Abstract
Campylobacter rectus is an understudied oral bacterium that contributes to periodontitis. Processes that contribute to the disease-causing capabilities of pathogens, such as chemotaxis, are largely unknown in C. rectus. The aim of this study was to better understand C. rectus chemotaxis, by examining the C. rectus genome for the presence of a cheY gene. CheY proteins play a part in chemotaxis by acting as two-component response regulators. Significantly, CheY proteins from several pathogens, including the related species Campylobacter jejuni, have been shown to contribute to bacterial virulence. Degenerate PCR, RT-PCR, sequence analyses, and structural modeling showed that C. rectus encodes a gene (Cr-CheY) which shares significant homology with previously characterized CheY proteins. Functional studies of a recombinant form of the protein supports a likely role of Cr-CheY in C. rectus chemotaxis. Cr-CheY is the first CheY characterized from the oral campylobacters.
Keywords: Campylobacter rectus, periodontitis, cheY, chemotaxis
INTRODUCTION
Periodontitis is an infectious disease of the supportive tissue of the teeth (Philstrom et al., 2005). It infects 10% of the population making it a relatively common infectious disease (Teng et al., 2002). Approximately 13% of patients develop severe forms of periodontitis, which, may result in tooth loss and systemic complications including an increased risk of pregnancy complication (Philstrom et al., 2005; Teng et al., 2002).
Campylobacter rectus is a Gram-negative, motile, oral bacterium that has been implicated as a cause of periodontitis (Columbo et al., 2006; Dzink et al., 1985; Lai et al., 1992). Women with periodontitis are seven times more likely to experience preterm labor than their healthy counterparts (Bobetsis et al., 2006; Offenbacher et al., 1996). Serological data has implied that C. rectus plays a role in the preterm labors of mothers with periodontitis (Madianos et al., 2001). Additionally, a pregnant mouse model has shown the association of C. rectus with a decreased survival of pups (Offenbacher et al., 2005; Yeo et al., 2005). Although these studies have established C. rectus as an agent of periodontitis, the mechanisms of C. rectus pathogenesis are not well known.
The genes important to the pathogenesis of C. rectus have not been well characterized. A few studies have identified potential virulence factors in C. rectus, including toxin genes (csxA and csxB; csxC and csxD), a potential cell invasion gene (ciaB), and a surface array protein (crsA gene) thought to play a role in avoiding the host immune system (Braun et al., 1999; LaGier and Threadgill, 2008; Wang et al., 2000; LaGier and Threadgill, 2014). Within the campylobacters, virulence factors have been best characterized from the gastrointestinal pathogen Campylobacter jejuni (Young et al., 2007). In C. jejuni, chemotaxis is important to establishing disease, by allowing the bacteria to locate an optimal site for infection (Zautner et al., 2012). Among the chemotaxis proteins in C. jejuni, CheY is believed to act as a response regulator that interacts with flagellar motor proteins (specifically FliM, when phosphorylated by the signal-transducing histidine kinase CheA) to influence chemotaxis (Marchant et al., 2002). In addition, mutant C. jejuni lacking CheY do not efficiently colonize mice, and do not induce campylobacteriosis in ferret models (Yao et al., 1997).
In this study, a cheY gene from C. rectus was discovered and characterized (Cr-cheY) using a combination of bioinformatics, PCR, reverse transcriptase PCR (RT-PCR), and functional complementation experiments in Escherichia coli. Interestingly, C. rectus appears to be capable of moving within a host from one site to a distant site of infection; specifically, from the subcutaneous tissue to the placenta of pregnant mice (Offenbacher et al., 2005; Yeo et al., 2005). C. rectus is also capable of chemotactic behavior in vitro (Paster and Gibbons, 1986). Hence, chemotaxis may contribute to the systemic complications of C. rectus-related periodontitis. Additional characterization of C. rectus chemotaxis, including Cr-CheY, has the potential to identify new ways to disrupt this process.
MATERIALS AND METHODS
Bacterial strains and DNA isolation
Campylobacter curvus (ATCC 33273) and Campylobacter rectus strains 314 and 33238 (LaGier and Threadgill, 2008) were grown under standard anaerobic conditions. C. curvus, like C. rectus, is an oral bacteria associated with periodontitis (Muchach and Tanner, 2000). Bacteria were grown on tryptic soy blood agar with sodium formate (0.3%). Genomic DNA was isolated from bacterial pellets using CTAB (Ausubel et al., 1990). For the complementation studies, an isogenic mutant strain of E. coli lacking CheY function (RP5232, ΔcheY) and the corresponding wild-type strain (RP437, WT) were used (Bourret et al., 1990).
