Abstract
RNA synthesis and replication of the members of the Flavivirus genus (including dengue, West Nile and Japanese encephalitis viruses) is regulated by a wide variety of mechanisms and actors. These include the sequestration of the RNA-dependent RNA polymerase (RdRp) for functions other than RNA synthesis, regulatory interactions with other viral and host proteins within the replication complex (RC), and regulatory elements within the RNA genome itself. In this review, we discuss our current knowledge of the multiple levels at which Flavivirus RNA synthesis is controlled. We aim to bring together two active research fields: the structural and functional biology of individual proteins of the RC and the impressive wealth of knowledge acquired regarding the viral genomic RNA.
1. Introduction
Flaviviruses (genus Flavivirus, family Flaviviridae) are single-strand positive-sense RNA (+RNA) viruses, which include important arthropod-borne human pathogens such as the dengue viruses (DENV, serotypes 1–4), West Nile virus (WNV), yellow fever virus (YFV) and the Japanese, St. Louis, and tick-borne encephalitis viruses (JEV, SLE, and TBEV, respectively). The Flavivirus genome bears a conserved type-1 RNA cap (m7GpppA2’OmG) at the 5’-end. Upon infection, the genome of ~11 kb is translated into a single polyprotein, which is processed into three structural and seven non-structural (NS) proteins. The NS proteins assemble with an array of host factors into membrane-bound replication complexes (RC) where viral RNA (vRNA) synthesis takes place. The main actor is the RNA-dependent RNA polymerase (RdRp) NS5, which replicates the genomic +RNA into the uncapped minus-strand RNA (−RNA). Within the resulting double-strand (ds) RNA replicative form (RF), the −RNA acts as a template to produce a large excess of +RNA [1–3]. In a replicative intermediate (RI), several nascent +RNA molecules are synthesized simultaneously from and on the same −RNA template. This synthesis is semi-conservative, i.e., the old +RNA is displaced by the nascent +RNA. The viral capping machinery harbored in NS3 and NS5 caps only newly-synthesized +RNAs [4–6]. Both the process of discrimination between −RNA and +RNA 5’-ends for capping and the precise timing of the capping process remain largely undefined. Recently, a model was proposed [3] where the low-level production of −RNA at early stages of RNA replication occurs in RCs associated with virus-induced membranes derived from the endoplasmic reticulum (ER) but not yet in fully formed vesicles. Then, at later stages of RNA replication, 100-nm vesicles that are connected to the cytosol via small pores [7,8] further protect the RC and the nascent +RNA. The correct initiation of RNA synthesis is essential for the integrity of the viral genome. The Flavivirus RdRp initiates RNA synthesis without a primer (primer-independent, or de novo initiation) over the last 3’-end nucleotide of the template. RNA synthesis always starts with the formation of the same short primer, pppAG, consistent with strict conservation of the 5’-and 3’-end sequences of all Flavivirus genomes as 5’AG…CU-3’ [9].
In this review, we discuss our current knowledge of the multiple levels at which viral RdRp activity and specificity are controlled to ensure faithful and efficient RNA synthesis and the vRNA elements that modulate vRNA replication. We focus on DENV but include when possible other members of the Flavivirus genus.
2. Regulation of RNA replication by NS5 adopting alternative functions
Unlike many other RNA viruses, such as +RNA viruses from the Nidovirales order or Togaviridae family, flaviviruses have no obvious stoichiometric control of non-structural versus structural protein production. The genome is translated as a single viral polyprotein that has to provide enough copies of structural proteins for viral assembly (180 copies of envelope protein in each DENV particle [10]). Active viral RdRp activity, embedded in the Flavivirus NS5 protein, is probably not required in so many copies. It is thus not surprising that additional functions have been and are still being discovered for NS5.
NS5 plays a direct role in viral defense mechanisms against the host innate immune response, which is triggered by recognition of vRNA as non-self RNA by intracellular RNA sensors [11]. This recognition in turn leads to the degradation of vRNA by cellular exonucleases [2,12] and triggers interferon (IFN) signaling pathways involving signal transducer and activator of transcription 2 (STAT2) and other defense mechanisms [6]. NS5 inhibits IFN type 1 signaling [13] through the promotion of STAT2 degradation [14,15].
NS5 proteins of YFV and DENV-2 are imported into the nucleus [16,17]. NS5 sequestration through nuclear import might represent a way to limit or regulate vRNA synthesis, which takes place in the cytoplasm [18]. At the same time this nuclear import of NS5 may serve to hijack cellular importins [18]. Nevertheless, substantial differences have been found in the percentage of NS5 that is directed to the nucleus between different flaviviruses and even among different DENV serotypes [19,20]. Clearly, more work is needed to clarify the role of nuclear import of NS5 in flaviviral defense mechanisms and/or regulation of vRNA synthesis [21,22].
Despite the alternative functions of NS5 that limit vRNA synthesis, Flavivirus RNA is produced in excess. One possible reason might be that it saturates the host exonuclease XRN1, which remains trapped and inactivated by the XRN1-resistant subgenomic Flavivirus RNA (sfRNA) [2].
3. Regulatory elements within the NS5 polymerase domain
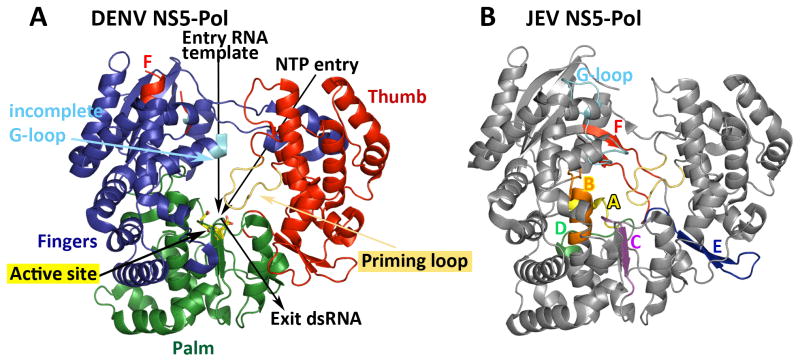
Flavivirus NS5 harbors the RdRp activity in its C-terminal domain, designated NS5-Pol in this review. NS5-Pol (74 kDa) is connected via a flexible short linker to the N-terminal methyltransferase (MTase) domain (30 kDa), which contains two MTase activities and possibly a guanylyltransferase activity (GTase), thus incorporating three of the four reactions necessary to form a type-1 RNA cap at the 5’-end of the nascent +RNA strand [4,5]). The NS5-Pol domain alone displays de novo RdRp activity [9,23–26]. NS5-Pol and NS5 follow the same overall RNA synthesis scheme observed for other de novo RNA polymerases: pppAG synthesis by de novo initiation, a transition phase involving a conformational change from an initiation to an elongation conformation, followed by processive elongation [9,27]. Structurally, the existence of two different conformations for initiation and elongation has been demonstrated for Flaviviridae Hepatitis C (HCV) RdRp [28]. The X-ray crystal structures of isolated NS5-Pol domains of DENV, WNV and JEV and of JEV NS5-Pol in the context of full-length NS5 have been determined [19,26,29–31]. They adopt the common right-hand fold of viral RdRps with three subdomains fingers, thumb and palm (Figure 1). A comparison to the initiation-state and elongation-state structures of HCV RdRp suggests that the existing Flavivirus RdRp structures are situated in between, since they present a closed priming loop essential for de novo initiation (see below) but a rather large distance between the fingers and thumb subdomains that could accommodate a template/primer duplex [32]. More structures in complex with NTP substrates, RNA templates or regulatory elements are needed that capture genuine initiation and elongation structures of Flavivirus RdRps.
