Abstract
XPD-like helicases constitute a prominent DNA helicase family critical for many aspects of genome maintenance. These enzymes share a unique structural feature, an auxiliary domain stabilized by an iron-sulphur (FeS) cluster, and a 5′-3′ polarity of DNA translocation and duplex unwinding. Biochemical analyses alongside two single-molecule approaches, total internal reflection fluorescence microscopy and high-resolution optical tweezers, have shown how the unique structural features of XPD helicase and its specific patterns of substrate interactions tune the helicase for its specific cellular function and shape its molecular mechanism. The FeS domain forms a duplex separation wedge and contributes to an extended DNA binding site. Interaction within this site position the helicase in an orientation to unwind the duplex, control the helicase rate, and verify the integrity of the translocating strand. Consistent with its cellular role, processivity of XPD is limited and is defined by an idiosyncratic stepping kinetics. DNA duplex separation occurs in single base pair steps punctuated by frequent backward steps and conformational rearrangements of the protein-DNA complex. As such, the helicase in isolation mainly stabilizes spontaneous base pair opening and exhibits a limited ability to unwind stable DNA duplexes. The presence of a cognate ssDNA binding protein converts XPD into a vigorous helicase by destabilizing the upstream dsDNA as well as by trapping the unwound strands. Remarkably, the two proteins can co-exist on the same DNA strand without competing for binding. The current model of the XPD unwinding mechanism will be discussed along with possible modifications to this mechanism by the helicase interacting partners and unique features of such bio-medically important XPD-like helicases as FANCJ (BACH1), RTEL1 and CHLR1 (DDX11).
1. Introduction
1.1. DNA helicases in maintenance of the genetic integrity
Helicases are essential components of many DNA repair machines. These vectorial enzymes produce ssDNA (single-stranded DNA) intermediates necessary for function of these machines, remodel non-canonical DNA structures or nucleoprotein complexes, and rearrange DNA repair intermediates. The cell uses helicases along with helicase-like DNA translocating motors and switches to maintain integrity of its genome, support DNA replication, and to rectify DNA damage caused by exogenous agents and byproducts of cellular metabolism [1–3]. Every day our genomes experience up to an astounding 200,000 DNA modifications [4, 5]. While the repertoire of repairable DNA lesions is extensive, their processing depends on a limited number of different helicases present in the cell. If not completely and accurately repaired, DNA lesions or their repair intermediates can cause genetic instability and chromosomal rearrangements [6], expedite the acquisition of mutations, and contribute to uncontrolled cell proliferation, tumorigenesis [7, 8], or cell senescence [9–11]. To properly respond to many possible lesions and intermediates, DNA helicases are often adapted to perform very different tasks when integrated into different macromolecular assemblies.
Helicases use ATP binding and hydrolysis to fuel two important biochemical activities (i) strand separation (also referred to as bona fide helicase activity), where dsDNA is unwound to produce transient single-stranded intermediates of DNA replication, recombination and repair; and (ii) translocation, a directional motion along the DNA molecule which can be coupled to remodeling of nucleoprotein complexes [2]. These two activities are related, but not identical: a helicase may unwind dsDNA, but may be stalled by bound proteins or by particular ssDNA secondary structures; on the other hand, it may displace proteins, but display no duplex separation activity whatsoever. The vast array of genome maintenance transactions requires the limited battery of helicases to switch between these activities, and to possess a range of tunable processivities and substrate specificities commensurate with the extensive number and variety of jobs to be done. Often, only one of these activities may be important for the cellular function of a particular helicase.
The majority of helicases involved in DNA repair belong to two major superfamilies, superfamily 1 (SF1) and superfamily 2 (SF2) [12–14]. Directional movement, a defining mechanistic feature of SF1 and SF2 helicases, is achieved through conformational transitions within the conserved motor core. Structural and biochemical evidence assembled to date suggests that the motor core of all these enzymes is comprised of the two RecA-like folds, 1A and 2A (in SF1), and HD1 and HD2 (in SF2), where domain 1A (HD1) contacts the 3′-end of the bound ssDNA, and domain 2A (HD2) faces the 5′-end (Figure 1) [12, 13]. The cleft which spans both domains 1A (HD1) and 2A (HD2) forms the bipartite primary DNA binding site. Residues within this DNA-binding cleft make extensive contacts with the strand on which the enzyme translocates. Notably, while the interactions within the helicase core domains are sufficient for enabling the directional translocation of the 3′-5′ moving helicases, additional contacts outside the canonical DNA binding site are indispensable in the helicases that move along DNA strand in the 5′ - 3′ direction. In the repeating mechano-chemical cycle ATP binding in the cleft between 1A (HD1) and 2A (HD2) domains brings the two domains together. ATP hydrolysis and release of ADP and inorganic phosphate allows the two domains to come apart. ATP binding and hydrolysis elicits conformational transitions, which are transferred through the helicase core to the DNA binding site and alter the contacts between the two RecA-domains of the helicase and the translocating strand. Cycling between the two metastable states enables directional translocation. Additional structural features unique to different helicase groups provide means for achieving duplex separation. Residues forming NTP (nucleotide triphosphate) and DNA binding sites as well as residues involved in communication between these two sites are highly conserved within each helicase superfamily and are referred to as the helicase signature motifs [14, 15]. The residues forming duplex separating structures display much less conservation or are unique to each helicase or to a helicase family.
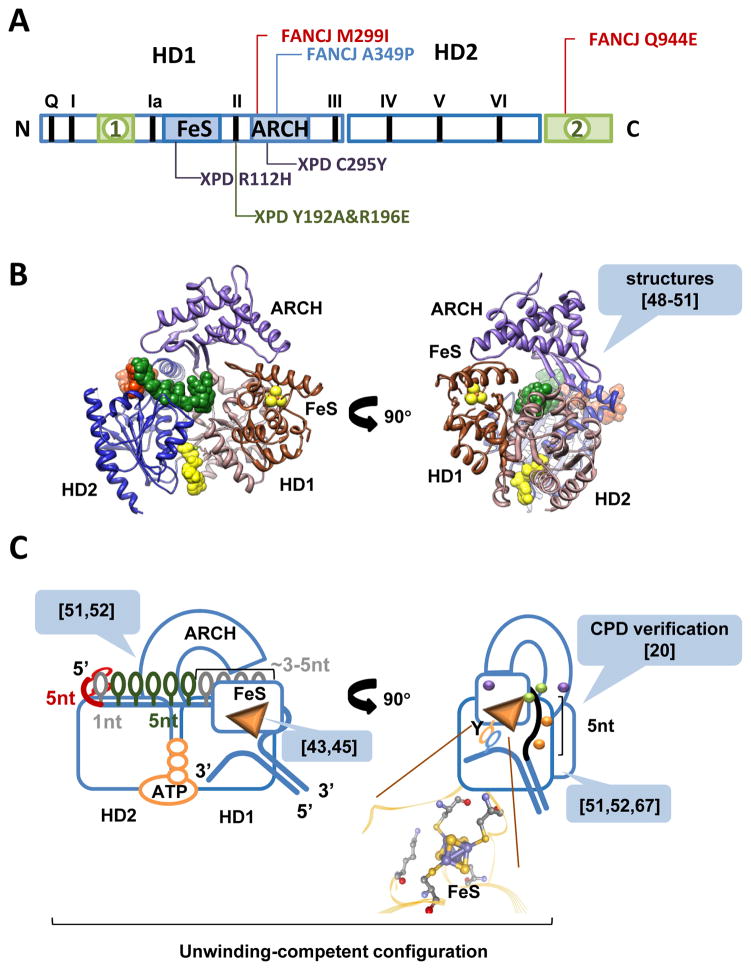
Figure 1. Modular organization of FeS DNA helicases.