Degenerate PCR
Genomic DNA from C. rectus 314 and 33238 were PCR scanned for cheY genes using oligonucleotides 5'-GATATGCCTATYATNATGGTWAC-3' (CheY-DF) and 5'-TTAAWACTTGNGGMGTAAAAGG-3' (CheY-DR). CheY-DF and CheY-DR were designed to amplify the entire cheY gene (Wren et al., 1992). Each 50 µl PCR reaction contained 0.5 µM of each primer. The cycling conditions used were as follows: 94°C for 1 minute, 45°C for 1 minute, and 72°C for 1 minute (10 cycles); followed by 30 cycles of 94°C for 1 minute, 50°C for 1 minute, and 72°C for 1 minute. Amplicons were purified (QIAquick gel extraction kit, Qiagen, USA) and cloned for sequencing into pCR2.1-TOPO (TOPO-TA kit, Invitrogen, USA).
RNA isolation and reverse transcription PCR
RNA was isolated from bacterial pellets using a QIAgen miniprep kit (Qiagen, USA). The RNA was then used for RT-PCR using the Access RT-PCR kit (Promega, USA). Primers CheY-RTF1 (5'-ATCCTAGGTTTATATCG-3') and CheY-RTR1 (5'-TAGGTAAAGCCGAGGTC-3') were used for amplification of cheY transcripts.
Recombinant plasmid construction and expression in E. coli
The complete open reading frame of C. rectus cheY (strain 314, GenBank Accession #EU119866) was cloned (BamHI and HindIII sites) into E. coli expression vector pQE80L (Invitrogen, USA) to create pQE80L-Cr-CheY. The engineered plasmid contains the complete cheY gene with an in-frame, N-terminus histidine tag. The fidelity of the construct was checked by DNA sequencing. The plasmid pQE80L-Cr-CheY was transformed into E. coli RP5232 and RP437. The expression of pQE80L-Cr-CheY was confirmed by SDS-PAGE analysis (Ausubel et al., 1990), after induction of the protein by the addition of 1mM IPTG to overnight cultures of E. coli growing in Luria broth; supplemented with 100 µg/mL ampicillin.
Swarming motility assays
E. coli swarming assays were carried out as previously described (Bourret et al., 1990; Li et al., 2006). Briefly, the WT (RP5232) and ΔcheY (RP437) strains transformed with pQE80L-Cr-CheY were grown overnight at 37°C in LB supplemented with 100 µg/mL ampicillin. A 5 µL sample (106 cells) of each culture was place in the center of motility agar plates (Li et al., 2006) supplemented with 100 µg/mL ampicillin and 0.0 or 10.0 µM IPTG. In pQE80L-Cr-CheY, the expression of the recombinant protein is under the control of an IPTG-inducible promoter. The plates were incubated at 30°C. After 24 hours, swarming was assessed by measuring the distance moved (radii measurements) by E. coli cells from the original site of inoculation. Three independent experiments were performed and the swarming radii were averaged. Each experiment contained replicate plates for each condition tested. From each plate, three radii measurements were taken.
PCR-based site directed mutagenesis
The QuikChange II site directed mutagenesis kit (Stratagene, USA) was used to change a residue important to the predicted functionality of Cr-CheY. The kit was used according to the manufacturer’s protocols, and the change in residue was from a conserved aspartate to an alanine. The PCR template used was pQE80L-Cr-CheY, and CheY-MUTF1 (5' - TGCTTATCACCAACTGGA -3') and CheY-MUTR1 (5' -GCATGTTCCAGTCGGTGATA-3') were used as primers to introduce the point mutation. The intended change was confirmed by sequencing the resultant plasmid, pQE80L-Cr-CheY-MUT. This plasmid was subjected to motility assays after transformation into E. coli RP5232 and RP437.
Sequence analysis and bioinformatics
DNA chromatographs were edited and assembled using Vector NTI software (Invitrogen, USA). Obtained sequences were then used as templates for sequence homology searches using BLAST. Searches were performed using the default parameters for BLASTx (Altschul et al., 1990). Alignments were generated using T-Coffee (Thompson et al., 1994). The resulting alignment was used to generate a WebLogo (Crooks et al., 2004).
I-TASSER was used to predict a tertiary structure for Cr-CheY (Zhang, 2008).
