Abstract
Aims
Nanoparticle conjugates have the potential for delivering siRNA, splice-shifting oligomers or nucleic acid vaccines, and can be applicable to anticancer therapeutics. This article compares tripartite conjugates with gold nanoparticles or synthetic methoxypoly(ethylene glycol)-block-polyamidoamine dendrimers.
Materials & methods
Interactions with model liposomes of a 1:1 molar ratio of tripalmitin:cholesterol or phospholipid:cholesterol were investigated by high-throughput absorbance, as well as fluorescence difference and cellular luminescence assays.
Results
Spectral differences and dynamic light-scattering spectroscopy shifts demonstrated the interaction of conjugates with liposomes. Biological activity was demonstrated by upregulation of gene expression via splice-shifting oligomers, delivery of anti-B-Raf siRNA in cultured human cancer cells or tuberculosis antigen 85B plasmid expression vector in a coculture model of antigen presentation.
Conclusion
The data suggests that gold nanoparticles and methoxypoly(ethylene glycol)-block-polyamidoamine dendrimer nanoconjugates may have potential for binding, stabilization and delivery of splice-shifting oligomers, siRNA and nucleic acid vaccines for preclinical trials.
Keywords: absorbance difference, B-Raf, fluorescence difference, gold nanoparticle, liposome, luciferase, mPEG–PAMAM, nanoconjugate, protamine
Absorbance difference spectroscopy (ADS) and fluorescence difference spectroscopy have been applied to characterize the cell-penetrating basic peptide, penatratin, in its penetration of a model liposome [1]. High-throughput screening (HTS) is one of the first steps in our drug-delivery research because it is essential to be able to characterize an interaction between nanoparticles and nucleic acids. Such an interaction would be evident in HTS techniques, including ADS and fluorescence difference spectroscopy, and the data can be presented in a quantitative manner, making them desirable techniques. The complexity of HTS can vary from innovative and complex technologies to ordinary technologies and simple assays. Despite its complexity, the use of HTS is advantageous, as it reduces the time and labor to perform assays and provides several data hits at one time [2]. Inthis study, ADS and fluorescence difference spectroscopy are applied for the first time in high throughput towards the delivery of splice-shifting oligomers (SSOs), siRNA, nucleic acid and protein-based biomolecular nanoconjugates.
The cell membrane is a limiting barrier that a nanoconjugate-bearing therapeutic nucleic acid must surmount for effective delivery. Gold nanoparticles (GNPs) are known to enter into the cellular membrane and are used as a model system because their physical and chemical properties can be easily manipulated [3]. GNPs can then bind nucleic acids, such as DNA or RNA, with the assistance of a delivery-enhancing vehicle such as protamine (Prot) or polyamidoamine (PAMAM) dendrimer [4]. The cationic peptide Prot is known for its powerful DNA condensation, cell membrane-penetrating and nuclear-trafficking properties [5,6]. Similar to Prot, PAMAM is a surface-active subnanomaterial, and approaches for delivering anticancer drugs using these polymeric molecules are of widespread interest. Physical characteristics of dendrimers, such as their monodispersity, water solubility, encapsulation ability and their large number of functionalizable peripheral groups, make them appropriate candidates for drug-delivery vehicles [7].
The process of PEGylation involves the modification of a protein or peptide by the linking of one or more poly(ethylene glycol) (PEG) chains. These molecules have several additional advantages as drug-delivery agents because they have a prolonged resistance in the body, decreased degradation by enzymes and a reduction of protein immunogenicity [8]. In this study, generation (G)3 methoxypoly(ethylene glycol) (mPEG)–PAMAM dendrimer is used as a delivery agent, along with Prot, as recent studies have shown that G3 mPEG–PAMAM can bind human serum albumin [9].
Multifunctional nanoparticles were recently tested in humans for the delivery of siRNA [10]. Delivery of nucleic acid vaccines or the dsRNA, polyinosine:polycytosine, have also shown promising preclinical immunogenic activity [11–13]. SSOs are another potential sequence-specific therapeutic strategy owing to their ability to correct errors in splicing, which are molecular-level aberrances underlying cancer and other human diseases [14,15].
The importance of this research and the underlying problem that was addressed is that nanoconjugates must interact with and penetrate the cell membrane. This study addresses this by HTS, as well as fluorescence difference and luminescence assays, to better enhance the assessment of interaction and delivery. To demonstrate preclinical potential, GNP conjugates with Prot or mPEG–PAMAM dendrimers were constructed to contain specific nucleic acid sequences of either tuberculosis antigen 85B (Ag85B) or B-Raf siRNA; two important molecular targets against tubercolosis and cancer, respectively [16,17].
Materials & methods
The anti-B-Raf siRNA and 705 SSO sequences used have previously been described [18] and were generously provided by the laboratories of R Juliano and R Kole at the University of North Carolina-Chapel Hill (NC, USA) and Lineberger Cancer Center core facility (NC, USA), respectively. SSOs and siRNA were dissolved in sterile water at 1 μM stock concentration prior to use. Prot, λ-phage DNA and tRNAphe used in the preparation of DNA:Prot or RNA:Prot (RNP) particle suspensions were obtained from Sigma-Aldrich (MO, USA) and dissolved in double-distilled deionized water at 0.5–1.0 mg/ml and stored frozen until use. Lipofectamine™ was obtained from Invitrogen (CA, USA). Bromophenol blue (BPB), fast green and Crocein Scarlet (acid red 73) were obtained from Matheson Coleman & Bell Laboratory Chemicals, Wilshire Chemical Co. Inc., in CA, USA. Amaranth red was obtained from Fisher Scientific (DE, USA). Trypan red and chromotrope 2R were obtained from Allied Chemical and Dye Corporation (NY, USA). All other dyes, including the alizarins, brilliant cresyl blue and Biebrich scarlet, were obtained from Allied Chemical and Dye Corporation. All dyes were used at a stock concentration of 1 mg/ml dissolved in double-distilled deionized water. GNPs were synthesized chemically or by pulsed laser deposition, by standard methodologiesin the laboratories of our collaborators A Wanekaya and K Ghosh in the Chemistry and Physics, Astronomy and Materials Science departments at Missouri State University (MO, USA). Characterization of these nanoparticles can be found in [19]. PEG-dendrimer was synthesized and purified as described in [20,21]. The synthesis of the PAMAM dendrimer block onto a mPEG-NH2 core consisted of two steps alternately repeated to achieve G4. In the first step, exhaustive Michael addition of methyl acrylate to the primary amine terminal groups of mPEG resulted in a tertiary amine branch point with methyl ester terminal groups. In the second step, reaction of methyl ester terminal groups with a large excess of ethylenediamine resulted in the primary amine terminal groups. In preparation of each generation, excess reactants (methyl acrylate and ethylenediamine) were removed under vacuum (p < 0.1 mmHg) after completion of the reactions. In addition, precipitation of each generation of the polymers in a large excess of ethyl ether resulted in separation of trace amounts of the reactants. Every generation of the polymers, including G4 mPEG–PAMAM, was analyzed by proton nuclear magnetic resonance spectroscopy. For example, the proton nuclear magnetic resonance spectrum of G4 mPEG–PAMAM in D2O was consistent with it being the only species present. Furthermore, the ratio of the terminal methoxy group of PEG (singlet, chemical shift [δ] = 3.38 ppm) to the protons next to -CH2CONH (multiplet, δ = 2.43 ppm) was 3:60 (or 1:20) as required by its structure. In addition, gel permeation chromatography analysis of the polymer further confirmed the purity of the product.