A. All XPD-like helicases contain the motor domains (HD1 and HD2) and auxiliary domains (FeS and ARCH). Helicase signature motifs are indicated by black bars and Roman numerals. Two additional domains found in FANCJ, CHLR1 and RTEL are shown in green. Most studied mutations in the auxiliary domains of XPD-like enzymes are shown in red (breast cancer causing mutations in FANCJ), blue (Fanconi anemia-associated mutation in FANCJ), purple (TTD-associated mutations in XPD) and green (mutations that disable the damage verification site). B. Structure of XPD (PDB: 4a15) is shown as ribbon diagram. Individual domains are colored as following: HD1 is salmon, HD2 is blue, FeS domain is brown and ARCH domain is purple. Five nucleotide ssDNA bound to the non-canonical site on HD2 resolved in the PDB: 4a15 structure are shown in red space filling model. From the structural superimposition of the motor cores XPD with another SF2 helicase (PDB: 2db3, Vasa DEAD-box RNA helicase bound to ssRNA) the ssDNA can be extended into the canonical DNA binding grove by additional 5 nt (green space filling model). AMP-PNP, an ATP analog is also from the PDB: 2db3 (yellow space filling model). C. Cartoon depiction of the XPD bound to DNA and ATP. Green and purple spheres correspond to mutations highlighted in (A); orange spheres correspond to the key residues within the extended DNA binding site that control helicase activity. References in the callouts indicated the manuscripts that laid the basis for each highlighted element of the model (see text for the details).
The versatility of helicases and related translocases raises an important question: how do these enzymes generate such a fantastic functional diversity using a limited set of structural features? What is especially intriguing is the complex relationship between NTP hydrolysis and regulated substrate remodeling. The free energy released due to the hydrolysis of one ATP molecule should be sufficient to separate several base pairs of the DNA duplex. On the other hand, disruption of the streptavidin-biotin interaction – a common reporter for translocase activity [16] – is energetically equivalent to multiple ATP molecules. Despite these facts, many helicases readily displace streptavidin from biotinylated DNA but cannot unwind duplex DNA. Helicase oligomerization, allosteric regulation by interacting partners, and specificity for particular substrates have been proposed as contextual determinants of unwinding and/or protein displacement activities [17]. The mechanism(s) underlying the specific regulation and dynamic behavior of these two activities, however, remains enigmatic.
1.2. XPD – a model system for FeS-containing SF2 DNA helicases
XPD (Xeroderma pigmentosum complementation group D) is a prototypical member of a prominent DNA helicase family whose deficiency or dysregulation is linked to human diseases ranging from cancer predisposition to hypertension (Figure 1A, Table 1 and references [18–40] within). Four human XPD orthologs (XPD, FANCJ, RTEL1 and CHLR1) are DNA helicases with roles in DNA repair and genome maintenance [41, 42]. Homologues of XPD helicase can also be found in archaea, bacteria and, all eukaryotes. Members of the XPD-like helicase family play distinct and only partially overlapping roles in the cell. All, however, are likely to share the overall structure and basic mechanism of DNA translocation and unwinding.
Table 1.
Human XPD-like helicases
| Helicase | activities | pathways | diseases |
|---|---|---|---|
| XPD/Rad3 (ERCC2) | Helicase [18, 19], damage verification [20] | Nucleotide excision repair (NER), transcription [27] | Xeroderma Pigmentosum (XP), Cockayne Syndrome (CS), Trichothiodystrophy (TTD), Cerebro-oculo-facial-skeletal developmental abnormalities (COFS) [33, 34] |
|
|
|||
| FANCJ (BACH1) | Helicase[21], tumor suppressor[21], anti-recombinase, translocase[22], resolution of G-quadruplexes [23] | Homologous recombination (HR)[21], interstrand cross-link (ICL) repair[28, 29] | Breast Cancer [21], Fanconi Anemia [28, 29] |
|
|
|||
| RTEL | Helicase, anti-recombinase[24], translocase, dismantles G4 structures at telomeres[25] | HR [24], telomere homeostasis[25], replication[30], regulation of meiotic cross-over events [31] | Cancer[35, 36], Dyskeratosis congenita[37], Crohn’s Disease[38], hypertension[39] |
|
|
|||
| CHLR1 (DDX11) | Helicase[26] | sister chromatid cohesion and DNA repair [32] | Warsaw Breakage Syndrome [40] |
Inserted in their HD1 domains, XPD-like helicases possess an auxiliary domain stabilized by a 4Fe-4S cluster [43]. Consequently, they are commonly referred to as FeS helicases. All characterized XPD-like enzymes are bona fide helicases; all move along ssDNA with 5′–3′ polarity [18, 43–47]; all are believed to share a similar structural organization [12, 48–50] (Figure 1), as well as a preference for branched DNA substrates. Current structural information on FeS helicases is limited to three apo structures of archaeal XPD homologs [48–50] (PDB: 2vsf; PDB: 3crv; PDB: 3crw; and PDB: 2vl7. Note that the FeS domain was not resolved in the two latter structures) and a more recent structure of XPD in a complex with a 5 nucleotide ssDNA fragment bound to the HD2 outside of the canonical DNA binding site [51] (Figure 1B; PDB: 4a15). To compensate for the paucity of structural information on the binary XPD-DNA and ternary XPD-ATP-DNA complex, we recently used a combination of fluorescence foot-printing, chemical protease incorporated into the DNA substrates, and site-directed mutagenesis to identify the orientation and placement of the DNA in the XPD helicase [52]. We determined that the HD2 domain contacts the 5′ end of the translocating strand [52]. This can be inferred also from the XPD-DNA structure [51]. In this conformation, the translocating strand is oriented in the primary DNA binding site so that HD1 is positioned proximal to the 3′ end while HD2 is near the 5′ end. Within this site, XPD-like FeS helicases are expected to interact with the phosphodiester backbone of ssDNA. This expectation is due to their ability to bypass damaged bases [53] and ssDNA binding proteins that specifically interact with the nucleobases [54] (see discussion below). The region of XPD helicase that interacts with the translocation strand is extended beyond the canonical DNA binding site spanning the surface of HD1 and HD2 [51, 52]. In fact, bound XPD occludes approximately 20 nucleotides of ssDNA [54] suggesting that the canonical DNA binding site constitutes only a part of the XPD surface that engages the DNA. As mentioned above, a patch of residues in the HD2 important for XPD helicase activity binds a 5-nucleotide stretch of the translocating strand [51]. The stability of this interaction suggests that the initial encounter with the DNA substrate likely occurs within this site. We imagine that the translocation strand is then placed into the canonical DNA binding site spanning across HD1 and HD2, and under the arch of the ARCH domain. It then continues through the hole formed by HD1, ARCH and FeS domains into the secondary DNA binding site between HD1 and FeS domains, which accommodates additional 5 nucleotides of the translocation strand. It has been shown that the integrity of the translocating strand is “verified” within the secondary site: the presence of a cyclobutane pyrimidine dimer (CPD), a prototypical UV-induced DNA lesion, stalls the helicase and triggers nucleotide excision repair [20]. Two point mutations in this site are the separation of function mutations that specifically affect damage verification and downstream signaling, but not the duplex unwinding per se, confirming that the two activities are distinct [20]. Damage verification was also corroborated by a recent atomic force microscopy (AFM) study which revealed a remarkable preference of the ATP-bound XPD for the damage containing DNA [55]. At the end of the secondary DNA binding site, the FeS domain contains a wedge structure that we believe separates the DNA duplex (Figure 1C) [52].
