Abstract
The mTOR complex 1 (mTORC1) protein kinase is a master growth regulator that responds to multiple environmental cues. Amino acids stimulate, in a Rag-, Ragulator-, and v-ATPase-dependent fashion, the translocation of mTORC1 to the lysosomal surface, where it interacts with its activator Rheb. Here, we identify SLC38A9, an uncharacterized protein with sequence similarity to amino acid transporters, as a lysosomal transmembrane protein that interacts with the Rag GTPases and Ragulator in an amino acid-sensitive fashion. SLC38A9 transports arginine with a high Km and loss of SLC38A9 represses mTORC1 activation by amino acids, particularly arginine. Overexpression of SLC38A9 or just its Ragulator-binding domain makes mTORC1 signaling insensitive to amino acid starvation but not to Rag activity. Thus, SLC38A9 functions upstream of the Rag GTPases and is an excellent candidate for being an arginine sensor for the mTORC1 pathway.
The mechanistic target of rapamycin complex 1 (mTORC1) protein kinase is a central controller of growth that responds to the nutritional status of the organism and is deregulated in several diseases, including cancer (1–3). Upon activation, mTORC1 promotes anabolic processes, including protein and lipid synthesis, and inhibits catabolic ones, such as autophagy (4). Environmental cues such as nutrients and growth factors regulate mTORC1, but how it senses and integrates these diverse inputs is unclear.
The Rag and Rheb GTPases have essential but distinct roles in mTORC1 pathway activation, with the Rags controlling the subcellular localization of mTORC1 and Rheb stimulating its kinase activity (5). Nutrients, particularly amino acids, activate the Rag GTPases, which then recruit mTORC1 to the lysosomal surface where they are concentrated (6, 7). Rheb also localizes to the lysosomal surface (6, 8–10) and, upon growth factor withdrawal, the tuberous sclerosis complex (TSC) tumor suppressor translocates there and inhibits mTORC1 by promoting GTP hydrolysis by Rheb (10). Thus, the Rag and Rheb inputs converge at the lysosome, forming two halves of a coincidence detector that ensures that mTORC1 activation occurs only when both are active.
There are four Rag GTPases in mammals and they form stable, obligate heterodimers consisting of RagA or RagB with RagC or RagD. RagA and RagB are highly similar and functionally redundant, as are RagC and RagD (1, 6). The function of each Rag within the heterodimer is poorly understood and their regulation is likely complex as many distinct factors play important roles. A lysosome-associated molecular machine containing the multi-subunit Ragulator and vacuolar ATPase (v-ATPase) complexes regulates the Rag GTPases and is necessary for mTORC1 activation by amino acids (11). Ragulator anchors the Rag GTPases to the lysosome and also has nucleotide exchange activity for RagA/B (12, 13), but the molecular function of the v-ATPase in the pathway is unknown. Two GTPase activating protein (GAP) complexes, which are both tumor suppressors, promote GTP hydrolysis by the Rag GTPases, with GATOR1 acting on RagA/B (14) and Folliculin-FNIP2 on RagC/D (15). Lastly, a distinct complex called GATOR2 negatively regulates GATOR1 through an unknown mechanism (14). Despite the identification of many proteins involved in signaling amino acid sufficiency to mTORC1, the actual amino acid sensors remain unknown.
SLC38A9 Interacts with Ragulator and the Rag GTPases
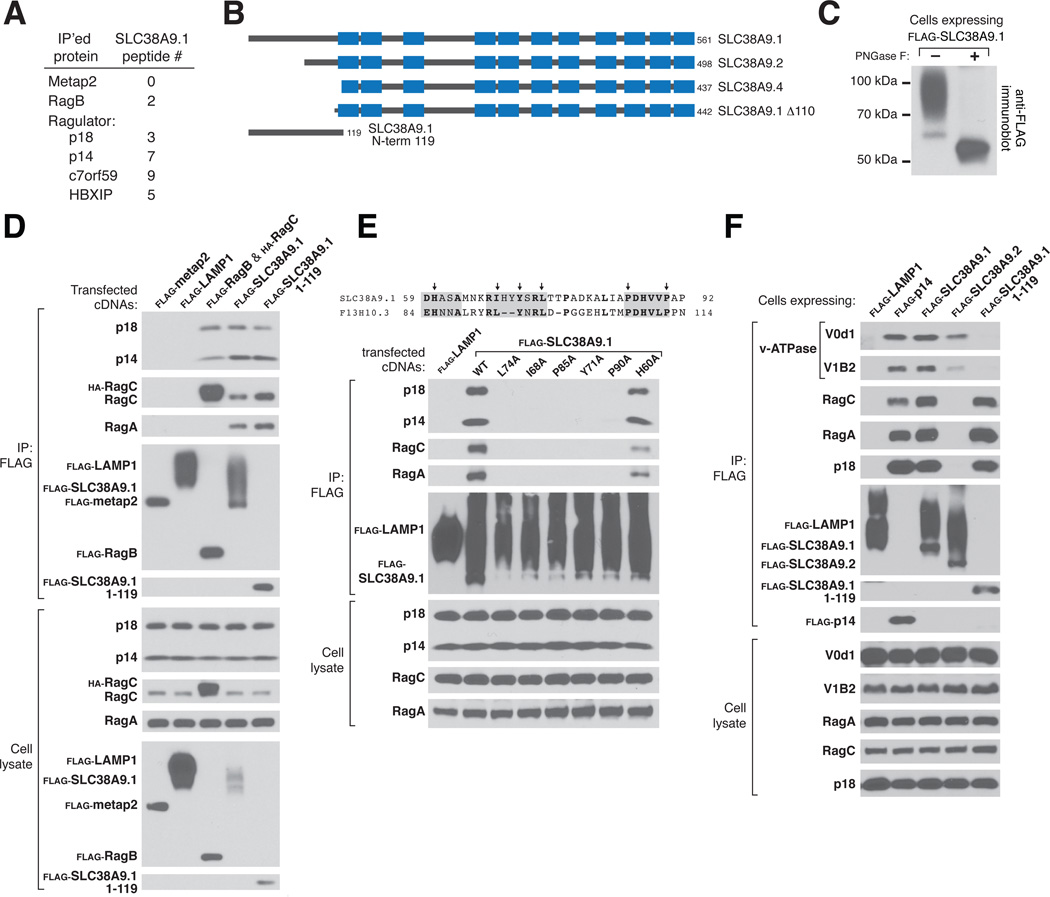
We have proposed that amino acid sensing initiates at the lysosome and requires the presence of amino acids in the lysosomal lumen (11). Thus, we sought to identify, as candidate sensors, proteins that interact with known components of the pathway and also have transmembrane domains. Mass spectrometric analyses of non-heated immunoprecipitates of several Ragulator components and, to a lesser extent, RagB, revealed the presence of isoform 1 of SLC38A9 (SLC38A9.1), a previously unstudied protein with sequence similarity to the SLC38 class of sodium-coupled amino acid transporters (16) (Fig. 1A). SLC38A9.1 is predicted to have eleven transmembrane domains, a cytosolic N-terminal region of 119 amino acids, and three N-linked glycosylation sites in the luminal loop between transmembrane domains 3 and 4 (Fig. 1B and fig. S1A and B). When stably expressed in human embryonic kidney (HEK)-293T cells, SLC38A9.1 migrated on SDS-PAGE as a smear that collapsed to near its predicted molecular weight of 63.8 kDa after treatment with Peptide-N-Glycosidase F (PNGase F) (Fig. 1C). Isoforms 2 (SLC38A9.2) and 4 (SLC38A9.4) lack the first 63 or 124 amino acids of SLC38A9.1, respectively (Fig. 1B).
