Abstract
JUN transcription factors bind DNA as part of the AP-1 complex, regulate many cellular processes, and play a key role in oncogenesis. The three JUN proteins (c-JUN, JUNB, and JUND) can have both redundant and unique functions depending on the biological phenotype and cell type assayed. Mechanisms that allow this dynamic switching between overlapping and distinct functions are unclear. Here we demonstrate that JUND has a role in prostate cell migration that is the opposite of c-JUN’s and JUNB's. RNA sequencing reveals that opposing regulation by c-JUN and JUND defines a subset of AP-1 target genes with cell migration roles. cis-regulatory elements for only this subset of targets were enriched for ETS factor binding, indicating a specificity mechanism. Interestingly, the function of c-JUN and JUND in prostate cell migration switched when we compared cells with an inactive versus an active RAS/extracellular signal-regulated kinase (ERK) signaling pathway. We show that this switch is due to phosphorylation and activation of JUND by ERK. Thus, the ETS/AP-1 sequence defines a unique gene expression program regulated by the relative levels of JUN proteins and RAS/ERK signaling. This work provides a rationale for how transcription factors can have distinct roles depending on the signaling status and the biological function in question.
INTRODUCTION
Most transcription factors can be grouped into families based on homologous DNA binding domains (1). These proteins often bind similar DNA sequences and therefore can compete to occupy genomic binding sites. Altering the balance of this competition toward factors with stronger, or weaker, transactivation functions is one mechanism of modulating gene expression. However, this complicates our understanding of individual transcription factor function, as a protein that activates transcription in one cell may attenuate transcription in a different cell depending upon the presence of other, stronger activators. Furthermore, signaling pathways can change the transactivation potential of transcription factors, thus dynamically altering the roles of competing factors. In the postgenomic era, where most cis-regulatory sequences are known, predicting the variable functions of a transcription factor binding site is a major new challenge.
The AP-1 transcription factor is an important regulator of cellular proliferation, survival, and locomotion and plays a central role in oncogenesis (2). AP-1 is a dimer of bZIP proteins from the JUN and FOS families that bind the consensus DNA sequence 5′-TGA(C/G)TCA-3′. JUN proteins can bind as either homo- or heterodimers, while FOS proteins can bind only DNA as heterodimers with JUN. Other bZIP families (ATF, MAF) can also pair with JUN, but this usually changes the DNA sequence preference. Once bound to DNA, each JUN family member has different and often antagonistic transcriptional functions (3, 4). Furthermore, the role of JUN members changes in different cell types. For example, c-JUN can promote apoptosis in fibroblasts and neurons but inhibits apoptosis in hepatocytes and keratinocytes (3). JUNB and JUND can either promote or inhibit cell cycle progression depending on the cellular background (5–7). The mechanisms that regulate these functional differences are not clear, making it difficult to predict the role of an AP-1 binding site in any particular cell type.
AP-1 can work together with a variety of other transcription factors, including those of the 28-member ETS family (8). Neighboring binding sites for ETS and AP-1 transcription factors were first identified in viral enhancers (9) and in the promoters of genes critical for cellular migration and invasion, including extracellular protease genes, such as PLAU, MMP1, and MMP9 (10–12). Together but not individually, the ETS and AP-1 binding sequences confer RAS/extracellular signal-regulated kinase (ERK) pathway responsiveness to neighboring genes (9, 13). ERK has been shown to mediate this function via either direct or indirect phosphorylation of various ETS proteins, such as ETS1, ETS2, ETV1, ETV4, and ETV5 (14, 15). This phosphorylation event can increase the affinity of ETS1 for the coactivator CBP/p300 and result in increased transcription of a neighboring gene (16). We recently identified another role for ETS proteins at these ETS/AP-1 sequences (17). In some prostate cancers, a chromosomal rearrangement results in the expression of one of four ETS genes (ERG, ETV1, ETV4, and ETV5) that are not normally expressed in prostate cells (18). These ETS proteins are oncogenic and promote prostate cell migration and invasion. We have shown that these oncogenic ETS proteins bind ETS/AP-1 sequences throughout the genome and activate transcription in the absence of RAS/ERK pathway activation (17). Therefore, we proposed two mechanisms of ETS function at ETS/AP-1 sequences. First, RAS/ERK-responsive ETS proteins can bind and activate transcription only when the RAS/ERK pathway is active. Second, oncogenic ETS proteins can bind and activate transcription in the absence of RAS/ERK signaling. While we have models for ETS function at ETS/AP-1 sequences, little is known about the role of AP-1 at this critical RAS/ERK response element.
Here we test the roles of the three JUN family members in regulating the cell migration transcriptional program mediated by ETS/AP-1 sequences. We find in prostate cell lines where RAS/ERK signaling is low that c-JUN promotes cell migration but that JUND inhibits migration. Interestingly, using transcriptome-wide RNA-sequencing (RNA-seq), we find that both c-JUN and JUND activate the majority of AP-1 target genes but that a subset of target genes are regulated in an opposite manner by c-JUN and JUND. This subset is enriched for cell migration genes and the ETS/AP-1 binding sequence. This indicates that the ETS/AP-1 sequence defines a gene expression program regulated in opposite directions by c-JUN and JUND. Surprisingly, the roles of JUN proteins are completely reversed in prostate cell lines that have an active RAS/ERK pathway. We show that the reason for this functional switch stems from the ability of ERK to bind and phosphorylate JUND, and we propose that this changes JUND from a weaker activator than c-JUN into a stronger activator than c-JUN. Therefore, these findings provide a mechanistic rationale for antagonistic functions of transcription factor family members that can be altered by RAS/ERK signaling.
MATERIALS AND METHODS
Cell culture and viral transduction.
All cell lines used in this study were authenticated by the University of Arizona Genetics Core using the PowerPlex 16HS assay (Promega) with a >80% match to eight core short tandom repeat (STR) loci (19). Cell lines were cultured according to ATCC recommendations as follows: 293 EBNA, HEK293T, and DU145 cells with Dulbecco's modified Eagle's medium (DMEM, Sigma) with 10% fetal bovine serum (FBS; Sigma); RWPE (RWPE1) and RWPE-KRAS (RWPE2) cells with keratinocyte serum-free medium (SFM) with growth supplements (Invitrogen); and PC3 cells with F12K medium (Mediatech-Cellgro) with 10% FBS. All media were supplemented with 1% penicillin-streptomycin (100× solution; Mediatech-Cellgro). Lentivirus for JUN knockdown experiments were produced by cotransfection of pLKO.1 constructs encoding short-hairpin RNAs (shRNAs) in HEK293T cells with pMDLg/pRRE, pRSV-Rev, and pMD2.G envelope plasmids (Addgene). shRNA sequences were luciferase, CTTACGCTGAGTACTTCGATTC; c-JUN, AAGGAAGCTGGAGAGAATCGC (20); JUNB, AAGCAACGGCGTGATCACGAC (21); and JUND, ACGCGAACCTGAGCAGCTATT (22). Full-length JUN open reading frames were cloned into the vector pQCXIH (Clontech) using oligonucleotides indicated in Table S2 in the supplemental material. JUN proteins were stably expressed in DU145 and PC3 cell lines via use of a retrovirus as described previously (17).
Transwell migration, in vitro scratch, and luciferase assays.
Transwell-migration assays were done as described previously (17). In brief, 5 × 104 cells were introduced to Transwells (8-μm pore size; BD Bioscience) and incubated for 48 h (DU145 and PC3) and 60 h (RWPE1, RWPE-KRAS). The mean of results for five representative fields per membrane was determined in each biological replicate. For in vitro scratch assays, cells were plated in 35-mm plates and grown to full confluence, and the cultures were scratched with a pipette tip. Migration into the open area was documented at 24 h postscratching by microscopy. Free area was measured using TScratch software (23; www.cse-lab.ethz.ch/software.html). Luciferase assays using wild-type and mutant ETS/AP-1 sequences were done in the cell lines indicated above using vectors and methods previously reported (24).
Measuring protein and RNA levels.
Total protein extract from equal numbers of cells were separated on 10% SDS-PAGE gels and transferred to nitrocellulose membranes by standard procedures (Bio-Rad). Membranes were blocked in 5% milk in TBS (10 mM Tris, pH 8.0, 150 mM NaCl), incubated with primary and secondary antibodies, and visualized by enhanced chemiluminescence (ECL) (Thermo Scientific) by using standard procedures. Antibodies for c-Jun (sc-45), JunB (sc-8051), and JunD (sc-74) were from Santa Cruz Biotechnology. Phospho-c-Jun (Ser 73, 9164) was from Cell Signaling. Antitubulin was bought from Sigma. RNA levels were measured by reverse transcription-quantitative PCR (RT-qPCR) as previously reported (17). Standard curves from diluted PCR products were used to measure absolute values for each gene product, and then each reading was standardized to the level of a housekeeping gene (EEF1A). Oligonucleotides used in RT-qPCR are described in Table S2 in the supplemental material.