Figure 1. X-ray crystal structures of DENV-3 NS5-Pol and JEV NS5-Pol.
Ribbon diagrams generated by PyMOL Molecular Graphics System (Version 1.7 Schrödinger, LLC). A) DENV-3 NS5-Pol (PDB code 4HHJ [31]) with fingers, palm and thumb subdomains colored in blue, green and red, respectively. The RNA template and NTP entry tunnels are indicated by arrows, as is the dsRNA exit. The priming loop is shown in light orange. The active site aspartic acids of motifs A and C are shown in sticks colored according to the atom type (carbon, yellow; oxygen, red). Motif F and incomplete G-loop are shown in red and cyan, respectively. B) JEV NS5-Pol (4HDG [23]) in complex with stabilizing GTP (not shown) showing Motifs A to F in yellow, orange, purple, light green, red and blue, respectively, and the G-loop in cyan. Motif F is stabilized by GTP but closes the NTP entry tunnel, and the G-loop aligns the entry of the RNA template tunnel.
RNA synthesis involves small regulatory conformational changes within the active center of NS5-Pol. The common viral RdRp active site motifs can be considered as basic intrinsic elements to control RNA synthesis. Flavivirus RdRp motifs A and C (Figure 1B) provide the two aspartic acid residues implicated in the nucleotidyl transfer catalyzed by two ions (in vitro Mg2+ or Mn2+; in vivo this remains an open question). Motif B seems to be involved in template binding and translocation [33]. Its second function, and also of motif D, is the control of NTP specificity [33–35]. Motif D may also provide the general acid for nucleotidyl transfer [36]. Motif F is not present in un-stabilized structures of DENV, WNV and JEV NS5-Pol domains (Figure 1A). Only when stabilized by GTP, ATP, crystal contacts or the NS5-MTase domain (Figure 1B), it forms the upper part of the NTP entry tunnel [23,29] as in other viral RdRps. Motif E is found at the end of the NTP tunnel and might be involved in the correct positioning of the priming nucleotide ATP [37]. Additionally, it is situated at the hinge between palm and thumb subdomains and might act as a pivot during conformational changes between de novo initiation and elongation states of NS5-Pol. Motif G was originally defined for primer-dependent RdRps [38]. De novo-initiating Flavivirus RdRps do not contain a sequence resemblance to motif G. The corresponding loop (G-loop) was absent in the first NS5-Pol structures [19,30] (Figure 1A) and was proposed to be involved in regulation of de novo RdRp activity [19], like the C-terminal extension of HCV RdRp [28]. In the JEV NS5-Pol structures stabilized by GTP, ATP or the MTase domain, the G-loop is well-defined and remains far from the active site (Figure 1B) [23,29]. Whether there is a biologically relevant regulatory conformational change of motifs F and G from a putative pre-initiation to an initiation conformation triggered by the incoming template or NTP substrates remains to be clarified [1,32]
Like other de novo-initiating viral RdRps, Flavivirus NS5-Pol has an elaborate thumb subdomain with a priming loop that closes the active site during initiation (Figure 1) [19,23,29,30]. This loop provides at least part of an ATP-specific priming site [9]. A conserved histidine in the priming loop (H798 in DENV-2) might act as a priming platform against which the ATP stacks upon de novo initiation; however, existing NS5-Pol structures do not provide enough information on the precise structure of this priming site in the de novo initiation complex. The ATP-specific priming site enables NS5-Pol to ensure the conservation of correct Flavivirus genome ends [9].
4. Regulation of RNA synthesis by the NS5 methyltransferase domain
A longstanding question has been whether the NS5-MTase domain regulates the RdRp activity of NS5-Pol. A prerequisite of regulation is interaction between the two domains. Genetic and in vitro interaction has indeed been shown for WNV and DENV NS5 MTase and RdRp domains (see references in [39]). The interaction appears weak and might use multiple transient interfaces. Indeed, Small Angle X-ray Scattering (SAXS) data showed that NS5 is mainly extended in solution with just a small subset of compact structures where NS5-Pol and NS5-MTase form an interface [40]. Concerning the regulation of the RdRp activity of NS5-Pol by NS5-MTase, in vitro studies comparing steady-state RdRp activities of recombinant DENV NS5 and NS5-Pol yielded contradictory results (summarized in [39]). Recently, we and others found consistent stimulatory effects by the DENV NS5-MTase domain on the overall steady-state RdRp activity of NS5-Pol [26,39]. This stimulation happens during initiation and elongation, while NS5-MTase does not seem to facilitate the conformational change between the two steps. Stimulation of de novo initiation is based on an increase in RNA and priming nucleotide affinity. During the elongation phase, NS5-MTase increases the affinity for the incoming nucleotide and its incorporation rate [39].