Figure 1.
Interaction of SLC38A9.1 with Ragulator and the Rag GTPases. (A) The spectral counts of SLC38A9-derived peptides detected by mass spectrometry in immunoprecipitates prepared from HEK-293T cells stably expressing the indicated FLAG-tagged proteins. (B) Schematic depicting SLC38A9 isoforms and truncation mutants. Transmembrane domains predicted by the TMHMM (transmembrane hidden Markov model) algorithm (http://www.cbs.dtu.dk/services/TMHMM/) are shown as blue boxes. (C) Effects of PNGase F treatment of SLC38A9.1 on its electrophoretic migration. (D) Interaction of full-length SLC38A9.1 or its N-terminal domain with endogenous Ragulator (p18 and p14) and RagA and RagC GTPases. HEK-293T cells were transfected with the indicated cDNAs in expression vectors and lysates were prepared and subjected to FLAG immunoprecipitation followed by immunoblotting for the indicated proteins. (E) Identification of key residues in the N-terminal domain of SLC38A9.1 required for it to interact with Ragulator and the Rag GTPases. Experiment was performed as in (D) using indicated SLC38A9.1 mutants. (F) Interaction of SLC38A9.1 with v-ATPase components V0d1 and V1B2. HEK-293T cells stably expressing the indicated FLAG-tagged proteins were lysed and processed as in (D).
As expected from the mass spectrometry results, immunoprecipitates of stably expressed FLAG-tagged SLC38A9.1, but not of three other lysosomal membrane proteins – LAMP1 (17), SLC36A1 (18), and SLC38A7 (19) – contained Ragulator (as detected by its p14 and p18 components), RagA, and RagC (Fig. 1D and fig. S2A). Indicative of the strength of the Ragulator-SLC38A9.1 interaction, the amounts of endogenous Ragulator that coimmunoprecipitated with SLC38A9.1 were similar to those associated with the RagB-RagC heterodimer (Fig. 1D). In contrast, SLC38A9.2, SLC38A9.4 or a mutant of SLC38A9.1 lacking its first 110 amino acids (SLC38A9.1 Δ110) did not associate with Ragulator (fig. S2B and C). The N-terminal region of SLC38A9.1 is sufficient for it to interact with Ragulator-Rag because on its own the first 119 amino acids of SLC38A9.1 coimmunoprecipitated similar amounts of Ragulator and Rag GTPases as did the full-length protein (Fig. 1D and fig. S2C). Using alanine scanning mutagenesis of residues in the N-terminal region conserved to the SLC38A9.1 homolog in C. elegans (F13H10.3), we identified I68, Y71, L74, P85, and P90 as required for the Ragulator-SLC38A9.1 interaction (Fig. 1E).
The v-ATPase and its activity are necessary for amino acid sensing by the mTORC1 pathway and, like SLC38A9.1, it coimmunoprecipitated with stably expressed FLAG-tagged Ragulator (11, 20, 21). Indicating the existence of a supercomplex, stably expressed SLC38A9.1, but not LAMP1, associated with endogenous components of the v-ATPase in addition to Ragulator and the Rag GTPases (Fig. 1F). Although SLC38A9.2 does not interact with Ragulator, it did co-immunoprecipitate the v-ATPase, albeit at lesser amounts than SLC38A9.1 (Fig. 1F). This suggests that the interaction between SLC38A9.1 and the v-ATPase is not mediated through Ragulator but directly or indirectly through the region of SLC38A9.1 that contains its transmembrane domains. Concordant with this interpretation, the N-terminal domain of SLC38A9.1, which interacts strongly with Ragulator, did not coimmunoprecipitate the v-ATPase (Fig. 1F).