Human XPD-like helicases FANCJ, RTEL, and CHLR1 are likely to share the basic molecular mechanism of DNA translocation and unwinding, as well as a common arrangement of the DNA binding site. An additional insertion in the HD1 found in these helicases, however, may provide them with the means of recognizing their preferred DNA substrates. It may also function in a similar way to RPA2, a small archaeal ssDNA-binding protein that assists XPD in dsDNA unwinding by melting the duplex ahead of the helicase [56]. I will focus this review on XPD, the most “simple” helicase within the family, because our structural and mechanistic understanding of this enzyme made it an ideal model system. Most of the mechanistic insights that I will describe are fruits of the single-molecule analyses placed in the context of biochemical and structural studies.
Two complementary single-molecule approaches have been applied to characterize XPD helicase: total internal reflection fluorescence microscopy (TIRFM) and high resolution optical tweezers. The latter gave us unprecedented insight into the mechanism of XPD-mediated duplex unwinding, while the former allowed us to monitor XPD nucleoprotein transactions in the presence of other proteins. In addition, a very recently published AFM study provided important insights into how XPD helicase verifies the presence of the DNA damage.
2. Taking advantage of the FeS cluster
2.1. FeS clusters in DNA helicases
FeS clusters are ubiquitous prosthetic groups whose biochemical utility usually rests upon their ability to accept and donate electrons, or upon a cationic feature that allows tight binding of the electron-rich oxygen and nitrogen atoms of organic substrates [57]. These features are influenced by solvent exposure and the local electrostatic environment, both of which may provide a means of regulation. The redox sensing function was proposed for the FeS cluster of XPD [58]. The FeS clusters also play structural roles by facilitating protein folding and by stabilizing the important structural elements. For example, FeS clusters organize the elements forming the DNA-interacting surface and position the key catalytic residues in several DNA glycosylases, stabilize the nuclease domain of AddAB helicase/nuclease and related RecB-family nucleases, and are also found in DNA primase and RNA polymerase (reviewed in [59]), as well as in B-family DNA polymerases [60]. The FeS domain in XPD forms a duplex separation wedge and contributes to a secondary DNA binding site, which positions the helicase in an orientation to unwind duplex, controls the helicase rate, and verifies the integrity of the translocating strand [20, 45, 51, 52]. Eukaryotic FeS containing helicases XPD, FANCJ, RTEL1 and DNA2 were shown to acquire their FeS clusters through interaction with MMS19, MIP18 and ANT2 proteins that are involved in cytoplasmic FeS cluster protein assembly [61].
Practical utility of the FeS clusters in these enzymes lays in their ability to serve as endogenous quenchers of a wide spectral range of fluorophores which can be used as reporters of the helicase-DNA transactions (Figure 2 A–C). We used this effect to develop new assays for ensemble and single-molecule analyses of XPD family helicases [45, 54, 56]. By altering the distance between bound helicase and DNA-tethered fluorescent dye (for example Cy3 or Cy5), the quenching signal can be calibrated: in the case of XPD, a change in the helicase position by 1 nucleotide (or 1-bp) corresponds to a 3% change in the Cy3 fluorescence intensity ([54] and our unpublished data) (Figure 2C). Cy3 quenching, therefore, can be used as a proximity indicator in analysis of XPD binding to and translocation along ssDNA, as well as in monitoring duplex separation or nucleoprotein remodeling by XPD helicase and related enzymes [62]. Furthermore, FeS cluster-mediated fluorescence quenching can be combined with Förster resonance energy transfer (FRET) [63, 64] between a pair of fluorophores thereby expanding the translocation and unwinding measurements to pseudo three color analyses of complex nucleoprotein interactions [54, 62].