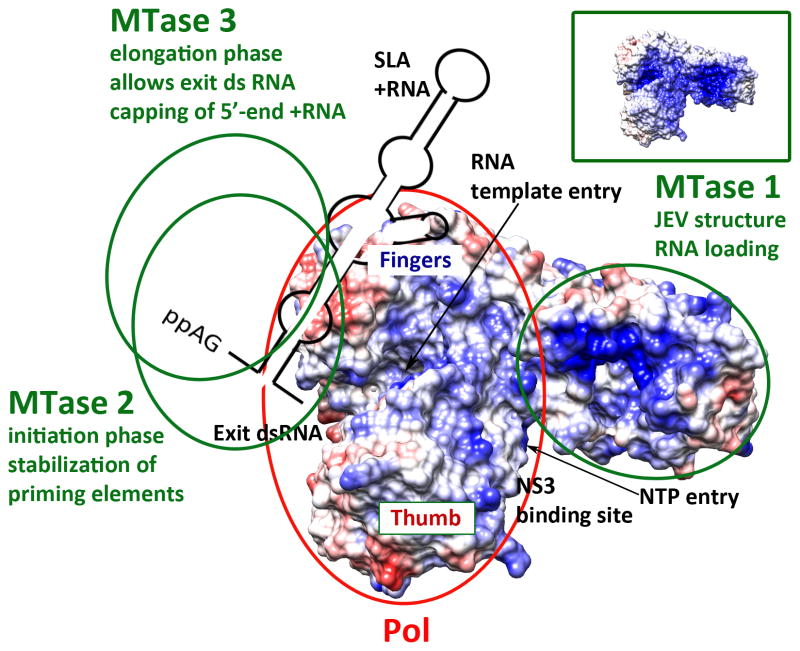
The observed stimulatory effect of NS5-MTase calls for two scenarios. First, the NS5-MTase may act via a single but flexible interface, which allows the necessary conformational changes of the NS5-Pol domain during RNA synthesis. This interface might correspond to the hydrophobic interface between NS5-MTase and NS5-Pol of the recent X-ray crystal structure of JEV NS5 [23]. In this conformation, the NS5-MTase (MTase 1 in Figure 2) appears to complete the RNA binding zone near the entrance of the RNA template tunnel. It might also support RNA binding by promoting the correct conformation of the RNA template tunnel. The optimal RNA template binding might then cause a long-range allosteric effect in the specific ATP-binding site involving the priming loop, thus stimulating de novo initiation. After pppAG synthesis, NS5-Pol is expected to undergo a conformational change to accommodate the growing primer-template dsRNA. The nascent dsRNA bears a 5’-triphosphate reaching into the solvent. Capping of nascent +RNA occurs with unknown choreography. In this scenario with a single interacting site, NS5-MTase is situated far from the dsRNA exit; and capping should involve an NS5-MTase domain from a second NS5 protein. This might indeed be possible since several NS5 proteins complete +RNA synthesis on the RI form [3]. The second scenario envisages several binding sites (Figure 2, MTase 1 to 3). The first one, facilitating RNA template binding, may correspond to the JEV NS5 structure discussed above (MTase 1). The structuring of the RNA template tunnel might then release the NS5-MTase from its binding site. The MTase domain would move to a second site (MTase 2) where it exerts an allosteric effect on the priming site for de novo initiation. The flexible linker allows for this binding site to be near the priming loop [28,29]. The priming loop later opens to allow the exit of the nascent dsRNA. The NS5-MTase might concurrently change position to a third site (MTase 3) to exert its effect on RNA elongation. In the NS5-Pol elongation-state interaction, the NS5-MTase could receive the 5’-end of +RNA after its passage towards NS3 (containing RNA triphosphatase activity) and the formation of the 5’SLA of around 70 nucleotides, which has been shown to be essential for N7-methylation [4]. In this scenario we may also consider that MTase 2 or 3 is from a second NS5 molecule, since apparent oligomerization of NS5 within the RC has been shown [41]. Structural and mutational data studying the existing interface of NS5 and other interfaces to be discovered in the future will hopefully clarify which scenario is correct.
Figure 2. Multi-interface model of Flavivirus NS5 during RNA synthesis, illustrating the stimulating effects of the MTase domain on the Pol domain.
The surface of JEV NS5 (PDB code 4K6M [29]) is shown, with the Pol domain on the left (red circle) and the MTase domain on the right (green circle labeled MTase 1). The polymerase domain is seen from above looking down the RNA template tunnel with the entry to the NTP tunnel on the right and the exit of dsRNA on the left. Fingers and thumb subdomains are labeled. The Coulombic surface coloring generated by Chimera (http://www.cgl.ucsf.edu/chimera) shows the positively charged nature of the surface (blue) of the template tunnel. This positive zone continues on the surface of the MTase domain. This is especially evident using the electrostatic surface coloring of NS5 (see inset on the right, also generated by Chimera) The existing structure of JEV-NS5 might represent a structure adopted by NS5 during RNA template loading, with the MTase extending the RNA binding area. The following is an attempt to accommodate reverse genetics experiments, observed conformations in solution, and the topology of RNA capping with novel structural data of JEV NS5 (see text). After a conformational change, the MTase may adopt a position at the other side of the Pol domain (MTase 2) near or at the exit of dsRNA and the priming loop, stimulating de novo initiation. Finally, MTase 3 might be the position of the MTase after a necessary conformational change of the Pol domain entering the elongation phase. The MTase would be near the exit of dsRNA. When +RNA is synthesized, the nascent 5’ end would dissociate from its template, the γ-phosphate would be hydrolyzed by NS3, the SLA structure formed and the diphosphate 5’-end could return to the active site of the MTase to be capped. The second and third positions of NS5-MTase are hypothetical but possible given the flexibility and length of the linker, and are consistent with genetic mapping data. However, MTase 2 and 3 contacts with NS5-Pol could also be provided by a second NS5 molecule. The binding site identified for NS3-Hel on NS5-Pol (see text) is indicated. It comprises NS5-Pol elements from the fingers and thumb subdomains near the RNA entry tunnel. In this position, NS3-Hel may dissociate the RF or remove dsRNA structures from the RNA genome before RNA template loading.
5. Regulation by viral or host proteins interacting with NS5
All viral NS proteins co-localize with dsRNA to ER membrane-embedded active RCs [7,8,41]. Among them, only NS3 seems to interact directly with NS5 [8,41], but several others, such as NS1 [42]), NS2A [43], NS4A [44–47], and NS4B [44,45,48], have been shown to regulate RNA synthesis, though the molecular mechanisms are still not completely defined. Apart from NS5, the second main player in the Flavivirus RC is NS3. NS3 harbors the NS2B-activated viral protease activity at its N-terminal domain (NS3-Pro) and the NTPase-driven helicase activity at its C-terminal domain (NS3-Hel) [49]. The latter also contains the 5’-RNA triphosphatase (RTPase) activity and is therefore thought to undertake the first step of vRNA cap formation at the 5’-end of the genomic RNA [6]. NS3 may thus influence Flavivirus RNA replication on several levels. First, its own activities are essential for viral replication [50]. Second, NS3 acts as a central hub within the RC, interacting via its membrane-bound co-factor NS2B with NS2A, NS4A and NS4B [41]. Third, it may influence RNA synthesis via its direct interactions with NS5, which have been demonstrated in vivo and in vitro. NS3-Hel and NS5-Pol interact [51–53] via a region of the fingers subdomain of NS5-Pol comprising its nuclear localization sequence (NLS) and reaching over to the thumb subdomain (Figure 2) [51,52]. There is evidence that binding between NS3-Hel and NS5 is enhanced by NS3-Pro [52], probably involving the NS5-MTase domain [53]. Within NS5-MTase, several negatively-charged residues were proposed to be involved in the interaction with NS3 [54]. Functionally, regulation of the polymerase activity of NS5 by NS3 has not been demonstrated. In contrast, NS5 seems to positively regulate the NTPase activity of NS3 [55] and possibly also its helicase and RTPase activities [56].
Flavivirus
RNA replication may also be modulated by host proteins, which interact with the viral RC components. Numerous studies screening interactions in the context of viral infection have yielded large numbers of putatively interacting host proteins, not only within the RC (see references in [57]). Among NS5-interacting host proteins modulating vRNA replication are GBF1, a guanine nucleotide exchange factor [57], cyclophilin A, a peptidyl-prolyl isomerase, [58], and the cellular protein kinase G, which has been shown to phosphorylate NS5 of mosquito-borne flaviviruses within the NS5-MTase and NS5-Pol domains [59,60]. Other identified host interaction partners of Flavivirus NS5 are heat shock protein Hsp70 [61] as well as eIF3L (eukaryotic translation initiation factor 3), Hdj2 (Hsp40 homolog) and PAZ proteins (reviewed in [3]). Host factors that interact with NS3 and influence RNA replication are fatty acid synthase (FASN) [62] and caveolin (Cav 1) [63]. The precise molecular mechanisms underlying the putative regulation of Flavivirus RNA synthesis by host proteins is under active investigation.