ChIP, protein purification, and in vitro kinase assays.
Chromatin immunoprecipitation (ChIP) from PC3 cells was done as previously reported (25) using a JUND antibody from Santa Cruz Biotechnology (sc-74). qPCR measurement of ChIP enrichment used the primers in Table S2 in the supplemental material. JUN proteins were cloned into the pet28a vector, which adds an N-terminal 6×His tag. After expression in BL-21 cells, protein was extracted and JUN proteins were purified by a Ni chromatography column. Each purified JUN protein could specifically bind the AP-1 sequence in a gel shift assay. c-JUN N-terminal kinase (JNK) and ERK2 kinases were from SignalChem. Reaction mixtures contained 0.5 ng/μl kinase and 25 ng/μl JUN protein and were incubated for 30 min at 30°C.
RNA sequencing.
Raw and processed RNA-seq files are available for download from NCBI's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) (see below). Total RNA from three biological replicates was isolated from PC3 cells transduced with lentiviral shRNA knockdown vectors using the RNeasy minikit (Qiagen) according to the manufacturer's instructions. Sequencing libraries for whole-transcriptome analysis were generated using a modified Illumina TruSeq sample preparation protocol. Total RNA was treated with TURBO DNase (Invitrogen). The DNase-treated RNA was poly(A) selected with oligo(dT) beads (Invitrogen). A Superscript III reverse transcriptase first-strand synthesis (Invitrogen) system was used to generate cDNA from the poly(A)-selected RNA with random hexamer primers (Invitrogen). After first-strand synthesis, a second strand was generated using Escherichia coli DNA ligase (New England BioLabs) and E. coli DNA polymerase I (New England BioLabs). The double-stranded cDNAs were sheared to ∼150 nucleotides using a Diagenode Bioruptor, and the size was confirmed by DNA gel electrophoresis. DNA end repair of the cDNA was performed using Klenow DNA polymerase (New England BioLabs), T4 DNA polymerase (New England BioLabs), and T4 DNA ligase (New England BioLabs) before the sample was subjected to QIAquick PCR purification (Qiagen). Adapters were ligated to DNA fragments using T4 DNA ligase (New England BioLabs). The product was run on a 2% agarose gel, size selected to be between 200 and 300 nucleotides, and then purified by a gel extraction kit (Qiagen). Universal and indexing adapters were taken from the TruSeq sample preparation kit (Illumina). Deep sequencing was performed on an Illumina HiSeq2000 instrument according to the manufacturer's protocol.
Informatics analysis of RNA-seq.
The Tuxedo Suite RNA sequencing pipeline was used to determine differential gene expression for PC3 Jun knockdown samples sent for deep sequencing as described previously (26), with some modifications. Raw FASTQ files were mapped to the human genome (UCSC release, version 19) using TopHat2 with Bowtie2. The annotated genome file used in the pipeline was modified to contain gene identifiers as well as transcript identifiers and to remove all noncoding RNA. Differential expression of genes and transcripts was determined using Cuffdiff2. The RNA-seq data were separated into quadrants based on gene expression changes in JUND and c-JUN knockdown cells in a comparison with that in luciferase knockdown cells. Only genes with nearest-neighbor JUND-bound regions in PC3 cells (17) were considered for further analysis. JUND-bound regions near genes in each quadrant were searched for the sequence MGGAAGW using the Regulatory Sequence Analysis Tools (RSAT) DNA pattern function (27; http://rsat.ulb.ac.be/) and were found to overlap ETV4-bound regions in PC3 cells (17). The genes from each quadrant were used as inputs for gene ontology using the program g:GOSt Gene Group Functional Profiling on g:Profiler (28; http://biit.cs.ut.ee/gprofiler/index.cgi). The default settings for g:GOSt were changed to search for only significant ontologies (P < 0.05). The significance threshold for each ontology category was determined using the Benjamini-Hochberg false-discovery rate. To eliminate broad uninformative categories, a maximum of 2,000 genes was set for the ontology category to be considered.
Microarray data accession number.
Raw and processed RNA-seq files are available for download from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE53470.
RESULTS
JUN family transcription factors have opposing functions in the regulation of cell migration.
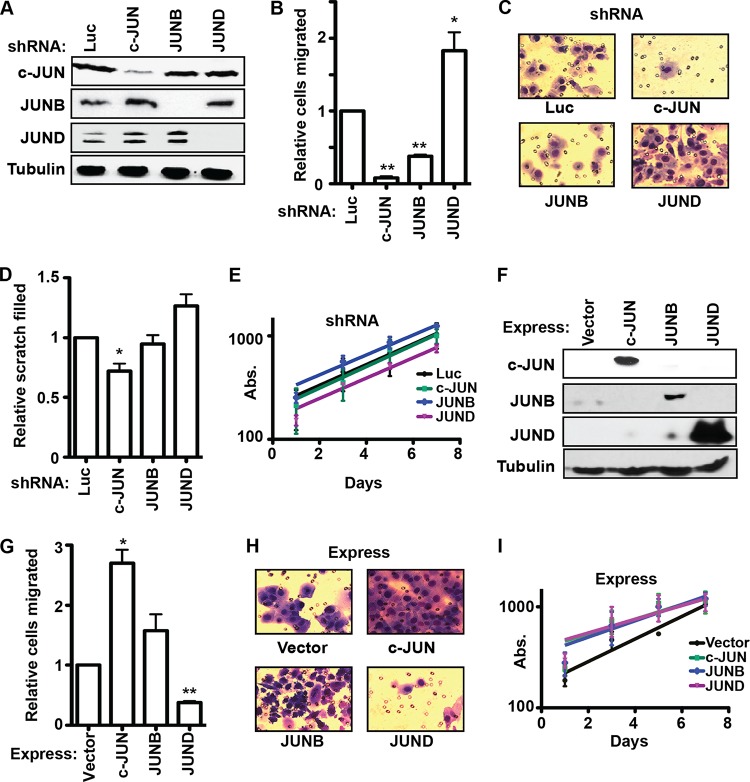
ETS/AP-1 binding sequences regulate cell migration genes and therefore play a role in the transition from neoplasia to invasive carcinoma. However, the function of individual AP-1 subunits at these sites is not known. Because JUN but not FOS family members have been shown to interact with various ETS proteins (29), we focused on the role of JUN family members on cell migration. Lentiviral constructs stably expressed shRNAs to knock down expression of each JUN family member in PC3 prostate cancer cells (Fig. 1A). Each knockdown specifically changed the expression of the target JUN protein but not those of the other family members. Loss of either c-JUN or JUNB resulted in a significant decrease in cell migration in a transwell assay (Fig. 1B and C), indicating that c-JUN and JUNB promote cell migration. Loss of JUND had the opposite effect, increasing cell migration (Fig. 1B and C), indicating that JUND is an inhibitor of cell migration. A second method of measuring cell migration, the scratch assay, revealed a similar trend (Fig. 1D). None of the JUN knockdown experiments significantly changed the proliferation rate of PC3 cells (Fig. 1E), indicating that differences in proliferation do not bias the migration assays. We next tested the effect of changing the relative levels of JUN proteins by overexpression. Each JUN protein was stably overexpressed in PC3 cells by a retroviral vector (Fig. 1F). Overexpression of c-JUN significantly increased cell migration, and JUNB overexpression resulted in a similar trend (Fig. 1G and H), consistent with an activating function. Overexpression of JUND significantly decreased cell migration (Fig. 1G and H), consistent with a repressive function. Unlike with their opposite roles in migration, overexpression of all three JUNs slightly decreased PC3 proliferation (Fig. 1I). In summary, both depletion and overexpression experiments indicate that, in PC3 cells, c-JUN and JUNB activate a cell migration gene expression program but that JUND has the opposite function.
FIG 1.