SLC38A9 is a Lysosomal Membrane Protein Required for mTORC1 Activation
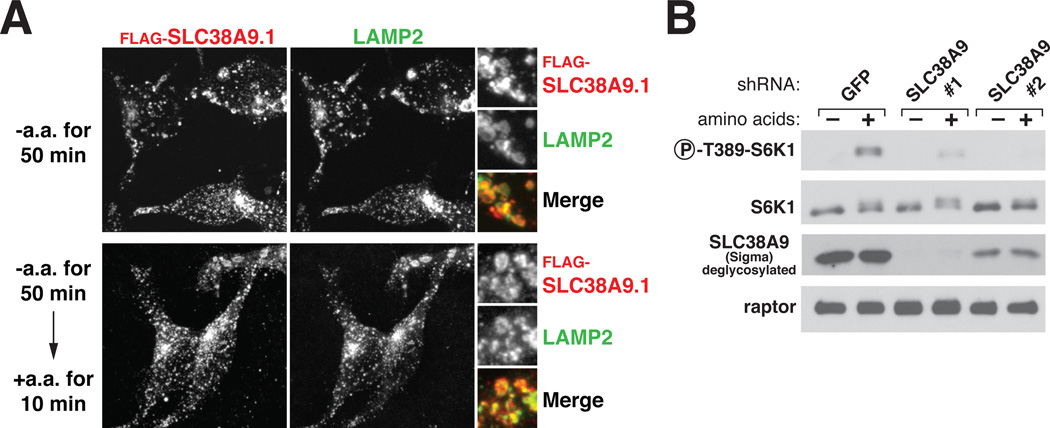
Well-characterized members of the SLC38 family of amino acid transporters (SLC38A1-5) localize to the plasma membrane (22) but at least one member, SLC38A7, is a lysosomal membrane protein (19). This is also the case for SLC38A9.1, SLC38A9.2, and SLC38A9.4 as in HEK-293T cells all three isoforms co-localized with LAMP2, an established lysosomal membrane protein (Fig. 2A and fig. S3A and B). Amino acids did not affect the lysosomal localization of SLC38A9.1 (Fig. 2A). As would be expected if SLC38A9.1 binds to Ragulator at the lysosome, a Ragulator mutant that does not localize to the lysosomal surface because its p18 component lacks lipidation sites (23) did not interact with SLC38A9.1 (fig. S3C).
Figure 2.
Localization of SLC38A9.1 to the lysosomal membrane in an amino acid-independent fashion and requirement of SLC38A9 for mTORC1 pathway activation by amino acids. (A) SLC38A9.1 localization in cells deprived of or replete with amino acids. HEK-293T cells stably expressing FLAG-SLC38A9.1 were starved and stimulated with amino acids for the indicated times. Cells were processed and immunostained for LAMP2 and FLAG-SLC38A9.1. (B) Requirement of SLC38A9 for the activation of the mTORC1 pathway by amino acids. HEK-293T cells expressing indicated short hairpin RNAs (shRNAs) were deprived of amino acids for 50 min or deprived of and then re-stimulated with amino acids for 10 min. Cell lysates were analyzed for the levels of indicated proteins and the S6K1 phosphorylation state.
ShRNA- or siRNA-mediated depletion of SLC38A9 in HEK-293T cells suppressed activation of mTORC1 by amino acids, as detected by the phosphorylation of its established substrate ribosomal protein S6 Kinase 1 (S6K1) (Fig. 2B and fig. S3D). Thus, like the five known subunits of Ragulator (12, 13), SLC38A9.1 is a positive component of the mTORC1 pathway. We conclude that SLC38A9.1 is a lysosomal membrane protein that interacts with Ragulator and the Rag GTPases through its N-terminal 119 amino acids (‘Ragulator-binding domain’) and is required for mTORC1 activation.
SLC38A9.1 Overexpression Makes mTORC1 Signaling Insensitive to Amino Acids
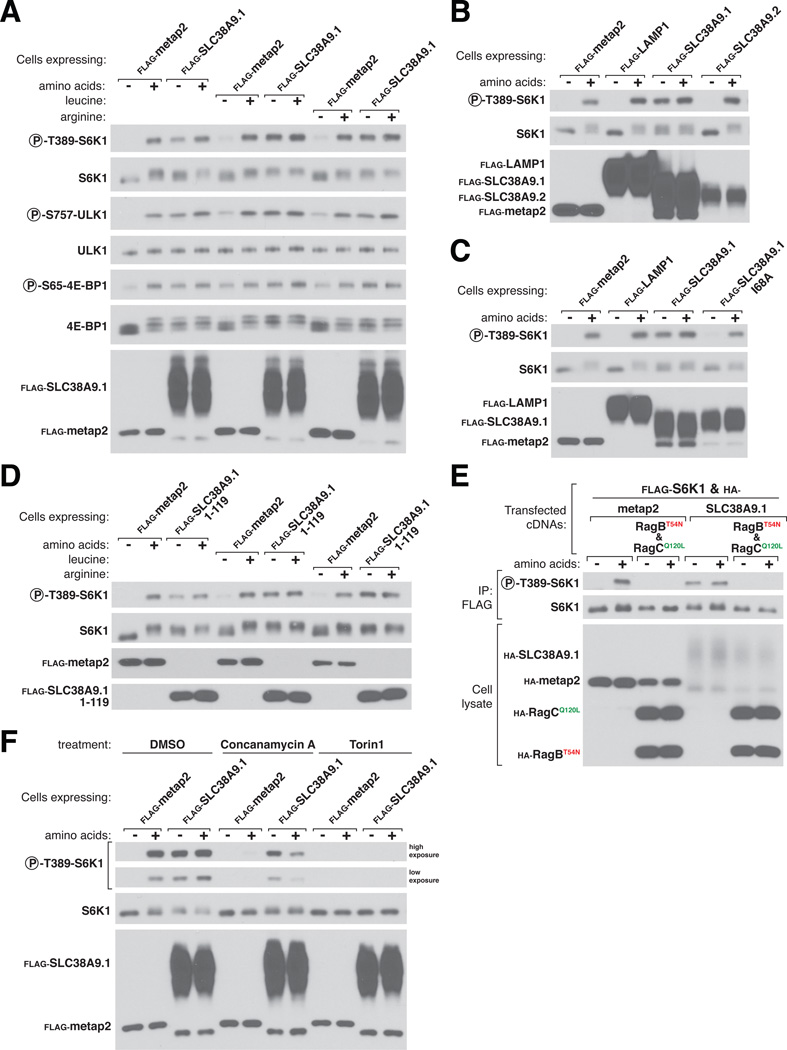
Given the similarity of SLC38A9.1 to amino acid transporters, we reasoned that it might act in conveying amino acid sufficiency to Ragulator and the Rag GTPases. In accord with this expectation, stable or transient overexpression in HEK-293T cells of SLC38A9.1, but not of several control proteins, rendered mTORC1 signaling resistant to total amino acid starvation or to just that of leucine or arginine, two amino acids that regulate mTORC1 activity in many cell types (24–26) (Fig. 3A and fig. S4A). Overexpression of SLC38A9.1 did not affect the regulation of mTORC1 by growth factor signaling (fig. S4D and E). Commensurate with its effects on mTORC1, SLC38A9.1 overexpression suppressed the induction of autophagy caused by amino acid starvation (fig. S4C), a phenotype shared with activated alleles of RagA and RagB (6, 7, 28). Overexpression of variants of SLC38A9 that do not interact with Ragulator and the Rag GTPases, including SLC38A9.2, SLC38A9.4, and the SLC38A9.1 Δ110 and SLC38A9.1 I68A mutants, failed to maintain mTORC1 signaling after amino acid withdrawal (Fig. 3B and Cand fig. S4A). Thus, even in cells deprived of amino acids, some of the overexpressed SLC38A9.1 protein appears to be in