pDNA expression vector
Ag85 DNA was isolated and amplified from Mycobacterium tuberculosis strain H37Rv genomic DNA (kindly supplied by M Bernstein, University of Noth Carolina-Chapel Hill, Department of Microbiology and Immunology) using a PCR with primers that the authors designed. The resulting antigen–DNA PCR product was then cloned into a NT-GFP Fusion TOPO® Expression vector (Invitrogen) carrying ampicillin resistance. Subsequently, the vector construct was utilized to transform TOP10 Escherichia coli. Ampicillin resistance was used as a selective screen on lysogeny broth agar plates containing 100 μg/ml ampicillin to verify transformation of E. coli. Surviving colonies were selected after 24 h and utilized to inoculate 2 ml 100 μg/ml ampicillin lysogeny broth liquid cultures. Plasmid isolation was performed on 16 pelleted liquid culture colonies. Samples from each colony were digested overnight at 37°C with the restriction endonuclease, Eco0109I, incubated and analyzed with agarose gel electrophoresis to produce a restriction map to confirm the presence and orientation of Ag85 in the vector. Two colonies that met the predicted criteria were sequenced at the University of Missouri (MO, USA) Core facility. Electronic sequence results were then analyzed with NCBI BLAST. Subsequent sequencing of the construct, as well as a control self-ligated NT-GFP Fusion TOPO Expression vector without insert, confirmed the presence and proper orientation of the Ag85 DNA insert in the antigen-containing vector construct.
Niosome & dioleoylphosphatidylcholine:cholesterol liposomes
Liposomes were prepared by coevaporation (Buchi Heidolph Instruments, Schwabach, Germany) of a 1:1 molar ratio of tripalmitin and cholesterol (Chol) obtained from Eastman Kodak (NY, USA) or dioleoylphosphatidylcholine (DOPC; Sigma-Aldrich). Lipid solids were dissolved in approximately 1 ml of chloroform in a round-bottom flask and evaporated to a thin film with gentle rotation at room temperature using a house vacuum and water as a coolant. The thin layer was then suspended in 10 ml of phophate-buffered saline (PBS), the flask was vortexed vigorously for 30 s and sonicated for approximately 5 min prior to storage at 4°C or direct use in the high-throughput chromophoric shift, high-throughput absorbance difference or dynamic light-scattering (DLS) spectroscopy assays described below. Fluorescent liposome was obtained by substitution of 10 μl of 1 mg/ml BODIPY® (Invitrogen)–cholesteryl (Molecular Probes, OR, USA) dissolved in chloroform into the DOPC:Chol or tripalmitin:Chol preparation prior to coevaporation.
Chromophoric shift
These experiments were conducted essentially as previously described in [19]. Briefly, 100 μl of each dye was pipetted into a 96-well plate and 100 μl of either PAMAM dendrimer G3 1:100 dilution of 5–10%(w/v) stock or RNP (0.1–0.5 mg/ml) suspension in double-distilled deionized water was added relative to double-distilled H2O itself as a control. Chromophoric shifts were observed and quantified on a FLUOstar Optima BMG Labtech instrument (BMG LABTECH Inc., NC, USA) monitored at 485, 544 and 595 nm or a wide-range spectrum obtained from 1–2 μl of the dye:RNP mixtures on a Thermo Fisher Scientific UV/Vis nanodrop. This series of experiments was performed in triplicate.
Fluorimetric shift & microscopy
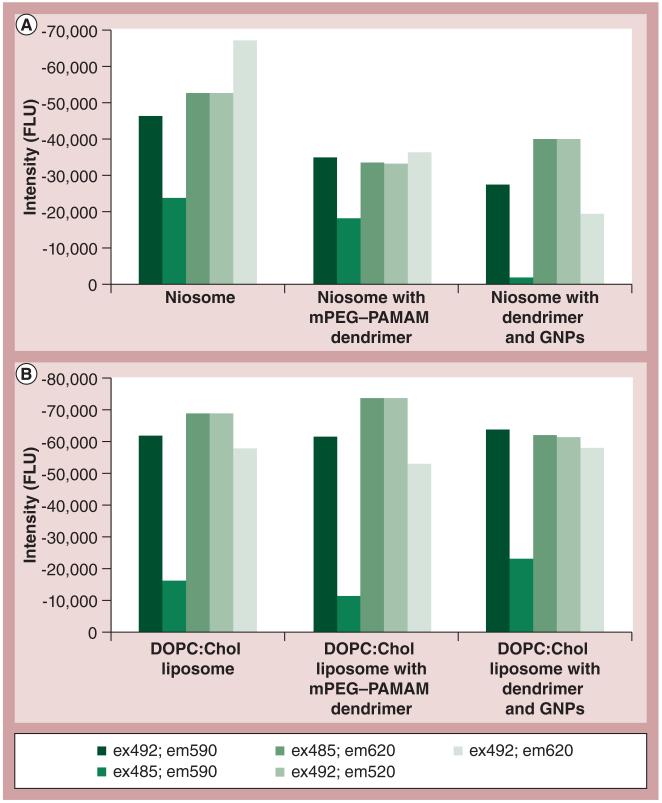
Fluorometric shifts were obtained similarly to the above, using uranine (1 mg/ml) incubated at a 1:1 (v/v) ratio with GNP, GNP–Prot or PEG-dendrimer. Excitation and emission wavelengths are indicated in Figure 1. Microscopy was performed on an Olympus BX60 Epifluorescence Microscope (Olympus, Center Valley, PA, USA) with built-in Digital Imaging Hardware and Software. Two BMG Labtech 96-well plates were used, labeled from A to H and 1 to 12, and used in comparison with two different types of liposome: niosome and DOPC:Chol. In A1 of the nine-well plate, there was 10 μl of uranine with a concentration of 1 mg/ml added to 90 μl of deionized water. In A2 of the 96-well plate, 10 μl of 1 mg/ml uranine was added to 50 μl of the corresponding liposome (niosome or DOPC:Chol) and 40 μl of deionized water. In A3, 10 μl of 1 mg/ml uranine was added to 50 μl of the corresponding liposome (niosome or DOPC:Chol), 20 μl of G4 mPEG–PAMAM dendrimer and 20 μl of deionized water. In A4, 10 μl of 1 mg/ml uranine was added to 50 μl of the corresponding liposome (niosome or DOPC:Chol), followed by the addition of 20 μl of G4 mPEG–PAMAM dendrimer and 20 μl of GNPs. The 96-well plates were then vortexed for 5 s each and incubated in a VWR Shelton Manufacturing Inc. (OR, USA) incubator at 37°C for 20 min. After incubation, the plates were inserted into the BMG Labtech manufactured FLUOstar OPTIMA Microplate Reader, and analyzed using OPTIMA 2.20 software in fluorescence intensity mode at various wavelengths: excitation 492 λ and emission 590 λ; excitation 485 λ and emission 590 λ; excitation 485 λ and emission 620 λ; excitation 492 λ and emission 520 λ and excitation 492 λ and emission 620 λ. All of the data were measured in fluorescence intensity units, then exported to Microsoft Excel 2007 and graphed accordingly. The A1 standard wells of uranine were subtracted out of the remaining columns of data.