JUND and c-JUN/JUNB have opposite functions in PC3 cell migration. (A) Immunoblots with the indicated antibodies (left) show c-JUN, JUNB, and JUND protein levels in PC3 cell lines expressing shRNAs targeting the indicated genes (top). Tubulin levels provide a loading control. (B) A transwell assay measured the cell migration of c-JUN-, JUNB-, and JUND-depleted PC3 cells. The relative numbers of cells that migrated are the means and standard errors of the means (SEM) from three biological replicates (each value is the mean of results from two technical replicates) relative to numbers in a control luciferase (Luc) knockdown experiment. (C) Representative images of transwell migration assays shown in panel B. (D) The fractions of scratches filled in 24 h by migrating PC3 cells with shRNAs with the indicated genes knocked down are shown as the means and SEM from five replicates relative to the fraction of the scratch filled in the control luciferase knockdown experiment. (E) A 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay measured proliferation of PC3 cells with the indicated genes knocked down by determining absorbance (Abs.) at 600 nM beginning 24 h after plating. The results are the means and SEM from two biological replicates (each value is the mean of results from three technical replicates). (F) Immunoblots show Jun protein levels in PC3 cells stably overexpressing the indicated genes (top). (G, H) A transwell assay measured cell migration in PC3 cells with the indicated overexpression of a JUN protein. The number of migrated cells is shown as in panels B and C. (I) Proliferation of PC3 cells overexpressing the indicated JUN protein is shown as in panel E. All P values were calculated by the t test (*, P < 0.05; **, P < 0.005).
JUN protein competition for ETS-AP-1 binding sites can explain opposing cell migration roles.
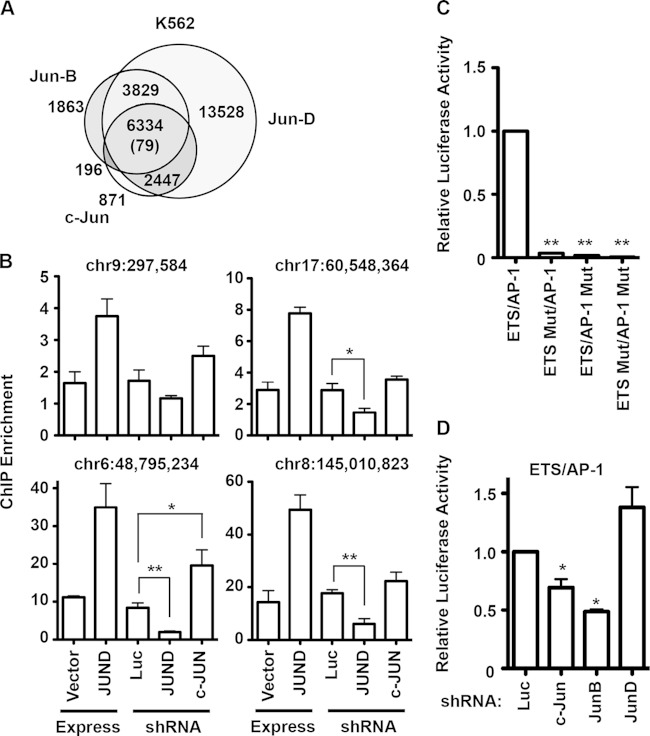
JUN family members bind the same DNA sequences in vitro (2) and therefore are predicted to compete for the same targets in vivo. To verify this, we compared genome-wide mapping findings from ENCODE (30) for c-JUN, JUND, and JUNB occupancy in the only cell line for which these data were available, K562 (Fig. 2A). Consistently with in vitro findings, the JUN proteins occupied a highly overlapping set of regions in vivo. To test whether JUN protein occupancy varies by expression level, chromatin was immunoprecipitated with a JUND antibody from PC3 cells with either JUND overexpression or shRNA depletion of JUND or c-JUN. Enrichment compared to that of unbound control loci was measured by quantitative PCR for four JUND target enhancers randomly selected from a JUND ChIP-DNA sequencing (ChIP-seq) data set (17). Enrichment at JUND target enhancers varied from 1.5- to 17-fold (Fig. 2B). In all four cases, overexpression of JUND increased JUND occupancy, indicating that JUND occupancy was not saturated at these sites (Fig. 2B). At all four targets, shRNA depletion of JUND resulted in a trend toward less enrichment. Also at all four targets, shRNA depletion of c-JUN resulted in a trend of increased JUND enrichment, consistent with competition between these factors. Therefore, we hypothesized that JUN proteins compete for occupancy of ETS/AP-1 sequences near cell migration genes. If c-JUN and JUNB are better activators than JUND in PC3 cells, this competition may account for their opposite roles observed in cell migration. To test whether ETS/AP-1 sequences could indeed mediate these opposite transcriptional functions, a reporter construct that has three copies of the ETS/AP-1 sequence controlling transcription of the firefly luciferase gene was tested. This reporter requires both the ETS and AP-1 sequences for activation (Fig. 2C). Expression of the reporter in PC3 cells with JUN knocked down mirrored the cell migration data, with loss of c-JUN and JUNB significantly reducing luciferase levels and loss of JUND resulting in a trend toward increased luciferase expression (Fig. 2D). Therefore, JUN family members have antagonistic functions at ETS/AP-1 sequences that match their cell migration functions.
FIG 2.
JUN protein transcriptional regulation through ETS/AP-1 sequences mirrors migration phenotypes. (A) A Venn diagram indicates overlaps of genomic regions bound by c-JUN (GSM1003609; Snyder Lab, Stanford University), GFP-JUNB, and GFP-JUND (GSM777638 and GSM777639; White Lab, University of Chicago) in K562 cells. The number in parentheses indicates the randomly predicted overlap, assuming that all open chromatin regions in K562 cells are potential accessible binding sites. (B) Chromatin immunoprecipitation (ChIP) of JUND in PC3 cells with the indicated overexpression or knockdown shRNA. The absolute quantity for each indicated locus was measured by quantitative PCR and is shown as a ratio to the mean quantity of two negative-control loci, such that values greater than 1 are considered enriched. The means and SEM from two independent replicates are shown for overexpression and from five replicates for knockdown shRNAs. P values were calculated by the t test (*, P < 0.05; **, P < 0.005). (C) A firefly luciferase reporter with either three copies of the ETS/AP-1 sequence or point mutations in the ETS or AP-1 sequences, as indicated, cloned upstream of a minimal promoter was transfected into PC3 cells. The ratio of firefly to renilla (control) luciferase is shown as a mean and SEM of results from three biological replicates relative to the wild-type ETS/AP-1 sequence. (D) The wild-type ETS/AP-1 reporter was transfected into PC3 cells with the indicated knockdown shRNAs, and reporter activity is shown as in panel C. P values were calculated by t test (*, P < 0.05; **, P < 0.001).
Genes regulated in opposite manners by c-JUN and JUND represent a subset of AP-1 targets with roles in cellular locomotion.
