Abstract
Signaling lymphocytic activation molecule F7 (SLAMF7) is a receptor present on immune cells, including natural killer (NK) cells. It is also expressed on multiple myeloma (MM) cells. This led to development of an anti-SLAMF7 antibody, elotuzumab, showing efficacy against MM. SLAMF7 mediates activating or inhibitory effects in NK cells, depending on whether cells express or do not express the adaptor EAT-2. Since MM cells lack EAT-2, we elucidated the inhibitory effectors of SLAMF7 in EAT-2-negative NK cells and tested whether these effectors were triggered in MM cells. SLAMF7-mediated inhibition in NK cells lacking EAT-2 was mediated by SH2 domain-containing inositol phosphatase 1 (SHIP-1), which was recruited via tyrosine 261 of SLAMF7. Coupling of SLAMF7 to SHIP-1 required Src kinases, which phosphorylated SLAMF7. Although MM cells lack EAT-2, elotuzumab did not induce inhibitory signals in these cells. This was at least partly due to a lack of CD45, a phosphatase required for Src kinase activation. A defect in SLAMF7 function was also observed in CD45-deficient NK cells. Hence, SLAMF7-triggered inhibition is mediated by a mechanism involving Src kinases, CD45, and SHIP-1 that is defective in MM cells. This defect might explain why elotuzumab eliminates MM cells by an indirect mechanism involving the activation of NK cells.
INTRODUCTION
Signaling lymphocytic activation molecule (SLAM) family receptors are hematopoietic-cell-specific receptors playing critical roles in normal immune regulation (1–4). They have also been firmly implicated in many human diseases, including immune deficiencies, autoimmunity, and hematological malignancies. SLAM family receptors can mediate either activating or inhibitory effects in immune cells, depending in part on whether they are coexpressed with members of the SLAM-associated protein (SAP) family of Src homology 2 (SH2) domain-only adaptors. Typically, SLAM family receptors activate in the presence of SAP family adaptors but are inhibitory in the absence of SAP family adaptors. Whereas much is known of the molecular mechanisms by which SLAM family receptors mediate activating effects, little is known about how they mediate inhibitory effects.
SLAMF7 (also named CS1 [CD2 subset 1], CRACC [CD2-like receptor-activating cytotoxic cell], and CD319) is a member of the SLAM family (1–4). The other members of the family are SLAM, 2B4, NK-T-B antigen (NTB-A)/Ly108, Ly-9, and CD84. Like most SLAM receptors, SLAMF7 is a self-ligand; i.e., it recognizes as ligand another SLAMF7 molecule on another cell. The only exception is 2B4, which recognizes CD48. SLAMF7 is found on natural killer (NK) cells, activated T cells, most B cells, including antibody-producing plasma cells, and myeloid cells (2, 5). It is also abundantly present in most cases of multiple myeloma (MM), a nearly universally fatal malignancy of plasma cells (either freshly isolated cells or cell lines) (3, 4).
In NK cells, SLAMF7 is usually a positive regulator of NK cell activation (5, 6). This activity requires expression of the SAP family adaptor Ewing's sarcoma-associated transcript 2 (EAT-2). SLAMF7 binds EAT-2 via phosphorylated tyrosine 281 (Y281) in its cytoplasmic segment, thereby triggering activating signals involving phospholipase C-γ (PLC-γ) (7). In the absence of EAT-2, SLAMF7 mediates inhibitory effects; these effects were documented in NK cells from EAT-2-deficient mice and normal activated T cells, which lack EAT-2 (5). However, the molecular basis of this inhibition is undetermined. Depending on the SLAM family receptor studied, it was suggested that inhibition might be mediated by SH2 domain-containing protein tyrosine phosphatase 1 (SHP-1), SHP-2, or SH2 domain-containing inositol phosphatase 1 (SHIP-1). However, firm genetic evidence in support of this idea has not been reported. Moreover, how any of the SLAM family receptors couples to its inhibitory effectors has not been addressed.
The nearly universal expression of SLAMF7 in MM led to development of a humanized anti-SLAMF7 monoclonal antibody (MAb), elotuzumab (3, 4). Preclinical studies using transplanted human MM cells in mice showed that elotuzumab caused MM cell elimination in vivo (8). The efficacy of elotuzumab in combination with lenalidomide was subsequently demonstrated in phase 1 and 2 trials of patients with refractory and relapsed MM (9–12). Phase 3 studies are ongoing. Surprisingly, elotuzumab had little or no direct inhibitory effects on MM cells in vitro, implying that its efficacy occurred through an indirect mechanism. Accordingly, maximal activity of elotuzumab in mice was NK cell dependent (8). Furthermore, elotuzumab directly activated NK cells through antibody-dependent cellular cytotoxicity (ADCC) via CD16 and triggering of SLAMF7 on NK cells (3, 4, 8, 13).
As MM cells lack EAT-2 (1, 13), we wanted to ascertain the molecular mechanism by which SLAMF7 mediates inhibition in NK cells and evaluate whether this mechanism is functional in MM cells. Our studies showed that the inhibitory function of SLAMF7 in EAT-2-negative NK cells was mediated by a molecular mechanism implicating lipid phosphatase SHIP-1, Src kinases, and protein tyrosine phosphatase CD45. Interestingly, coupling of SLAMF7 to SHIP-1 was severely compromised in MM cells. This defect correlated with a lack of CD45, which is required for activation of Src family kinases in hematopoietic cells and was needed for initiation of SLAMF7 inhibitory signaling.
MATERIALS AND METHODS
Mice.
C57BL/6J, 129S1/Sv, and CD45-deficient (B6.129-Ptprctm1Holm/J) mice were from The Jackson Laboratory (Bar Harbor, ME). EAT-2-deficient mice (in 129/Sv) were described previously (14). In all experiments, normal littermates were used as a control. The IRCM Animal Care Committee approved all experimentation in accordance with the Canadian Council for Animal Care.
Cells.
YT-S, Cos-1, HeLa, B16, and DT40 cells were described previously (5, 15, 16). B16 cells expressing mouse SLAMF7 (mSLAMF7) or mCD48 were described previously (5). IM-9, U266, and NCI-H929 cells were from Jerry Pelletier, McGill University (Montreal, Quebec, Canada). MM1S cells were from the American Type Culture Collection (Manassas, VA). OPM2 and KMS-12-PE cells were from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). Isolation of NK cells from mice primed with poly(I·C) and generation of interleukin-2 (IL-2)-expanded mouse NK cells were described previously (5, 17). YT-S cells expressing wild-type (WT) or mutant mSLAMF7, human SLAMF7 (hSLAMF7), or hEAT-2 were reported previously (5, 7). HeLa or DT40 cells expressing green fluorescent protein (GFP) alone or in combination with SLAMF7 were produced by retroviral infection, as detailed in reference 5. SHIP-1-deficient DT40 cells were reconstituted with mSHIP-1, as outlined previously (15). For expression of epidermal growth factor receptor (EGFR)-CD45, OPM2 cells and MM1S cells were electroporated with pAW-EGFR-CD45 (18) (provided by Arthur Weiss, University of California, San Francisco, San Francisco, CA) and selected in G418-containing medium. Cells expressing EGFR-CD45 were sorted using anti-EGFR antibody. Cos-1 transfections were performed as detailed previously (19).
Antibodies.
Antibodies recognizing m2B4 (eBio244F4), mCD48 (HM48-1), mNKG2D (CX5), mDNAM-1 (10E5), mCD45 (30F11), h2B4 (C1.7), hNTB-A (NT-7), hNKp46 (9E2), hCD45 (2D1 and HI30), hCD84 (CD84.1.21), hLy-9 (HLy9.1.25), or hCD148 were from eBioscience (San Diego, CA) or BioLegend (San Diego, CA). Anti-hEGFR antibody was from BD Biosciences (Mississauga, Ontario, Canada). Mouse anti-chicken IgM (M-1) antibody was from Southern Biotech (Birmingham, AL). Polyclonal rabbit anti-rat IgG, rabbit anti-mIgG, and rabbit anti-hIgG antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). Antibodies recognizing mSLAMF7 (4G2), mCD84 (1D3), mLy-9 (Ly9.2), mLy108 (3E11), mSLAM (12F12), phosphotyrosine (4G10), hEAT-2 (10F7), mEAT-2 (8F12), and SAP (12C4) were produced as reported previously (5, 7, 17). Hybridomas producing anti-hSLAMF7 (162) and anti-hDNAM-1 (11A8) were provided by Marco Colonna, Washington University (St. Louis, MO) (6, 20). Elotuzumab (HuLuc63) was provided by Bristol-Myers Squibb (Lawrenceville, NJ) (BMS) (3). Polyclonal rabbit antibodies recognizing SHIP-1, Shc, Vav-1, Fyn, Lyn, Src, Lck, Syk, ZAP-70, and c-Cbl were produced in our laboratory. Flow cytometry was performed using a BD Biosciences FACSCalibur flow cytometer and CellQuest software (BD Biosciences). Immunoprecipitations and immunoblotting were performed as detailed elsewhere (21).