an active conformation that confers amino acid insensitivity on mTORC1 signaling in a manner dependent on its capacity to bind Ragulator and Rags. SLC38A9.1 overexpression also activated mTORC1 in the absence of amino acids in HEK-293E, HeLa, and LN229 cells, as well as in mouse embryonic fibroblasts (MEFs), with the degree of activation proportionate to the amount of SLC38A9.1 expressed (fig. S4B). Interestingly, overexpression of just the Ragulator-binding domain of SLC38A9.1 mimicked the effects of the full-length protein on mTORC1 signaling (Fig. 3D), indicating that it can adopt an active state when separated from the transmembrane portion of SLC38A9.1.
Figure 3.
Stable overexpression of full-length SLC38A9.1 or its N-terminal Ragulator-binding domain makes the mTORC1 pathway insensitive to amino acid deprivation. (A) Stable overexpression of FLAG-SLC38A9.1 largely restores mTORC1 signaling during total amino acid starvation and completely restores it upon deprivation of leucine or arginine. HEK-293T cells transduced with lentiviruses encoding the specified proteins were deprived for 50 min of all amino acids, leucine, or arginine and, where indicated, re-stimulated for 10 min with the missing amino acid(s). Cell lysates were analyzed for the levels of the specified proteins and the phosphorylation states of S6K1, ULK1, and 4E-BP1. (B and C) Overexpression of neither SLC38A9.2 nor a point mutant of SLC38A9.1 that fails to bind Ragulator rescues mTORC1 signaling during amino acid starvation. Experiment was performed as in (A) except that cells were stably expressing SLC38A9.2 (B) or SLC38A9.1 I68A (C). (D) Stable overexpression of the Ragulator-binding domain of SLC38A9.1 largely restores mTORC1 signaling during total amino acid starvation and completely rescues it upon deprivation of leucine or arginine. Experiment was performed as in (A) except cells were stably expressing FLAG-SLC38A9.1 1-119. (E) The ability of SLC38A9.1 overexpression to rescue mTORC1 signaling during amino acid starvation is eliminated by co-expression of RagBT54N-RagCQ120L, a Rag heterodimer locked in the nucleotide configuration associated with amino acid deprivation. Effects of expressing the indicated proteins on mTORC1 signaling were monitored by the phosphorylation state of co-expressed FLAG-S6K1. (F) Effects of concanamycin A and Torin1 on mTORC1 signaling in cells stably expressing SLC38A9.1. HEK-293T cells stably expressing the indicated FLAG-tagged proteins were treated with the DMSO vehicle or the specified small molecule inhibitor during the 50 min starvation for and, where indicated, the 10 min stimulation with amino acids.
The gain of function phenotype caused by SLC38A9.1 overexpression offered an opportunity to test its relation to the Rag GTPases, mTORC1, and the v-ATPase. The Rag GTPases and mTORC1 both clearly function downstream of SLC38A9.1 as expression of the dominant negative Rag heterodimer (RagBT54N-RagCQ120L) or treatment with the mTOR inhibitor Torin1 (29) completely inhibited mTORC1 activity, irrespective of whether SLC38A9.1 was overexpressed or not (Fig. 3E and F). In contrast, the v-ATPase has a more complex relationship with SLC38A9.1. Its inhibition with concanamycin A eliminated mTORC1 signaling in the control cells but only partially blocked it in cells overexpressing SLC38A9.1 (Fig. 3F). These results suggest a model in which SLC38A9.1 and the v-ATPase represent parallel pathways that converge upon the Ragulator-Rag GTPase complex.