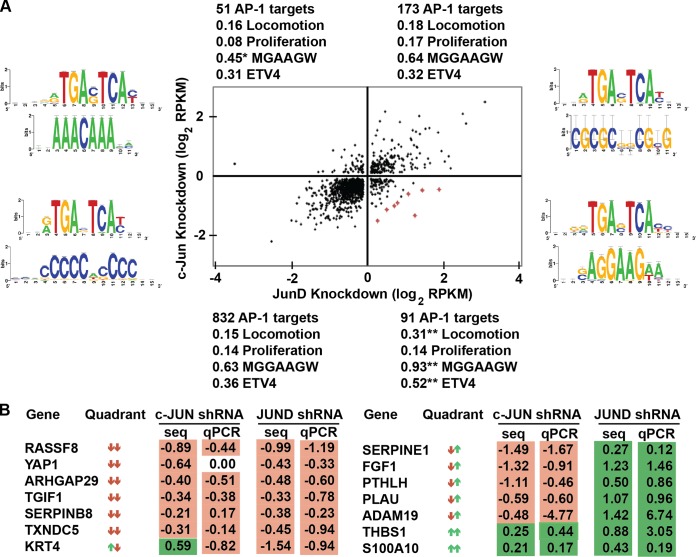
Previous mapping of AP-1 targets by JUND ChIP-seq in PC3 cells identified enrichment of an AP-1 binding sequence but not an ETS sequence (17). Ontology analysis of all AP-1 direct target genes does not identify cell migration functions. These data indicate that ETS/AP-1 cis-regulatory elements and cell migration genes represent a small subset of all AP-1 targets. Our cell migration and reporter data (Fig. 1 and 2) are consistent with a model in which c-JUN (and, to a lesser extent, JUNB) can activate cell migration genes via ETS/AP-1 sequences and in which JUND represses, or attenuates, transcription of the same genes. To identify the gene expression program that follows this pattern, mRNA levels in PC3 cell lines with c-JUN or JUND knocked down by shRNA were compared to mRNA levels in control cells (shRNA targeting luciferase) using next-generation sequencing. The Tuxedo Suite (26) was used to quantify mRNA levels from three biologically independent RNA-seq replicates for each sample. To identify potential direct AP-1 target genes, we analyzed only genes for which our previous PC3 ChIP-seq data (17) identified a neighboring JUND-bound region (see Table S1 in the supplemental material). These direct AP-1 target genes were separated into four categories, downregulated in both c-JUN and JUND knockdown experiments, upregulated in both, or downregulated in one and upregulated in the other (Fig. 3A). As a control, expression changes for 14 target genes selected for cell migration and roles in epithelial-to-mesenchymal transition (EMT) were tested by reverse transcription-quantitative PCR (RT-qPCR), and 26 of 28 changes identified by RNA-seq were confirmed (Fig. 3B). The majority of genes (73%) were in the quadrant downregulated by both c-JUN and JUND knockdown (Fig. 3A, lower left), suggesting a common activating function of these two JUN proteins at most AP-1 targets. However, we predicted that genes that promote cell migration would decrease in the c-JUN knockdown experiment and increase in the JUND knockdown experiment. There were 91 predicted direct target genes in this quadrant (Fig. 3A, lower right). The eight genes with the largest expression changes in this quadrant are marked in red in Fig. 3A and listed in Table 1. Strikingly, these eight genes encode three matrix metalloproteases, PLAU, MMP9, and ADAM19, a metalloprotease inhibitor, SERPINE1, and signaling ligands FGF1 and PTHLH, all factors involved in cell migration and/or communication with the tumor microenvironment (31–33). To test whether the 91 genes in this quadrant share functions consistent with the cell migration phenotype, unbiased gene ontology analysis was used (Table 2). The four most overrepresented functional categories of genes in this quadrant were “extracellular matrix organization,” “extracellular structure organization,” “locomotion,” and “cellular component movement,” consistent with cell migration and invasion roles. No other quadrant was enriched for these categories. A directed search found that 31% of the genes in the lower right quadrant have a role in cellular locomotion, representing a significant (P < 0.01) enrichment over the 15 to 18% of genes in each of the other quadrants (Fig. 3A). In comparison, there was no enrichment of genes with a role in cellular proliferation, consistent with the lack of a proliferation phenotype (Fig. 1E). Therefore, we conclude that opposite regulation by c-JUN and JUND can define a unique subset of AP-1 target genes that function to control cellular migration and reorganization of the extracellular matrix.
FIG 3.
Genes activated by c-JUN and repressed by JUND have ETS/AP-1 motifs and migration roles. (A) A dot plot shows 1,147 genes with nearest-neighbor JUND-bound regions and with a log2 fold change of >0.1 read per kilobase per million (RPKM) in both c-JUN- and JUND-depleted PC3 cells. Genes listed in Table 1 are marked in red in the dot plot. JUND-bound regions from each quadrant were used to query RSAT (52) to identify overrepresented sequence motifs. Position weight matrices for the first (above)- and second (below)-most-significant motifs returned are shown as logos on either side of the dot plot. Genes in each quadrant were compared to a list of genes involved in locomotion, chemotaxis, and extracellular matrix organization (Locomotion) or genes involved in proliferation as categorized by AmiGO v1.8 (53). The fraction of genes in these ontologies compared to the total number of genes in the quadrant is shown. Each JUND-bound region was searched for the ETS binding motif MGGAAGW (M = A/C; W = A/T). The fraction of bound regions containing this site relative to the total number of genes in each quadrant is shown. JUND-bound regions were compared to ETV4-bound regions in the same cell line (PC3). The fraction of overlapping regions compared to the total number of genes in each quadrant is shown. P values are calculated by chi-square testing (*, P < 0.01 [depleted]; **, P < 0.001 [enriched]). (B) Reverse transcription-quantitative PCR (RT-qPCR) confirmation of selected RNA-seq (seq) expression changes. Log2 fold changes in the activation of the indicated knockdown shRNAs from the activation of the knockdown control (luciferase) are shown as means from at least two replicates.
TABLE 1.
Extracellular proteases and growth factors are activated by c-JUN and repressed by JUND in PC3 cells
| Ranka | Gene identifier | Name | Presence of: |
|
|---|---|---|---|---|
| ETV4b | MGGAAGWc | |||
| 1 | FGF1 | Fibroblast growth factor 1 | Yes | Yes |
| 2 | PTPN5 | Tyrosine protein phosphatase nonreceptor type 5 | No | Yes |
| 3 | ADAM19 | Disintegrin and metalloproteinase domain protein 19 | Yes | Yes |
| 4 | SERPINE1 | Plasminogen activator inhibitor 1 | Yes | Yes |
| 5 | CXCR7 | C-X-C chemokine receptor type 7 | No | No |
| 6 | MMP9 | Matrix metalloproteinase 9 | No | No |
| 7 | PLAU | Urokinase-type plasminogen activator | Yes | Yes |
| 8 | PTHLH | Parathyroid hormone-related protein | Yes | Yes |
The top eight genes with the largest combined decreases in c-JUN repression and increases in JUND repression in PC3 cells are shown.
The presence of a neighboring ETV4-bound region was determined by ChIP-seq and reported by Hollenhorst et al. (17).
Presence of the ETS consensus sequence in a neighboring JUND-bound region.
TABLE 2.
AP-1 target genes activated by c-JUN and repressed by JUND in PC3 cells are enriched for ontologies associated with cell migration and invasion
| Quadranta | No. of genes | Most overrepresented ontology | P value |
|---|---|---|---|
| c-JUN act, JUND rep | 91 | Extracellular-matrix organization | 4.96E−08 |
| Extracellular-structure organization | 5.12E−08 | ||
| Locomotion | 2.70E−06 | ||
| Cellular-component movementc | 3.67E−06 | ||
| Epithelium development | 5.03E−05 | ||
| Tissue morphogenesis | 5.44E−05 | ||
| Positive regulation of phosphorus metabolic process | 1.36E−04 | ||
| Morphogenesis of an epithelium | 2.40E−04 | ||
| c-JUN act, JUND act | 832 | Phospholipid binding | 2.80E−16 |
| Lipid binding | 6.13E−15 | ||
| Positive regulation of the RNA metabolic process | 8.97E−15 | ||
| Positive regulation of transcription (DNA dependent) | 1.10E−14 | ||
| Positive regulation of gene expression | 1.11E−14 | ||
| Positive regulation of the nitrogen compound metabolic process | 2.89E−13 | ||
| Positive regulation of the nucleobase compound metabolic process | 3.30E−13 | ||
| Positive regulation of the macromolecule biosynthetic process | 7.15E−13 | ||
| c-JUN rep, JUND actb | 51 | PI3K-Akt signaling pathway | 3.94E−03 |
| Regulation of the actin cytoskeleton | 3.40E−02 | ||
| c-JUN rep, JUND rep | 173 | Anatomical-structure formation involved in morphogenesis | 2.11E−07 |
| Morphogenesis of an epithelium | 6.43E−07 | ||
| Tissue morphogenesis | 7.73E−07 | ||
| Tube development | 7.78E−07 | ||
| Tube morphogenesis | 1.31E−06 | ||
| Odontogenesis | 1.96E−06 | ||
| Chordate embryonic development | 2.52E−06 | ||
| Morphogenesis of embryonic epithelium | 2.56E−06 |
The top eight most enriched ontologies for genes either activated (act) or repressed (rep) by c-JUN and JUND are shown.
This category had only two ontologies, for which the P value was less than 0.1.
Cellular-component movement encompasses cell migration and neuronal projections.
Opposite regulation by c-JUN and JUND identifies AP-1 targets with ETS/AP-1 binding sequences.
PLAU and MMP9 are among the best-documented ETS/AP-1 target genes (10, 11). To test whether the 89 genes that coregulated with PLAU and MMP9 in the RNA-seq data represent a subset of AP-1 target genes that are distinguished by ETS/AP-1 binding sequences, an unbiased search identified the most overrepresented sequence motifs in all JUND-bound regions (Fig. 3A). As expected, the first motif identified in all four quadrants matched an AP-1 consensus binding sequence (TGA[C/G]TCA). However, the second motif identified in each quadrant differed. Significantly, a motif matching an ETS binding sequence (AGGAAG) was the second-most overrepresented in the quadrant associated with the cell migration phenotype (Fig. 3A, lower right), and this motif was not identified as significantly enriched in any of the other three quadrants (Fig. 3A and data not shown). The AGGAAG motif is consistent with an enhancer-enriched binding motif that can be bound by multiple ETS factors (17, 34). A directed search for the ETS sequence (MGGAAGW) found it to be significantly enriched among genes in the lower right quadrant compared to the other three quadrants (Fig. 3A). We have previously reported that PC3 cells overexpress the oncogenic ETS protein ETV4 and that ETV4 binds ETS/AP-1 sequences (17, 35). To test whether ETV4 binding was enriched at genes that were activated by c-JUN and repressed by JUND, we examined our previously published PC3 cell ETV4 ChIP-seq data (17). ETV4-bound regions overlapped JUND-bound regions significantly more often when the JUND-bound region was near a gene in the lower right quadrant than near the other three quadrants (Fig. 3A). Together, the RNA-seq analysis data indicate that JUN proteins regulate multiple gene expression programs and that the specific program that is activated by c-JUN and attenuated by JUND in PC3 cells correlates with the presence of ETS/AP-1 sequences and occupancy of an oncogenic ETS transcription factor.