Antibody-mediated stimulation.
For immunoblot analyses, 5 × 106 cells were incubated with 6 μg anti-SLAMF7 antibody for 5 min at room temperature. After being washed, cells were stimulated with 10 μg of the relevant secondary antibody for the periods of time indicated in the figures at 37°C. After lysis, SLAMF7 and SHIP-1 were immunoprecipitated. Unstimulated cells were processed similarly, except that the primary antibody was added after cell lysis. For inhibition of Src kinases, cells were incubated with PP2 (dissolved in dimethyl sulfoxide [DMSO] and purchased from Calbiochem, Darmstadt, Germany) for 1 h at 37°C and then stimulated as outlined above. To study Ca2+ fluxes, cells were loaded with Indo-1 (Invitrogen, Burlington, Ontario, Canada) for 20 min at 37°C. After being washed, 2 × 106 cells were incubated with 2 μg of anti-BCR antibody, with or without 2 μg anti-SLAMF7 antibody, for 5 min at room temperature and then loaded on a BD LSR cell analyzer. After the relevant secondary antibody was added, changes in intracellular Ca2+ were monitored over time at 37°C using the Fluo-4 (FL4) to Fluo-5 (FL5) ratio. Control cells were stimulated with only the secondary antibody.
NK cell cytotoxicity.
NK cell-mediated cytotoxicity was assayed as detailed elsewhere (17). Duplicates were used in each experiment.
RNA interference.
Scrambled small interfering RNA (siRNA) (1027280) and SHIP-1-specific siRNAs (SI00078575 and SI03029005) were from Qiagen (Valencia, CA). Transfection of YT-S cells was performed using Amaxa cell line Nucleofector kit R (VCA-1001) from Lonza (Basel, Switzerland) according to the manufacturer's protocol and the program O-017. Transfected cells were grown for 36 to 48 h and processed for cytotoxicity or immunoblot assays.
Reverse transcription (RT)-PCR.
RNA was purified with TRIzol (Invitrogen) according to the manufacturer's protocol and reverse transcribed into cDNA using random primers and Superscript II (Invitrogen). PCRs were performed using single-stranded cDNAs, gene-specific primers, and Taq polymerase (Invitrogen). The primers to distinguish the human SLAMF7 isoforms were CS1 F727 (5′-TCTCTTTGTACTGGGGCTATTTC-3′) and CS1 R955 (5′-TTTTCCATCTTTTTCGGTATTT-3′), as described previously (22). The primers to detect human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were 5′-AGGTCGGAGTCAACGGATTTG-3′ and 5′-GTGATGGCATGGACTGTGGT-3′.
Statistical analysis and quantitation.
Unpaired Student's t tests (two-tailed) were performed using the Prism software program. Bands in autoradiograms were quantified with the Image J software program.
RESULTS
SLAMF7-mediated inhibition in NK cells is accompanied by tyrosine phosphorylation of SHIP-1.
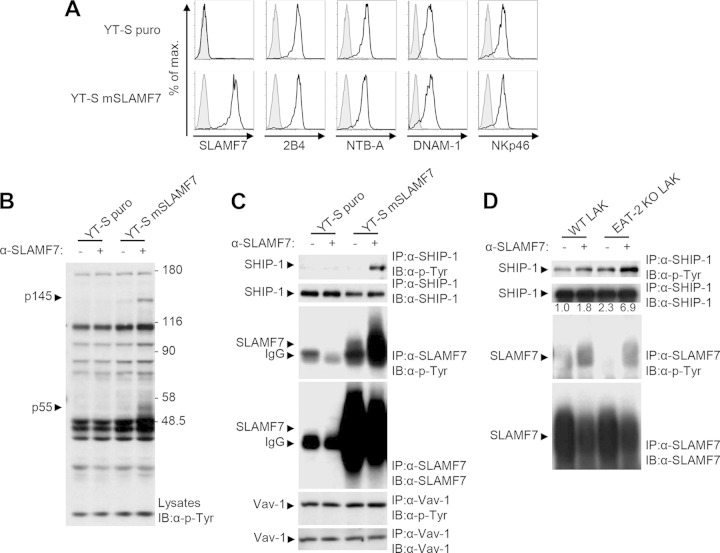
It was proposed that inhibition by SLAM family receptors might be mediated by various effectors, including SHP-1, SHP-2, SHIP-1, and Csk (23, 24). To identify the effectors of SLAMF7-mediated inhibition, we used the human NK cell line YT-S ectopically expressing or not expressing wild-type (WT) mSLAMF7 (Fig. 1A). As YT-S lacks EAT-2, SLAMF7 is inhibitory in these cells (5). Since SLAM family signaling is initiated by protein tyrosine phosphorylation (5, 15, 17, 23, 25, 26), we focused on tyrosine phosphorylation signals (Fig. 1B). Engagement of SLAMF7 by anti-SLAMF7 antibodies resulted in tyrosine phosphorylation of two polypeptides of 55 and 145 kDa in total cell lysates. These effects were not seen in YT-S cells lacking SLAMF7.
FIG 1.
SHIP-1 is tyrosine phosphorylated in response to the engagement of SLAMF7. (A to C) YT-S cells were transfected with a vector encoding the puromycin resistance marker (puro) alone or in combination with mouse SLAMF7 (mSLAMF7). Polyclonal populations were used for analyses. (A) Expression of SLAMF7, 2B4, NTB-A, DNAM-1, and NKp46 (black lines with no shading) was analyzed by flow cytometry. Isotype controls are shown as gray lines with shading. Results are representative of 3 experiments. (B) Cells were stimulated or not stimulated for 3 min with the anti-SLAMF7 MAb 4G2 and rabbit anti-rat IgG. After lysis, overall protein tyrosine phosphorylation was determined by immunoblotting (IB) of total cell lysates with antiphosphotyrosine (α-p-Tyr) antibodies. The positions of prestained molecular mass markers (in kilodaltons) are shown on the right. (C) Tyrosine phosphorylation of SHIP-1, SLAMF7, and Vav-1 was determined by immunoprecipitating (IP) the indicated proteins with specific antibodies and subsequent immunoblotting with antiphosphotyrosine antibodies. The abundance of SHIP-1, SLAMF7, and Vav-1 in the immunoprecipitates was verified by probing parallel immunoprecipitates or reprobing the immunoblot membrane with substrate-specific antibodies. It is noteworthy that the amount of SLAMF7 recovered from cells stimulated with anti-SLAMF7 was frequently lower than that obtained from unstimulated cells. This was due to the fact that a proportion of SLAMF7 becomes detergent insoluble upon SLAMF7 stimulation with antibodies. The position of the heavy chain of IgG, which is recognized by the immunoblotting antibodies, is shown by an arrowhead on the left. Results are representative of 3 experiments. (D) NK cells from wild-type (WT) or EAT-2-deficient (knockout; KO) mice were stimulated for 3 min with anti-SLAMF7, and tyrosine phosphorylation of SHIP-1 and SLAMF7 was analyzed as detailed for panels A to C. A quantitation of the relative tyrosine phosphorylation of SHIP-1 is shown. Results are representative of 2 experiments.
Based on their apparent masses, p55 and p145 were consistent with SLAMF7 and SHIP-1, respectively. To test this idea, immunoprecipitation experiments were conducted (Fig. 1C). Although SHIP-1 was not detectably tyrosine phosphorylated in unstimulated YT-S cells, it became tyrosine phosphorylated following engagement of SLAMF7. Likewise, although some tyrosine phosphorylation of SLAMF7 was detected in unstimulated YT-S cells, this phosphorylation was augmented in stimulated cells. Vav-1, a positive regulator of NK cell activation (15), did not undergo tyrosine phosphorylation upon SLAMF7 triggering. SLAMF7 migrated as a broad band or multiple bands in protein gels, presumably reflecting differential glycosylation.
Similar experiments were performed using NK cells from WT or EAT-2-deficient mice (Fig. 1D). While stimulation of SLAMF7 on WT NK cells increased SHIP-1 tyrosine phosphorylation, this increase was more prominent in EAT-2-deficient cells. In contrast, levels of tyrosine phosphorylation of SLAMF7 were equivalent, in keeping with the report that EAT-2 is not required for SLAMF7 tyrosine phosphorylation (5).