6. Regulation by host proteins interacting with viral RNA
Investigations of host protein-vRNA interactions improve our understanding of the contributions of host proteins to viral infection. However, mechanisms detailing the role of host proteins in flaviviral RNA replication are poorly defined and to date, only five host-derived factors have been demonstrated to directly impact Elavivirus RNA replication through interacting with vRNA.
Eukaryotic translation elongation factor eEF1A binds to the 3’SL, the large terminal stem-loop of the 3’UTR (Figure 3B) of several flaviviruses [64], co-localizes to the RC of WNV and DENV, and was shown to facilitate WNV negative-strand synthesis [65]. Polypyrimidine tract-binding protein (PTB) translocates from the nucleus to the cytoplasm during DENV infection [66]. Besides interacting with DENV RNA [67], PTB also binds DENV NS4A and modulates negative-strand RNA synthesis [68]. Another host factor NF90 interacts with the DENV 3’SL and positively regulates DENV replication [69]. The DEAD-box RNA helicase DDX6, a component of P-bodies and stress granules, has been shown to interact with DENV2 RNA via binding to the 3’UTR DB structures (Figure 3B), which are required for viral translation and replication [70]. Finally, p100 interacts with the DENV 3’SL,and p100 knock-down reduced viral RNA and protein levels in DENV-infected cells [71].
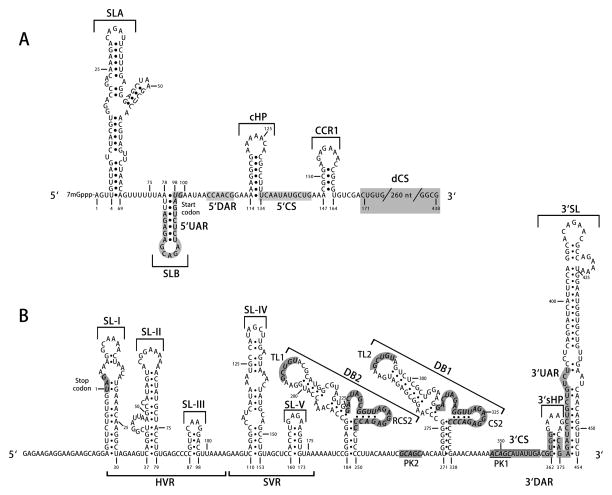
Figure 3. Schematic representation of conserved sequences and structures in the DENV untranslated regions (UTRs) and capsid-coding region.
(A) 5’UTR and capsid-coding region. The DENV 5’UTR contains one large stem-loop (SLA) and one short stem-loop (SLB). The 5’ upstream AUG region (5’UAR) is located within SLB. Downstream of the start codon, within the capsid-coding region, there are several cis-acting RNA elements, including a short 5’ downstream of AUG region (5’DAR), capsid-coding region hairpin (cHP), 5’ cyclization sequences (5’CS), conserved capsid-coding region 1 (CCR1), and downstream of the DENV 5’ cyclization sequence (dCS) element. (B) 3’UTR. The 3’UTR begins with a variable region (VR) containing the hypervariable region (HVR) and semi-variable region (SVR), followed by two dumbbell structures in tandem (DB2 and DB1), 3’ cyclization sequences (3’CS), 3’ downstream of AUG region (3’DAR), and 3’ upstream AUG region (3’UAR). The latter two RNA structures overlap with the 3’ stem-loop (3’SL), which includes a small 3’-end hairpin structure (3’sHP) and a large stem-loop. The top loops (TLs) of the dumbbells are proposed to interact with downstream pseudoknot (PK) regions. The 3’UTR nucleotides are numbered starting from and including the stop codon. Nucleotide numbering is according to DENV2 strain 16681.
Additional proteins that bind the DENV 3’-UTR, such as YB1 [72] and PABP [73], have been shown to play roles in other stages of the viral life cycle. Several other host proteins, including La, hnRNP-A1, hnRNP-A2/B1, and hnRNP-Q [67,72,73], bind to the DENV 3’UTR, but their exact roles in regulation of vRNA synthesis remain elusive [74].
7. Regulation by cis-acting RNA elements
Efficient viral RNA replication is regulated by conserved sequences and structural elements in the 5’UTR, capsid-coding region, and 3’UTR (Figure 3). DENV has a relatively short 5’UTR (96 nucleotides [nt] for DENV2) (Figure 3A) consisting of two stem-loops (SLA and SLB) separated by an oligoU spacer. The DENV NS5 RdRp specifically binds to the viral genome by interacting with a promoter element in the 5’-end SLA (~70nt), which is conserved across flaviviruses [75]. Circularization of the genome via long-range RNA-RNA interactions enables delivery of the NS5 RdRp to the 3’-end of the genome to initiate negative-strand synthesis [75]. Binding to the SLA alone is not sufficient to induce RdRp activity, and specific nucleotides in the SLA are necessary to activate the polymerase-promoter complex for RNA synthesis [76].
The other short stem-loop in the 5’-UTR, SLB (21nt), contains the 5’ upstream of AUG region 5’UAR (16nt) [77]. DENV RNA replication requires long-range RNA-RNA-mediated cyclization of the viral genome, which involves at least three pairs of complementary RNA sequences at both ends of the genome: the 5’- and 3’ cyclization sequences (5’-3’CS; 11nt) [78], 5’ and 3’ upstream of AUG region (5’-3’UAR; 16nt) [77,79,80], and the 5’ and 3’downstream of AUG region (5’-3’DAR; 6nt) [81,82]. These form a “zipper” along the panhandle of the circularized vRNA. In addition, the composition of the sequence downstream of the DENV 5’CS (dCS) element (268nt) in the capsid gene also influences circularization of the viral genome and hence affects vRNA replication [83]. Furthermore, a capsid-coding region hairpin element (cHP) (21nt) is required for efficient vRNA synthesis in a sequence-independent manner [84], while it also has an independent role in translation start site selection [85]. Detailed analysis of the interplay of these 5’ RNA elements with respect to RNA replication revealed that the position of SLA is restricted to the 5’-terminus of the DENV genome, whereas the 5’UAR, 5’DAR, cHP, and 5’CS form a second unit that is more flexible in position and likely act jointly to facilitate formation of the higher-order RNA structure of the circularized DENV genome [82]. Most 5’UTR and capsid-coding region RNA structures shown in Figure 3A are required for DENV RNA synthesis, with one exception the conserved capsid-coding region 1 (CCR1) (20nt), an RNA sequence element that likely modulates assembly [86].
DENV has a relatively long 3’UTR (454nt for DENV2) (Figure 3B), which contains several important RNA sequences/structures in addition to the three complementary sequences (3’-CS, 3’-DAR, and 3’-UAR) that are needed for genome circularization and negative-strand RNA synthesis. The variable region (VR), whose size varies (<50nt to >127nt) depending on the serotype [87], is located at the very 5’-terminus of the 3’UTR. VR consists of the hypervariable region (HVR), including three stem-loop structures (SL-I, SL-II, and SL-III), and the semi-variable region (SVR) containing two stem-loop structures (SL-IV and SL-V). The length of HVR and the specific sequence of the SVR impact RNA synthesis [88]. SL-II forma a pseudoknot structure that stalls the cellular 5’-3’ exonuclease XRN1 to form the sfRNA [89]. WNV sfRNA interferes with the RNAi antiviral pathway by inhibiting Dicer [90] and contributes to viral evasion of the type I interferon response [91]. Recent data also suggest that sfRNA inhibits XRN1 activity and stabilizes cellular mRNA [12].