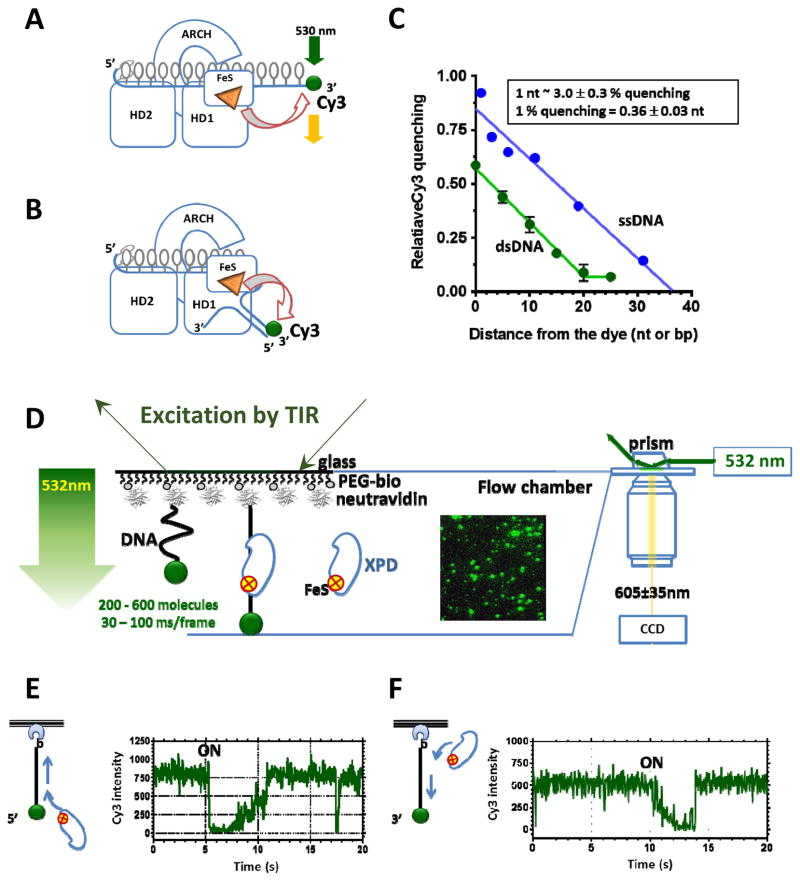
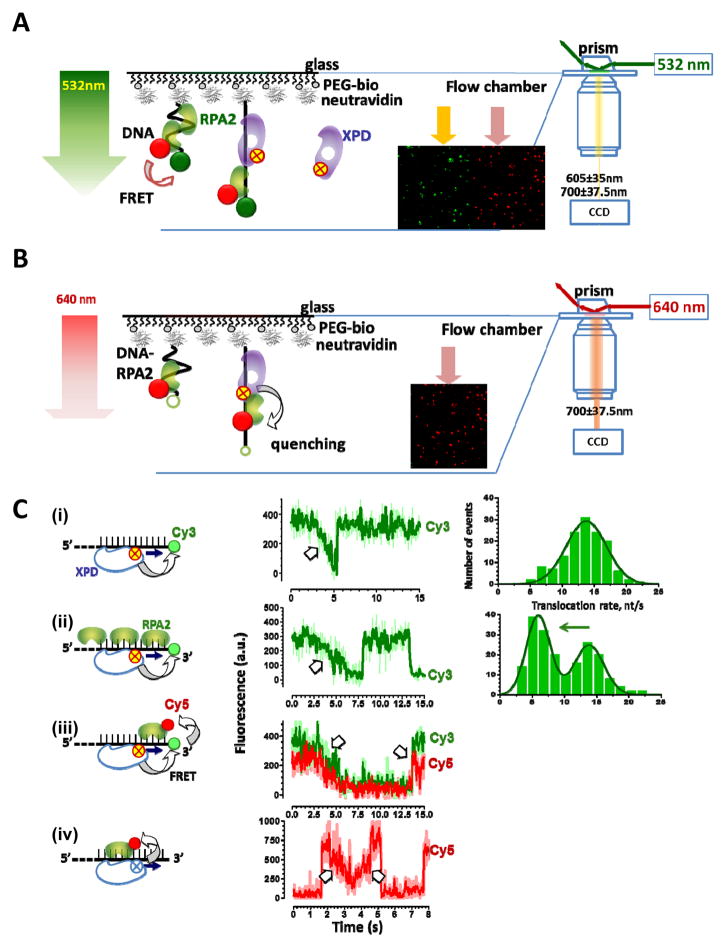
Figure 2. FeS cluster is a built-in proximity indictor.
A&B. Schematic depiction of utilization of the FeS cluster in XPD to monitor ssDNA translocation and unwinding, respectively. C. Distance-dependence of the fluorescence quenching magnitude. D. One-color TIRFM experimental setup for monitoring XPD translocation. Excitation by TIR and resulting evanescent wave (green arrow) illuminates a thin layer of the flow cell and excites Cy3-labeled DNA molecules tethered to its surface. Fragment of the movie frames is shown as the insert. In it, each green spot corresponds to a single surface-tethered DNA molecule. E&F. Fragments of the representative fluorescence trajectories for the helicase moving away and towards the label, respectively. The data shown were adapted from [54].
2.2. Visualizing directional translocation
Single-molecule fluorescence imaging by TIRFM is ideally suited for visualizing movement of slow and marginally processive XPD helicase [54, 62]. The minimal single-color TIRFM experiment is sufficient to follow, via the FeS-mediated fluorescence quenching, XPD binding to and movement on the surface-tethered fluorescently-labeled DNA molecules [54, 62] (Figure 2D). Similarly, DNA translocation and unwinding by other FeS helicases are readily observed using single-color TIRFM-based approach (our unpublished data).
To visualize XPD translocation, Cy3-labeled ssDNA is immobilized on the surface of the microscope flow cell through interaction between biotin moiety incorporated into the DNA molecule and neutravidin or neutravidin coating the flow cell surface. Hundreds of surface-tethered molecules are monitored simultaneously, each yielding a fluorescence trajectory, i.e. fluorescence intensity recorded from a particular location of the flow cell over time. Individual binding and translocation events are observed as changes in the Cy3 fluorescence intensity followed by the fluorescence recovery to the baseline when XPD dissociates [54] (Figure 2E&F). XPD is a 5′-3 helicase: it requires a 5′ ssDNA overhang to unwind DNA duplex [18, 19] and displaces streptavidin from ssDNA biotinylated near the 3′ end [45]. The 5′-3′ directionality of the XPD movement is confirmed in single-molecule experiments by placing the dye at either 3′ or 5′ end of the tethered DNA molecule: trajectories originating from 3′-labeled DNA depict gradual Cy3 quenching indicative of the helicase movement towards the dye followed by abrupt recovery of the fluorescence signal, which corresponds to the helicase dissociation. In contrast, trajectories recorded using 5′-labeled ssDNA show abrupt quenching followed by a gradual fluorescence recovery validating the 5′-3′ directional translocation of XPD (Figures 2&3) [54]. In contrast to other DNA helicases that shuttle on ssDNA [65, 66], the translocation behavior of XPD is simple: it binds ssDNA, translocates in 5′-3′ direction, and either translocates off the free ssDNA end or dissociates after encountering the neutravidin-bound end [54] (Figure 2E&F). Notably, a fraction of the latter encounters results in disruption of the neutravidin-biotin interaction and the subsequent detachment of the DNA molecule from the TIRFM slide.
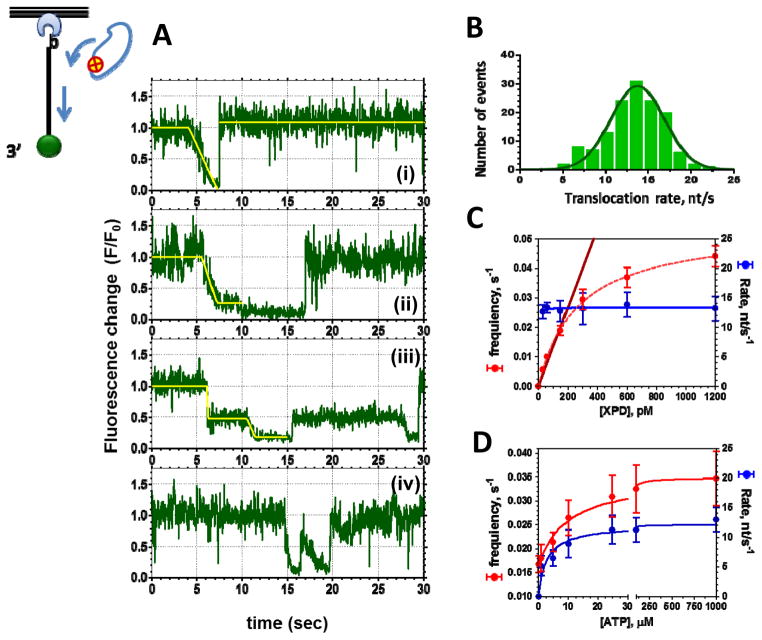
Figure 3. Analysis of XPD translocation trajectories.