RESULTS
Identification and expression of C. rectus cheY
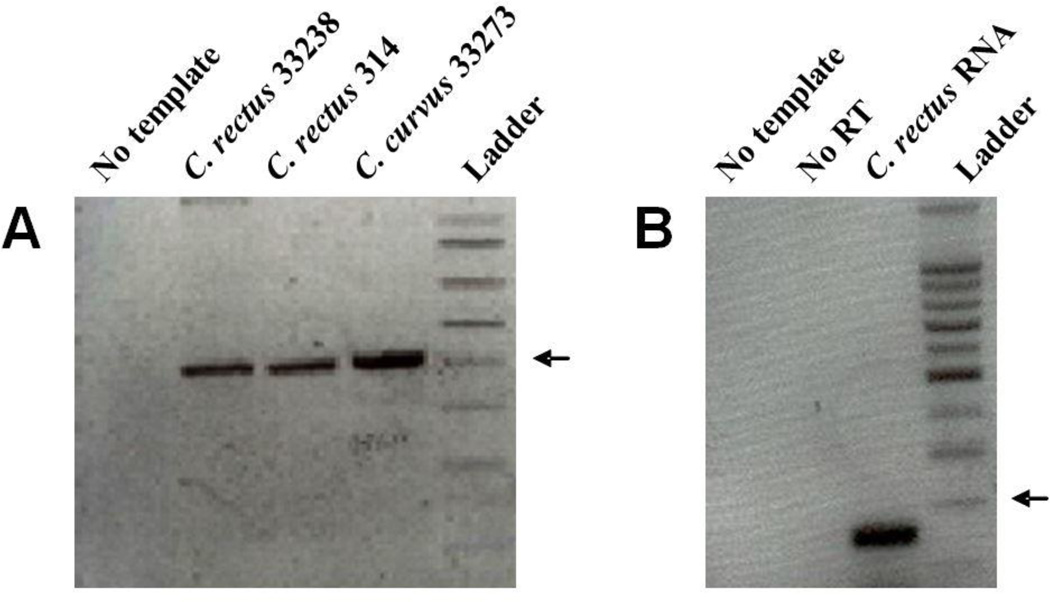
Degenerate PCR was used to amplify a potential cheY gene from C. rectus genomic DNA. PCR yielded an amplicon of the expected size (366 base pairs, Figure 1A). Amplicons of the correct size were also detected from C. rectus 33238 and Campylobacter curvus 33273. Cr-CheY from strain 314, which is the most widely studied strain of C. rectus, was used in all subsequent experiments. RT-PCR showed the expression of Cr-cheY (Figure 1B), with an expected amplicon of 151 base pairs generated from isolated C. rectus RNA. DNA sequencing of the degenerate PCR amplicon revealed a Cr-cheY ORF of 366 base pairs. The 366 base pair sequence is available at NCBI (#EU119867.1). The open reading frame (ORF) included a start codon (GTG), stop codon (TAA), and translation of the ORF reveals a protein of 121 amino acids, with a predicted molecular weight of 13.7 Kilodaltons.
Figure 1.
A. Agarose gel of C. rectus or Campylobacter curvus DNA amplified by degenerate PCR. An arrow marks 400 base pairs. B. Agarose gel of C. rectus strain 314 cDNA amplified with RT-PCR. No template = No template control, No RT = No Reverse-Transcriptase control. An arrow marks 200 base pairs.
In silico characterization of Cr-CheY
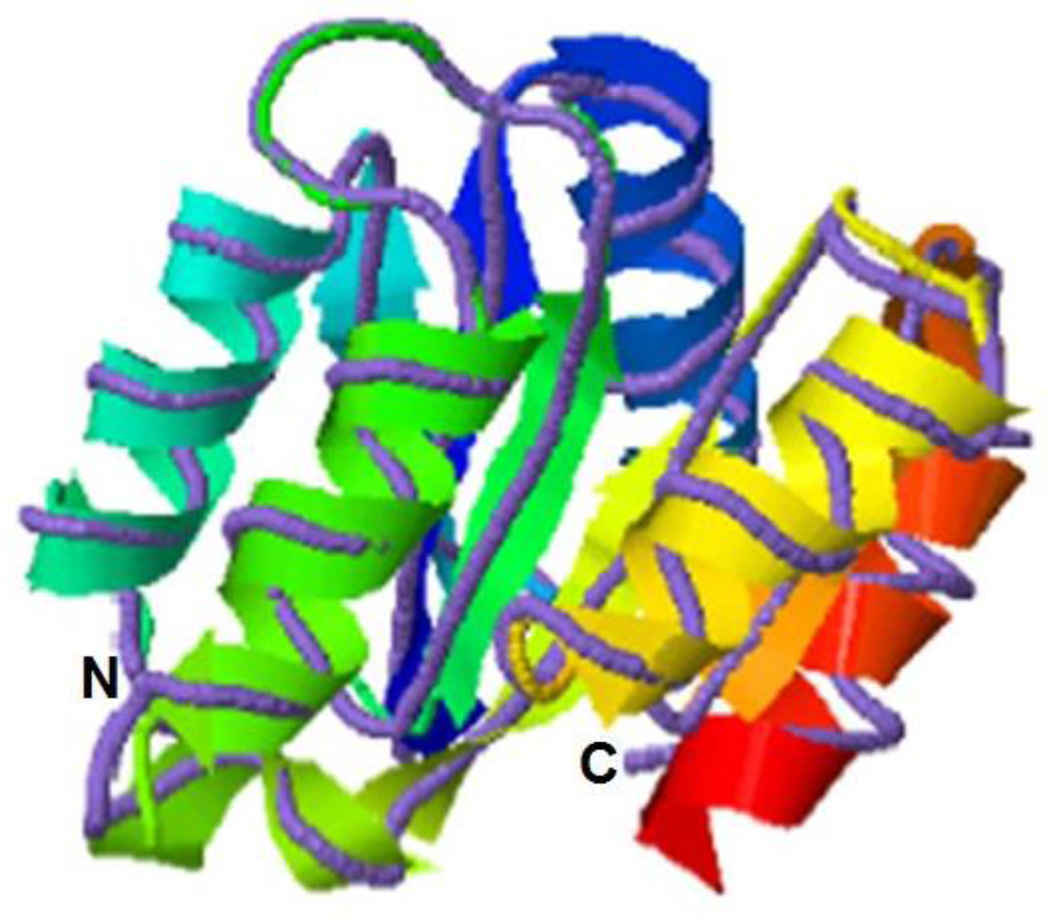
BLASTx analysis of Cr-CheY revealed a C. rectus gene sharing significant sequence identity and similarity to CheY proteins from campylobacters, including C. jejuni (NCBI #YP_179249.1; 88% identity, and E-value of 5e-71). A WebLogo (Figure 2, based on a T-Coffee alignment) of Cr-CheY showed homology with CheY proteins, including the presence of conserved residues that contribute to the function of CheY as a response regulator. In the model bacterium E. coli, CheY interacts with the flagellar motor switch protein FliM (when