 9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 36.Davies JD, Carpenter KL, Challis IR, Figg NL, McNair R, Proudfoot D, Weissberg PL, Shanahan CM. 2005. Adipocytic differentiation and liver X receptor pathways regulate the accumulation of triacylglycerols in human vascular smooth muscle cells. J Biol Chem 280:3911–3919. doi: 10.1074/jbc.M410075200. [DOI] [PubMed] [Google Scholar]
- 37.Huuskonen J, Fielding PE, Fielding CJ. 2004. Role of p160 coactivator complex in the activation of liver X receptor. Arterioscler Thromb Vasc Biol 24:703–708. doi: 10.1161/01.ATV.0000121202.72593.da. [DOI] [PubMed] [Google Scholar]
- 38.Labalette C, Renard CA, Neuveut C, Buendia MA, Wei Y. 2004. Interaction and functional cooperation between the LIM protein FHL2, CBP/p300, and beta-catenin. Mol Cell Biol 24:10689–10702. doi: 10.1128/MCB.24.24.10689-10702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.