Figure 1. Relative fluorescence difference.
(A) Niosome and (B) DOPC:Chol liposome in the presence or absence of mPEG–PAMAM dendrimer and GNPs. Samples were run at different emission and excitation wavelengths.
Chol: Cholestrol; DOPC: Dioleoylphosphatidylcholine; em: Emission; ex: Excitation; FLU: Fluorescence unit; GNP: Gold nanoparticle; mPEG–PAMAM: Methoxypoly(ethylene glycol)-block-polyamidoamine.
Dynamic laser light scatter shift & zeta potential
DLS was conducted on a Malvern Zetasizer Nano ZS-90 instrument. Prot or RNA solutions (0.1–1.0 mg/ml) were prepared in double-distilled deionized water and vortexed for approximately 10 s to dissolve and mix the solutions. An estimated 1:1 (v/v) Prot:RNA solution was prepared and 200 μl of Prot and 200 μl of RNA were added to low-volume cuvettes, provided by the manufacturer, to be vortexed for 10 s. A total of three independent DLS size measurements were taken, with each solution being vortexed for 10 s between each measurement. Various mass ratios were tested (1:2, 1:1, 2:1 and 4:1) concomitantly with the splice-shifting assay. Samples containing GNP, PEG-dendrimer conjugates with liposome were mixed at equal volume ratios.
UV/Vis spectroscopy
In some cases the concentrations of the biomolecules were read on a Perkin Elmer (MA, USA) Lambda 650 UV/Vis spectrometer. UV/Vis spectrometer measurements were taken and blanked from double-distilled deionized water from 200 to 800 nm wavelength. Background absorbance was corrected and the biomolecules’ interaction monitored by following the loss of Prot and PAMAM at 214–220 nm and DNA or RNA at 260–280 nm from the supernatant upon their sedimentation by microcentrifugation at 13,000 RPM for 1–2 min.
Gel electrophoresis
Gel analysis was performed essentially as previously described in [18]. Prot:RNA complexes were prepared by mixing equal volumes of 0.1–1.0 mg/ml Prot or PEG–G3 dendrimer with 0.1–1.0 mg/ml RNA to a final volume of 50–100 μl and 10–20 μl of the complexes or controls were loaded directly in the presence of 0.5–1.0 μl ethidium bromide. Loading samples were made by mixing 5 μl of Prot:RNA complex with 5 μl of the prescreened dyes and 4 μl of 40–60% sucrose. Samples were run through electrophoresis on a 2% agarose gel at 100 V for approximately 1 h. Images were taken on a Gel Logic 200 (Kodak, Eastman Chemical Company, TN, USA) imaging station.
Luciferase delivery
Fully confluent A375 cells were plated in six wells of a clear 96-well plate in quadruplicate, with 150 μl per well, and were then incubated overnight at 37°C in a NuAire (MN, USA) CO2 Water-Jacketed Incubator. The following day, Prot, GNPs and luciferase (Luc; stock solution from Promega Corporation, WI, USA) were made using 2 mg/ml Prot in double-distilled H2O, 1 mg/ml GNPs in double-distilled H2O and 0.1 mg/ml Luc in PBS, and were incubated at room temperature for 10 min. The first well remained A375-only and 10 μl of each solution was added in different combinations at equal volumes to each additional well in the order of: GNP-only; GNP + Prot; Prot only; GNP + Prot + Luc; and Luc-only. The plate was then incubated again overnight at 37°C in the CO2 incubator. The following day, the solutions were removed from the cells and the cells were washed twice with PBS. Then, 100 μl of 1× passive lysis buffer (Promega Corporation) was added from a stock concentration of 5× and the plate was vortexed for 20 s. The entire sample of the lysed cells was then transferred to a white OptiPlate 96-well plate and 150 μl of Luc substrate/buffer system was added to each well. The plate was incubated for 10 min at room temperature and then the luminescence was analyzed on the FLUOstar OPTIMA Microtiter Plate Reader.
HeLa pLuc 705 SSO & B-Raf siRNA systems
The complete protocol for this assay has been previously reported in [18] and performed in quadruplicate. Briefly, 103–104 HeLa-705cells were inoculated into 96-well plates (Nunclon, USA Scientific Inc., FL, USA) and allowed to attach overnight. Complexes of Prot:SSO were added to serum-free Opti-MEM media (100–200 μl) for 6–8 h and then the media was withdrawn, indicator-free 10% fetal bovine serum (FBS)/Dulbecco’s Modified Eagle medium was added and the cells allowed to grow undisturbed until the following day (36–48 h). Thereafter, the cells were washed twice with PBS and relative light units per well were quantified using Luc assay kit (Promega Corporation) on a TopCount1 Top Count NXT Perkin Elmer top-count machine (average of n = 4 wells) following manufacturers recommendations. For B-Raf, 7500 A375 cells per well were plated in a 24-well plate using culture medium containing FBS and antibiotics. The cells were left to adhere overnight. The following day each well was treated with nanoconjugates, as described above, or lipofectamine, per manufacturers’ recommendations (Invitrogen), to inhibit the production of B-Raf protein via siRNA. Note that as the nanoconjugates are triconjugates (GNP–Prot:siRNA), the siRNA stock concentration was approximately one-third less than that of the lipofectamine:siRNA (14.4 ng/μl). The western blot for B-Raf protein followed procedures similar to those that have been reported previously [18], whereby the crude A375 protein lysate was run on an SDS-PAGE gradient gel, blotted, incubated with primary antibody and visualized upon development with a horseradish peroxidase-conjugated secondary antibody. Only one putative B-Raf band was found in the correct size range for B-Raf proteinwhen examined next to a molecular weight ladder [16]. This experiment was standardized to total cell protein loaded via bicinchonic acid assay.