The middle part of the DENV 3’UTR forms two dumbbell structures in tandem (DB2 and DB1; 68nt), which are required for optimal vRNA synthesis [92]. The top loops (TLs) (5nt) of these dumbbells are proposed to interact with downstream pseudoknot (PK) (5nt) regions, and both TLs are needed for optimal translation [93]. Recently, the TL1/PK2 interaction was predicted to form an H-type pseudoknot that may participate in regulation of vRNA synthesis [94]. The repeated conserved sequences (CS2 and RCS2) (22nt) located within DB1 and DB2 are highly conserved between flaviviruses [95].
Finally, the 3’-end stem-loop (3’SL) (93nt) is essential for RNA replication and translation [96–98]. This is the most conserved part of the 3’UTR and includes a small 3’-end hairpin structure (3’sHP) (14nt), which is essential for replication [81,99], and an adjacent long stem-loop (79nt). The 3’SL has overlapping sequence with the 3’DAR, and it contains the entire sequence of the 3’UAR. The 3’sHP is present only in the linear form of the genome; alternatively, it can form a duplex with complementary sequences in the 5’UTR via long-range 5’-3’ interactions [81]. A recent systematic mutational analysis in the DENV 3’sHP revealed a stricter requirement for specific sequences in the 3’sHP for viral replication in mosquito cells than in mammalian cells [99]. Maintaining a balance between the circular and linear forms of the DENV genome is crucial for viral replication [100].
8. Conclusion and perspectives
The aim of this review is to bring together two research fields in Flavivirus RNA replication. The structural biology of the individual proteins NS3 and NS5 has reached a stage where further leaps in knowledge will be achieved only when RNAs are included into structural models. Conversely, the impressive wealth of knowledge of Flavivirus RNA biochemistry will significantly expand when NS3 and NS5 are included in biochemical/biophysical assays. The time is ripe to integrate the two approaches to decipher Flavivirus RNA synthesis. The abundance of potential molecular switches in the 5’ and 3’UTRs has to be matched with the forthcoming multiple structures and conformational changes of NS3, NS5, and both assembled in an NS3/NS5 complex. Since exposure of vRNA synthesis intermediates can trigger the host innate immune response, the timing and coordination of RNA synthesis and capping must be under constant evolutionary pressure, and experiments in the near future will need to address the timing of events during RNA synthesis. Flavivirus proteins involved in RNA synthesis together with RNA regulatory elements now represent a mature field, merging structural biology of macromolecular assembly, RNA structure and biochemistry, and virology. Importantly, these efforts are directed towards emerging pathogens, such as DENV, for which preventive and therapeutic options remain challenging, and the knowledge gained by these studies should contribute to development of innovative antiviral strategies.
Highlights.
Detailed structural models are available for Flavivirus NS5 and NS3 proteins
Complex structures of NS5 and NS3 with NTP and RNA substrates and regulatory elements are needed
The precise sequence and in many cases functions of Flavivirus RNA elements are known
Interaction maps of NS3 and/or NS5 with 5’ and 3’ UTR RNA are within reach
Future challenges involve deciphering the timing and coordination of RNA synthesis and capping
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* of special interest
** of outstanding interest
- 1.Malet H, Massé N, Selisko B, Romette J-L, Alvarez K, Guillemot JC, Tolou H, Yap TL, Vasudevan S, Lescar J, et al. The flavivirus polymerase as a target for drug discovery. Antiviral Res. 2008;80:23–35. doi: 10.1016/j.antiviral.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Roby JA, Pijlman GP, Wilusz J, Khromykh AA. Noncoding subgenomic flavivirus RNA: multiple functions in West Nile virus pathogenesis and modulation of host responses. Viruses. 2014;6:404–427. doi: 10.3390/v6020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3*.Brinton MA. Replication cycle and molecular biology of the West Nile virus. Viruses. 2014;6:13–53. doi: 10.3390/v6010013. This recent comprehensive review summarizes our knowledge of WNV replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong H, Fink K, Züst R, Lim SP, Qin C-F, Shi P-Y. Flavivirus RNA methylation. J Gen Virol. 2014;95:763–778. doi: 10.1099/vir.0.062208-0. [DOI] [PubMed] [Google Scholar]
- 5.Saeedi BJ, Geiss BJ. Regulation of flavivirus RNA synthesis and capping. Wiley Interdiscip Rev RNA. 2013;4:723–735. doi: 10.1002/wrna.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatel-Chaix L, Bartenschlager R. Dengue virus- and hepatitis C virus-induced replication and assembly compartments: the enemy inside--caught in the web. J Virol. 2014;88:5907–5911. doi: 10.1128/JVI.03404-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CKE, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9*.Selisko B, Potisopon S, Agred R, Priet S, Varlet I, Thillier Y, Sallamand C, Debart F, Vasseur J-J, Canard B. Molecular basis for nucleotide conservation at the ends of the dengue virus genome. PLoS Pathog. 2012;8:e1002912. doi: 10.1371/journal.ppat.1002912. This study shows the decisive role of the priming loop of DENV NS5-Pol for ATP-specific de novo initiation of RNA synthesis, and how it translates into the conservation of the first and last nucleotide of the viral RNA genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, Mukhopadhyay S, Baker TS, Strauss JH, Rossmann MG, Kuhn RJ. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol. 2003;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasirudeen AMA, Wong HH, Thien P, Xu S, Lam K-P, Liu DX. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl Trop Dis. 2011;5:e926. doi: 10.1371/journal.pntd.0000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, Khromykh AA, Wilusz J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA N Y N. 2012;18:2029–2040. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrison J, García-Sastre A. STAT2 signaling and dengue virus infection. JAK-STAT. 2014;3:e27715. doi: 10.4161/jkst.27715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashour J, Laurent-Rolle M, Shi P-Y, García-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol. 2009;83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazzon M, Jones M, Davidson A, Chain B, Jacobs M. Dengue virus NS5 inhibits interferon-alpha signaling by blocking signal transducer and activator of transcription 2 phosphorylation. J Infect Dis. 2009;200:1261–1270. doi: 10.1086/605847. [DOI] [PubMed] [Google Scholar]
- 16.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 17.Buckley A, Gaidamovich S, Turchinskaya A, Gould EA. Monoclonal antibodies identify the NS5 yellow fever virus non-structural protein in the nuclei of infected cells. J Gen Virol. 1992;73 ( Pt 5):1125–1130. doi: 10.1099/0022-1317-73-5-1125. [DOI] [PubMed] [Google Scholar]
- 18.Rawlinson SM, Pryor MJ, Wright PJ, Jans DA. Dengue virus RNA polymerase NS5: a potential therapeutic target? Curr Drug Targets. 2006;7:1623–1638. doi: 10.2174/138945006779025383. [DOI] [PubMed] [Google Scholar]