phosphorylated by the histidine kinase CheA) to influence chemotaxis (Bourret et al., 1990). Cr-CheY contains a conserved site for phosphorylation (Zautner et al., 2012) by CheA (aspartate, D53 in Cr-CheY; D71 in the WebLogo), and residues involved with binding FliM and CheA (Figure 2). The CheY from the related species C. jejuni contains the same conserved sites (Yao et al., 1997); and preliminary BLASTx analysis has identified potential FliM (NCBI #WP_002945945.1) and CheA (WP_002943755.1) homologs from C. rectus. The structure of Cr-CheY protein was modeled using the I-TASSER server. As shown in Figure 3, the predicted tertiary structure of Cr-CheY shares significant homology with the known structure of a CheY protein from the related epsilon-proteobacterium Helicobacter pylori (protein database #3H1F, http://www.rcsb.org/pdb/home/home.do); including a conserved arrangement and number of alpha helices and beta strands.
Figure 2.
WebLogo of Cr-CheY and related CheY proteins. The identity and position of conserved amino acids within Cr-CheY are indicated. A score of 4.0 bits indicates 100% conservation of the indicated amino acid among the 24 CheY proteins originally aligned with T-Coffee (Materials and Methods). The circled aspartate (D) is phosphorylated by CheA in related CheY proteins. The residues surrounded by rectangles are involved with binding FliM. Residues involved with binding CheA are surrounded by triangles.
Figure 3.
Model of Cr-CheY structure. The predicted tertiary structure of Cr-CheY (ribbon diagram) shares conserved structural features with the solved structure of a CheY from Helicobacter pylori (purple line, protein database #3H1F). The structural similarities are significant according to I-TASSER, with a TM-score of 0.939 (Zhang, 2008). A TM-score of greater than 0.5 is significant. The N and C-termini are indicated.