A375 tumorigenesis assay
Taken from a fully confluent T25 flask, A375 cells were trypsinized in 0.5 ml of trypsin and resuspended in 4 ml of Dulbecco’s Modified Eagle medium media (10% FBS and 1% penicillin/streptomycin). The cells were treated via a tumorigenesis assay with the following solutions: 20 μl of GNP only at a concentration of 1 mg/ml; 20 μl of dendrimer only at a concentration of 1 mg/ml; 40 μl of a B-Raf + dendrimer mix (B-Raf concentration was 28.6 ng/μl); and 40 μl of a GNP + Prot + B-Raf mix at a concentration of 347.9 ng/μl. The two mixed samples were incubated at room temperature for 20 min. Then, 200 μl of cells were plated in triplicate in a 24-well plate followed by a 6:4 ratio of an agar/media solution (Carolina nutrient agar [Carolina Biological Supply Company, NC, USA], item #821045 and Dulbecco’s Modified Eagle medium media with 10% FBS, 1% penicillin/streptomycin), in which 0.5 ml was added to each well. Finally, the cells were treated with the various mixtures, as described above, and the plate was incubated at 37°C in a NuAire CO2 Water-Jacketed Incubator for 14 days and the number of tumor spheroids formed per quadrant of each well were counted via microscopy.
Antigen presentation CD4 coculture assay
The assay involved detection and quantification, by ELISA, of IL-2 expression from CD4 T-hybridoma (DB-1) cells that had been added to human acute monocytic leukemia cell line (THP-1) cellstreated with nanoparticles containing DNA construct [22]. Firstly, THP-1 cells were incubated in a 96-well flat-bottom plate (1.5 × 105 cells/well) with 10 ng/ml phobol myristate acetate in infection medium for 24 h to fix the cells. Cells were washed once with infection medium and incubated with 100 IU/ml of recombinant human IFN-γ for a further 24 h. Cells were then washed twice with infection medium and nanoparticle preparations containing DNA were added to duplicate wells. After 48 h incubation DB-1 cells were added (2 × 105 cells/well) and plates were incubated for a further 72 h. Cells were separated by centrifugation and the supernatant was harvested and assayed by ELISA for IL-2 expression. Positive controls (three wells), where rAg85B protein was added instead of nanoparticles, were also prepared. Nanoparticles were prepared as follows: a DNA solution of approximately 150 μg/ml in water was saturated with Prot. A total of 150 μl of this was added to manganese (~1 mg or less needed), zinc (~1 mg or less needed) and gold (~100 μl of a 20 nM commercially prepared solution), respectively, and vortex mixed. A total of 20 μl volumes of these preparations were added to wells. The positive controls were prepared by adding 20 μl of an approximately 1 mg/ml solution of rAg85B protein to wells. The ELISA, to determine IL-2 expression from the DB-1 cells, was conducted using a BioSource™ mouse IL-2 kit (Invitrogen) with an eight-point calibration curve.
Results
HTS ADS
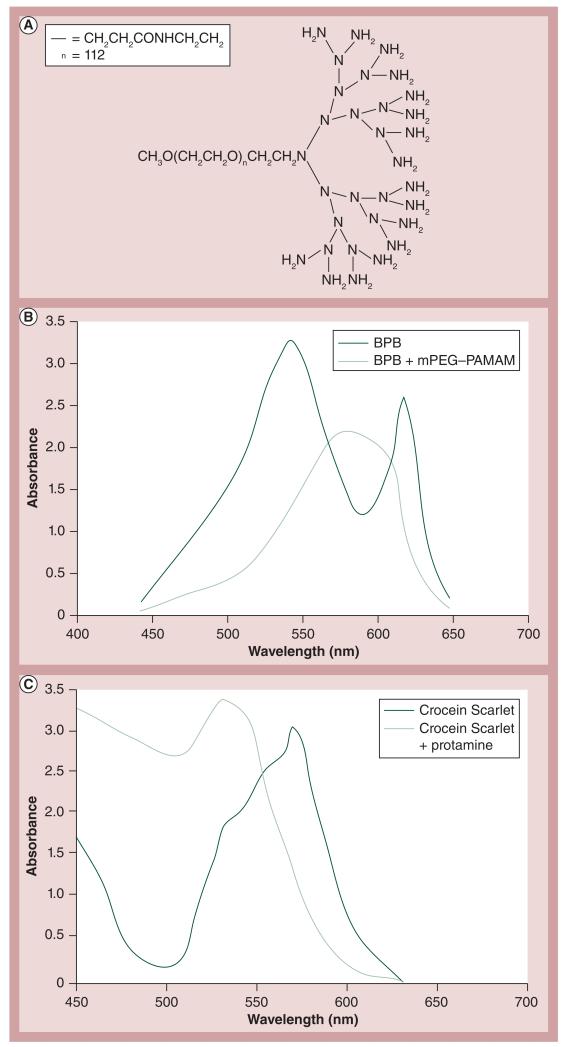
PAMAM’s interaction with coomassie blue has previously been described in [23] and mPEG–PAMAM has been stained by BPB and its interaction demonstrated by gel shift [24]. By means of absorbance shift, the authors wanted to demonstrate a similar interaction between G3 mPEG–PAMAM (whose structure is found in Figure 2A ) and BPB (Figure 2B) , and compare this absorbance shift to that of Crocein Scarlet and Prot (Figure 2C).
Figure 2. Methoxypoly(ethylene glycol)-block-polyamidoamine and protamine absorbance difference.
(A) mPEG–PAMAM dendrimer structure (generation 3). (B) Absorbance difference of BPB in the presence of generation 3 mPEG–PAMAM dendrimer. (C) Absorbance difference of Crocein Scarlet in the presence of protamine
BPB: Bromophenol blue; mPEG–PAMAM: Methoxypoly(ethylene glycol)-block-polyamidoamine.
ADS can also be applied in order to detect the interaction of Prot with nucleic acids, such as DNA or RNA. Several dyes were tested for their affect on the absorbance difference and to see if a positive or negative absorbance shift occurred when the dye was complexed with RNPs. Over 100 dyes were screened in a HTS assay and those in TABlE 1 were the ones with the best results. The dyes that underwent the greatest absorbance shift were BPB, quinoline yellow, alazarine yellow and uranine.