Modulation of the SLC38A9-Rag-Ragulator Interactions by Amino Acids
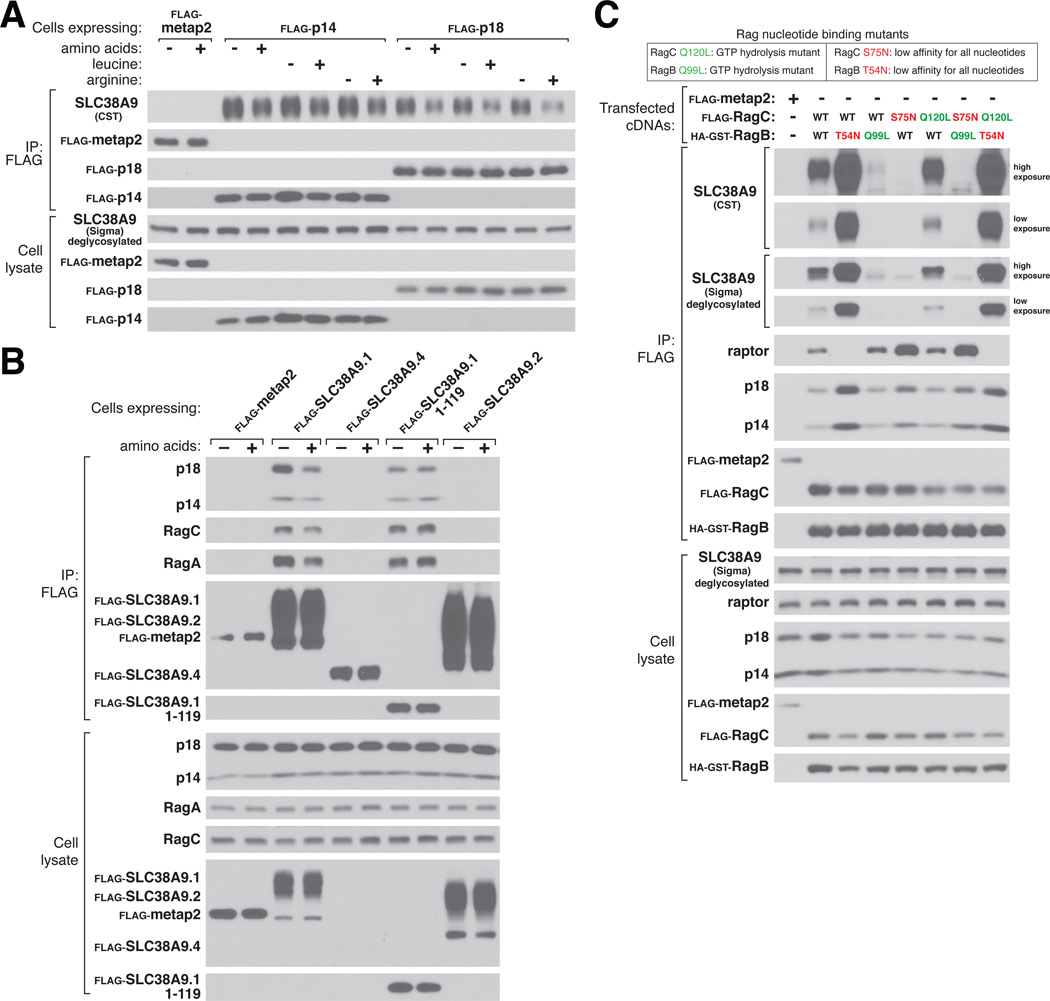
Amino acids modulate the interactions between many of the established components of the amino acid sensing pathway, so we tested if this was also the case for the SLC38A9.1-Ragulator-Rag complex. Indeed, amino acid starvation strengthened the interaction between stably expressed or endogenous Ragulator and endogenous SLC38A9 (Fig. 4A, fig. S5) and between stably expressed SLC38A9.1 and endogenous Ragulator and Rags (Fig. 4B). We obtained similar results when cells were deprived of and stimulated with just leucine or arginine (Fig. 4A). Curiously, although the N-terminal domain of SLC38A9.1 readily bound Ragulator, the interaction was insensitive to amino acids (Fig. 4B), suggesting that the transmembrane region is required to confer amino acid responsiveness.
Figure 4.
Modulation of the interaction between SLC38A9 and Ragulator and the Rag GTPases by amino acids. (A) Effects of amino acids on interaction between the Ragulator complex and endogenous SLC38A9. HEK-293T cells stably expressing the indicated FLAG-tagged Ragulator components were deprived of total amino acids, leucine, or arginine for 1 hour and, where indicated, re-stimulated with amino acids, leucine, or arginine for 15 min. After lysis, samples were subject to FLAG immunoprecipitation and immunoblotting for the indicated proteins. Quantification of SLC38A9 levels in the stimulated state relative to starved state, p14 IP: 0.75 (+AA), 0.79 (+L), 0.74 (+R); p18 IP: 0.56 (+AA), 0.57 (+L), 0.49 (+R). (B) Effects of amino acids on the interaction between full-length or truncated SLC38A9.1 and endogenous Ragulator and the Rag GTPases. Experiment was performed as in (A) except that cells stably expressed the indicated SLC38A9 isoforms or its N-terminal domain (SLC38A9.1 1-119). Quantification of indicated protein levels in the stimulated state relative to starved state, SLC38A9.1 IP: 0.43 (p18), 0.51 (p14), 0.61 (RagC), 0.58 (RagA); SLC38A9.1 1-119 IP: 0.99 (p18), 1.05 (p14), 1.04 (RagC), 1.09 (RagA). (C) Effects of the RagBT54N mutation on association with endogenous SLC38A9. HEK-293T cells were transfected with the indicated cDNAs in expression vectors and lysates were prepared and subjected to FLAG immunoprecipitation followed by immunoblotting for the indicated proteins. Two different antibodies were used to detect endogenous SLC38A9.
As amino acid starvation alters the nucleotide state of the Rag GTPases (6, 7), we tested whether SLC38A9 interacted differentially with mutants of the Rags that lock their nucleotide state. Heterodimers of epitope-tagged RagB-RagC containing RagBT54N, which mimics the GDP-bound state (6, 7), were associated with more endogenous SLC38A9 than heterodimers containing wild-type RagB (Fig. 4C). In contrast, heterodimers containing RagBQ99L, which lacks GTPase activity and so is bound to GTP (6, 7, 15), interacted very weakly with SLC38A9 (Fig. 4C). Thus, like Ragulator, SLC38A9 interacts best with Rag heterodimers in which RagA/B is GDP-loaded, which is consistent with SLC38A9 binding to Ragulator and with Ragulator being a GEF for RagA/B. These results suggest that amino acid modulation of the interaction of SLC38A9.1 with Rag-Ragulator largely reflects amino acid-induced changes in the nucleotide state of the Rag GTPases. Because the RagB mutations had greater effects on the interaction of the Rag GTPases with SLC38A9 than with Ragulator (in Figure 4C compare the SLC38A9 blots with those for p14 and p18), it is very likely that the Rag heterodimers make Ragulator-independent contacts with SLC38A9 that affect the stability of Rag-SLC38A9 interaction.