- 19*.Malet H, Egloff M-P, Selisko B, Butcher RE, Wright PJ, Roberts M, Gruez A, Sulzenbacher G, Vonrhein C, Bricogne G, et al. Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J Biol Chem. 2007;282:10678–10689. doi: 10.1074/jbc.M607273200. This paper reports the first crystal structure of a flavivirus polymerase domain comprising the nuclear localization sequence; with a reverse genetics study of NS5-Pol and MTase interaction. [DOI] [PubMed] [Google Scholar]
- 20*.Hannemann H, Sung P-Y, Chiu H-C, Yousuf A, Bird J, Lim SP, Davidson AD. Serotype-specific differences in dengue virus non-structural protein 5 nuclear localization. J Biol Chem. 2013;288:22621–22635. doi: 10.1074/jbc.M113.481382. This study demonstrates the somewhat surprising fact that the percentage of NS5 going to the nucleus varies considerably between DENV serotypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawlinson SM, Pryor MJ, Wright PJ, Jans DA. CRM1-mediated nuclear export of dengue virus RNA polymerase NS5 modulates interleukin-8 induction and virus production. J Biol Chem. 2009;284:15589–15597. doi: 10.1074/jbc.M808271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar A, Bühler S, Selisko B, Davidson A, Mulder K, Canard B, Miller S, Bartenschlager R. Nuclear localization of dengue virus nonstructural protein 5 does not strictly correlate with efficient viral RNA replication and inhibition of type I interferon signaling. J Virol. 2013;87:4545–4557. doi: 10.1128/JVI.03083-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Surana P, Satchidanandam V, Nair DT. RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state. Nucleic Acids Res. 2014;42:2758–2773. doi: 10.1093/nar/gkt1106. Structures of JEV NS5-Pol stabilized or not by bound ATP and GTP are presented and analyzed exhaustively in comparison with existing NS5 and NS5-Pol structures of flaviviruses and other RdRp structures. Innovative activity assays are presented that propose a special role of GTP in de novo initiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Latour DR, Jekle A, Javanbakht H, Henningsen R, Gee P, Lee I, Tran P, Ren S, Kutach AK, Harris SF, et al. Biochemical characterization of the inhibition of the dengue virus RNA polymerase by beta-d-2’-ethynyl-7-deaza-adenosine triphosphate. Antiviral Res. 2010;87:213–222. doi: 10.1016/j.antiviral.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Iglesias NG, Filomatori CV, Gamarnik AV. The F1 motif of dengue virus polymerase NS5 is involved in promoter-dependent RNA synthesis. J Virol. 2011;85:5745–5756. doi: 10.1128/JVI.02343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim SP, Koh JHK, Seh CC, Liew CW, Davidson AD, Chua LS, Chandrasekaran R, Cornvik TC, Shi P-Y, Lescar J. A crystal structure of the dengue virus non-structural protein 5 (NS5) polymerase delineates interdomain amino acid residues that enhance its thermostability and de novo initiation activities. J Biol Chem. 2013;288:31105–31114. doi: 10.1074/jbc.M113.508606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem. 2001;276:39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- 28.Caillet-Saguy C, Lim SP, Shi P-Y, Lescar J, Bressanelli S. Polymerases of hepatitis C viruses and flaviviruses: structural and mechanistic insights and drug development. Antiviral Res. 2014;105:8–16. doi: 10.1016/j.antiviral.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 29**.Lu G, Gong P. Crystal Structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 2013;9:e1003549. doi: 10.1371/journal.ppat.1003549. The long-awaited first structure of a full-length Flavivirus NS5 (of JEV) revealed a hydrophobic interface and the MTase domain near entry of the RNA template tunnel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Yap TL, Xu T, Chen Y-L, Malet H, Egloff M-P, Canard B, Vasudevan SG, Lescar J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J Virol. 2007;81:4753–4765. doi: 10.1128/JVI.02283-06. The first crystal structure of a DENV polymerase domain as apoenzyme and bound to 3’dGTP with a detailed active site analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noble CG, Lim SP, Chen Y-L, Liew CW, Yap L, Lescar J, Shi P-Y. Conformational Flexibility of the Dengue Virus RNA-Dependent RNA Polymerase Revealed by a Complex with an Inhibitor. J Virol. 2013;87:5291–5295. doi: 10.1128/JVI.00045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potisopon S, Priet S, Selisko B, Canard B. Comparison of dengue virus and HCV: from impact on global health to their RNA-dependent RNA polymerase. Future Virol. 2014;9:53–67. [Google Scholar]
- 33.Garriga D, Ferrer-Orta C, Querol-Audí J, Oliva B, Verdaguer N. Role of motif B loop in allosteric regulation of RNA-dependent RNA polymerization activity. J Mol Biol. 2013;425:2279–2287. doi: 10.1016/j.jmb.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Smidansky ED, Maksimchuk KR, Lum D, Welch JL, Arnold JJ, Cameron CE, Boehr DD. Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure. 2012;20:1519–1527. doi: 10.1016/j.str.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verdaguer N, Ferrer-Orta C. Conformational changes in motif D of RdRPs as fidelity determinant. Structure. 2012;20:1448–1450. doi: 10.1016/j.str.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, Götte M, Konigsberg W, Cameron CE. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lang DM, Zemla AT, Zhou CLE. Highly similar structural frames link the template tunnel and NTP entry tunnel to the exterior surface in RNA-dependent RNA polymerases. Nucleic Acids Res. 2013;41:1464–1482. doi: 10.1093/nar/gks1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorbalenya AE, Pringle FM, Zeddam J-L, Luke BT, Cameron CE, Kalmakoff J, Hanzlik TN, Gordon KHJ, Ward VK. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol. 2002;324:47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Potisopon S, Priet S, Collet A, Decroly E, Canard B, Selisko B. The methytransferase domain of the dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain. Nucleic Acids Res. 2014 Sep 10; doi: 10.1093/nar/gku666. pii: gku666. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bussetta C, Choi KH. Dengue virus nonstructural protein 5 adopts multiple conformations in solution. Biochemistry (Mosc) 2012;51:5921–5931. doi: 10.1021/bi300406n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu L, Takeda K, Markoff L. Protein-protein interactions among West Nile non-structural proteins and transmembrane complex formation in mammalian cells. Virology. 2013;446:365–377. doi: 10.1016/j.virol.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Muller DA, Young PR. The flavivirus NS1 protein: molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antiviral Res. 2013;98:192–208. doi: 10.1016/j.antiviral.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 43.Xie X, Gayen S, Kang C, Yuan Z, Shi P-Y. Membrane topology and function of dengue virus NS2A protein. J Virol. 2013;87:4609–4622. doi: 10.1128/JVI.02424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nemésio H, Palomares-Jerez F, Villalaín J. NS4A and NS4B proteins from dengue virus: membranotropic regions. Biochim Biophys Acta. 2012;1818:2818–2830. doi: 10.1016/j.bbamem.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 45.Khromykh AA, Sedlak PL, Westaway EG. cis- and trans-acting elements in flavivirus RNA replication. J Virol. 2000;74:3253–3263. doi: 10.1128/jvi.74.7.3253-3263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller S, Kastner S, Krijnse-Locker J, Bühler S, Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem. 2007;282:8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 47.Shiryaev SA, Chernov AV, Aleshin AE, Shiryaeva TN, Strongin AY. NS4A regulates the ATPase activity of the NS3 helicase: a novel cofactor role of the non-structural protein NS4A from West Nile virus. J Gen Virol. 2009;90:2081–2085. doi: 10.1099/vir.0.012864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Umareddy I, Chao A, Sampath A, Gu F, Vasudevan SG. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J Gen Virol. 2006;87:2605–2614. doi: 10.1099/vir.0.81844-0. [DOI] [PubMed] [Google Scholar]