Table 1.
High-throughput screening absorbance difference of dyes in the presence H of RNA:protamine at wavelengths 485, 545 and 595 nm.
| Dye | Wavelength | ||
|---|---|---|---|
| 485 nm | 545 nm | 595 nm | |
| Amaranth red | Decrease | Decrease | Increase |
| Bromophenol blue | Decrease | Increase | Increase |
| Bromophenol purple | No change | Increase | Increase |
| Congo red | Decrease | Decrease | Increase |
| Biebrich scarlet | Decrease | Decrease | Increase |
| Alazarin yellow | Increase | Increase | Increase |
| Quinoline yellow | Increase | Increase | Increase |
| Uranine | Increase | Increase | Increase |
| Trypan red | No change | No change | Increase |
| Alazarin red | No change | Increase | Increase |
| Fast green | Increase | Decrease | Decrease |
| Crocein Scarlet | Decrease | Decrease | Increase |
Liposomes as model cell membranes
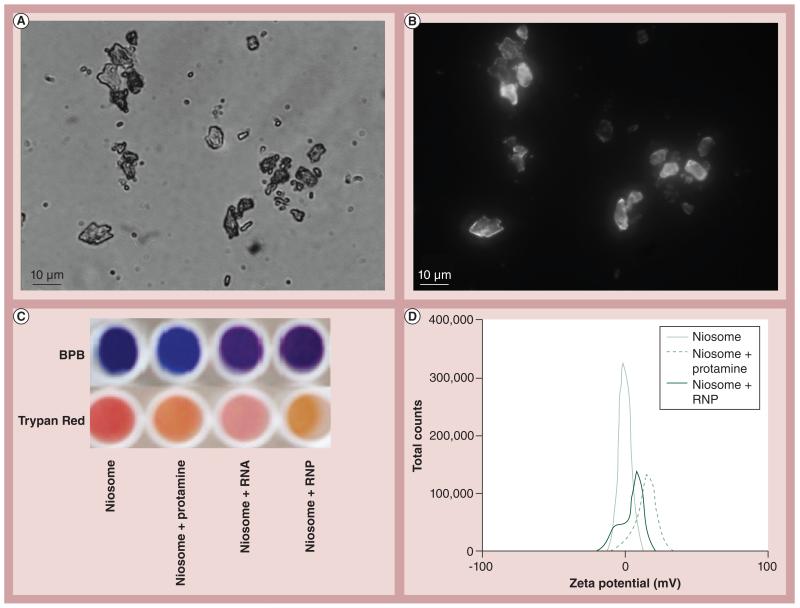
To apply the chromophoric and ADS assays to the nanoconjugate’s interaction with the membrane, two different model liposome membranes were created. A first-generation neutral liposome contained a 1:1 molar rio of tripalmitin and Chol (niosome). These were shown by light microscopy (Figure 3A) . Alternatively, as shown in Figure 3B , when a fluorescently derivitized version of Chol containing BODIPY conjugate is incorporated in the niosome, fluorescent images of the niosome could be obtained by fluorescence microscopy. Furthermore, DLS analysis of these niosomes show that they are homogeneous and 1–2 μm in size [DeLong RK, Kanomata M, Unpublished Data]. Chromophoric shift of the niosome in the presence of BPB or trypan red as a function of treatment with cell penetrating Prot, RNA or RNP is shown in Figure 3C . Further analysis of this interaction is characterized in Figure 3D , where a surface charge shift can be observed when the niosome is in the presence of Prot or RNPs.
Figure 3. Niosome as a model cell membrane.
Niosomes can be viewed by (A) light microscopy or (B) fluorescence microscopy when BODIPY®–cholesterol was substituted into the membrane. (C) Chromophoric shift of niosome in the presence of protamine, RNA or RNP nanoparticles. (D) Shift in the dynamic light-scattering spectroscopy zeta potential spectrum as a consequence of interaction with protamine or RNP with tripalmitin:cholesterol niosome.
BPB: Bromophenol blue; RNP: RNA:protamine.
Collectively, the previous data strongly suggest that the attachment of proteins and nucleic acids as nanoconjugates mediate binding, if not penetration of the model membranes. GNP surface activity and binding at the cell membrane is critical[20] and functionalization of the PAMAM dendrimer is also known to affect its interactions [21]. To further compare the effect dendrimer has on the efficiency of GNP surface activity, the authors tested the two liposomes by fluorescence difference, whereby the fluorescent uranine dye previously identified in the ADS assay was incubated with niosome or DOPC:Chol liposome in the presence or absence of mPEG–PAMAM dendrimer and GNP. These data are summarized in Figure 1.
The data show that the fluorescence difference was wavelength and liposome-dependent. Excitation and emission at 485 and 590 nm, respectively, revealed the most sensitive range, particularly when GNP was in the presence of mPEG–PAMAM and overall the DOPC:Chol liposome exhibited the greatest fluorescence difference.
Prot:RNA stoichiometry
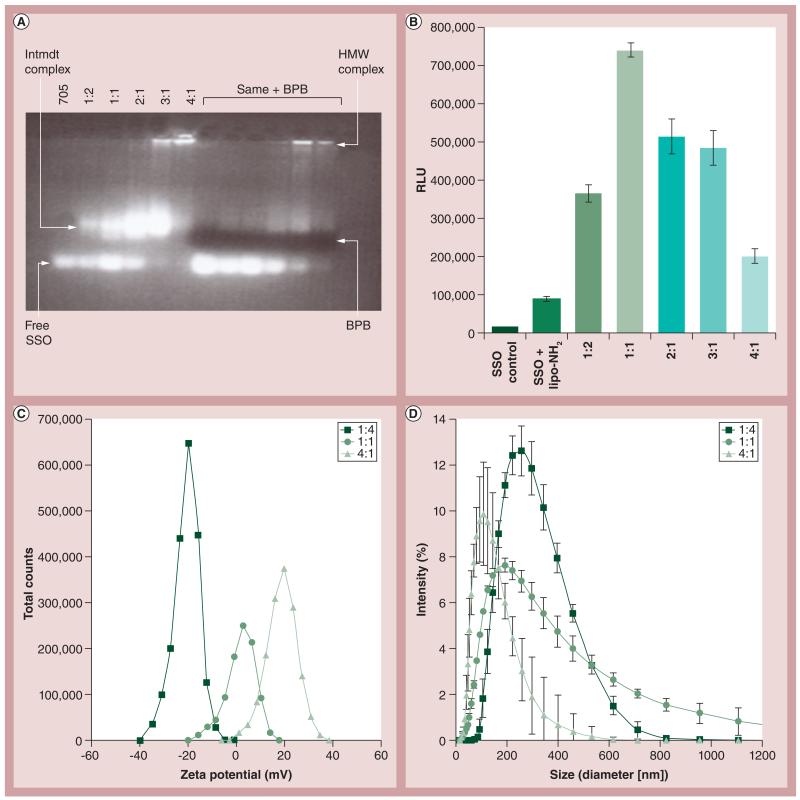
Previous data demonstrate that Prot interacts with nucleic acids and also has cell membrane interacting and penetrating capabilities. Several assays were executed in order to determine the proper ratio at which these interactions are most efficient. As shown in Figure 4 , similar to other nucleic acid-polymer nanoconjugates [25], the ratio of Prot:SSO impacted the gel shift pattern and splice-shifting, demonstrating that the optimal ratio between Prot:SSO for highest levels of activity is 1:1 molar ratio. Figure 4 also shows the expected effect of Prot:SSO stochiometry on the particle’s surface charge and size.