SLC38A9.1 is an Amino Acid Transporter
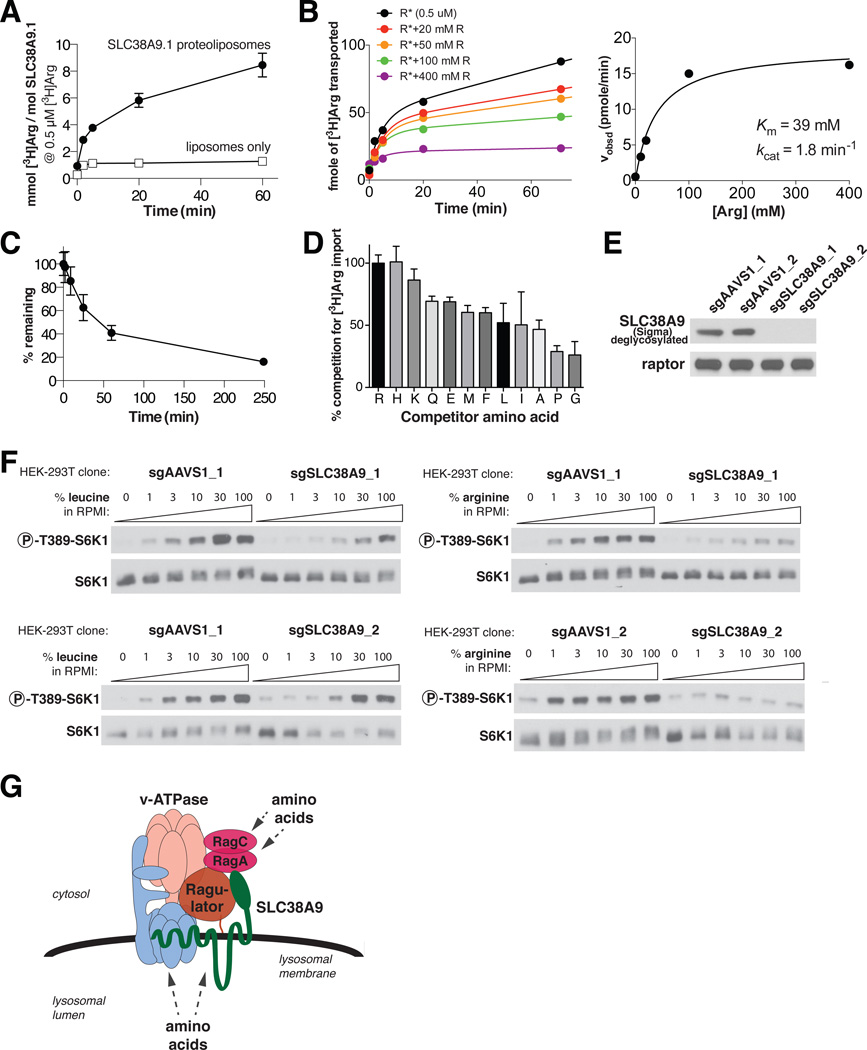
We failed to detect SLC38A9.1-mediated amino acid transport or amino acid-induced sodium currents in live cells in which SLC38A9.1 was so highly overexpressed that some reached the plasma membrane (fig. S6A-E). Because these experiments were confounded by the presence of endogenous transporters or relied on indirect measurements of transport, respectively, we reconstituted SLC38A9.1 into liposomes to directly assay the transport of radiolabelled amino acids. Affinity-purified SLC38A9.1 inserted unidirectionally into liposomes so that its N-terminus faced outward in an orientation analogous to that of the native protein in lysosomes (fig. S6F-H). We could not use radiolabelled L-leucine in transport assays because it bound non-specifically to liposomes so we focused on the transport of L-arginine, which had low background binding (fig. S6I). The SLC38A9.1-containing proteoliposomes exhibited time-dependent uptake of radiolabelled arginine while those containing LAMP1 interacted with similar amounts of arginine as liposomes (Fig. 5A, fig. S6I). Steady-state kinetic experiments revealed that SLC38A9.1 has a Michaelis constant (Km) of ~39 mM and a catalytic rate constant (kcat) of ~1.8 min−1 (Fig. 5B), indicating that SLC38A9.1 is a low-affinity amino acid transporter. SLC38A9.1 can also efflux arginine from the proteoliposomes (Fig. 5C), but its orientation in liposomes makes it impossible to obtain accurate Km and kcat measurements for this activity. It is likely that by having to assay the transporter in the ‘backwards’ direction we are underestimating its affinity for amino acids during their export from lysosomes.
Figure 5.
SLC38A9.1 is a low affinity amino acid transporter and is necessary for mTORC1 pathway activation by arginine. (A) Time-dependent uptake of [3H]arginine at 0.5 μM by proteoliposomes containing 22.4 pmol of SLC38A9.1. To recapitulate the pH gradient across the lysosomal membrane, the lumen of the proteoliposomes is buffered at pH 5.0, while the external buffer is pH 7.4. (B) Steady-state kinetic analysis of SLC38A9.1 uptake activity reveals a Michaelis constant (Km) of ~39mM and catalytic rate constant (kcat) of ~1.8min−1. (Left) Time course of [3H]arginine (R*) uptake, given fixed [3H]arginine (0.5 μM) and increasing concentrations of unlabeled arginine. (Right) Velocity, calculated from left panel, as a function of total arginine concentration. Data were fitted to the Michaelis-Menton equation. Experiment was repeated over 4 times with similar results and a representative one is shown. (C) Time-dependent efflux of SLC38A9.1 proteoliposomes following 1.5 hr loading with 0.5 μM [3H]arginine. (D) Competition of 0.5 μM [3H]arginine transport by SLC38A9.1 using 100 mM of indicated unlabeled amino acids. In A-D, error bars represent standard deviation derived from at least 3 measurements. (E) HEK-293T cells null for SLC38A9 were generated using CRISPR-Cas9 genome editing using two different guide sequences and isolated by single cell cloning. The AAVS1 locus was targeted as a negative control. (F) Impairment of arginine-induced activation of the mTORC1 pathway in SLC38A9-null HEK-293T cells. Cells were starved of the indicated amino acid for 50 minutes and stimulated for 10 minutes using the indicated amino acid concentrations. The leucine and arginine concentrations in RPMI are, respectively, 381 μM and 1.14 mM. (G) Model for distinct amino acid inputs to the Rag GTPases in signaling amino acid sufficiency to mTORC1.