The roles of c-JUN and JUND in cell migration switch when the RAS/ERK pathway is activated.
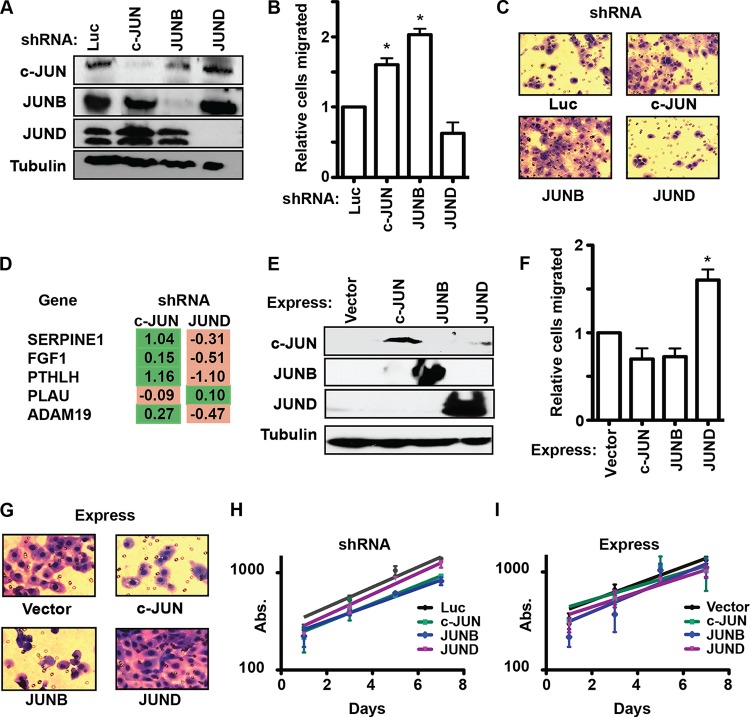
To test whether the ability of c-JUN and JUNB to activate, and JUND to attenuate, cell migration is specific to the PC3 cell line, each JUN family member was knocked down in a second prostate cancer cell line, DU145 (Fig. 4A). Surprisingly, the transwell assay with DU145 cells showed a result exactly opposite to that with PC3 cells. In DU145 cells, JUND was an activator of cell migration (a trend toward loss of migration in the knockdown experiment), and both c-JUN and JUNB were repressors of cell migration (Fig. 4B and C). Furthermore, compared to findings with PC3 cells, knockdown of c-JUN and JUND in DU145 cells had the opposite effect on the expression of four out of five target genes tested (compare Fig. 3B to 4D). As in PC3 cells, overexpression of each JUN protein in DU145 cells (Fig. 4E) confirmed the knockdown results, with JUND increasing cell migration and c-JUN and JUNB trending toward decreased migration (Fig. 4F and G). Changing JUN levels in DU145 cells also did not affect proliferation (Fig. 4H and I). So, in DU145 cells, the role of JUN transcription factors in the cell migration phenotype is completely reversed from that in PC3 cells (Fig. 1).
FIG 4.
The role of JUN proteins in cell migration is cell line dependent. (A) Immunoblots of DU145 cells expressing shRNAs targeting the indicated genes, with antibodies listed on left. (B, C) A transwell assay measured the migration of c-JUN, JUNB, and JUND knockdown in DU145 cells as described for Fig. 1B and C. (D) RT-qPCR of changes in the expression of the indicated genes after c-JUN or JUND knockdown in DU145 cells as described for Fig. 3B. (E) Immunoblots using the indicated antibodies of DU145 cells overexpressing the indicated JUN proteins by means of a retroviral vector. (F, G) A transwell assay measured the migration of JUN overexpression in DU145 cells as described for Fig. 1B and C. (H, I) Proliferation of DU145 cells with JUN shRNA repressed (H) or overexpressed (I) is measured as described for Fig. 1E. P values were calculated by the t test (*, P < 0.05).
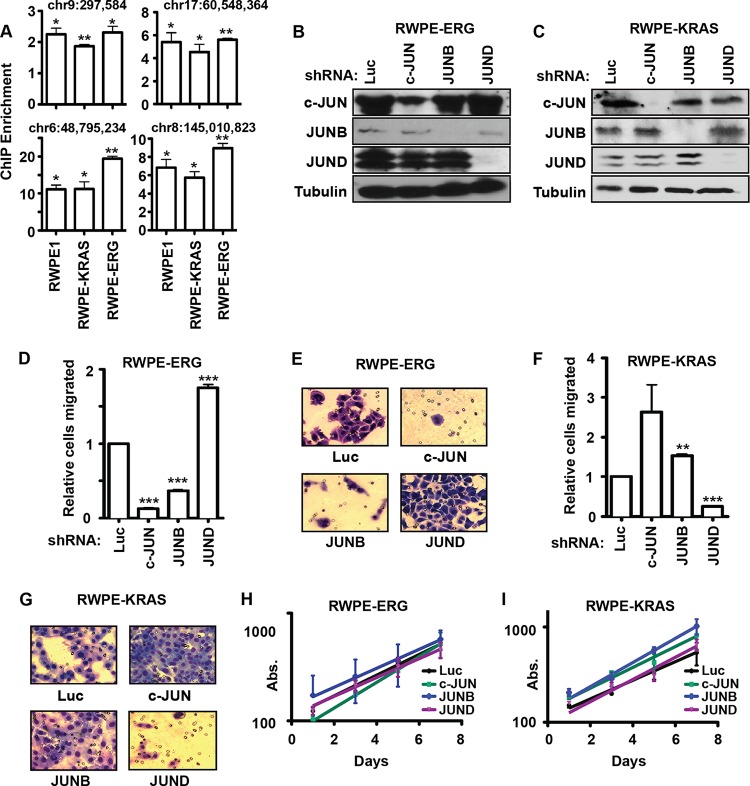
We have previously shown that both RAS/ERK pathway activation and oncogenic ETS expression can promote cell migration, but not synergistically (24). PC3 cells express an oncogenic ETS protein (ETV4) but have low levels of ERK activation, while DU145 cells have high ERK activity and lack an oncogenic ETS protein (24). To determine if this difference alone could explain the altered role of the JUN family transcription factors, we sought to compare JUN functions in otherwise isogenic cell lines where the only difference was oncogenic ETS expression or RAS/ERK activation. RWPE1 cells are derived from normal prostate cells and exhibit little cell migration (36). Stable overexpression of either the oncogenic Kirsten RAS (kiRAS) (RWPE-KRAS) or the oncogenic ETS protein ERG (RWPE-ERG) induces cell migration (24). ChIP indicated that JUND bound target sites in each of these cell lines (Fig. 5A). We knocked down all three JUN proteins in both RWPE-ERG and RWPE-KRAS cells (Fig. 5B and C) and measured cell migration (Fig. 5D to G). The changes in cell migration in RWPE-ERG cells were the same as those in PC3 cells (compare Fig. 5D to 1B), and changes in RWPE-KRAS cells mirrored those in DU145 cells (compare Fig. 5F to 4B). As with the other cell lines, no significant change in proliferation was observed (Fig. 5H and I). Therefore, c-JUN and JUNB increase migration in cells with oncogenic ETS expression (PC3 and RWPE-ERG), while JUND increases migration in cells with RAS/ERK pathway activation (DU145 and RWPE-KRAS).
FIG 5.
Oncogenic ETS proteins and RAS/ERK signaling define distinct roles for JUN proteins. (A) ChIP of JUND (as described for Fig. 2B) in the indicated cell lines, except that means and SEM from three independent replicates are shown. P values compare enrichment to the hypothetical mean of 1 (no enrichment) by a one-value t test (*, P < 0.05; **, P < 0.005). (B, C) Immunoblots of RWPE-ERG (B) and RWPE-KRAS (C) cells expressing shRNAs targeting the indicated JUN genes. (D to G) Transwell assays measured the migration of RWPE-ERG (D, E) and RWPE-KRAS (F, G) cells with c-JUN, JUNB, and JUND knocked down as described for Fig. 1B and C, except that the means from four replicates are shown. (H, I) Proliferation of RWPE-ERG (H) or RWPE-KRAS (I) cells with JUN shRNA knocked down is measured as described for Fig. 1E. P values were calculated by the t test (**, P < 0.005; ***, P < 0.0005).