Effect of expressing Cr-CheY on swarming motility of E. coli
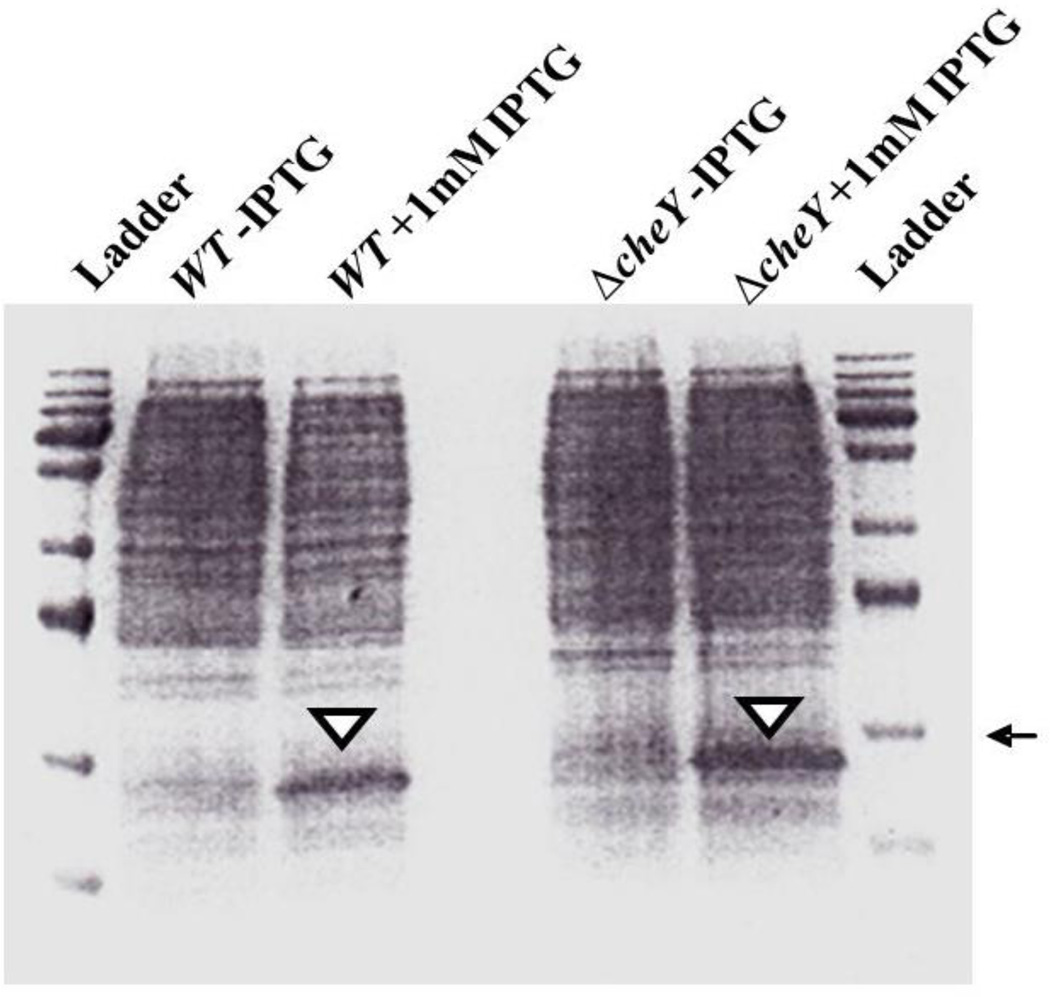
The potential function of Cr-CheY was further characterized by functional complementation studies in E. coli (Materials and Methods). The expression of recombinant Cr-CheY in E. coli was confirmed by SDS-PAGE analysis. Following induction of Cr-CheY expression with IPTG, a protein of the expected size (16 Kilodaltons; predicted molecular weight of Cr-CheY plus the weight of the N-terminus histidine tag) was visualized in both WT (RP437) and mutant E. coli (ΔcheY) lacking CheY (RP5232, Figure 4).
Figure 4.
Coomassie-stained SDS-PAGE gel (15%) showing expression of recombinant Cr-CheY (from C. rectus) in E. coli. Equal amounts of cell lysate were loaded per well. The arrow indicates 17 Kilodaltons and the arrowheads denote IPTG-induced Cr-CheY at the expected molecular weight (16 Kilodaltons). Ladder = EZ-Run protein standard (Fisher Scientific, USA); WT = wild type E. coli; ΔcheY = isogenic mutant E. coli.
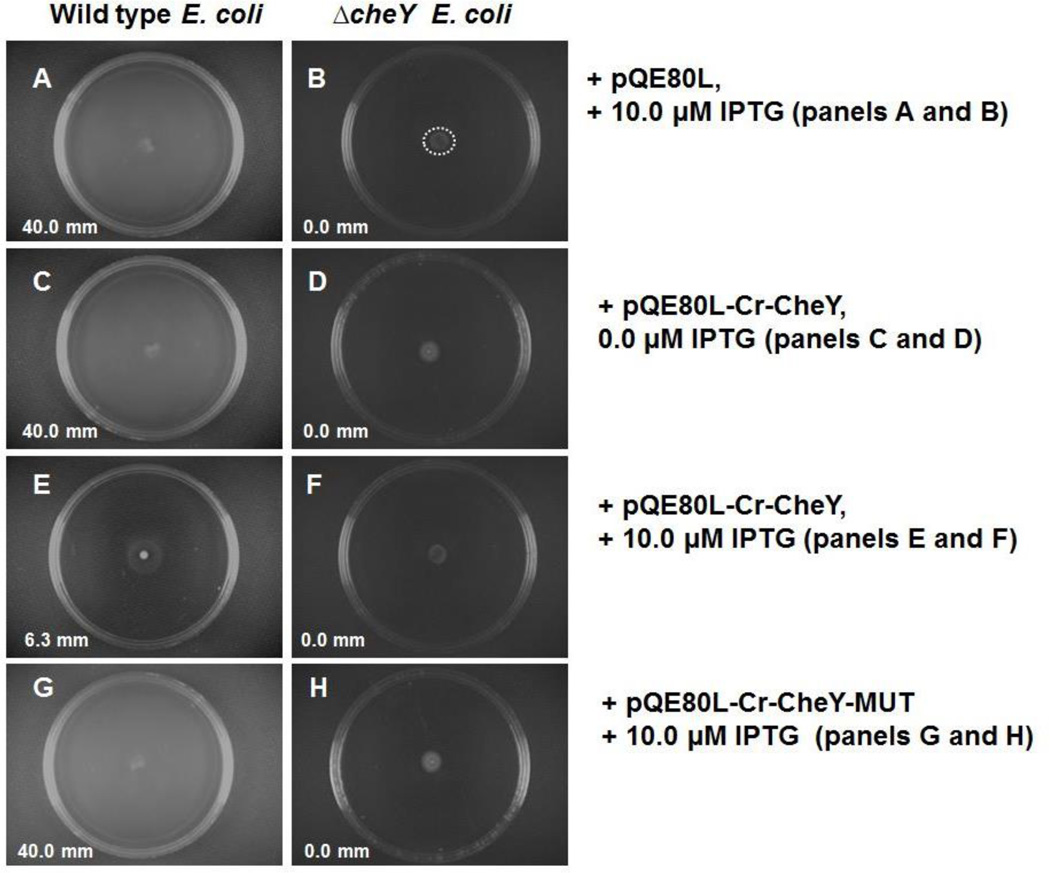
The effect of Cr-CheY in WT and ΔcheY E. coli was assessed by observing changes in swarming motility that requires CheY functionality, and thus requires chemotaxis. While WT E. coli display swarming motility, ΔcheY E. coli lack swarming motility after 24 hours of growth (Figure 5, panels A, B, C, D). Cr-CheY expressed in ΔcheY E. coli did not restore swarming, and therefore does not directly replace the function of E. coli cheY (Figure 5, panel F). However, the expression of Cr-CheY in WT E. coli inhibited the swarming ability of WT cells (Figure 5, panel E). The mean radii swarming distance decreased from 40.0 mm to 6.3 mm (Figure 5, compare panels C and E). The inhibition of swarming in WT E. coli was not due to differences in growth rates. More specifically, tracking the optical density (600 nanometer readings) of WT E. coli carrying Cr-CheY in the presence or absence of 10 µM IPTG showed no difference in growth rate. After 24 hours of incubation, the mean optical densities for samples minus or plus IPTG were 1.81 and 1.80, respectively. As expected, a mutated version of Cr-CheY; in which a key aspartate residue (D53) has been changed to an alanine, restores 100% of swarming motility when expressed in WT E. coli (Figure 5, panel G). The expression of mutated Cr-CheY in WT E. coli was confirmed by SDS-PAGE (data not shown).
Figure 5.