Thus, an increase in SLAMF7-mediated inhibition was consistently paralleled by an enhancement of SLAMF7-triggered tyrosine phosphorylation of SHIP-1.
SHIP-1 is critical for SLAMF7-mediated inhibition.
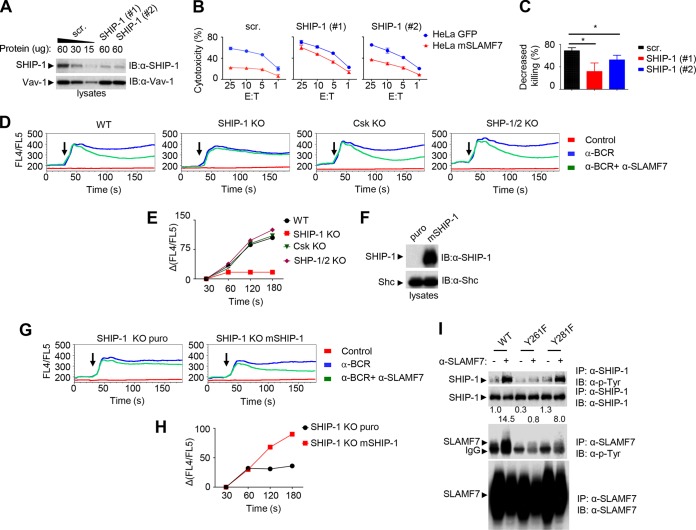
To examine whether SHIP-1 was critical for inhibition, the impact of downregulating SHIP-1 expression in YT-S cells was tested (Fig. 2A to C). YT-S cells expressing mSLAMF7 were independently transfected with two small interfering RNAs (siRNAs) against SHIP-1 or with irrelevant siRNAs. SHIP-1-specific siRNAs resulted in an ∼70% reduction of SHIP-1 levels (Fig. 2A). Cells were then tested for their ability to kill HeLa targets expressing or not expressing the ligand of SLAMF7, i.e., SLAMF7 itself. SHIP-1-specific siRNAs resulted in an increased ability to kill HeLa cells (Fig. 2B). In keeping with the involvement of SHIP-1 in multiple inhibitory pathways (15, 27, 28), this effect was observed whether HeLa cells expressed SLAMF7 or not. Nonetheless, calculation of the extent of inhibition conferred by expression of SLAMF7 on targets showed that SLAMF7-mediated inhibition was attenuated by loss of SHIP-1 (Fig. 2C). This finding represented the first direct genetic evidence for a specific effector of SLAM family receptor-mediated inhibition in an immune cell function.
FIG 2.
SHIP-1 is required for SLAMF7-mediated inhibition in NK cells and DT40 B cells. (A to C) Expression of SHIP-1 in YT-S cells expressing mSLAMF7 was downregulated using two different SHIP-1-specific small interfering RNAs (siRNAs), #1 and #2. (A) Expression of SHIP-1 was determined by immunoblotting of total cell lysates with anti-SHIP-1 antibodies. Vav-1 was used as loading control. For cells transfected with scrambled siRNAs (scr.), various amounts of lysates were loaded to confirm the linearity of the immunoblot assays. (B) Cytotoxicity to HeLa target cells expressing green fluorescent protein (GFP) alone or in combination with mSLAMF7 was determined using a 51Cr release assay. The effector-to-target ratios (E:T) are shown at the bottom. Average values of duplicates are shown, while error bars depict standard deviations. Results are representative of 3 experiments. (C) The percentage of decreased killing was calculated as [(cytotoxicity to GFP+ targets − cytotoxicity to SLAMF7+ targets)/cytotoxicity to GFP+ targets] × 100 at the 25:1 E/T ratio. Average values from 3 independent experiments with standard deviations are represented. *, P < 0.05. (D and E) Wild-type (WT) DT40 cells or variants lacking the indicated inhibitory molecules were transduced with a cDNA encoding mouse SLAMF7. Cells were loaded with the Ca2+ indicator dye Indo-1 and then stimulated with anti-B cell receptor (α-BCR) antibodies alone or in combination with anti-SLAMF7. (D) Changes in intracellular Ca2+ were analyzed over time by determining the FL4/FL5 ratio using flow cytometry. Arrows indicate the time at which receptor cross-linking was triggered. Stimulation with only the secondary antibody was used as a control. (E) A quantitation of inhibition of Ca2+ levels in cells stimulated with anti-BCR plus anti-SLAMF7, in comparison to anti-BCR alone, at different times is represented graphically. Results are representative of 3 experiments. (F to H) SHIP-1-deficient DT40 cells were transfected with a plasmid encoding the puromycin resistance marker (puro) alone or in combination with mouse SHIP-1. (F) SHIP-1 expression was determined by immunoblotting of total cell lysates with anti-SHIP-1 antibodies. (G and H) Ca2+ fluxes were analyzed as detailed for panels D and E. (I) YT-S cells expressing WT, Y261F SLAMF7, or Y281F SLAMF7 were stimulated with anti-SLAMF7. Tyrosine phosphorylation of SHIP-1 and SLAMF7 was examined as detailed for Fig. 1C. A quantitation of the relative tyrosine phosphorylation of SHIP-1 is shown. Results are representative of 3 experiments.
We also tested the role of SHIP-1 using the DT40 system. DT40 is a chicken B cell line that has variants lacking SHIP-1 or other inhibitory effectors, such as SHP-1, SHP-2, or Csk (29). It lacks EAT-2 (15). Therefore, parental or mutant DT40 cells expressing mSLAMF7 were generated. These cells expressed equivalent amounts of SLAMF7 and B cell antigen receptor (BCR) (data not shown).
As engagement of SLAMF7 resulted in inhibition of BCR-triggered Ca2+ fluxes in DT40 cells (data not shown), we focused our studies on Ca2+ fluxes. In contrast to parental DT40 cells, cells lacking SHIP-1 displayed a near abolition of the ability of SLAMF7 to inhibit BCR-evoked Ca2+ fluxes (Fig. 2D). Cells devoid of Csk or SHP-1 and SHP-2 had no defect. A quantitation of the extent of inhibition is depicted in Fig. 2E. To confirm that this defect was due to a lack of SHIP-1, SHIP-1-deficient cells were reconstituted with SHIP-1 (Fig. 2F; data not shown). Reexpression of SHIP-1 restored SLAMF7-mediated inhibition (Fig. 2G and H).
SLAMF7 contains two conserved intracytoplasmic tyrosines, Y261 and Y281. Y261 is critical for SLAMF7-mediated inhibition when NK cells lack EAT-2, whereas Y281 is needed for SLAMF7-mediated activation when NK cells express EAT-2 (5). Therefore, the roles of Y261 and Y281 in tyrosine phosphorylation of SHIP-1 were also assessed (Fig. 2I). Engagement of SLAMF7 resulted in robust tyrosine phosphorylation of SHIP-1 in cells expressing the WT protein or a protein with a tyrosine 281-to-phenylalanine 281 (Y281F) mutation but not a Y261F mutation (SLAMF7). Mutation of either Y261 or Y281 resulted in reduced SLAMF7 tyrosine phosphorylation, implying that both sites contribute to SLAMF7 phosphorylation. Hence, Y261, but not Y281, was needed for tyrosine phosphorylation of SHIP-1 in response to SLAMF7 engagement. Therefore, SHIP-1, but not SHP-1, SHP-2, or Csk, was responsible for SLAMF7-mediated inhibition.
Src kinases are critical for SLAMF7-induced inhibitory signals.
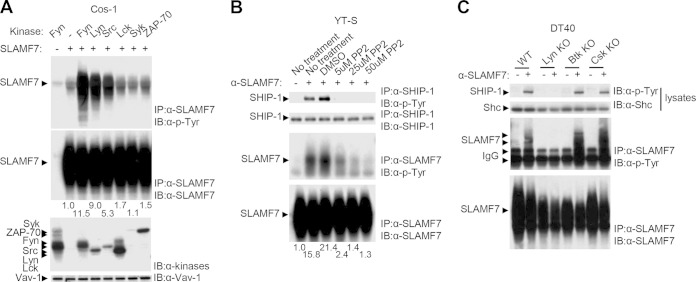
The kinase(s) responsible for initiation of SLAMF7-induced tyrosine phosphorylation was identified. Using transient transfection in Cos-1 cells, we found that the Src family kinases Fyn, Lyn, and Src triggered an increase in SLAMF7 tyrosine phosphorylation (Fig. 3A). No effect was seen with Lck, Syk, and ZAP-70. We also examined the impact of a pharmacological inhibitor of Src kinases, PP2, in YT-S cells (Fig. 3B). PP2 completely eliminated the ability of SLAMF7 to evoke tyrosine phosphorylation of SHIP-1 in these cells. A full effect was seen at 5 μM. PP2 also caused a reduction of SLAMF7 tyrosine phosphorylation. Although only a minimal effect was seen at 5 μM, a nearly complete elimination was seen at 50 μM.