- 49.Li K, Phoo WW, Luo D. Functional interplay among the flavivirus NS3 protease, helicase, and cofactors. Virol Sin. 2014;29:74–85. doi: 10.1007/s12250-014-3438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lescar J, Luo D, Xu T, Sampath A, Lim SP, Canard B, Vasudevan SG. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from Dengue virus as a target. Antiviral Res. 2008;80:94–101. doi: 10.1016/j.antiviral.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 51.Johansson M, Brooks AJ, Jans DA, Vasudevan SG. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-beta and the viral helicase, NS3. J Gen Virol. 2001;82:735–745. doi: 10.1099/0022-1317-82-4-735. [DOI] [PubMed] [Google Scholar]
- 52**.Zou G, Chen Y-L, Dong H, Lim CC, Yap LJ, Yau YH, Shochat SG, Lescar J, Shi P-Y. Functional analysis of two cavities in flavivirus NS5 polymerase. J Biol Chem. 2011;286:14362–72. doi: 10.1074/jbc.M110.214189. A comprehensive mutational exploration of two cavities of DENV NS5-Pol using in vitro polymerase assays of full-length NS5 and viral replication assays. Cavity B is important for de novo initiation and is thus a candidate for rational drug design. Interestingly, it is also important for binding to NS3-Hel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi H, Takahashi C, Moreland NJ, Chang Y-T, Sawasaki T, Ryo A, Vasudevan SG, Suzuki Y, Yamamoto N. Establishment of a robust dengue virus NS3-NS5 binding assay for identification of protein-protein interaction inhibitors. Antiviral Res. 2012;96:305–314. doi: 10.1016/j.antiviral.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 54.Kroschewski H, Lim SP, Butcher RE, Yap TL, Lescar J, Wright PJ, Vasudevan SG, Davidson AD. Mutagenesis of the dengue virus type 2 NS5 methyltransferase domain. J Biol Chem. 2008;283:19410–19421. doi: 10.1074/jbc.M800613200. [DOI] [PubMed] [Google Scholar]
- 55.Cui T, Sugrue RJ, Xu Q, Lee AK, Chan YC, Fu J. Recombinant dengue virus type 1 NS3 protein exhibits specific viral RNA binding and NTPase activity regulated by the NS5 protein. Virology. 1998;246:409–417. doi: 10.1006/viro.1998.9213. [DOI] [PubMed] [Google Scholar]
- 56.Yon C, Teramoto T, Mueller N, Phelan J, Ganesh VK, Murthy KHM, Padmanabhan R. Modulation of the nucleoside triphosphatase/RNA helicase and 5’-RNA triphosphatase activities of Dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J Biol Chem. 2005;280:27412–27419. doi: 10.1074/jbc.M501393200. [DOI] [PubMed] [Google Scholar]
- 57.Carpp LN, Rogers RS, Moritz RL, Aitchison JD. Quantitative proteomic analysis of host-virus interactions reveals a role for GBF1 in dengue infection. Mol Cell Proteomics MCP. 2014 doi: 10.1074/mcp.M114.038984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qing M, Yang F, Zhang B, Zou G, Robida JM, Yuan Z, Tang H, Shi P-Y. Cyclosporine inhibits flavivirus replication through blocking the interaction between host cyclophilins and viral NS5 protein. Antimicrob Agents Chemother. 2009;53:3226–3235. doi: 10.1128/AAC.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhattacharya D, Mayuri, Best SM, Perera R, Kuhn RJ, Striker R. Protein kinase G phosphorylates mosquito-borne flavivirus NS5. J Virol. 2009;83:9195–9205. doi: 10.1128/JVI.00271-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keating JA, Bhattacharya D, Lim P-Y, Falk S, Weisblum B, Bernard KA, Sharma M, Kuhn RJ, Striker R. West Nile virus methyltransferase domain interacts with protein kinase G. Virol J. 2013;10:242. doi: 10.1186/1743-422X-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye J, Chen Z, Zhang B, Miao H, Zohaib A, Xu Q, Chen H, Cao S. Heat shock protein 70 is associated with replicase complex of Japanese encephalitis virus and positively regulates viral genome replication. PloS One. 2013;8:e75188. doi: 10.1371/journal.pone.0075188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.García Cordero J, León Juárez M, González-Y-Merchand JA, Cedillo Barrón L, Gutiérrez Castañeda B. Caveolin-1 in lipid rafts interacts with dengue virus NS3 during polyprotein processing and replication in HMEC-1 cells. PloS One. 2014;9:e90704. doi: 10.1371/journal.pone.0090704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blackwell JL, Brinton MA. Translation elongation factor-1 alpha interacts with the 3’ stem-loop region of West Nile virus genomic RNA. J Virol. 1997;71:6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davis WG, Blackwell JL, Shi P-Y, Brinton MA. Interaction between the cellular protein eEF1A and the 3’-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J Virol. 2007;81:10172–10187. doi: 10.1128/JVI.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Agis-Juárez RA, Galván I, Medina F, Daikoku T, Padmanabhan R, Ludert JE, del Angel RM. Polypyrimidine tract-binding protein is relocated to the cytoplasm and is required during dengue virus infection in Vero cells. J Gen Virol. 2009;90:2893–2901. doi: 10.1099/vir.0.013433-0. [DOI] [PubMed] [Google Scholar]
- 67.De Nova-Ocampo M, Villegas-Sepúlveda N, del Angel RM. Translation elongation factor-1alpha, La, and PTB interact with the 3’ untranslated region of dengue 4 virus RNA. Virology. 2002;295:337–347. doi: 10.1006/viro.2002.1407. [DOI] [PubMed] [Google Scholar]
- 68.Jiang L, Yao H, Duan X, Lu X, Liu Y. Polypyrimidine tract-binding protein influences negative strand RNA synthesis of dengue virus. Biochem Biophys Res Commun. 2009;385:187–192. doi: 10.1016/j.bbrc.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gomila RC, Martin GW, Gehrke L. NF90 binds the dengue virus RNA 3’ terminus and is a positive regulator of dengue virus replication. PloS One. 2011;6:e16687. doi: 10.1371/journal.pone.0016687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2–3’ UTR structures. RNA Biol. 2011;8:1173–1186. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lei Y, Huang Y, Zhang H, Yu L, Zhang M, Dayton A. Functional interaction between cellular p100 and the dengue virus 3’ UTR. J Gen Virol. 2011;92:796–806. doi: 10.1099/vir.0.028597-0. [DOI] [PubMed] [Google Scholar]
- 72.Paranjape SM, Harris E. Y box-binding protein-1 binds to the dengue virus 3’-untranslated region and mediates antiviral effects. J Biol Chem. 2007;282:30497–30508. doi: 10.1074/jbc.M705755200. [DOI] [PubMed] [Google Scholar]
- 73.Polacek C, Friebe P, Harris E. Poly(A)-binding protein binds to the non-polyadenylated 3’ untranslated region of dengue virus and modulates translation efficiency. J Gen Virol. 2009;90:687–692. doi: 10.1099/vir.0.007021-0. [DOI] [PubMed] [Google Scholar]