To assess the substrate specificity of SLC38A9.1, we performed competition experiments using unlabeled amino acids (Fig. 5D). The positively charged amino acids histidine and lysine competed radiolabelled arginine transport to similar degrees as arginine, while leucine had a modest effect and glycine was the least effective competitor. Thus, it appears that SLC38A9.1 has a relatively non-specific substrate profile with a preference for polar amino acids.
Given the preference of SLC38A9.1 for the transport of arginine and that arginine is highly concentrated in rat liver lysosomes (30) and yeast vacuoles (31), we asked whether SLC38A9.1 may have an important role in transmitting arginine levels to mTORC1. Towards this end we examined how mTORC1 signaling responded to a range of arginine or leucine concentrations in HEK-293T cells in which we knocked out SLC38A9 using CRISPR-Cas9 genome editing (Fig. 5E). Interestingly, activation of mTORC1 by arginine was strongly repressed at all arginine concentrations while the response to leucine was only blunted so that high leucine concentrations activated mTORC1 equally well in null and control cells (Fig. 5F).
Conclusions
Several properties of SLC38A9.1 are consistent with it functioning as an amino acid sensor for the mTORC1 pathway. Purified SLC38A9.1 transports and therefore directly interacts with amino acids. Overexpression of SLC38A9.1 or just its Ragulator-binding domain activates mTORC1 signaling even in the absence of amino acids. The activation of mTORC1 by amino acids, particularly arginine, is defective in cells lacking SLC38A9. Given these results and that arginine is highly enriched in lysosomes from at least one mammalian tissue (30), we suggest that SLC38A9.1 is a strong candidate for being a lysosome-based arginine sensor for the mTORC1 pathway. To substantiate this possibility it will be necessary to determine the actual concentrations of arginine and other amino acids in the lysosomal lumen and cytosol and compare them to the affinity of SLC38A9.1 for amino acids. If high arginine levels are a general feature of mammalian lysosomes it could explain why SLC38A9.1 appears to have a relatively broad amino acid specificity; perhaps no other amino acid besides arginine is in the lysosomal lumen at levels that approach its Km.
The notion that proteins with sequence similarity to transporters function as both transporters and receptors (transceptors) is not unprecedented (32, 33). The transmembrane region of SLC38A9.1 might undergo a conformational change upon amino acid binding that is then transmitted to Ragulator through its N-terminal domain. What this domain does is unknown but it could regulate Ragulator nucleotide exchange activity or access to the Rag GTPases by other components of the pathway. To support a role as a sensor, it will be necessary to show that amino acid binding regulates the biochemical function of SLC38A9.1.
Even if SLC38A9.1 is an amino acid sensor, additional sensors, even for arginine, are almost certain to exist as we already know that amino acid-sensitive events exist upstream of Folliculin (15, 34) and GATOR1 (35), which, like Ragulator, also regulate the Rag GTPases. An attractive model is that distinct amino acid inputs to mTORC1 converge at the level of the Rag GTPases with some initiating at the lysosome through proteins like SLC38A9.1 and others from cytosolic sensors that remain to be defined (Fig. 5G). Indeed, such a model would explain why the loss of SLC38A9.1 specifically affects arginine sensing but its overexpression makes mTORC1 signaling resistant to arginine or leucine starvation: hyperactivation of the Rag GTPases through the deregulation of a single upstream regulator is likely sufficient to overcome the lack of other positive inputs. A similar situation may occur upon loss of GATOR1, which, like SLC38A9.1 overexpression, causes mTORC1 signaling to be resistant to total amino acid starvation (14).
Modulators of mTORC1 have clinical utility in disease states associated with or caused by mTORC1 deregulation. The allosteric mTOR inhibitor rapamycin is used in cancer treatment (36) and transplantation medicine (37). However, to date, there have been few reports on small molecules that activate mTORC1 by engaging known components of the pathway. The identification of SLC38A9.1—a protein that is a positive regulator of the mTORC1 pathway and has an amino acid binding site—provides an opportunity to develop small molecule agonists of mTORC1 signaling. Such molecules should promote mTORC1-mediated protein synthesis and could have utility in combatting muscle atrophy secondary to disuse or injury. Lastly, there is reason to believe that a selective mTORC1 pathway inhibitor may have better clinical benefits than rapamycin, which in long-term use inhibits both mTORC1 and mTORC2 (38). SLC38A9.1 may be an appropriate target to achieve this.
Supplementary Material
Acknowledgements
We thank all members of the Sabatini Lab for helpful insights, E. Spooner for the mass spectrometric analyses, and G. Superti-Furga and M. Rebsamen for suggesting the use of the Sigma antibody to detect SLC38A9. This work was supported by grants from the NIH (R01 CA103866 and AI47389) and Department of Defense (W81XWH-07-0448) to D.M.S., and fellowship support from the NIH to Z.T. (F30 CA180754), to S.W. (T32 GM007753 and F31 AG044064), to L.C. (F31 CA180271), and to R.W. (T32 GM007753); an NDSEG Fellowship to G.A.W.; an NSF Graduate Research Fellowship to T.W.; an American Cancer Society - Ellison Foundation Postdoctoral Fellowship to W.C. (PF-13-356-01-TBE); a German Academic Exchange Service/DAAD Fellowship to C.S., and support from the Howard Hughes Medical Institute to T.D.J., C.K. and J.P. D.M.S. and B.L.S. are investigators of the Howard Hughes Medical Institute.