FIG 3.
Src family kinases are responsible for SLAMF7-dependent tyrosine phosphorylation of SHIP-1. (A) Cos-1 cells were transiently transfected with cDNAs encoding the indicated protein tyrosine kinases, without or with SLAMF7. Tyrosine phosphorylation of SLAMF7 was determined by immunoblotting of SLAMF7 immunoprecipitates with antiphosphotyrosine antibodies. A quantitation of the relative tyrosine phosphorylation of SLAMF7 is shown. Results are representative of 3 experiments. (B) YT-S cells expressing mSLAMF7 were pretreated with the indicated concentrations of PP2 (a Src family kinase inhibitor) or with vehicle (dimethyl sulfoxide [DMSO]) alone. Cells were then stimulated with anti-SLAMF7, and tyrosine phosphorylation of SHIP-1 and SLAMF7 was determined as detailed for Fig. 1C. A quantitation of the relative tyrosine phosphorylations of SLAMF7 is shown. Results are representative of 2 experiments. (C) WT or kinase-deficient DT40 cells expressing SLAMF7 were stimulated or not stimulated with anti-SLAMF7 antibody. Tyrosine phosphorylation of SHIP-1 and SLAMF7 was analyzed as detailed for Fig. 1C. Results are representative of 5 experiments.
We also tested the roles of various protein tyrosine kinases (PTKs) in the DT40 cell system by expressing hSLAMF7 in DT40 variants lacking the kinases Lyn (the only Src kinase in DT40 cells), Btk, and Csk. All cells expressed equivalent amounts of hSLAMF7 (data not shown). Stimulation with anti-SLAMF7 MAb revealed that tyrosine phosphorylation of SLAMF7 and SHIP-1 was eliminated in cells lacking Lyn but not in cells lacking Btk or Csk. Thus, Src kinases, in particular Fyn and Lyn, are responsible for SLAMF7-triggered protein tyrosine phosphorylation.
Elotuzumab fails to trigger tyrosine phosphorylation of SHIP-1 in multiple myeloma cells.
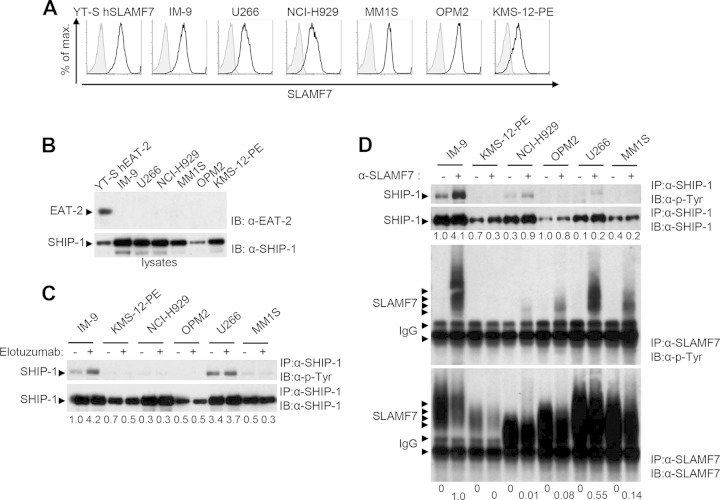
MM cells express high levels of SLAMF7 but lack EAT-2 (1, 3, 4, 8, 13) (Fig. 4A and B). Hence, we tested whether SLAMF7-mediated inhibitory signals identified in EAT-2-negative NK cells also occurred in MM cells. Several human MM cell lines were stimulated with the therapeutic anti-SLAMF7 MAb elotuzumab, and SHIP-1 tyrosine phosphorylation was examined (Fig. 4C and D). IM-9, an Epstein-Barr virus-transformed B cell line also expressing SLAMF7 but lacking EAT-2, was used as a control.
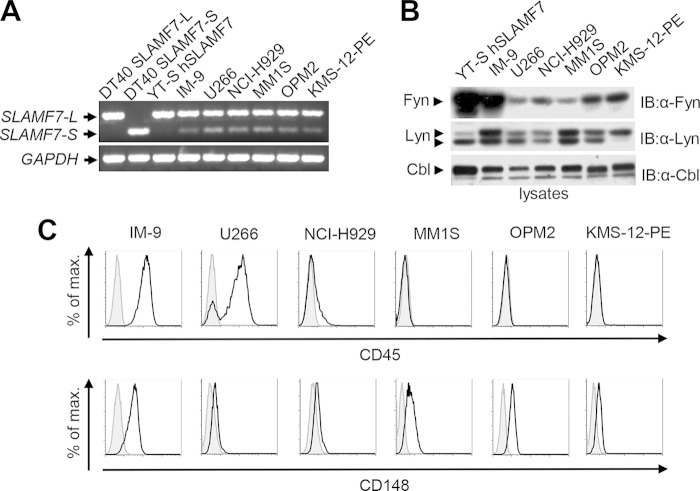
FIG 4.
Engagement of SLAMF7 on multiple myeloma cells fails to induce tyrosine phosphorylation of SHIP-1 and, to a lesser extent, SLAMF7. (A and B) Expression of SLAMF7, EAT-2, and SHIP-1 in several multiple myeloma (MM) cell lines, in addition to YT-S cells ectopically expressing human SLAMF7 (hSLAMF7) or hEAT-2 and IM-9 cells, was examined by flow cytometry (A) or immunoblotting of total cell lysates (B). (A) Staining with anti-SLAMF7 is depicted as black lines with no shading, whereas staining with isotype controls is shown as gray lines with shading. Results are representative of 3 experiments. (C and D) Cells were stimulated with elotuzumab (C) or anti-SLAMF7 MAb 162 (D), followed by the relevant secondary antibody. Tyrosine phosphorylation of SHIP-1 and SLAMF7 was determined as outlined for Fig. 1C. Note that as elotuzumab is a humanized antibody, it was inefficient at immunoprecipitating SLAMF7 with the currently available secondary reagents. As a result, SLAMF7 could be efficiently immunoprecipitated only from cells stimulated with MAb 162. The differences in electrophoretic mobility of SLAMF7 between cell lines were presumably due to differential glycosylation. Quantitations of the relative tyrosine phosphorylation of SHIP-1 (C and D) and SLAMF7 (D) are shown. Results are representative of 3 experiments.
Elotuzumab triggered prominent tyrosine phosphorylation of SHIP-1 in IM-9 cells (Fig. 4C). However, it failed to evoke a similar response in MM cells, although SHIP-1 was constitutively tyrosine phosphorylated in some cell lines (in particular U266). Similar findings were made with anti-SLAMF7 MAb 162 (Fig. 4D). In contrast to the defect in SLAMF7-evoked SHIP-1 tyrosine phosphorylation, the compromise in tyrosine phosphorylation of SLAMF7 was variable (Fig. 4D). The latter was severely reduced in KMS-12-PE, NCI-H929, OPM2, and MM1S cells but not in U266 cells. Hence, unlike NK cells and the B cell line IM-9, MM cells had a defect in SLAMF7-evoked tyrosine phosphorylation of SHIP-1 and, to a lesser extent, SLAMF7.
Multiple myeloma cells express SLAMF7-L and Src kinases but lack the phosphatase CD45.
We tested whether any of the intermediates linking SLAMF7 to SHIP-1 in NK cells were defective in MM cells (Fig. 5). Human SLAMF7 exists as two isoforms produced by alternative splicing: the long isoform (SLAMF7-L), which contains all tyrosines, including Y261, and the short isoform (SLAMF7-S), which bears an alternative intracytoplasmic domain lacking Y261 (22, 30, 31). In DT40 cells, SLAMF7-L, but not SLAMF7-S, was inhibitory (data not shown). Therefore, we ascertained whether MM cells expressed the long isoform of SLAMF7. RT-PCR analyses showed that, like YT-S and IM-9 cells, all MM cells expressed predominantly SLAMF7-L (Fig. 5A). We also verified expression of Fyn and Lyn, which were most effective at triggering SLAMF7 tyrosine phosphorylation in Cos-1 cells (Fig. 3A). Immunoblot analyses showed that all MM cells contained Fyn and Lyn (Fig. 5B). Of note, however, levels of Fyn tended to be lower in MM cells than in IM-9 cells.
FIG 5.