ERK phosphorylation switches JUND to an activator of cell migration.
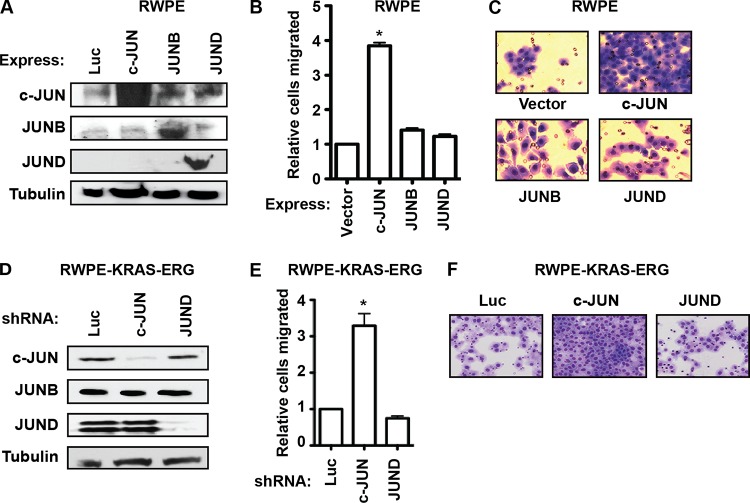
To explain the differences between c-JUN and JUND, we hypothesized that either c-JUN requires the presence of an oncogenic ETS to be an activator of migration or JUND requires an activated RAS/ERK pathway to activate migration. To differentiate these models, we overexpressed each JUN family member in a cell line that does not express an oncogenic ETS protein or have an activated RAS/ERK pathway: RWPE1 (Fig. 6A). c-JUN, but not JUND, increased the cell migration of RWPE1 cells (Fig. 6B and C). This indicates that c-JUN does not require the presence of an oncogenic ETS protein, but JUND does require an activated RAS/ERK pathway to increase migration. In a reciprocal experiment, c-JUN and JUND were depleted by shRNA in RWPE cells expressing both oncogenic KRAS and ERG (Fig. 6D), and migration assays were performed by transwell migration (Fig. 6E and F). The RWPE-KRAS-ERG cells showed a migration response that was similar to that of RWPE-KRAS cells and the opposite of that of RWPE-ERG cells (compare Fig. 6 and 5), indicating that KRAS signaling, and not the oncogenic ETS, is the important factor differentiating c-JUN and JUND functions.
FIG 6.
The switch in JUN functions is due to KRAS signaling. (A) Immunoblots of RWPE1 cells transduced with retroviral vectors expressing the indicated JUN members. (B, C) A transwell assay measured the migration of RWPE1 cells overexpressing c-JUN, JUNB, and JUND as described for Fig. 1B and C, except that means and SEM from two replicates are shown relative to values for cells expressing the empty vector. (D) Immunoblots of JUN proteins from RWPE-KRAS-ERG cells expressing the indicated shRNA. (E, F) Relative migration of cell lines in panel D by transwell assay. All P values were calculated by the t test (*, P < 0.05).
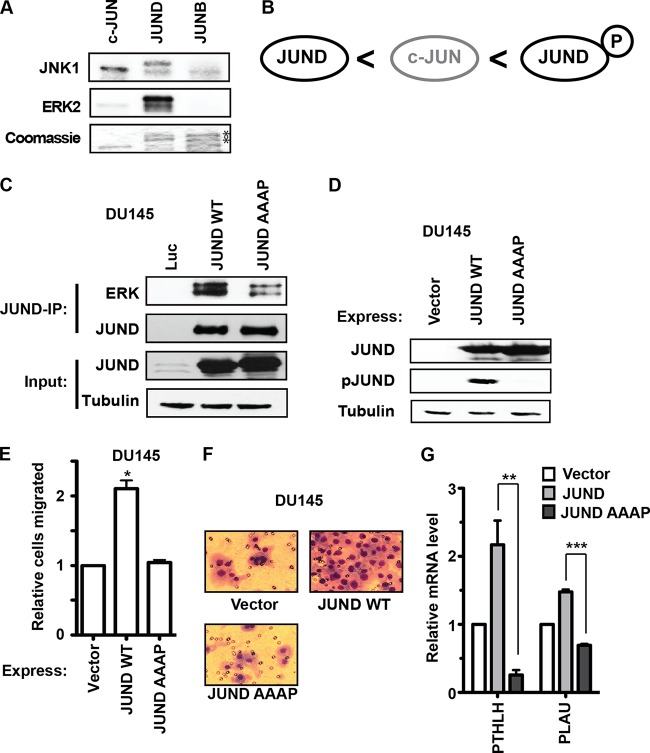
All three JUN proteins have D domains that mediate binding and phosphorylation by the mitogen-activated protein kinase (MAPK) JNK. Consistently with this, we found that JNK could phosphorylate purified recombinant versions of each JUN protein (Fig. 7A). However, JUND also has a docking site for ERK, the FXFP (DEF) domain (sequence FLYP). A previous study showed that this DEF domain allowed JUND but not c-JUN to be efficiently phosphorylated by ERK in response to epidermal growth factor (EGF) signaling in HEK293 cells (37). Furthermore, ERK phosphorylation of JUND allows increased transcriptional activation downstream of EGF signaling (37). Consistently with this previous finding, we found that JUND, but not c-JUN or JUNB, was phosphorylated by ERK in vitro (Fig. 7A). Therefore, we proposed a model in which c-JUN is a better activator than JUND in the absence of RAS/ERK pathway activation but that when the pathway is active, ERK phosphorylates JUND via the DEF domain and JUND becomes an even stronger activator than c-JUN (Fig. 7B). To test whether ERK phosphorylation of JUND is required for JUND activation of cell migration, we created a mutant version of JUND where the ERK binding motif was changed from FLYP to AAAP (JUND AAAP). Endogenous ERK was immunoprecipitated with a JUND antibody in DU145 cells with overexpressed JUND, but the AAAP mutation reduced the interaction of JUND and ERK (Fig. 7C). Both JUND and JUND AAAP were overexpressed at similar levels in DU145 cells, but only JUND was detected by an anti-phospho-JUN antibody, indicating that the loss of ERK binding results in a loss of phosphorylation (Fig. 7D). Importantly, overexpression of JUND in DU145 cells increased cell migration, but overexpression of JUND AAAP did not (Fig. 7E and F). Furthermore, JUND could activate expression of two target genes, PTHLH and PLAU, but JUND AAAP could not (Fig. 7G). Therefore, we conclude that ERK binding is required for the role of JUND in cell migration. Together, these data indicate that RAS/ERK signaling can switch the roles of JUND and c-JUN at the ETS/AP-1 sequences that regulate cell migration. Therefore, both the relative expression levels of JUN proteins and the level of RAS/ERK signaling combine to dynamically regulate a gene expression program critical to cancer progression.
FIG 7.
JUND interaction with ERK is required to promote cell migration. (A) His-tagged c-JUN, JUND, and JUNB were expressed and purified from bacteria. An autoradiogram of kinase assays (top two blots) used radiolabeled ATP and purified JNK1 and ERK2. Coomassie staining (bottom blot) shows input levels of JUN proteins. Like endogenous JUND, recombinant JUND runs as a doublet. There are two copurifying bands with JUNB, but these are not phosphorylated (asterisks). (B) Model of the relative transactivation activities of JUND, c-JUN, and phosphorylated JUND at ETS/AP-1 sequences. (C) JUND or JUND AAAP was overexpressed in DU145 cells and immunoprecipitated with a JUND antibody. Proteins were visualized by immunoblotting, as indicated. Note that the input-JUND blot is overexposed to show the overexpression level relative to the level of expression of endogenous protein. (D) Immunoblots of DU145 cell extracts with overexpression of wild-type (WT) and mutant (AAAP) JUND proteins. JUND phosphorylation was detected by using phospho-c-Jun (Ser 73) antibody, which recognizes phosphoserine 100 of JUND (37). (E, F) A transwell assay measured the migrations of wild-type JUND and JUND AAAP in DU145 cells. Means and SEM of results from three biological replicates relative to values for cells expressing the empty vector are shown. (G) Expression of PTHLH or PLAU mRNA in DU145 cells expressing the indicated construct as determined by RT-qPCR, with values normalized to those for the housekeeping gene EEF1A and relative to values with the vector only. P values were calculated by the t test (*, P < 0.05; **, P < 0.01; ***, P < 0.0001).