Swarming motility of WT and ΔcheY E. coli following IPTG-induced expression of Cr-CheY. Panels A-H show representative swarming motility agar plates. Panels denoted as +pQE80L (A and B) are E. coli transformed with plasmid alone (lacking the cr-cheY gene from C. rectus) and exposed to 10.0 µM IPTG. The number presented in each panel is the mean of swarming radii collected from three independent experiments. Growth at the original inoculation site, as exemplified by the dotted circle in panel B is not considered swarming motility (Bourret et al., 1990; Li et al., 2006). In panels A, C, and G, the entire plate surface is coated by swarming E. coli.
DISCUSSION
In this study a cheY gene from C. rectus, an understudied periodontal pathogen, was identified and characterized. Degenerate PCR, RT-PCR and bioinformatics (Figures 1, 2, and 3) supported the notion that C. rectus, which is motile and capable of chemotaxis (Paster and Gibbons, 1986), expresses a gene (Cr-CheY) that shares significant homology with known CheY proteins. E. coli strains without CheY lack swarming motility, which is dependent on chemotaxis (Bourett et al., 1990). To begin to characterize the function of Cr-CheY, a recombinant version of the protein was expressed in an E. coli strain lacking CheY. Although Cr-CheY did not complement the function of E. coli (Figure 5, panel F), the expression of Cr-CheY in wild-type E. coli inhibited swarming motility (Figure 5, panel E). The lack of complementation may be due to insufficient functional similarity. In E. coli, chemotaxis is dependent on CheY being phosphorylated by a histidine kinase, CheA (Bourret et al., 1990). The mechanism by which Cr-CheY inhibits E. coli swarming is unknown. However, the same effect has been observed in E. coli complementation experiments using CheY proteins from Leptospira interrogans and Helicobacter pylori (Li et al., 2006; Foynes et al., 2000). Interestingly, a mutated version of Cr-CheY expressed in wild-type E. coli, which lacks an aspartate residue that is phosphorylated during E. coli chemotaxis, does not inhibit swarming (Figure 5, panel G). This observation supports speculation that Cr-CheY expressed in wild-type E. coli competes with E. coli CheY for phosphates from CheA; and thus inhibits swarming (Foynes et al., 2000; Pittman et al., 2001). Overall, the ability of Cr-CheY to inhibit swarming of E. coli suggests that Cr-CheY likely plays a role in C. rectus chemotaxis.