Multiple myeloma cells express the long isoform of SLAMF7 and Src family kinases but not CD45. (A) Cells were examined for expression of the long (SLAMF7-L) and short (SLAMF7-S) isoforms of SLAMF7 and GAPDH using RT-PCR, as detailed in Materials and Methods. DT40 transfectants expressing either SLAMF7-L or SLAMF7-S and YT-S transfectants expressing SLAMF7-L were used as controls. Results are representative of 3 experiments. (B) Expression of the Src family kinases Fyn and Lyn was analyzed in cells shown in panel A. It should be noted that Lyn exists as two isoforms, p53 and p56, which are generated by alternative splicing. Results are representative of 3 experiments. (C) Expression of CD45 and CD148 was examined on cells shown in panel A. Staining with anti-CD45 or anti-CD148 is depicted as black lines with no shading, whereas staining with isotype controls is shown as gray lines with gray shading. Results are representative of 3 experiments.
These data suggested that the activity, rather than the expression, of Src kinases might be defective in MM cells. In B cells, activation of Src kinases requires two transmembrane protein tyrosine phosphatases (PTPs), CD45 and CD148 (32–34). These PTPs dephosphorylate the inhibitory tyrosine of Src kinases. Whereas IM-9 cells expressed both CD45 and, to a lesser extent, CD148, MM cells expressed CD148 but not CD45 (Fig. 5C). The only exception was U266 cells, which contained both CD45-positive and CD45-negative populations. It should also be noted that, whereas MM cells expressed CD148, levels were lower than in IM-9 cells. Therefore, the defect in SLAMF7-triggered tyrosine phosphorylation of SHIP-1 and, to a lesser extent, SLAMF7 in MM cells correlated at least in part with a lack of CD45.
CD45 is required for SLAMF7-triggered tyrosine phosphorylation of SHIP-1.
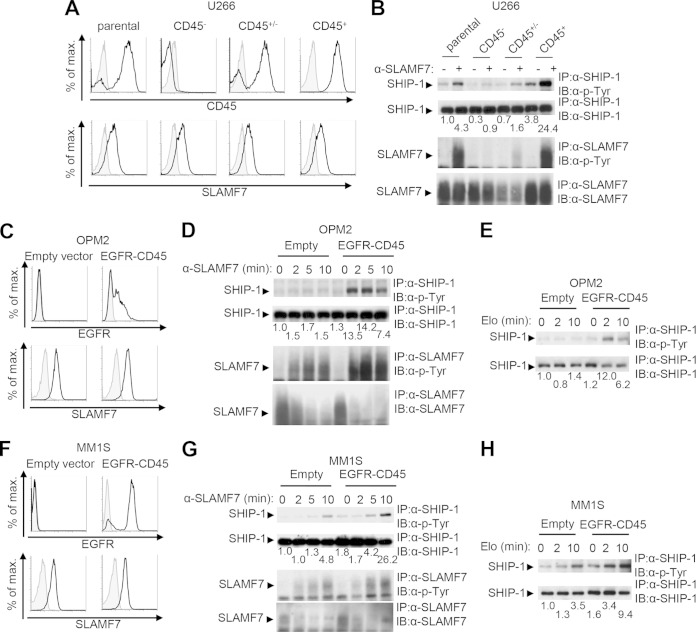
To assess whether a lack of CD45 was responsible for defective SLAMF7-evoked protein tyrosine phosphorylation in MM cells, two approaches were taken (Fig. 6). First, limiting dilution was used to generate CD45-positive and CD45-negative variants of U266 cells, which contain a mixture of the two populations (Fig. 6A). Stimulation of a CD45-negative variant of U266 cells with anti-SLAMF7 resulted in minimal tyrosine phosphorylation of SHIP-1 and SLAMF7 (Fig. 6B). In contrast, triggering of a CD45-positive variant resulted in prominent tyrosine phosphorylation of SHIP-1 and SLAMF7. Intermediate responses were observed in a variant containing CD45-positive and CD45-negative cells. This variant also expressed smaller amounts of SLAMF7.
FIG 6.
CD45 is critical for SHIP-1 tyrosine phosphorylation in multiple myeloma cells. (A and B) Subclones of U266 cells were tested for CD45 and SLAMF7 expression. (A) Staining with anti-CD45 or anti-SLAMF7 is depicted as black lines with no shading, whereas staining with isotype controls is shown as gray lines with shading. (B) Cells were then stimulated with anti-SLAMF7 MAb 162, and tyrosine phosphorylation of SHIP-1 and SLAMF7 was tested as detailed for Fig. 1C. A quantitation of the relative tyrosine phosphorylation of SHIP-1 is shown. Results are representative of 2 experiments. (C to H) OPM2 cells (C to E) or MM1S cells (F to H) were stably transfected with a vector encoding the epidermal growth factor receptor (EGFR)-CD45 chimera or an empty vector. Expression of surface EGFR and SLAMF7 was detected by flow cytometry. (C and F) Staining with anti-EGFR or anti-SLAMF7 is depicted as black lines with no shading, whereas staining with isotype controls is shown as gray lines with shading. (D, E, G, H) Cells were then stimulated for the indicated times with anti-SLAMF7 MAb 162 (D and G) or elotuzumab (Elo) (E and H), and tyrosine phosphorylation of SHIP-1 and SLAMF7 was tested as detailed for Fig. 1C. It is noteworthy that the amount of SLAMF7 recovered from cells stimulated with anti-SLAMF7 was lower than that obtained from unstimulated cells. This was due to the fact that a proportion of SLAMF7 becomes detergent insoluble upon SLAMF7 stimulation with antibodies. Quantitations of the relative tyrosine phosphorylation of SHIP-1 are shown. Results are representative of 4 experiments.
Second, expression of CD45 was reconstituted in the CD45-negative cell lines OPM2 and MM1S (Fig. 6C to H). As full-length CD45 is notoriously difficult to express in transfected cells, we used a chimeric CD45 in which the extracellular and transmembrane domains of the epidermal growth factor receptor (EGFR) are fused to the CD45 cytoplasmic region, which contains the phosphatase domain (18). After transfection with a cDNA encoding EGFR-CD45, ∼50% of OPM2 and ∼80% of MM1S cells expressed the chimeric receptor (Fig. 6C and F). Expression of EGFR-CD45 in OPM2 and MM1S cells resulted in a marked increase in SHIP-1 tyrosine phosphorylation in response to SLAMF7 stimulation by MAb 162 or elotuzumab (Fig. 6D, E, G, and H). A smaller effect was seen on the tyrosine phosphorylation of SLAMF7. Thus, the lack of CD45 was responsible at least in part for the defect in SHIP-1 tyrosine phosphorylation upon SLAMF7 engagement in MM cells. The absence of CD45 also reduced the ability to evoke tyrosine phosphorylation of SLAMF7, although this effect was less pronounced.
CD45 is required for the function of SLAMF7 in NK cells.
To obtain evidence that CD45 was needed for SLAMF7 function, we tested the impact of CD45 on the ability of SLAMF7 to control NK cell activation, using NK cells from CD45-deficient mice. Since these mice express EAT-2, the activating function of SLAMF7 was analyzed (Fig. 7). It was not possible to analyze the inhibitory function of SLAMF7 using NK cells lacking both CD45 and EAT-2, since the mice lacking CD45 or EAT-2 suitable for these studies were generated in a different genetic background (C57BL/6 and 129/Sv, respectively). NK cell functions are drastically different between the two strains.
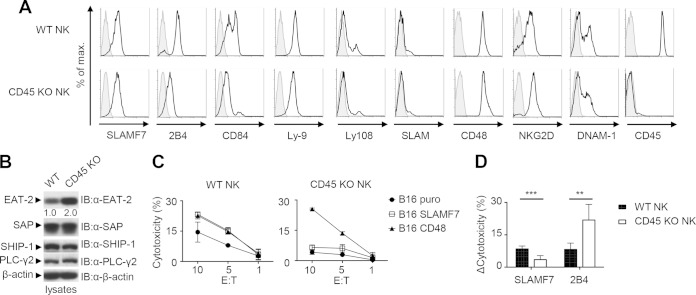
FIG 7.