A. Four distinct classes of unwinding events routinely observed in XPD translocation experiments. The top three classes could be fitted with multi-segment lines to determine the translocation rate. The fourth trajectory depicts two translocation events that occurred too close to one another to be analyzed. B. The rates of individual translocation events are binned and plotted as a histogram whose Gaussian fit yields average translocation rate. C. XPD concentration dependence of the frequency of the observed events and XPD translocation rates suggests that at the XPD concentrations below 200 nM we were observing translocation by individual XPD monomers. D. Both the translocation rate and the frequency of the observed events display Michaelis-Menten dependence on ATP concentration. The data shown were adapted from [54].
As expected for a motor protein, the average translocation rate displayed a Michaelis-Menten dependence on ATP concentration (Figure 3D) [54]. This is because the overall cycle of such a motor contains two steps that can be partially kinetically limiting. At low ATP concentrations, ATP binding limits the translocation rate. In contrast, at saturating ATP concentrations, the forward stepping becomes rate limiting. Under latter conditions, archaeal XPD translocates along ssDNA with an average rate of approximately 12 nucleotides per second (Figure 3 B–D). Translocation rates of individual XPD molecules, however, vary significantly around mean displaying a broad bell-shaped distribution (Figure 3B) – a feature commonly observed in single-molecule studies of motor proteins. The frequency of observed binding events also depends on ATP concentration suggesting that ATP bound XPD has higher affinity for ssDNA than the ATP-free enzyme (Figure 3 D) [54].
3. DNA unwinding mechanism
3.1. XPD stepping behavior via high resolution optical tweezers
While it is generally accepted that all SF1 and SF2 helicases translocate on ssDNA by a so-called inch-worm mechanism whereby the conformational transitions in the helicase motor core are biased to achieve a ratchet-like directional progression of the helicase in single nucleotide steps [67], it remains unclear whether there exists a unified duplex unwinding mechanism that can be universally applied to SF1 and SF2 helicases [17]. Groups of related helicases within each superfamily are, however, likely to share fundamental unwinding mechanisms. This makes XPD an excellent model for all helicases in the FeS group of SF2B helicases.
Application of a powerful single-molecule technique, high resolution optical tweezers [68, 69], has revealed the detailed mechanism of XPD-mediated dsDNA unwinding [70]. The construct unwound by XPD consisted of a dumbbell structure with DNA between two polystyrene beads and a dsDNA hairpin containing a single-stranded region at its 5′ end in the middle (Figure 4A). The ssDNA region served as a loading site from which XPD initiated hairpin unwinding (Figure 4B). Each unwound base pair increased the dumbbell length by the length of two released nucleotides of ssDNA. Capable of angstrom-level resolution, the high resolution optical tweezers system was adequate for monitoring discrete steps of the helicase as small as a single base pair [70] (Figure 4C). The steps revealed by this method correspond to the kinetic steps size, or the presence of a repeating rate limiting step in duplex unwinding mechanism [71], either of which may or may not relate to either the physical step size of the motor, or the duplex length traversed in a single ATPase cycle. The kinetic step size of 4–6 base pairs (a half-turn of B-form dsDNA) is commonly observed for the 3′-5′ SF1 and SF2 helicases [71–74], that were also shown to move on ssDNA in single nucleotide steps and hydrolyze one ATP for each step [75–77]. More complex stepping behavior has been observed for some helicases. The unwinding mechanism of NS3 helicase from Hepatitis C Virus, for example, combines nested steps of 1-, 3- and 11-bps [78–80] with the asynchronous release of separated ssDNA [79].
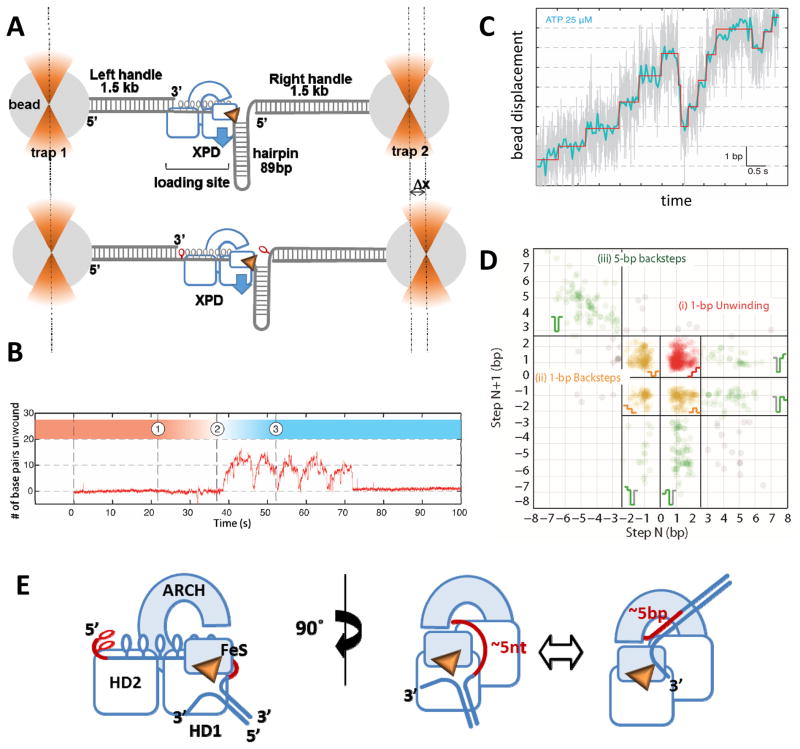
Figure 4. Analysis of XPD stepping kinetics in the high-resolution optical tweezers experiment.
A. The construct for monitoring XPD helicase activity consisted of a dumbbell DNA structure, in which two 1.5 kb handles were tethered to the polystyrene beads held in the optical traps, and an 89 bp hairpin structure flacked by a ssDNA region that served as a loading site for XPD. Blue arrow depicts direction of the XPD movement along the hairpin. Each unwound base pair lengthens the construct by 2 nucleotides (shown in red). The substrate is constructed in a way that allows only the hairpin to be unwound and not the handles (see [67] for details). B. A representative unwinding trajectory. After dumbbell incubation in the XPD containing channel (1) the construct is transferred into the ATP-containing channel (2) where unwinding is detected under the constant applied force as the change in the length of the dumbbell structure resulting in the change in the position of the right bead. This particular trajectory shows 5 consecutive attempts at hairpin unwinding by a single XPD molecule. Each unwinding burst is followed by sliding of the helicase back to the hairpin base. C. A fragment of the representative unwinding trajectory with actual extension data (grey) overlaid with filtered data (blue) and steps determined from fitting the data (red). D. Representative scatter plot of step pairs. Each data points represent the size of every adjacent pair of steps. Consecutive forward steps at highlighted in red, the pairs consisting of at least one backward step are in orange, and 5-bp steps are in green. The data shown were adapted from [67]. E. Proposed mechanics of 5-bp stepping.