Figure 4. Effect of protamine:RNA ratio on physicochemical characteristics and splice-shifting oligomers activity.
(A) Gel shift demonstrating binding abilities of protamine:SSO at different ratios. (B) Effect of protamine:RNA stoichiometry on SSO upregulation of luciferase, in the HeLa pLuc splice-switching oligomer delivery system. The y-axis represents RLU/5000 seeded cells post 24-h treatment with protamine:SSO. (C) Charge and (D) size of particles also as a function of protamine:RNA stoichiometry (molar ratio). Error bars represent standard deviation values.
BPB: Bromophenol blue; HMW: High molecular weight; Intmdt: Intermediate band; lipo: Lipofectamine™; RLU: Relative luminescence units; SSO: Splice-shifting oligomer.
Nanoconjugates as delivery agents
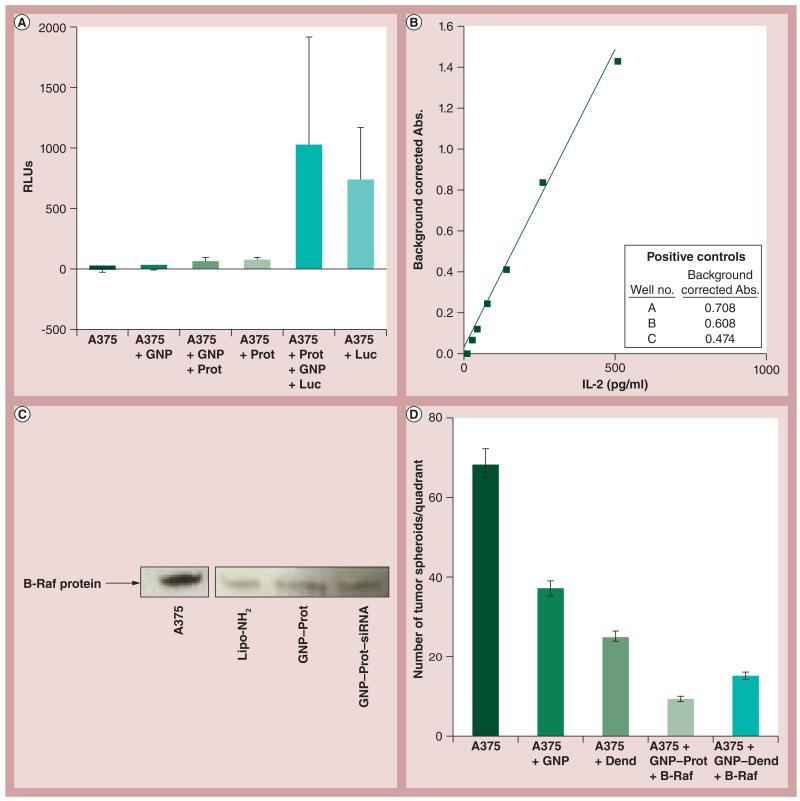
In order to assay for biological activity, several tripartite conjugates were formed, based on the data obtained thus far, and added to cellular systems. Tripartite conjugate complexes formed with GNP, cell-penetrating Prot and Luc protein enzyme were incubated with human melanoma cells (A375) grown in culture and assayed for luminescence. These data show cell-associated luminescence values several log orders higher than background (Figure 5A) . The authors also compared DNA nanoconjugates of either GNP or mPEG–PAMAM dendrimer for a pDNA expression vector of tuberculosis Ag85B. These data are shown in Figure 5B , in a presentation coculture assay system, in which effective antigen presentation is assessed by induction of IL-2 expression [22]. Finally, the author and colleagues tested the GNP–Prot conjugates for delivery of an anti-B-Raf siRNA into the same human melanoma cell line in culture (A375) (Figures 5C & 5D). Figure 5D shows results from a tumorgenicity assay in which A375 cells were grown in soft agar and treated with nanoconjugates. The data is consistent with that of Figure 5C , such that the tripartite conjugate of GNP–Prot–B-Raf siRNA or dendrimer complexes had a profound effect on the expression of oncogenic properties of the A375 cells.
Figure 5. Biological activity of tripartite nanoconjugates.
(A) The effect of protamine:GNPs incubated with luciferase protein, which were then exposed to A375 cells, and then the cells were rinsed. The cell-associated luminescence activity was quantified. (B) Tuberculosis antigen 85B presentation coculture assay positive controls. For details, see the experimental methods section in [22]. (C) Western blot of B-Raf expression in A375 cells after anti-B-Raf siRNA treatment with delivery by Lipofectamine™ (lipo-NH2)-siRNA, GNP–Prot control or the GNP–Prot–siRNA nanoconjugate. (D) Comparison of methoxypoly(ethylene glycol)-block-polyamidoamine dendrimer and GNP nanoconjugate effect on delivery of B-Raf siRNA. Represents the number of A375 tumor ‘spheroids’ formed after treatment of nanoconjugates. Error bars represent standard deviation values.
Abs.: Absorbance; B-Raf: B-Raf siRNA; Dend: Dendrimer; GNP: Gold nanoparticle; Luc: Luciferase; Prot: Protamine; RLU: Relative light unit.
Discussion
HTS, such as ADS, can play a major role in the characterization and detection of nanoconjugate surface interactions. A common method used in our research is the utilization of different dyes to observe these interactions, either via a shift in the absorbance or a color change. As seen in Figure 2, an absorbance shift does occur for both BPB and Crocein Scarlet in the presence of mPEG–PAMAM and Prot, respectively, within the wavelengths of 500–650 nm. The absorbance of BPB in the presence of mPEG–PAMAM increased, while the absorbance of Crocein Scarlet in the presence of Prot decreased, which can also be seen in TABlE 1 . To the best of our knowledge this represents the first time ADS has been applied to bionanomaterials.