CD45 is required for the function of SLAMF7. (A and B) NK cells from the spleens of WT or CD45-deficient (knockout [KO]) mice were analyzed for expression of various cell surface molecules by flow cytometry. (A) Staining with specific antibodies is depicted as black lines with no shading, whereas staining with isotype controls is shown as gray lines with shading. (B) NK cells were also assessed for expression of several intracellular signaling molecules by immunoblotting of total cell lysates with the indicated antibodies. A quantitation of the relative expression of EAT-2 is shown. (A and B) Results are representative of 3 experiments. (C) NK cells from the indicated mice were tested for their ability to kill B16 melanoma cells expressing the puromycin resistance marker (puro) alone or in combination with SLAMF7 or CD48, as described for Fig. 2B. The effector-to-target ratios (E:T) are shown at the bottom. Average values of duplicates are shown, while error bars depict standard deviations. Results are representative of 5 experiments. (D) The enhancement of cytotoxicity by the expression of the ligands of SLAMF7 or 2B4 (CD48) on targets was quantitated for multiple independent experiments. Enhanced killing was calculated as cytotoxicity to SLAMF7 or CD48+ targets minus cytotoxicity to puro targets at the 10:1 E/T ratio. Average values from 3 independent experiments with standard deviations are represented. **, P < 0.01; ***, P < 0.001.
As reported elsewhere (35–37), the lack of CD45 had little impact on NK cell development (data not shown). Moreover, it had no effect on the expression of SLAMF7, the activating NK cell receptors NKG2D and DNAM-1, or the effectors of SLAMF7 signaling PLC-γ2 and SHIP-1 (Fig. 7A and B). Rather, expression of EAT-2 was enhanced in CD45-deficient NK cells. Paradoxically, expression of the SLAM family receptors 2B4 and CD84 was reduced. The basis for the latter alterations remains to be clarified.
NK cells were incubated with B16 melanoma cells, expressing or not expressing SLAMF7, and cytotoxicity was measured (Fig. 7C). B16 cells expressing CD48, the ligand of 2B4, were analyzed as a control. Unlike SLAMF7, 2B4 mediates its activating function through EAT-2 and its relative SAP (17). NK cells lacking CD45 displayed reduced killing of B16 cells lacking SLAMF7 or CD48. Since cytotoxicity to B16 cells is mediated largely by DNAM-1 (38–40), this finding suggested that the function of DNAM-1 might be compromised. More importantly, enhancement of cytotoxicity caused by expression of SLAMF7 on B16 cells was reduced in CD45-deficient NK cells. This was not the case for expression of CD48 on B16 cells, which still augmented cytotoxicity by CD45-deficient NK cells. A quantitation of these findings is shown in Fig. 7D. Hence, CD45 was required for the activating function of SLAMF7 but not of 2B4 in NK cells.
DISCUSSION
Although the ability of SLAM family receptors, including SLAMF7, to mediate inhibitory effects in immune cells is well established, the molecular mechanism triggering these effects is largely undetermined. To identify the effectors of SLAMF7-mediated inhibition, we first used the human EAT-2-negative NK cell line YT-S. Stimulation of SLAMF7 on YT-S cells with anti-SLAMF7 resulted in tyrosine phosphorylation of two proteins, SLAMF7 and SHIP-1. Tyrosine phosphorylation of SHIP-1 was also observed upon triggering of SLAMF7 on mouse NK cells, and this effect was enhanced in EAT-2-deficient mice. Thus, SLAMF7 was coupled to SHIP-1 in NK cells, and this effect was enhanced in cells lacking EAT-2.
Several findings firmly supported the idea that SHIP-1 was the critical effector of SLAMF7-mediated inhibition. First, downregulation of SHIP-1 expression using siRNAs attenuated SLAMF7-mediated inhibition in YT-S cells. Second, the ability of SLAMF7 to inhibit BCR-triggered Ca2+ fluxes in DT40 cells was eliminated in cells lacking SHIP-1 but not in cells lacking SHP-1, SHP-2, or Csk. This defect was corrected by reintroduction of SHIP-1. Third, the tyrosine required for SLAMF7-mediated inhibition in YT-S cells, Y261, was needed for coupling to SHIP-1.
Although these data implied that SHIP-1 was the inhibitory effector of SLAMF7 in EAT-2-negative NK cells, this may not be true for all SLAM receptors or all cell types. In studies using the heterologous system DT40, it was suggested that the inhibitory effectors of 2B4 were SHIP-1 and, to a lesser extent, SHP-1 and SHP-2 (15). Conversely, experiments with a pharmacological inhibitor suggested that the inhibitory function of SLAMF6 (Ly108) in CD8+ T cells and NK-T cells is SHP-1 (41, 42). Hence, the inhibitory effectors of SLAM receptors may vary from one family member to another or from one cell type to another.
In YT-S cells, Y261, but not Y281, of SLAMF7 was needed for SLAMF7-triggered tyrosine phosphorylation of SHIP-1. As SHIP-1 contains an SH2 domain, one scenario is that, upon SLAMF7 engagement, phosphorylated Y261 recruits SHIP-1 via its SH2 domain. Intriguingly, however, a synthetic SLAMF7 peptide phosphorylated at Y261 did not interact with SHIP-1 in vitro (data not shown), suggesting that SLAMF7 does not bind directly to SHIP-1. Possibly, an intermediate molecule, such as another adaptor like Grb2, Grap, or Shc, mediates binding of SLAMF7 to SHIP-1. Alternatively, Y261 may recruit the PTK that phosphorylates SHIP-1.
To understand further how SLAMF7 coupled to its inhibitory effector, we elucidated the PTKs responsible for SLAMF7 tyrosine phosphorylation. Cotransfection experiments with Cos-1 cells, experiments with DT40 cells lacking PTKs, and studies with pharmacological inhibitors indicated that Src kinases, in particular Fyn and Lyn, were responsible for the tyrosine phosphorylation of SLAMF7. No effect was observed with Lck, ZAP-70, Syk, Btk, or Csk. Therefore, Src kinases initiated SLAMF7-mediated inhibitory signaling. Since SLAMF7 tyrosine phosphorylation also triggers the activating function of SLAMF7 (5, 6), Src kinases are probably required as well for initiation of SLAMF7-mediated activation.
Although MM cells lack EAT-2 (4), engagement of SLAMF7, including by elotuzumab, failed to trigger tyrosine phosphorylation of SHIP-1. Tyrosine phosphorylation of SLAMF7 was also partially reduced. This was in contrast to what occurred with the EBV-transformed B cell line IM-9, in which prominent tyrosine phosphorylation of SHIP-1 and SLAMF7 was evoked. The defect in SHIP-1 tyrosine phosphorylation in MM cells was not due to a lack of SLAMF7-L, Fyn and Lyn, or SHIP-1. Rather, it correlated with a lack of CD45, a PTP required for Src kinase activation in hematopoietic cells. Although we documented the lack of EAT-2 in all MM cell lines tested, including U266, it should be noted that another group stated that U266 cells expressed EAT-2, although the data to that effect were not reported (13). The basis for this distinction is not known but may relate to clonal variation between the U266 lines used.
Subcloning of CD45-positive and CD45-negative variants of U266 cells, as well as transfection of EGFR-CD45 in CD45-negative OPM2 and MM1S cells, indicated that the absence of CD45 explained at least in part the defect in SLAMF7 signaling in MM cells. In all likelihood, CD45 was needed for activation of Src kinases in these cells. However, CD45 had a more variable impact on the tyrosine phosphorylation of SLAMF7. Perhaps, the pool of Src kinases responsible for SLAMF7 phosphorylation was less dependent on CD45 for activation.
Elotuzumab mediates its anti-MM cell effect by activating immune cells like NK cells, rather than by having direct effects on MM cells (3, 4, 8). Our finding that elotuzumab failed to trigger inhibitory signaling in MM cells might explain at least in part why elotuzumab has no direct effect in these cells. Nonetheless, it should be mentioned that EGFR-CD45 was unable to render MM cells susceptible to a direct antitumor effect from elotuzumab (data not shown). Possibly, the levels of CD45 achieved by EGFR-CD45 were insufficient. Alternatively, additional defects might contribute to the resistance of MM cells to a direct effect from elotuzumab. The lower levels of CD148 and Fyn observed in these cells may contribute.
MM cells, both primary samples from patients and cell lines, frequently lack CD45 (43, 44). This is also true for normal plasma cells. Compared to CD45-positive MM cells, CD45-negative MM cells have been linked to disease progression and poor treatment response (45). Xenotransplantation experiments also suggested that CD45-negative MM cells contain a greater proportion of long-lived and/or myeloma-initiating cells (46). Interestingly, these cells tended to have higher PI3′K activity than CD45-positive MM cells (47). Considering our finding that SLAMF7-mediated inhibition involved SHIP-1, a negative regulator of PI3′K, it is tempting to speculate that increased PI3′K activity in MM cells lacking CD45 was due at least in part to reduced SLAMF7-dependent activation of SHIP-1.