DISCUSSION
AP-1 is a critical regulator of many aspects of cellular behavior; however, the myriad of subunits that can constitute this dimeric transcription factor make it difficult to dissect functional mechanisms. We have focused on the role of AP-1 subunits from the JUN family and identified a gene expression program controlling cell migration that is regulated in opposite manners by JUND and c-JUN/JUNB. This program consists of a subset of AP-1-regulated genes that are defined by the presence of neighboring ETS and AP-1 binding sequences in cis-regulatory elements. The RAS/ERK pathway can alter the role of JUN family transcription factors at these ETS/AP-1 sequences by changing JUND from an attenuator of the gene expression program to an activator. Together, these data provide a novel mechanism for how AP-1 transcription factor functions can vary depending on the relative levels of JUN family members, the particular gene expression program being assayed, and the cellular signaling background.
cis-regulatory elements in DNA are collections of transcription factor binding sequences that together mediate the proper regulation of neighboring genes. JUND and c-JUN had opposite functions at a subset of AP-1 target genes controlled by cis-regulatory elements with neighboring AP-1 and ETS binding sequences (Fig. 3A, lower right). In contrast, JUND and c-JUN share an activating function when there is an AP-1, but no ETS, binding sequence (Fig. 3A, lower left). This indicates that the neighboring ETS protein alters the function of AP-1 and provides an interesting example of a combinatorial code where the function of neighboring transcription factors is different from the simple addition of each individual function. Many transcription factors have been shown to activate some targets and repress others in the same cell (38, 39); however, the cis-regulatory sequences that differentiate these functions are largely unknown. Here we show that the ETS/AP-1 sequence motif defines a distinct subset of AP-1 targets where c-JUN and JUND function antagonistically.
Our data also suggest that the ability of ERK to phosphorylate JUND and increase transactivation occurs in the context of ETS/AP-1 targets. ERK can phosphorylate, in addition to JUND, a number of ETS proteins, such as ETS1 and ETS2 (14). ERK phosphorylation increases the affinity of ETS1 and ETS2 for the coactivator, CBP/p300, resulting in increased transactivation (16, 40). Interestingly, an isolated ETS binding sequence does not confer ERK responsiveness to a neighboring gene. RAS-responsive elements consist of either two adjoining ETS binding sequences, such as those found in the MMP3 promoter (41), or neighboring ETS and AP-1 binding sequences, such as those found in the MMP9 promoter and PLAU enhancer (10, 11). It is interesting to speculate that a cooperative interaction with two phosphorylated transcription factors (either ETS/ETS or ETS/JUND) is required to efficiently recruit a coactivator, such as CBP/p300, to an enhancer in a RAS-responsive manner. Although RAS/ERK signaling can increase the transactivation function of JUND (37, 42), JUND is known to negatively regulate RAS-mediated cellular transformation by downregulating cell growth (43). Our findings provide an explanation for this discrepancy by showing that JUND controls multiple gene expression programs, and the ETS/AP-1 program that JUND activates in response to RAS/ERK signaling is associated with cell migration and not cell proliferation.
We also identify an ETS/JUN combination that allows activation of cell migration gene expression in the absence of RAS/ERK signaling. In cells lacking RAS/ERK activity, c-JUN and oncogenic ETS factors (ERG, ETV1, ETV4, and ETV5) can cooperate to activate cell migration genes through ETS/AP-1 sequences. This finding is consistent with several recent reports that indicate an important role for c-JUN in both ERG- and ETV1-positive prostate cancer (44, 45). It is interesting that the E3 ubiquitin ligase COP1 targets ETV1, ETV4, ETV5, and c-JUN (46–48). Loss of COP1 would allow both oncogenic ETS and c-JUN proteins to be stabilized and synergistically activate ETS/AP-1 target genes. The fact that the COP1 gene is deleted or downregulated in various types of cancer, including prostate cancer (46, 47), provides a mechanism for upregulation of both oncogenic ETS proteins and c-JUN. We have recently shown that the ability of oncogenic ETS proteins, such as ERG, to activate through ETS/AP-1 sequences requires the phosphoinositol 3-kinase (PI3K)/AKT pathway (24). A recent report indicates that PI3K/AKT activation and overexpression of ERG in megakaryocytes increases dependence on c-JUN (49), indicating that synergism between these factors may extend to other cell types. The JNK signaling pathway is also known to be a major regulator of c-JUN function (50), and JNK signaling can cooperate with PI3K signaling during prostate cancer progression (51). Interesting foci of future work will be the mechanism that allows c-JUN to work with oncogenic ETS proteins and the contributions of both JNK and PI3K/AKT signaling to this function.
Taken together, these findings suggest that the ETS/AP-1 sequence element defines a cell migration gene expression program that is regulated by ETS and AP-1 transcription factor competition and mitogenic signaling pathways.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a research scholar award to P.C.H. (RSG-13-215-01-DMC) from the American Cancer Society. This work was also supported by funds from the Walther Cancer Foundation and the Showalter Trust.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00982-14.
REFERENCES
- 1.Messina DN, Glasscock J, Gish W, Lovett M. 2004. An ORFeome-based analysis of human transcription factor genes and the construction of a microarray to interrogate their expression. Genome Res 14:2041–2047. doi: 10.1101/gr.2584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eferl R, Wagner EF. 2003. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 3.Hess J, Angel P, Schorpp-Kistner M. 2004. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci 117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 4.Mechta-Grigoriou F, Gerald D, Yaniv M. 2001. The mammalian Jun proteins: redundancy and specificity. Oncogene 20:2378–2389. doi: 10.1038/sj.onc.1204381. [DOI] [PubMed] [Google Scholar]
- 5.Weitzman JB, Fiette L, Matsuo K, Yaniv M. 2000. JunD protects cells from p53-dependent senescence and apoptosis. Mol Cell 6:1109–1119. doi: 10.1016/S1097-2765(00)00109-X. [DOI] [PubMed] [Google Scholar]
- 6.Meixner A, Karreth F, Kenner L, Wagner EF. 2004. JunD regulates lymphocyte proliferation and T helper cell cytokine expression. EMBO J 23:1325–1335. doi: 10.1038/sj.emboj.7600133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Passegue E, Jochum W, Schorpp-Kistner M, Mohle-Steinlein U, Wagner EF. 2001. Chronic myeloid leukemia with increased granulocyte progenitors in mice lacking junB expression in the myeloid lineage. Cell 104:21–32. doi: 10.1016/S0092-8674(01)00188-X. [DOI] [PubMed] [Google Scholar]
- 8.Chinenov Y, Kerppola TK. 2001. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi Y, Satake M, Ito Y. 1989. Two overlapping sequence motifs within the polyomavirus enhancer are independently the targets of stimulation by both the tumor promoter 12-O-tetradecanoylphorbol-13-acetate and the Ha-ras oncogene. J Virol 63:1040–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nerlov C, Rorth P, Blasi F, Johnsen M. 1991. Essential AP-1 and PEA3 binding elements in the human urokinase enhancer display cell type-specific activity. Oncogene 6:1583–1592. [PubMed] [Google Scholar]
- 11.Gum R, Lengyel E, Juarez J, Chen JH, Sato H, Seiki M, Boyd D. 1996. Stimulation of 92-kDa gelatinase B promoter activity by ras is mitogen-activated protein kinase kinase 1-independent and requires multiple transcription factor binding sites including closely spaced PEA3/ets and AP-1 sequences. J Biol Chem 271:10672–10680. doi: 10.1074/jbc.271.18.10672. [DOI] [PubMed] [Google Scholar]
- 12.Gutman A, Wasylyk B. 1990. The collagenase gene promoter contains a TPA and oncogene-responsive unit encompassing the PEA3 and AP-1 binding sites. EMBO J 9:2241–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wasylyk B, Wasylyk C, Flores P, Begue A, Leprince D, Stehelin D. 1990. The c-ets proto-oncogenes encode transcription factors that cooperate with c-Fos and c-Jun for transcriptional activation. Nature 346:191–193. doi: 10.1038/346191a0. [DOI] [PubMed] [Google Scholar]