Building on their knowledge of these nanoconjugate interactions, the author’s group wanted to see if they could characterize these surface and/or penetration properties with model cell membranes in the form of niosomes or liposomes. The chromophoric shift (Figure 3C) suggests an interaction of the niosome when it is in the presence of nanoconjugates, and the DLS data seen in Figure 3D show that Prot or RNPs had an effect on the surface charge of the niosome consistent with binding and/or penetration of the membrane. The niosome is a neutral biostructure and thus in the presence of Prot it is expected that it will bear a positive charge and will then become more negative with the Prot:RNA nanoconjugate. The data seen in Figure 3D are consistent with this hypothesis. A second-generation liposome containing DOPC and Chol at a 1:1 molar ratio was then created to better reflect a simple model membrane, whereby phospholipid rather than triacylglyceride would be more present [26].
The data shown in Figure 1 compares how effective the mPEG–PAMAM dendrimer is in increasing the surface activity of GNPs on both the niosome and the liposome. Data show that there are enhanced surface effects when the GNPs are in the presence of mPEG–PAMAM and that there is also an enhanced surface effect with the DOPC:Chol liposome, as compared with the niosome based on the fluorescence difference intensity between the two. The overall decrease in fluorescence compared with the free dye confirms uranine’s interaction with either liposome. However, there is a greater negative difference in the DOPC:Chol liposome, suggesting the possibility that the nanoconjugate complexes bind to or penetrate the membrane of the liposome and the uranine dye is no longer as readily available to give off fluorescence.
As previously stated, Prot is known for its powerful DNA condensation, cell membrane-penetrating and nuclear-trafficking properties. The latter activity of Prot would be critical for DNA vaccine expression and SSO activity, and can be applied when assaying for biological activity. We determined that a 1:1 molar ratio of Prot and SSO is the most effective for expression and stabilization (Figure 4). Figure 4A shows a gel shift that demonstrates the binding abilities of Prot:SSO at different ratios and Figure 4B displays that there is significantly more activity of SSO when the SSO:Prot ratio is 1:1. Figures 4C & 4D show the expected effect of Prot:SSO stochiometry on the particle’s surface charge and size. Since RNA bears a negative charge and Prot bears a positive charge, it is expected that when they are at equal concentrations, they will create a complex that has a neutral charge. Similar size comparisons are also shown. Knowing that this 1:1 ratio is the most effective for expression and stabilization, the authors were then able to apply tripartite nanoconjugates of Prot, GNPs and biomolecules to cellular systems and uncover various biological applications of these bionanoconjugates. The data shown in Figure 5A suggest interaction or penetration of the nanoconjugates bearing Luc onto or into the cells. In order to assess whether the Luc was intracellular or extracellular, a similar experiment was conducted in which lysed versus unlysed cells were compared, and results showed that there was an overall higher relative light unit value in lysed cells [Risor A, DeLong RK, Unpublished Data]. A modest amount of bioactivity was shown with the siRNA delivery system, where 40–50% B-Raf knock-down was achieved. While activity of the DNA expression vector was not shown, the delivery of the vector was demonstrated by observing the nanoparticles entering the assay system test cells (THP-1 cells). The potential for use of the coculture assay system was demonstrated by detection of a large amount of IL-2 expression in the positive controls(Figure 5B) . The last cellular system that was considered was that of the B-Raf protein that is expressed in A375 cells. The author and colleagues complexed anti-B-Raf siRNA with GNP and Prot. The B-Raf protein was significantly repressed when viewed on a western blot (Figure 5C). Figure 5D shows quantitative results obtained from a tumorigenesis assay in which tumor spheroids from A375 cells were counted after being treated with these conjuguates. Results suggest that the nanoconjugates bound or penetrated through the cells because there was a significant decrease in the presence of tumors consistent with, and to be expected for, reduced B-Raf protein expression shown previously.
Conclusion
In summary, chromophoric shift, absorbance and fluorescence differences have been shown for the firsttime in high throughput, as applied to protein and nucleic acid nanoconjugates, and their interactions with model cell membranes. It is important to characterize physicochemical attributes, which are likely to underscore biological activity of such nanoconjugates, especially as the preclinical development of these systems continue to evolve. Intracellular delivery of SSOs, siRNA and nucleic acid vaccines requires their binding and penetration through the cell membrane. In this study, the authors have presented a promising approach to probe these critical requirements as a function of the surface activity of the mPEG–PAMAM dendrimer, Prot and GNP nanoconjugates.
Future perspective
The fields of biomolecular science and biomedical nanotechnology are beginning to converge. Nanomaterials with biomolecules, such as protein and RNA bound to them, present the ultimate opportunity in being able to re-engineer cells; for example, the delivery of a functionally expressive RNA or an active protein enzyme into cells via a nanoparticle. Nanomaterials from almost every bioelement in the periodic chart are now being synthesized and when these are attached to biomolecules to create ‘nanoconjugates’, they have seemingly limitless opportunities as nanomedical agents. Each nanomaterial bound to a biomolecule potentially represents a new molecular entity and some of these are very likely to have powerful biological activity and specificity. During the next 5–10 years one can also speculate that GNPs, dendrimer and surface chemistries will also evolve, expand and be adapted to other nanomaterials and polymers. Thus, in conclusion, we are witnessing the emergence of a new field called biomolecular nanotechnology. Rapid advancement of such biomolecular nanoconjugates hinges critically on better characterization techniques, which this study has explored by high-throughput absorbance, fluorescence difference or cellular delivery.
Executive summary.
■A surface interaction occurs between RNA and proteins, and can be characterized via the absorbance difference between several different dyes.
■Liposomes act as model cell membranes and when in the presence of nanoconjugates, a surface interaction and/or penetrating mechanism can be observed.
■Methoxypoly(ethylene glycol)-block-polyamidoamine dendrimer increases the surface interaction efficiency of gold nanoparticles, especially in the presence of dioleylphosphatidylcholine:cholestrol liposomes.
■The optimal molar ratio of protamine and splice-shifting oligomers for effective splice-shifting oligomer activity is 1:1.
■Protamine and gold nanoparticle conjugates significantly improve the delivery mechanism of several proteins and nucleic acids, such as B-Raf siRNA and tubercolosis antigen 85B, which have significant future applications for drug therapeutics.
Acknowledgements
The authors wish to express their gratitude for the early preliminary work of former students Kindra Grozinger, Vince Butano, Renee Ehrenstrom, Lisa Cillessen and Stephanie Barber, leading the way to this report. They also wish to thank Adam Wanekaya for the synthesis and provision of gold nanoparticles utilized herein.