Chemotaxis, or directed movement in response to chemical stimulants, has been shown to play a role in the disease-causing capabilities of many bacterial pathogens including C. jejuni, Helicobacter pylori, Vibrio cholerae, and Listeria monocytogenes (Yao et al., 1997; Foynes et al., 2000; Butler and Camili 2004; Dons et al., 2004). The protein network in model bacteria that function to direct chemotaxis includes methyl-accepting chemoreceptors (MCPs) that sense changes in extracellular chemical stimulant levels, and cytoplasmic proteins (e.g., cheY, cheA, and cheW) which transmit signals from the MCPs to the flagellum via a series of phosphotransfers (Baker et al., 2006). Although this study focused on CheY, preliminary analysis of the draft C. rectus genome suggests that C. rectus contains several genes, including MCPs and phosphotransfer proteins, known to play a role in chemotaxis (https://img.jgi.doe.gov/cgi-bin/w/main.cgi?section=KeggMap&page=keggMap&map_id=map02030&gene_oid=644263128&myimg=0). In addition, this study is the first effort to characterize a cheY gene from any of the oral campylobacters.
In the related pathogen C. jejuni, cells lacking CheY do not swarm, colonize mice, and fail to induce disease in ferrets (Yao et al., 1997). C. rectus appears to be capable of moving within a host from one site to a distant site of infection; from the subcutaneous tissue to the placenta of pregnant mice (Offenbacher et al., 2005; Yeo et al., 2005). C. rectus is also capable of chemotaxis in vitro (Paster and Gibbons, 1986). Most significantly, C. rectus is attracted to a potential energy source, formate (Rams et al., 1993), which is also found in periodontal pockets (Paster and Gibbons, 1986). Hence, chemotaxis may contribute to both the systemic complications of C. rectus-related periodontitis and assist the bacterium in colonizing subgingival pockets during the development of periodontal disease. Additional studies of C. rectus chemotaxis have the potential to discover new approaches for disrupting this process.
ACKNOWLEDGEMENTS
Michael J. LaGier was supported in part by FGCU Grant #09101. This study was also supported in part by the NSF-POWRE MCB-9973861 to Deborah S. Threadgill.
Contributor Information
Michael J. LaGier, Department of Biology, Grand View University, Des Moines, Iowa, USA
Ihor Bilokopytov, Department of Biological Sciences, Florida Gulf Coast University, Fort Myers, Florida, USA.
Bradley Cockerill, Department of Biological Sciences, Florida Gulf Coast University, Fort Myers, Florida, USA.
Deborah S. Threadgill, Department of Veterinary Pathology, Texas A&M University, College Station, Texas, USA
REFERENCES
- 1.Altschul S, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F, et al. Current protocols in molecular biology. New York: John Wiley and Sons; 1990. [Google Scholar]
- 3.Baker, et al. Signal transduction in bacterial chemotaxis. BioEssays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 4.Bobetsis YA, Barros SP, Offenbacher S. Exploring the relationship between periodontal disease and pregnancy complications. J Am Dent Assoc. 2006;137:7S–13S. doi: 10.14219/jada.archive.2006.0403. [DOI] [PubMed] [Google Scholar]
- 5.Bourret RB, et al. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natal Acad Sci USA. 1990;87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun M, et al. Cloning and characterization of two bistructural S-layer-RTX proteins from Campylobacter rectus. J Bacteriol. 1999;181:2501–2506. doi: 10.1128/jb.181.8.2501-2506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler SM, Camili A. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc Natl Acad Sci USA. 2004;101:5018–5023. doi: 10.1073/pnas.0308052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colombo AV, et al. Identification of oral bacteria associated with crevicular epithelial cells from chronic periodontitis lesions. J Med Microbiol. 2006;55:609–615. doi: 10.1099/jmm.0.46417-0. [DOI] [PubMed] [Google Scholar]