- 14.Yang BS, Hauser CA, Henkel G, Colman MS, Van Beveren C, Stacey KJ, Hume DA, Maki RA, Ostrowski MC. 1996. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol Cell Biol 16:538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh S, Shin S, Janknecht R. 2012. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta 1826:1–12. doi: 10.1016/j.bbcan.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foulds CE, Nelson ML, Blaszczak AG, Graves BJ. 2004. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol Cell Biol 24:10954–10964. doi: 10.1128/MCB.24.24.10954-10964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollenhorst PC, Ferris MW, Hull MA, Chae H, Kim S, Graves BJ. 2011. Oncogenic ETS proteins mimic activated RAS/MAPK signaling in prostate cells. Genes Dev 25:2147–2157. doi: 10.1101/gad.17546311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. 2008. Recurrent gene fusions in prostate cancer. Nat Rev Cancer 8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capes-Davis A, Reid YA, Kline MC, Storts DR, Strauss E, Dirks WG, Drexler HG, MacLeod RA, Sykes G, Kohara A, Nakamura Y, Elmore E, Nims RW, Alston-Roberts C, Barallon R, Los GV, Nardone RM, Price PJ, Steuer A, Thomson J, Masters JR, Kerrigan L. 2013. Match criteria for human cell line authentication: where do we draw the line? Int J Cancer 132:2510–2519. doi: 10.1002/ijc.27931. [DOI] [PubMed] [Google Scholar]
- 20.Caffarel MM, Moreno-Bueno G, Cerutti C, Palacios J, Guzman M, Mechta-Grigoriou F, Sanchez C. 2008. JunD is involved in the antiproliferative effect of Delta9-tetrahydrocannabinol on human breast cancer cells. Oncogene 27:5033–5044. doi: 10.1038/onc.2008.145. [DOI] [PubMed] [Google Scholar]
- 21.Gurzov EN, Bakiri L, Alfaro JM, Wagner EF, Izquierdo M. 2008. Targeting c-Jun and JunB proteins as potential anticancer cell therapy. Oncogene 27:641–652. doi: 10.1038/sj.onc.1210690. [DOI] [PubMed] [Google Scholar]
- 22.Delestre L, Berthon C, Quesnel B, Figeac M, Kerckaert JP, Galiegue-Zouitina S, Shelley CS. 2011. Repression of the RHOH gene by JunD. Biochem J 437:75–88. doi: 10.1042/BJ20100829. [DOI] [PubMed] [Google Scholar]
- 23.Geback T, Schulz MM, Koumoutsakos P, Detmar M. 2009. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques 46:265–274. [DOI] [PubMed] [Google Scholar]
- 24.Selvaraj N, Budka JA, Jerde TJ, Ferris MW, Hollenhorst PC. 2014. Prostate cancer ETS rearrangements switch a cell migration gene expression program from RAS/ERK to PI3K/AKT regulation. Mol Cancer 13:61. doi: 10.1186/1476-4598-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. 2007. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev 21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas-Chollier M, Darbo E, Herrmann C, Defrance M, Thieffry D, van Helden J. 2012. A complete workflow for the analysis of full-size ChIP-seq (and similar) data sets using peak-motifs. Nat Protoc 7:1551–1568. doi: 10.1038/nprot.2012.088. [DOI] [PubMed] [Google Scholar]
- 28.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. 2007. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res 35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bassuk AG, Leiden JM. 1995. A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity 3:223–237. doi: 10.1016/1074-7613(95)90092-6. [DOI] [PubMed] [Google Scholar]
- 30.The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page-McCaw A, Ewald AJ, Werb Z. 2007. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol 8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beenken A, Mohammadi M. 2009. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov 8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foley J, Nickerson N, Riese DJ II, Hollenhorst PC, Lorch G, Foley AM. 2012. At the crossroads: EGFR and PTHrP signaling in cancer-mediated diseases of bone. Odontology 100:109–129. doi: 10.1007/s10266-012-0070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ. 2009. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet 5:e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hollenhorst PC, Paul L, Ferris MW, Graves BJ. 2011. The ETS gene ETV4 is required for anchorage-independent growth and a cell proliferation gene expression program in PC3 prostate cells. Genes Cancer 1:1044–1052. doi: 10.1177/1947601910395578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bello D, Webber MM, Kleinman HK, Wartinger DD, Rhim JS. 1997. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis 18:1215–1223. doi: 10.1093/carcin/18.6.1215. [DOI] [PubMed] [Google Scholar]
- 37.Vinciguerra M, Vivacqua A, Fasanella G, Gallo A, Cuozzo C, Morano A, Maggiolini M, Musti AM. 2004. Differential phosphorylation of c-Jun and JunD in response to the epidermal growth factor is determined by the structure of MAPK targeting sequences. J Biol Chem 279:9634–9641. doi: 10.1074/jbc.M308721200. [DOI] [PubMed] [Google Scholar]
- 38.Ma J. 2005. Crossing the line between activation and repression. Trends Genet 21:54–59. doi: 10.1016/j.tig.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson ML, Kang HS, Lee GM, Blaszczak AG, Lau DK, McIntosh LP, Graves BJ. 2010. Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to coactivator CBP. Proc Nat Acad Sci U S A 107:10026–10031. doi: 10.1073/pnas.0915137107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jayaraman G, Srinivas R, Duggan C, Ferreira E, Swaminathan S, Somasundaram K, Williams J, Hauser C, Kurkinen M, Dhar R, Weitzman S, Buttice G, Thimmapaya B. 1999. p300/cAMP-responsive element-binding protein interactions with ets-1 and ets-2 in the transcriptional activation of the human stromelysin promoter. J Biol Chem 274:17342–17352. doi: 10.1074/jbc.274.24.17342. [DOI] [PubMed] [Google Scholar]
- 42.Gallo A, Cuozzo C, Esposito I, Maggiolini M, Bonofiglio D, Vivacqua A, Garramone M, Weiss C, Bohmann D, Musti AM. 2002. Menin uncouples Elk-1, JunD and c-Jun phosphorylation from MAP kinase activation. Oncogene 21:6434–6445. doi: 10.1038/sj.onc.1205822. [DOI] [PubMed] [Google Scholar]
- 43.Pfarr CM, Mechta F, Spyrou G, Lallemand D, Carillo S, Yaniv M. 1994. Mouse JunD negatively regulates fibroblast growth and antagonizes transformation by ras. Cell 76:747–760. doi: 10.1016/0092-8674(94)90513-4. [DOI] [PubMed] [Google Scholar]
- 44.Cai C, Hsieh CL, Shemshedini L. 2007. c-Jun has multiple enhancing activities in the novel cross talk between the androgen receptor and Ets variant gene 1 in prostate cancer. Mol Cancer Res 5:725–735. doi: 10.1158/1541-7786.MCR-06-0430. [DOI] [PubMed] [Google Scholar]
- 45.Zong Y, Xin L, Goldstein AS, Lawson DA, Teitell MA, Witte ON. 2009. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Nat Acad Sci U S A 106:12465–12470. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vitari AC, Leong KG, Newton K, Yee C, O'Rourke K, Liu J, Phu L, Vij R, Ferrando R, Couto SS, Mohan S, Pandita A, Hongo JA, Arnott D, Wertz IE, Gao WQ, French DM, Dixit VM. 2011. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature 474:403–406. doi: 10.1038/nature10005. [DOI] [PubMed] [Google Scholar]
- 47.Migliorini D, Bogaerts S, Defever D, Vyas R, Denecker G, Radaelli E, Zwolinska A, Depaepe V, Hochepied T, Skarnes WC, Marine JC. 2011. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J Clin Invest 121:1329–1343. doi: 10.1172/JCI45784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao J, Teng Y, Padia R, Hong S, Noh H, Xie X, Mumm JS, Dong Z, Ding HF, Cowell J, Kim J, Han J, Huang S. 2013. COP1 and GSK3beta cooperate to promote c-Jun degradation and inhibit breast cancer cell tumorigenesis. Neoplasia 15:1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stankiewicz MJ, Crispino JD. 2013. AKT collaborates with ERG and Gata1s to dysregulate megakaryopoiesis and promote AMKL. Leukemia 27:1339–1347. doi: 10.1038/leu.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 51.Hubner A, Mulholland DJ, Standen CL, Karasarides M, Cavanagh-Kyros J, Barrett T, Chi H, Greiner DL, Tournier C, Sawyers CL, Flavell RA, Wu H, Davis RJ. 2012. JNK and PTEN cooperatively control the development of invasive adenocarcinoma of the prostate. Proc Nat Acad Sci U S A 109:12046–12051. doi: 10.1073/pnas.1209660109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 53.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, Ami GOH, Web Presence Working Group. 2009. AmiGO: online access to ontology and annotation data. Bioinformatics 25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.