A clear impact of CD45 on SLAMF7 function was documented in mouse NK cells. Whereas SLAMF7 was activating in WT NK cells, this activating effect was lost in CD45-deficient NK cells. This was presumably because CD45-dependent activation of Src kinases was needed for tyrosine phosphorylation of SLAMF7 or, possibly, EAT-2 and PLC-γ2. Surprisingly, this was not the case for another SLAM receptor, 2B4, which was still activating in CD45-deficient NK cells. Perhaps, this distinction relates to the fact that 2B4 is also coupled to SAP (15), which binds Fyn via the Fyn SH3 domain and, thus, may activate Fyn in a CD45-independent manner (23).
In summary, our data indicated that SLAMF7-mediated inhibition in EAT-2-negative NK cells was mediated by the lipid phosphatase SHIP-1. SLAMF7 was coupled to SHIP-1 by way of Y261, which was phosphorylated by Src family kinases. This mechanism was defective in MM cells, at least in part due to a lack of PTP CD45, which is needed for the activation of Src kinases in hematopoietic cells and for initiation of SLAMF7 inhibitory signaling. In addition to clarifying the mechanism of inhibition by SLAMF7 and, possibly, other SLAM family receptors, these data help us understand why elotuzumab has no direct effect on MM cells but rather causes elimination of these cells by activating normal immune cells, such as NK cells.
ACKNOWLEDGMENTS
This work was supported by grants from the Canadian Institute of Health Research, the Canadian Cancer Society Research Institute, and Bristol-Myers Squibb (BMS) to A.V. H.G. held an Alma Mater Fellowship from McGill University and a Joseph-Paulin doctoral scholarship from IRCM. M.-E.C.-M. held fellowships from the Terry Fox Foundation and Cancer Research Society Inc., and N.W. holds a fellowship from the Fonds de Recherche du Québec—Santé. M.R. is an employee of BMS. A.V. holds the Canada Research Chair in Immune Cell Signaling.
A.V. served on the Scientific Advisory Board for elotuzumab at BMS.
REFERENCES
- 1.Veillette A, Guo H. 2013. CS1, a SLAM family receptor involved in immune regulation, is a therapeutic target in multiple myeloma. Crit Rev Oncol Hematol 88:168–177. doi: 10.1016/j.critrevonc.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Bouchon A, Cella M, Grierson HL, Cohen JI, Colonna M. 2001. Activation of NK cell-mediated cytotoxicity by a SAP-independent receptor of the CD2 family. J Immunol 167:5517–5521. doi: 10.4049/jimmunol.167.10.5517. [DOI] [PubMed] [Google Scholar]
- 3.Hsi ED, Steinle R, Balasa B, Szmania S, Draksharapu A, Shum BP, Huseni M, Powers D, Nanisetti A, Zhang Y, Rice AG, van Abbema A, Wong M, Liu G, Zhan F, Dillon M, Chen S, Rhodes S, Fuh F, Tsurushita N, Kumar S, Vexler V, Shaughnessy JD Jr, Barlogie B, van Rhee F, Hussein M, Afar DE, Williams MB. 2008. CS1, a potential new therapeutic antibody target for the treatment of multiple myeloma. Clin Cancer Res 14:2775–2784. doi: 10.1158/1078-0432.CCR-07-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tai YT, Dillon M, Song W, Leiba M, Li XF, Burger P, Lee AI, Podar K, Hideshima T, Rice AG, van Abbema A, Jesaitis L, Caras I, Law D, Weller E, Xie W, Richardson P, Munshi NC, Mathiot C, Avet-Loiseau H, Afar DE, Anderson KC. 2008. Anti-CS1 humanized monoclonal antibody HuLuc63 inhibits myeloma cell adhesion and induces antibody-dependent cellular cytotoxicity in the bone marrow milieu. Blood 112:1329–1337. doi: 10.1182/blood-2007-08-107292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruz-Munoz ME, Dong Z, Shi X, Zhang S, Veillette A. 2009. Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function. Nat Immunol 10:297–305. doi: 10.1038/ni.1693. [DOI] [PubMed] [Google Scholar]
- 6.Tassi I, Colonna M. 2005. The cytotoxicity receptor CRACC (CS-1) recruits EAT-2 and activates the PI3K and phospholipase Cgamma signaling pathways in human NK cells. J Immunol 175:7996–8002. doi: 10.4049/jimmunol.175.12.7996. [DOI] [PubMed] [Google Scholar]
- 7.Perez-Quintero LA, Roncagalli R, Guo H, Latour S, Davidson D, Veillette A. 2014. EAT-2, a SAP-like adaptor, controls NK cell activation through phospholipase Cgamma, Ca++, and Erk, leading to granule polarization. J Exp Med 211:727–742. doi: 10.1084/jem.20132038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Rhee F, Szmania SM, Dillon M, van Abbema AM, Li X, Stone MK, Garg TK, Shi J, Moreno-Bost AM, Yun R, Balasa B, Ganguly B, Chao D, Rice AG, Zhan F, Shaughnessy JD Jr, Barlogie B, Yaccoby S, Afar DE. 2009. Combinatorial efficacy of anti-CS1 monoclonal antibody elotuzumab (HuLuc63) and bortezomib against multiple myeloma. Mol Cancer Ther 8:2616–2624. doi: 10.1158/1535-7163.MCT-09-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zonder JA, Mohrbacher AF, Singhal S, van Rhee F, Bensinger WI, Ding H, Fry J, Afar DE, Singhal AK. 2012. A phase 1, multicenter, open-label, dose escalation study of elotuzumab in patients with advanced multiple myeloma. Blood 120:552–559. doi: 10.1182/blood-2011-06-360552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakubowiak AJ, Benson DM, Bensinger W, Siegel DS, Zimmerman TM, Mohrbacher A, Richardson PG, Afar DE, Singhal AK, Anderson KC. 2012. Phase I trial of anti-CS1 monoclonal antibody elotuzumab in combination with bortezomib in the treatment of relapsed/refractory multiple myeloma. J Clin Oncol 30:1960–1965. doi: 10.1200/JCO.2011.37.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lonial S, Vij R, Harousseau JL, Facon T, Moreau P, Mazumder A, Kaufman JL, Leleu X, Tsao LC, Westland C, Singhal AK, Jagannath S. 2012. Elotuzumab in combination with lenalidomide and low-dose dexamethasone in relapsed or refractory multiple myeloma. J Clin Oncol 30:1953–1959. doi: 10.1200/JCO.2011.37.2649. [DOI] [PubMed] [Google Scholar]
- 12.Lonial S, Kaufman J, Laubach J, Richardson P. 2013. Elotuzumab: a novel anti-CS1 monoclonal antibody for the treatment of multiple myeloma. Expert Opin Biol Ther 13:1731–1740. doi: 10.1517/14712598.2013.847919. [DOI] [PubMed] [Google Scholar]
- 13.Collins SM, Bakan CE, Swartzel GD, Hofmeister CC, Efebera YA, Kwon H, Starling GC, Ciarlariello D, Bhaskar S, Briercheck EL, Hughes T, Yu J, Rice A, Benson DM Jr. 2013. Elotuzumab directly enhances NK cell cytotoxicity against myeloma via CS1 ligation: evidence for augmented NK cell function complementing ADCC. Cancer Immunol Immunother 62:1841–1849. doi: 10.1007/s00262-013-1493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roncagalli R, Taylor JE, Zhang S, Shi X, Chen R, Cruz-Munoz ME, Yin L, Latour S, Veillette A. 2005. Negative regulation of natural killer cell function by EAT-2, a SAP-related adaptor. Nat Immunol 6:1002–1010. doi: 10.1038/ni1242. [DOI] [PubMed] [Google Scholar]
- 15.Dong Z, Davidson D, Perez-Quintero LA, Kurosaki T, Swat W, Veillette A. 2012. The adaptor SAP controls NK cell activation by regulating the enzymes Vav-1 and SHIP-1 and by enhancing conjugates with target cells. Immunity 36:974–985. doi: 10.1016/j.immuni.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Weber M, Treanor B, Depoil D, Shinohara H, Harwood NE, Hikida M, Kurosaki T, Batista FD. 2008. Phospholipase C-gamma2 and Vav cooperate within signaling microclusters to propagate B cell spreading in response to membrane-bound antigen. J Exp Med 205:853–868. doi: 10.1084/jem.20072619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong Z, Cruz-Munoz ME, Zhong MC, Chen R, Latour S, Veillette A. 2009. Essential function for SAP family adaptors in the surveillance of hematopoietic cells by natural killer cells. Nat Immunol 10:973–980. doi: 10.1038/ni.1763. [DOI] [PubMed] [Google Scholar]