References and Notes
- 1.Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nature reviews. Molecular cell biology. 2013 Mar;14:133. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howell JJ, Manning BD. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends in endocrinology and metabolism: TEM. 2011 Mar;22:94. doi: 10.1016/j.tem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature reviews. Molecular cell biology. 2011 Jan;12:21. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Molecular cell. 2010 Oct 22;40:310. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012 Apr 13;149:274. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008 Jun 13;320:1496. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nature cell biology. 2008 Aug;10:935. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saito K, Araki Y, Kontani K, Nishina H, Katada T. Novel role of the small GTPase Rheb: its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. Journal of biochemistry. 2005 Mar;137:423. doi: 10.1093/jb/mvi046. [DOI] [PubMed] [Google Scholar]
- 9.Buerger C, DeVries B, Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochemical and biophysical research communications. 2006 Jun 9;344:869. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- 10.Menon S, et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014 Feb 13;156:771. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011 Nov 4;334:678. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012 Sep 14;150:1196. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010 Apr 16;141:290. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bar-Peled L, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013 May 31;340:1100. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsun ZY, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Molecular cell. 2013 Nov 21;52:495. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sundberg BE, et al. The evolutionary history and tissue mapping of amino acid transporters belonging to solute carrier families SLC32, SLC36, and SLC38. Journal of molecular neuroscience : MN. 2008 Jun;35:179. doi: 10.1007/s12031-008-9046-x. [DOI] [PubMed] [Google Scholar]
- 17.Carlsson SR, Roth J, Piller F, Fukuda M. Isolation and characterization of human lysosomal membrane glycoproteins, h-lamp-1 and h-lamp-2. Major sialoglycoproteins carrying polylactosaminoglycan. The Journal of biological chemistry. 1988 Dec 15;263:18911. [PubMed] [Google Scholar]
- 18.Sagne C, et al. Identification and characterization of a lysosomal transporter for small neutral amino acids. Proceedings of the National Academy of Sciences of the United States of America. 2001 Jun 19;98:7206. doi: 10.1073/pnas.121183498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapel A, et al. An extended proteome map of the lysosomal membrane reveals novel potential transporters. Molecular & cellular proteomics : MCP. 2013 Jun;12:1572. doi: 10.1074/mcp.M112.021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, et al. Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 (MTORC1) activity. The Journal of biological chemistry. 2013 Dec 13;288:35769. doi: 10.1074/jbc.M113.511212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, et al. Epidermal growth factor-induced vacuolar (H+)-atpase assembly: a role in signaling via mTORC1 activation. The Journal of biological chemistry. 2012 Jul 27;287:26409. doi: 10.1074/jbc.M112.352229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackenzie B, Erickson JD. Sodium-coupled neutral amino acid (System N/A) transporters of the SLC38 gene family. Pflugers Archiv : European journal of physiology. 2004 Feb;447:784. doi: 10.1007/s00424-003-1117-9. [DOI] [PubMed] [Google Scholar]
- 23.Nada S, et al. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. The EMBO journal. 2009 Mar 4;28:477. doi: 10.1038/emboj.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ban H, et al. Arginine and Leucine regulate p70 S6 kinase and 4E-BP1 in intestinal epithelial cells. International journal of molecular medicine. 2004 Apr;13:537. [PubMed] [Google Scholar]
- 25.Hara K, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. The Journal of biological chemistry. 1998 Jun 5;273:14484. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 26.Yao K, et al. Dietary arginine supplementation increases mTOR signaling activity in skeletal muscle of neonatal pigs. The Journal of nutrition. 2008 May;138:867. doi: 10.1093/jn/138.5.867. [DOI] [PubMed] [Google Scholar]
- 27.Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends in molecular medicine. 2012 Sep;18:524. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Efeyan A, et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013 Jan 31;493:679. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. The Journal of biological chemistry. 2009 Mar 20;284:8023. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harms E, Gochman N, Schneider JA. Lysosomal pool of free-amino acids. Biochemical and biophysical research communications. 1981 Apr 15;99:830. doi: 10.1016/0006-291x(81)91239-0. [DOI] [PubMed] [Google Scholar]
- 31.Kitamoto K, Yoshizawa K, Ohsumi Y, Anraku Y. Dynamic aspects of vacuolar and cytosolic amino acid pools of Saccharomyces cerevisiae. Journal of bacteriology. 1988 Jun;170:2683. doi: 10.1128/jb.170.6.2683-2686.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holsbeeks I, Lagatie O, Van Nuland A, Van de Velde S, Thevelein JM. The eukaryotic plasma membrane as a nutrient-sensing device. Trends in biochemical sciences. 2004 Oct;29:556. doi: 10.1016/j.tibs.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Hyde R, Cwiklinski EL, MacAulay K, Taylor PM, Hundal HS. Distinct sensor pathways in the hierarchical control of SNAT2, a putative amino acid transceptor, by amino acid availability. The Journal of biological chemistry. 2007 Jul 6;282:19788. doi: 10.1074/jbc.M611520200. [DOI] [PubMed] [Google Scholar]
- 34.Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. The Journal of cell biology. 2013 Sep 30;202:1107. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chantranupong L, et al. The Sestrins Interact with GATOR2 to Negatively Regulate the Amino-Acid-Sensing Pathway Upstream of mTORC1. Cell reports. 2014 Sep 24; doi: 10.1016/j.celrep.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nature reviews. Drug discovery. 2011 Nov;10:868. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 37.Kahan BD, Camardo JS. Rapamycin: clinical results and future opportunities. Transplantation. 2001 Oct 15;72:1181. doi: 10.1097/00007890-200110150-00001. [DOI] [PubMed] [Google Scholar]
- 38.Lamming DW, Ye L, Sabatini DM, Baur JA. Rapalogs and mTOR inhibitors as anti-aging therapeutics. The Journal of clinical investigation. 2013 Mar 1;123:980. doi: 10.1172/JCI64099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.