- 74.Paranjape SM, Harris E. Control of dengue virus translation and replication. Curr Top Microbiol Immunol. 2010;338:15–34. doi: 10.1007/978-3-642-02215-9_2. [DOI] [PubMed] [Google Scholar]
- 75**.Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. A 5’ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006;20:2238–2249. doi: 10.1101/gad.1444206. This elegant landmark study demonstrates that binding of NS5 to the 5’ end of the DENV genome enables priming of negative-strand RNA synthesis at the 3’ end of the RNA via circularization of the viral genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Filomatori CV, Iglesias NG, Villordo SM, Alvarez DE, Gamarnik AV. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J Biol Chem. 2011;286:6929–6939. doi: 10.1074/jbc.M110.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alvarez DE, Lodeiro MF, Ludueña SJ, Pietrasanta LI, Gamarnik AV. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shurtleff AC, Beasley DW, Chen JJ, Ni H, Suderman MT, Wang H, Xu R, Wang E, Weaver SC, Watts DM, et al. Genetic variation in the 3’ non-coding region of dengue viruses. Virology. 2001;281:75–87. doi: 10.1006/viro.2000.0748. [DOI] [PubMed] [Google Scholar]
- 79.Alvarez DE, Filomatori CV, Gamarnik AV. Functional analysis of dengue virus cyclization sequences located at the 5’ and 3’UTRs. Virology. 2008;375:223–235. doi: 10.1016/j.virol.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 80.Polacek C, Foley JE, Harris E. Conformational changes in the solution structure of the dengue virus 5’ end in the presence and absence of the 3’ untranslated region. J Virol. 2009;83:1161–1166. doi: 10.1128/JVI.01362-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Friebe P, Shi P-Y, Harris E. The 5’ and 3’ downstream AUG region elements are required for mosquito-borne flavivirus RNA replication. J Virol. 2011;85:1900–1905. doi: 10.1128/JVI.02037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Friebe P, Harris E. Interplay of RNA elements in the dengue virus 5’ and 3’ ends required for viral RNA replication. J Virol. 2010;84:6103–6118. doi: 10.1128/JVI.02042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Friebe P, Peña J, Pohl MOF, Harris E. Composition of the sequence downstream of the dengue virus 5’ cyclization sequence (dCS) affects viral RNA replication. Virology. 2012;422:346–356. doi: 10.1016/j.virol.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Clyde K, Barrera J, Harris E. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology. 2008;379:314–323. doi: 10.1016/j.virol.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85*.Clyde K, Harris E. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J Virol. 2006;80:2170–2182. doi: 10.1128/JVI.80.5.2170-2182.2006. This study characterizes the first RNA structural element identified within the Flavivirus coding region and shows that it plays two distinct roles -- in translation start site selection and in RNA replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Groat-Carmona AM, Orozco S, Friebe P, Payne A, Kramer L, Harris E. A novel coding-region RNA element modulates infectious dengue virus particle production in both mammalian and mosquito cells and regulates viral replication in Aedes aegypti mosquitoes. Virology. 2012;432:511–526. doi: 10.1016/j.virol.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gebhard LG, Filomatori CV, Gamarnik AV. Functional RNA elements in the dengue virus genome. Viruses. 2011;3:1739–1756. doi: 10.3390/v3091739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tajima S, Nukui Y, Takasaki T, Kurane I. Characterization of the variable region in the 3’ non-translated region of dengue type 1 virus. J Gen Virol. 2007;88:2214–2222. doi: 10.1099/vir.0.82661-0. [DOI] [PubMed] [Google Scholar]
- 89*.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi P-Y, Hall RA, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. This paper presents the characterization of the Flavivirus sfRNA and demonstrates a role in pathogenesis. [DOI] [PubMed] [Google Scholar]
- 90*.Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, Goldbach RW, Vlak JM, Kohl A, Khromykh AA, Pijlman GP. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J Virol. 2012;86:13486–13500. doi: 10.1128/JVI.01104-12. This study demonstrates that WNV subgenomic flavivirus RNA (sfRNA) efficiently suppressed RNAi pathways by inhibiting in vitro cleavage of double-stranded RNA by Dicer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schuessler A, Funk A, Lazear HM, Cooper DA, Torres S, Daffis S, Jha BK, Kumagai Y, Takeuchi O, Hertzog P, et al. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J Virol. 2012;86:5708–5718. doi: 10.1128/JVI.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. Role of RNA structures present at the 3’UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005;339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 93.Manzano M, Reichert ED, Polo S, Falgout B, Kasprzak W, Shapiro BA, Padmanabhan R. Identification of cis-acting elements in the 3’-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. J Biol Chem. 2011;286:22521–22534. doi: 10.1074/jbc.M111.234302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sztuba-Solinska J, Teramoto T, Rausch JW, Shapiro BA, Padmanabhan R, Le Grice SFJ. Structural complexity of Dengue virus untranslated regions: cis-acting RNA motifs and pseudoknot interactions modulating functionality of the viral genome. Nucleic Acids Res. 2013;41:5075–5089. doi: 10.1093/nar/gkt203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Markoff L. 5’- and 3’-noncoding regions in flavivirus RNA. Adv Virus Res. 2003;59:177–228. doi: 10.1016/S0065-3527(03)59006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zeng L, Falgout B, Markoff L. Identification of specific nucleotide sequences within the conserved 3’-SL in the dengue type 2 virus genome required for replication. J Virol. 1998;72:7510–7522. doi: 10.1128/jvi.72.9.7510-7522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Holden KL, Stein DA, Pierson TC, Ahmed AA, Clyde K, Iversen PL, Harris E. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3’ stem-loop structure. Virology. 2006;344:439–452. doi: 10.1016/j.virol.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 98.Tilgner M, Deas TS, Shi P-Y. The flavivirus-conserved penta-nucleotide in the 3’ stem-loop of the West Nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology. 2005;331:375–386. doi: 10.1016/j.virol.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 99.Villordo SM, Gamarnik AV. Differential RNA sequence requirement for dengue virus replication in mosquito and mammalian cells. J Virol. 2013;87:9365–9372. doi: 10.1128/JVI.00567-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100*.Villordo SM, Alvarez DE, Gamarnik AV. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA NY N. 2010;16:2325–2335. doi: 10.1261/rna.2120410. Functional analysis of RNA sequences at its best: formation of a small hairpin controls the linearity of the genome and plays a key role in the balance between linear and circular forms of the DENV genome. [DOI] [PMC free article] [PubMed] [Google Scholar]