- 9.Crooks GE, et al. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dons L, et al. Role of flagellin and the cheA/cheY system of Listeria monocytogenes in host cell invasion and virulence. Infect Immun. 2004;72:3237–3244. doi: 10.1128/IAI.72.6.3237-3244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dzink JL, et al. Gram negative species associated with active destructive periodontal lesions. J Clin Periodontol. 1985;5:648–659. doi: 10.1111/j.1600-051x.1985.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 12.Foynes S, et al. Helicobacter pylori possesses two cheY response regulators and a histidine kinase regulator, cheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect Immun. 2000;68:2016–2023. doi: 10.1128/iai.68.4.2016-2023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LaGier MJ, Threadgill DS. Identification of novel genes in the oral pathogen Campylobacter rectus. Oral Microbiol Immunol. 2008;23:406–412. doi: 10.1111/j.1399-302X.2008.00443.x. 2008. [DOI] [PubMed] [Google Scholar]
- 14.LaGier MJ, Threadgill DS. Identification and characterization of an invasion antigen B gene from the oral pathogen Campylobacter rectus. Indian J Microbiol. 2014;54:33–40. doi: 10.1007/s12088-013-0406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai CH, et al. Wolinella recta in adult gingivitis and periodontitis. J Periodontal Res. 1992;27:8–14. doi: 10.1111/j.1600-0765.1992.tb02079.x. [DOI] [PubMed] [Google Scholar]
- 16.Li ZH, et al. Characterization of the cheY genes from Leptospira interrogens and their effects on the behavior of Escherichia coli. Biochem Biophys Res Commun. 2006;345:858–866. doi: 10.1016/j.bbrc.2006.04.159. [DOI] [PubMed] [Google Scholar]
- 17.Madianos PN, et al. Maternal periodontitis and prematurity Part II: Maternal infection and fetal exposure. Ann Periodontol. 2001;6:175–182. doi: 10.1902/annals.2001.6.1.175. [DOI] [PubMed] [Google Scholar]
- 18.Marchant J, Wren B, Ketley J. Exploiting genome sequence: predictions for mechanisms of campylobacter chemotaxis. Trends Microbiol. 2002;10:155–159. doi: 10.1016/s0966-842x(02)02323-5. [DOI] [PubMed] [Google Scholar]
- 19.Mucach P, Tanner AC. Campylobacter species in health gingivitis and periodontitis. J Dent Res. 2000;79:785–792. doi: 10.1177/00220345000790021301. [DOI] [PubMed] [Google Scholar]
- 20.Offenbacher S, et al. Periodontal infection as a possible risk factor for preterm low birth weight. J Periodontol. 1996;67:1103–1113. doi: 10.1902/jop.1996.67.10s.1103. [DOI] [PubMed] [Google Scholar]
- 21.Offenbacher S, et al. Effects of maternal Campylobacter rectus infection on murine placenta fetal and neonatal survival and brain development. J Periodontol. 2005;76:2133–2143. doi: 10.1902/jop.2005.76.11-S.2133. [DOI] [PubMed] [Google Scholar]
- 22.Paster BJ, Gibbons RJ. Chemotactic response to formate by Campylobacter concisus and its potential role in gingival colonization. Infect Immun. 1986;52:378–382. doi: 10.1128/iai.52.2.378-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 24.Pittman MS, et al. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for cheW and the three cheV paralogues, and evidence for cheV2 phosphorylation. Microbiol. 2001;147:2493–2504. doi: 10.1099/00221287-147-9-2493. [DOI] [PubMed] [Google Scholar]
- 25.Rams TE, et al. Campylobacter rectus in human periodontitis. Oral Microbiol Immunol. 1993;8:230–235. doi: 10.1111/j.1399-302x.1993.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 26.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng YT, et al. Periodontal health and systemic disorders. J Can Dent Assoc. 2002;68:188–192. [PubMed] [Google Scholar]
- 28.Wang B, Kraig E, Kolodrubetz D. Use of defined mutants to assess the role of the Campylobacter rectus S-layer in bacterium-epithelial cell interactions. Infect Immun. 2000;68:1465–1473. doi: 10.1128/iai.68.3.1465-1473.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wren BW, et al. Degenerate PCR primers for the amplification of fragments from genes encoding response regulators from a range of pathogenic bacteria. FEMS Microbiol Lett. 1992;78:287–291. doi: 10.1016/0378-1097(92)90042-m. [DOI] [PubMed] [Google Scholar]
- 30.Yao R, Burr DH, Guerry P. CheY-modulation of Campylobacter jejuni virulence. Mol Microbiol. 1997;23:1021–1031. doi: 10.1046/j.1365-2958.1997.2861650.x. [DOI] [PubMed] [Google Scholar]
- 31.Yeo A, et al. Campylobacter rectus mediates growth restriction in pregnant mice. J Periodontol. 2005;76:551–557. doi: 10.1902/jop.2005.76.4.551. [DOI] [PubMed] [Google Scholar]
- 32.Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol. 2007;5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 33.Zuatner AE, et al. Chemotaxis in Campylobacter jejuni. Eur J Microbiol (Bp) 2012;2:24–31. doi: 10.1556/EuJMI.2.2012.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]