- 18.Desai DM, Sap J, Schlessinger J, Weiss A. 1993. Ligand-mediated negative regulation of a chimeric transmembrane receptor tyrosine phosphatase. Cell 73:541–554. doi: 10.1016/0092-8674(93)90141-C. [DOI] [PubMed] [Google Scholar]
- 19.Latour S, Fournel M, Veillette A. 1997. Regulation of T-cell antigen receptor signalling by Syk tyrosine protein kinase. Mol Cell Biol 17:4434–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs A, Cella M, Giurisato E, Shaw AS, Colonna M. 2004. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). J Immunol 172:3994–3998. doi: 10.4049/jimmunol.172.7.3994. [DOI] [PubMed] [Google Scholar]
- 21.Veillette A, Bookman MA, Horak EM, Bolen JB. 1988. The CD4 and CD8 T cell surface antigens are associated with the internal membrane tyrosine-protein kinase p56lck. Cell 55:301–308. doi: 10.1016/0092-8674(88)90053-0. [DOI] [PubMed] [Google Scholar]
- 22.Lee JK, Boles KS, Mathew PA. 2004. Molecular and functional characterization of a CS1 (CRACC) splice variant expressed in human NK cells that does not contain immunoreceptor tyrosine-based switch motifs. Eur J Immunol 34:2791–2799. doi: 10.1002/eji.200424917. [DOI] [PubMed] [Google Scholar]
- 23.Veillette A. 2010. SLAM-family receptors: immune regulators with or without SAP-family adaptors. Cold Spring Harb Perspect Biol 2:a002469. doi: 10.1101/cshperspect.a002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cannons JL, Tangye SG, Schwartzberg PL. 2011. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol 29:665–705. doi: 10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- 25.Veillette A, Latour S. 2003. The SLAM family of immune-cell receptors. Curr Opin Immunol 15:277–285. doi: 10.1016/S0952-7915(03)00041-4. [DOI] [PubMed] [Google Scholar]
- 26.Sayos J, Martin M, Chen A, Simarro M, Howie D, Morra M, Engel P, Terhorst C. 2001. Cell surface receptors Ly-9 and CD84 recruit the X-linked lymphoproliferative disease gene product SAP. Blood 97:3867–3874. doi: 10.1182/blood.V97.12.3867. [DOI] [PubMed] [Google Scholar]
- 27.Galandrini R, Tassi I, Mattia G, Lenti L, Piccoli M, Frati L, Santoni A. 2002. SH2-containing inositol phosphatase (SHIP-1) transiently translocates to raft domains and modulates CD16-mediated cytotoxicity in human NK cells. Blood 100:4581–4589. doi: 10.1182/blood-2002-04-1058. [DOI] [PubMed] [Google Scholar]
- 28.Banh C, Miah SM, Kerr WG, Brossay L. 2012. Mouse natural killer cell development and maturation are differentially regulated by SHIP-1. Blood 120:4583–4590. doi: 10.1182/blood-2012-04-425009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurosaki T. 1999. Genetic analysis of B cell antigen receptor signaling. Annu Rev Immunol 17:555–592. doi: 10.1146/annurev.immunol.17.1.555. [DOI] [PubMed] [Google Scholar]
- 30.Lee JK, Mathew SO, Vaidya SV, Kumaresan PR, Mathew PA. 2007. CS1 (CRACC, CD319) induces proliferation and autocrine cytokine expression on human B lymphocytes. J Immunol 179:4672–4678. doi: 10.4049/jimmunol.179.7.4672. [DOI] [PubMed] [Google Scholar]
- 31.Kim JR, Mathew SO, Patel RK, Pertusi RM, Mathew PA. 2010. Altered expression of signalling lymphocyte activation molecule (SLAM) family receptors CS1 (CD319) and 2B4 (CD244) in patients with systemic lupus erythematosus. Clin Exp Immunol 160:348–358. doi: 10.1111/j.1365-2249.2010.04116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shrivastava P, Katagiri T, Ogimoto M, Mizuno K, Yakura H. 2004. Dynamic regulation of Src-family kinases by CD45 in B cells. Blood 103:1425–1432. doi: 10.1182/blood-2003-03-0716. [DOI] [PubMed] [Google Scholar]
- 33.Zhu JW, Brdicka T, Katsumoto TR, Lin J, Weiss A. 2008. Structurally distinct phosphatases CD45 and CD148 both regulate B cell and macrophage immunoreceptor signaling. Immunity 28:183–196. doi: 10.1016/j.immuni.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhee I, Veillette A. 2012. Protein tyrosine phosphatases in lymphocyte activation and autoimmunity. Nat Immunol 13:439–447. doi: 10.1038/ni.2246. [DOI] [PubMed] [Google Scholar]
- 35.Hesslein DG, Takaki R, Hermiston ML, Weiss A, Lanier LL. 2006. Dysregulation of signaling pathways in CD45-deficient NK cells leads to differentially regulated cytotoxicity and cytokine production. Proc Natl Acad Sci U S A 103:7012–7017. doi: 10.1073/pnas.0601851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin SM, Mehta IK, Yokoyama WM, Thomas ML, Lorenz RG. 2001. Development of intestinal intraepithelial lymphocytes, NK cells, and NK 11+ T cells in CD45-deficient mice. J Immunol 166:6066–6073. doi: 10.4049/jimmunol.166.10.6066. [DOI] [PubMed] [Google Scholar]
- 37.Yamada H, Kishihara K, Kong YY, Nomoto K. 1996. Enhanced generation of NK cells with intact cytotoxic function in CD45 exon 6-deficient mice. J Immunol 157:1523–1528. [PubMed] [Google Scholar]
- 38.Lakshmikanth T, Burke S, Ali TH, Kimpfler S, Ursini F, Ruggeri L, Capanni M, Umansky V, Paschen A, Sucker A, Pende D, Groh V, Biassoni R, Hoglund P, Kato M, Shibuya K, Schadendorf D, Anichini A, Ferrone S, Velardi A, Karre K, Shibuya A, Carbone E, Colucci F. 2009. NCRs and DNAM-1 mediate NK cell recognition and lysis of human and mouse melanoma cell lines in vitro and in vivo. J Clin Invest 119:1251–1263. doi: 10.1172/JCI36022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilfillan S, Chan CJ, Cella M, Haynes NM, Rapaport AS, Boles KS, Andrews DM, Smyth MJ, Colonna M. 2008. DNAM-1 promotes activation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors. J Exp Med 205:2965–2973. doi: 10.1084/jem.20081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iguchi-Manaka A, Kai H, Yamashita Y, Shibata K, Tahara-Hanaoka S, Honda S, Yasui T, Kikutani H, Shibuya K, Shibuya A. 2008. Accelerated tumor growth in mice deficient in DNAM-1 receptor. J Exp Med 205:2959–2964. doi: 10.1084/jem.20081611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, Schwartzberg PL, Crotty S. 2012. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity 36:986–1002. doi: 10.1016/j.immuni.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao F, Cannons JL, Dutta M, Griffiths GM, Schwartzberg PL. 2012. Positive and negative signaling through SLAM receptors regulate synapse organization and thresholds of cytolysis. Immunity 36:1003–1016. doi: 10.1016/j.immuni.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Rajkumar SV, Kimlinger T, Greipp PR, Witzig TE. 2005. CD45 expression by bone marrow plasma cells in multiple myeloma: clinical and biological correlations. Leukemia 19:1466–1470. doi: 10.1038/sj.leu.2403823. [DOI] [PubMed] [Google Scholar]
- 44.Moreau P, Robillard N, Avet-Loiseau H, Pineau D, Morineau N, Milpied N, Harousseau JL, Bataille R. 2004. Patients with CD45 negative multiple myeloma receiving high-dose therapy have a shorter survival than those with CD45 positive multiple myeloma. Haematologica 89:547–551. [PubMed] [Google Scholar]
- 45.Pellat-Deceunynck C, Bataille R. 2004. Normal and malignant human plasma cells: proliferation, differentiation, and expansions in relation to CD45 expression. Blood Cells Mol Dis 32:293–301. doi: 10.1016/j.bcmd.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Kim D, Park CY, Medeiros BC, Weissman IL. 2012. CD19−CD45low/−CD38high/CD138+ plasma cells enrich for human tumorigenic myeloma cells. Leukemia 26:2530–2537. doi: 10.1038/leu.2012.140. [DOI] [PubMed] [Google Scholar]
- 47.Descamps G, Pellat-Deceunynck C, Szpak Y, Bataille R, Robillard N, Amiot M. 2004. The magnitude of Akt/phosphatidylinositol 3′-kinase proliferating signaling is related to CD45 expression in human myeloma cells. J Immunol 173:4953–4959. doi: 10.4049/jimmunol.173.8.4953. [DOI] [PubMed] [Google Scholar]