Not all helicases display complex kinetic mechanisms. Using high resolution optical tweezers, we showed that XPD unwinds DNA in single base pair steps [70]. In addition to the 1 bp forward steps, frequent 1 bp backward steps and occasional large (~5-bp) backward and forward steps were observed (Figure 4D). The size of the forward and backward steps was uniform across all tested ATP concentrations. Albeit very rare, back-steps were previously observed in high-resolution optical tweezers analysis of NS3 helicase [79]. Yeast Pif1, which like XPD is a 5′-3′ helicase, unwinds dsDNA in single base pair steps [81]. The kinetics of XPD 1 bp steps displayed Michaelis-Menten-like dependence on ATP with the dwell times between individual steps as well as the frequency of backward steps decreasing with increasing ATP concentration. The 5 bp steps were unique to XPD and were ATP-independent [70]. Moreover, a backward 5 bp step was always observed first and was always followed by a 5-bp forward step before XPD could resume single base pair stepping. In contrast, single base pair forward and backward steps displayed no correlation with past behavior [70]. The frequency of backward and forward stepping by XPD displayed a significant sequence dependence: back-stepping was more frequent when the upstream sequence was stable. When the probability to find the terminal base pair open was small, the rate of forward stepping was suppressed and the frequency of back-stepping increased [70]. Frequent back-stepping might not be unique to XPD helicase. Other SF2 helicases and helicase-like translocases may display similar stepping kinetics, especially when analyzed in isolation from their respective supramolecular machines. If analyzed using bulk pre-steady state kinetics, distributions of unwinding times obtained in single-molecule TIRFM measurements, or low-resolution force-extension approaches, the presence of frequent backward steps is likely to result in an erroneous assignment of the step size.
Why are these details important? Accurate characterization of a helicase’s stepping behavior is critical for understanding which attributes of its mechanism are affected by interacting partners, posttranslational modifications and mutations associated with human diseases. Furthermore, this level of analysis is required in order to determine whether all related helicases (for example the four human XPD-like FeS helicases) unwind their respective substrates using the same mechanism. This information may prove invaluable when placing a key helicase in the context of its cellular function or disease-associated malfunction. It may also be useful if/when specific inhibitors are desired to target a single helicase family member without affecting structurally and functionally related enzymes.
3.2. The source of the 5 bp steps
While 1-bp steps can be attributed to the movement of motor domains during the mechanochemical cycle, 5-bp steps can only be explained in the context of translocation strand placement in the secondary DNA binding site: approximately 5 base pairs can be accommodated between the aperture of the hole between HD1, ARCH and FeS domains, which defines the entrance into the secondary DNA binding site, and the wedge feature on the back of the FeS domain, which splits the duplex (Figures 1C & 4E). When the translocating strand dissociates from this secondary DNA binding site, the released 5 nucleotides of the translocating strand re-anneal with the complementary strand, manifesting in a 5-bp back step. This, however, results in the XPD-DNA complex incapable of duplex unwinding. To re-initiate unwinding, the 5 re-annealed base pairs need to reopen, so that the ss-dsDNA junction can reengage correctly (Figure 4E). Not surprisingly, a 5-bp forward step is always observed following a 5-bp backward step and is never detected on its own. The dwell times of the 5-bp steps are exponentially distributed suggesting a single rate-limiting step and are independent of ATP concentration which corroborates the model where the secondary DNA binding site is distant from the canonical bipartite ssDNA binding grove of SF2 helicase [70]. XPD-like helicases FANCJ, RTEL1 and CHLR1 differ from XPD by an insertion in the HD1, which may alter either the architecture of the secondary DNA binding site, or the duplex separation feature. This may affect the presence or the size of large backward steps in their kinetic mechanisms. This said, the 1-bp forward and backward steps are likely represent a mechanistic feature common for all helicases in this family.
3.3 XPD is a partially “active” helicase
Although all helicases use ATP to fuel duplex unwinding and are therefore active motors, these enzymes are designated as “active” or “passive” based on the interaction potential between the helicase and the ss-dsDNA junction [82–84]. “Passive” helicases hydrolyze ATP only to rectify the directionality of translocation. These enzymes take advantage of the transient base pair opening by trapping thermally frayed base pairs. Consequently, the forward rate of a fully passive helicase should be directly proportional to the probability of the upstream duplex to be open. The number of the upstream base pairs that has to be thermally frayed for a passive helicase to advance depends on the physical step size of this helicase. For example, if the helicase unwinds duplex in 1-bp steps, fraying of a single base pair should be sufficient to enable its forward motion. If the helicase makes 2-bp steps, its progression will depend on the probability to find 2 upstream base pairs frayed. In contrast, “active” helicases drive duplex melting and accelerate base pair opening at the ss-dsDNA junction. One distinguishing feature of a fully “active” helicase is that the rates of its translocation on ssDNA and duplex unwinding are equal or nearly equal. Only a few cases are known where the translocation and unwinding rates of a helicase were measured under the same conditions and were found identical. RecBC, Pif1 and UvrD helicases, for example, have also been classified as active helicases whose unwinding and translocation rates are very similar [73, 81, 85]. This said, there exists a broad range of interaction potentials between the helicase and the ss-dsDNA junctions resulting in different degrees to which an individual helicase may destabilize upstream duplex. The sensitivity and resolution of instrumentation, as well as a prior mechanistic knowledge of the assayed enzyme often limit how accurately the interaction potential can be defined. Of a particular importance here are the physical step size and the presence or absence of the back-stepping in the mechanism of a particular enzyme. In the case of yeast Pif1 helicase, which unwinds dsDNA with 1 bp steps and seems to exhibit a tight mechano-chemical coupling by hydrolyzing approximately 1 ATP molecule per 1 bp unwound, a bulk pre-steady state measurement was sufficient to identify it as an active helicase [81]. UvrD translocation and unwinding rates were inferred from the DNA hairpin unzipping and re-zipping rates recorded in the single-molecule force-extension experiment [73]. Arbitrarily, it was proposed to consider a helicase active if it displays >0.25 ratio between unwinding and translocation rates [86]. This classification will place in the same category the helicases whose interaction with the upstream duplex is relatively weak, and those exhibiting idiosyncratic stepping behavior. Only when the stepping kinetics of a particular helicase is disambiguated, can its degree of activity be properly and unambiguously assessed. We showed recently that XPD utilizes a “partially” active unwinding mechanism. Its forward stepping rate during duplex unwinding never matches the 12 nt/s ssDNA translocation rate, but should achieve 12 bp/s at the limit of base pair opening probability of 1. In fact, the rate of its forward stepping is sequence dependent and can be correlated to the base pair opening probability. XPD, however, is not a fully passive helicase: interaction between XPD and ss-dsDNA junction contributes approximately 1.9 kBT to the duplex destabilization [70].