Footnotes
Financial & competing interests disclosure
RK DeLong and R Garrad are supported by an AREA/R15 grant from the National Cancer Institute entitled, ‘Anticancer RNA Nanoconjugates’ (1 R15 CA139390–01). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
■of interest
■■of considerable interest
- 1.Bárány-Wallje E, Keller S, Serowy S, et al. A critical reassessment of penetratin translocation across lipid membranes. Biophys. J. 2005;89(4):2513–2521. doi: 10.1529/biophysj.105.067694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shoemaker RH, Scudiero DA, Melillo G, et al. Application of high-throughput, molecular-targeted screening to anticancer drug discovery. Curr. Top. Med. Chem. 2002;2(3):229–246. doi: 10.2174/1568026023394317. [DOI] [PubMed] [Google Scholar]
- 3.Chithrani DB. Intracellular uptake, transport and processing of gold nanostructures. Mol. Membr. Biol. 2010;27(7):299–311. doi: 10.3109/09687688.2010.507787. [DOI] [PubMed] [Google Scholar]
- 4.Vijayanathan V, Thomas T, Thomas TJ. DNA nanoparticles and development of DNA delivery vehicles for gene therapy. Biochemistry. 2002;41(48):14085–14094. doi: 10.1021/bi0203987. ■ Describes the importance of polyamines and polymers as DNA delivery vehicles.
- 5.Brewer LR, Corzett M, Balhorn R. Protamine-induced condensation and decondensation of the same DNA molecule. Science. 1999;286(5437):120–123. doi: 10.1126/science.286.5437.120. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds F, Weissleder R, Josephson L. Protamine as an efficient membrane-translocating peptide. Bioconjug. Chem. 2005;16(5):1240–1245. doi: 10.1021/bc0501451. [DOI] [PubMed] [Google Scholar]
- 7.Cheng Y, Xu Z, Ma M, Xu T. Dendrimers as drug carriers: applications in different routes of drug administration. J. Pharm. Sci. 2008;97(1):123–143. doi: 10.1002/jps.21079. [DOI] [PubMed] [Google Scholar]
- 8.Veronese FM, Pasut G. PEGylation, successful approach to drug delivery. Drug Discov. Today. 2005;10(21):1451–1458. doi: 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- 9.Froehlich E, Mandeville JS, Jennings CJ, Segadhat-Herati R, Tajmir-Riahi HA. Dendrimers bind human serum albumin. J. Phys. Chem. B. 2009;113(19):6986–6993. doi: 10.1021/jp9011119. ■■ Supports the author’s reasoning for using generation 3 methoxypoly(ethylene glycol)-block-polyamidoamine dendrimer in their studies.
- 10.Davis ME, Zuckerman JE, Choi CHJ, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kutzler MA, Weiner DB. DNA vaccines: ready for the prime time. Nat. Gen. 2008;9(10):776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navabi H, Jasani B, Reece A, et al. A clinical grade poly I:C-analogue (Ampligen) promotes optimal DC maturation and Th1-type T cell responses of healthy donors and cancer patients in vitro. Vaccine. 2009;27(1):107–115. doi: 10.1016/j.vaccine.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 13.Liu MA. DNA vaccines: a historical perspective and view to the future. Immunol. Rev. 2011;239(1):62–84. doi: 10.1111/j.1600-065X.2010.00980.x. [DOI] [PubMed] [Google Scholar]
- 14.Bauman JA, Li SD, Yang A, Huang L, Kole R. Anti-tumor activity of splice-switching oligonucleotides. Nucleic Acids Res. 2010;38(22):8348–8356. doi: 10.1093/nar/gkq731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22(5):535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 16.Hingorani SR, Jacobetz MA, Robertson GP, Herlyn M, Tuveson DA. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res. 2003;63(17):5198–5202. [PubMed] [Google Scholar]
- 17.Aagaard C, Hoang TT, Izzo A, et al. Protection and polyfunctional T cells induced by Ag85B-TB10.4/IC31 against Mycobacterium tuberculosis is highly dependent on the antigen dose. PLoS ONE. 2009;4(6):e5930. doi: 10.1371/journal.pone.0005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLong RK, Akhtar U, Sallee M, et al. Characterization and performance of nucleic acid nanoparticles combined with protamine and gold. Biomaterials. 2009;30(32):6451–6459. doi: 10.1016/j.biomaterials.2009.07.067. ■■ Former research carried out by our group, which underlies the basis of the current study.
- 19.DeLong RK, Cillessen L, Reynolds C, et al. Biomolecular triconjugates formed between gold, protamine and nucleic acid: comparative characterization on the nanoscale. J. Nanotechnol. 2012 doi:10.1155/2012/954601. Epub ahead of print. [Google Scholar]
- 20.Arvizo RR, Miranda OR, Thompson MA, et al. Effect of nanoparticle surface charge at the plasma membrane and beyond. Nano Lett. 2010;10(7):2543–2548. doi: 10.1021/nl101140t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer J, Fleming K, Hammond PT. Synthesis and solution properties of new linear-dendritic diblock copolymers. Macromolecules. 1998;31:8757–8765. [Google Scholar]
- 22.Ramachandra L, Smialek JL, Shank SS, et al. Phagosomal processing of Mycobacterium tuberculosis Antigen85B is modulated independently of mycobacterial viability and phagosome maturation. Infect. Immun. 2005;73(2):1097–1105. doi: 10.1128/IAI.73.2.1097-1105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma A, Desai A, Ali R, Tomalia D. Polyacrylamide gel electrophoresis separation and detection of polyamidoamine dendrimers possessing various cores and terminal groups. J. Chromatogr. A. 2005;1081(2):238–244. doi: 10.1016/j.chroma.2005.05.074. [DOI] [PubMed] [Google Scholar]
- 24.Luo D, Haverstick K, Belcheva N, Han E, Saltzman WM. Poly(ethylene glycol)-conjugated PAMAM dendrimer for biocompatible, high-efficiency DNA delivery. Macromolecules. 2002;35(9):3456–3462. [Google Scholar]
- 25.Chittimalla C, Zammut-Italiano L, Zuber G, Behr JP. Monomolecular DNA nanoparticles for intravenous delivery of genes. J. Am. Chem. Soc. 2005;127(32):11436–11441. doi: 10.1021/ja0522332. [DOI] [PubMed] [Google Scholar]
- 26.Tombaccini D, Fallani A, Mugnai G, Ruggieri S. Lipid composition of Balb/c3T3, SV3T3, and concanavalin A-selected revertant cells grown in media containing lipid-depleted serum. J. Lip. Res. 1981;22(4):590–597. [PubMed] [Google Scholar]