3.4 XPD is a marginally processive helicase
Sequence dependence of the XPD stepping kinetics also affects its processivity: the rate constants of forward and backward stepping are equal when the probability of base pair opening is 0.1, resulting in helicase stalling, slipping or dissociation. Even under applied tension that assists unwinding, XPD processivity on random sequences is limited to tens of base pairs; this modest processivity, however, is consistent with is proposed cellular function(s). While the exact role of archaeal XPD helicase remains unclear, structurally and mechanistically it serves as a good approximation of eukaryotic XPD and Rad3 helicases that participate in nucleotide excision repair and transcription as integral components of the TFIIH complex. Within eukaryotic TFIIH, the bona fide helicase activity of XPD is essential for the nucleotide excision repair, but is dispensable for transcription initiation [87], where it may play a scaffolding role [88] and is positioned away from the DNA [89]. The helicase activity of XPD, or at least the integrity of its FeS domain, is required for transcription associated recombination in mammalian cells [90]. Together with XPB helicase, XPD is also involved the cellular response to retroviral infections [91]. It is unclear, however, whether XPD participates in these processes by itself or as a component of TFIIH. XPD also has additional TFIIH-independent roles that may not require duplex separation. As a part of MMXD (MMS19-MIP19-XPD) complex, XPD localizes to the mitotic spindle where it participates in proper chromosome segregation [92, 93].
Our analysis of XPD stepping kinetics and unwinding mechanism specifically focused on isolated XPD monomers. When two or more XPD monomers were allowed to load onto the same DNA molecule, duplex unwinding processivity was dramatically increased [70]. This enhanced processivity is likely due to reducing the back-stepping probability of the leading XPD monomer by the trailing monomer. In the cell, low processivity derived from intrinsic back-stepping kinetics may provide a means to regulate helicase activity. Although this remains to be confirmed, it is likely that ssDNA binding proteins control back-stepping by archaeal XPD and possibly by other XPD-like helicases. Similarly, the processivity of viral NS3 helicase, a 3′-5′ SF2 enzyme, is enhanced by even a non-cognate ssDNA binding protein (SSB) [94]. Notably, stimulation of both XPD and NS3 helicases does not require physical interaction between the helicase and the SSB [56, 94]. Interacting partners or integration into larger macromolecular machines (such as eukaryotic TFIIH complex) may also modulate the intrinsic back-stepping and ensure that the helicase is processive only when its activity is required. In FANCJ, REL1, and CHLR1, an additional modular domain incorporated into HD1 may be involved in duplex destabilization resulting in the helicases that are both faster and more processive.
4. Interplay between XPD and ssDNA binding proteins
4.1. XPD translocation on protein-coated DNA
In addition to increasing processivity of a helicase by preventing back-stepping, protein partners may directly participate in duplex separation. Ferroplasma acidarmanus XPD helicase, for example, acts in conjunction with RPA2, one of the two cognate archaeal SSBs [56]. RPA2 targets XPD to the fork DNA structures and enhances its helicase activity enabling efficient duplex unwinding by XPD monomers. Based on footprinting analysis, we proposed that not only does RPA2 trap the DNA strands released downstream of the advancing XPD monomers, but that it also actively destabilizes the ss-dsDNA junction ahead of XPD [56]. The two activities may be performed by the same or by different molecules of RPA2. In either case, XPD should be able to navigate RPA2-coated DNA lattice.
Using TIRFM we monitored XPD translocation on individual molecules of protein-free DNA and on DNA coated with ssDNA binding proteins RPA1 or RPA2. A marked decrease in XPD translocation velocity in the presence of RPA2 was observed. We envisioned two possible explanations for the slower translocation: XPD and RPA2 may compete for or co-exist on the same DNA molecule. To distinguish between these possibilities, we visualized the two proteins simultaneously. The presence of Cy5-labeled RPA2 on the DNA was monitored either through fluorescence resonance energy transfer (FRET) from Cy3 on DNA or via direct excitation by red laser light (Figures 5A&B). At the same time, XPD helicase was tracked by following FeS-dependent quenching of Cy3 on DNA or quenching of Cy5 on RPA2 bound to ssDNA. Simultaneous visualization of both the helicase and its obstacle revealed that XPD can translocate over bound RPA2 without dissociating from the lattice and without displacing RPA2. In contrast, the second F. acidarmanus SSB, FacRPA1 competed with XPD for ssDNA access [54]. Considering the ubiquity of SSB and their known functional and physical interactions with DNA helicases, the observed protein bypass by XPD may represent a feature as common as protein displacement by helicases (reviewed in [95]).
Figure 5. Obstacle bypass by XPD helicase visualized in pseudo-tricolor TIRFM experiment.
A & B. Schematic representation of the experimental setups for simultaneous detection of XPD and Cy5-labeled RPA2. In (A), the presence of RPA2 on ssDNA is detected due to FRET between Cy3 on DNA and Cy5 on RPA2, while XPD binding and translocation due to FeS mediated fluorescence quenching of both Cy3 and Cy5. In (B), Cy5-RPA2 fluorescence is excited directly. Note that despite of Cy5-RPA2 abundance in the solution, only molecule bound to the surface-tethered DNA produce fluorescence signal (inset). C. Four schemes for detecting XPD translocation (i) on protein-free DNA, (ii) on the protein-coated ssDNA but with only XPD detection, (iii) on the protein coated ssDNA with simultaneous detection of both ssDNA and RPA2, and (iv) by directly exciting RPA2. Representative fluorescence trajectories and rate distributions are shown on the right. The data shown were adapted from [54].
4.2. A large conformational transition in XPD is expected to coincide with RPA2 bypass
XPD translocation on RPA2-coated ssDNA further broadened the range of known lattices on which helicases and helicase-related motors can translocate. It also raised the question of how do these two proteins manage to share the lattice. RPA2 is a monomer consisting of a single OB fold, similar to that of eukaryotic ssDNA binding proteins [96]. It therefore is expected to interact with the DNA bases [97]. XPD, on the other hand, likely tracks along the phosphodiester backbone of DNA making minimal contacts, if any, with the bases. This would allow an arrangement where XPD contacts the DNA backbone while RPA2 interacts with the bases of the same region of ssDNA simultaneously. Moving over bound RPA2 presents another challenge: the predicted path of the translocating strand within XPD [51, 52] traverses through the narrow hole between the FeS and ARCH domains. In all XPD structures solved to date, this hole is topologically closed and provides an opening only 15–20Å wide, which is too narrow to thread through the ssDNA plus OB fold (about 30Å wide). Due to its domain arrangement, XPD will have to undergo a conformational change to bypass RPA2. ARCH and FeS domain have to be sufficiently separated to accommodate the ssDNA-RPA2 complex (Figure 6A). The same conformational transition is a prerequisite for the ssDNA binding by XPD as a part of TFIIH, which binds to the internal sites on ssDNA, and does not thread the DNA end through the hole, unwinding bubble structures in vitro [98] and in vivo [99]. Another argument for the presence of the conformational transition and its importance in the XPD mechanism has been provided by the recent AFM study which compared the geometries of the XPD-DNA complexes in the presence and in the absence of two distinct types of DNA damage [55]. Both CPD and fluorescein (a mimic of a bulky adduct) were recognized albeit in a different manner by XPD, and both resulted in the complexes containing bent DNA substrate.
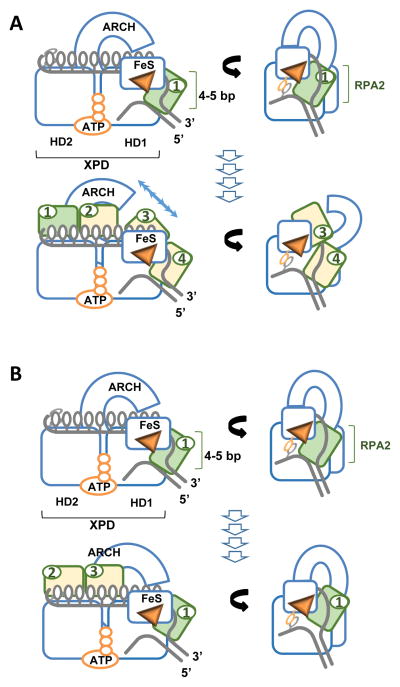
Figure 6. Building an active helicase.
Two models for cooperation between XPD and RPA2 A. Scenario 1: RPA2 (depicted in green) binds at the ss-dsDNA junction, destabilizes 4–5 bp dsDNA upstream of the helicase. XPD then advances into the melted region trapping the released strands. Bound RPA2 moves with the translocating strand through the motor core of XPD. The cycle repeats with the next RPA2 molecule (yellow 2&3) binding at the junction. The prerequisite for this scenario is opening of the ARCH domain sufficient to accommodate RPA2-ssDNA complex. B. Alternatively, The same molecule of RPA2 may remain associated with the helicase jammed between the junction and the hole through which the translocating strand passes. Additional RPA2 molecules (2&3) may trap the unwound strands behind the helicase.
The requirement for the conformational change affecting the ARCH domain of XPD is a plausible explanation for the slower translocation rate on RPA2-coated ssDNA relative to XPD movement on a protein free lattice [54]. Human FeS helicases may have similar “partnerships” with one or several of the small SSBs containing just one or two OB folds, whose numbers continue to grow [100]. Visualization of the XPD conformational dynamics awaits new breakthroughs in structural analyses by X-ray crystallography or SAXS that would trap the protein in a different conformation, or direct visualization of the conformational transitions in the fluorescently-labeled XPD.
4.3. Building a processive helicase
Knowing the precise stepping mechanics of XPD helicase [70], the arrangement of the DNA fork bound to XPD [51, 52], and the functional interaction between XPD and RPA2 [54, 56], can we propose a realistic model of how the two proteins cooperate? Considering that XPD can translocate over RPA2 bound to ssDNA, and that the combination of RPA2 and XPD produces an efficient helicase where RPA2 acts to destabilize duplex, two mechanisms are possible (Figure 6): (i) after RPA2 melts DNA at the junction, XPD translocates over RPA2, which remains associated with ssDNA and now functions to prevent XPD back-stepping; the second RPA2 molecule then binds at the junction and melts the next 4 base pairs; (ii) alternatively, interaction with the ss-dsDNA junction may modulate the ability of XPD to bypass RPA2; RPA in this situation would remain lodged between the pore in the helicase and the junction continuously melting the duplex ahead of the helicase while additional RPA2 molecules trap the separated DNA strands. The former mechanism would necessitate high mobility of the ARCH domain, while the latter is more consistent with the proposed “gripping” function of the pore [101]. To resolve which of the two models is correct will require development of new experimental approaches.
5. Concluding remarks
To date XPD helicase has proven an invaluable model for understanding the structure and mechanism of bio-medically important FeS helicases. It has also spurred the development of cutting edge single-molecule methodologies for analyzing these enzymes. Application of the technologies developed using XPD as a model helicase, such as fluorescence quenching [62], high resolution optical tweezers [70], or simultaneous application of both force and fluorescence detection [102], to human XPD, FANCJ, RTEL1 and CHLR1 is the next challenge to the field. Comprehensive analyses of these helicases acting alone and in the presence of their numerous interacting partners using these methods will provide powerful mechanistic insight into the activities of this prominent helicase family. It will permit us to see how these structurally similar helicases are tuned to perform drastically distinct cellular functions, and how the disease associated mutations in these proteins alter their mechanisms.
High resolution structures of ternary XPD-DNA-ATP complexes may further advance our understanding of this helicase family. The value added benefit of combining the knowledge of helicase mechanism derived from single-molecule analyses with high resolution structures of the helicase “caught” at different steps of its mechanochemical cycle is immense. Besides parsing the explicit features of helicase mechanism and regulation that allow an enzyme from the XPD family to function as a bona fide helicase or as a DNA translocase, it will permit the rational development of novel therapeutic interventions against diseases associated with malfunctions of XPD-like helicases.
Acknowledgments
I would like to express my gratitude to the American Cancer Society (RSG-09-182-01-DMC), Howard Hughes Medical Institute (Early Career Scientist Award), NIH (1 R01 GM101167-01A1) and University of Iowa Carver College of Medicine for the financial support and to Mohamed Ghoneim, Sarah Hengel, David Beyer, Dr. Colin Wu, Dr. Masayoshi Honda and Dr. M. Todd Washington for critically reading the manuscript.
References
- 1.Suhasini AN, Brosh RM., Jr DNA helicases associated with genetic instability, cancer, and aging. Adv Exp Med Biol. 2013;767:123–144. doi: 10.1007/978-1-4614-5037-5_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu CG, Spies M. Overview: what are helicases? Adv Exp Med Biol. 2013;767:1–16. doi: 10.1007/978-1-4614-5037-5_1. [DOI] [PubMed] [Google Scholar]
- 3.Daley JM, Niu H, Sung P. Roles of DNA helicases in the mediation and regulation of homologous recombination. Adv Exp Med Biol. 2013;767:185–202. doi: 10.1007/978-1-4614-5037-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billen D. Spontaneous DNA damage and its significance for the “negligible dose” controversy in radiation protection. Radiat Res. 1990;124:242–245. [PubMed] [Google Scholar]
- 5.Ciccia A, Elledge SJ. The DNA Damage Response: Making It Safe to Play with Knives. Molecular Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanaar R, Hoeijmakers JH, van Gent DC. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 7.Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Campisi J. Cancer, aging and cellular senescence. In Vivo. 2000;14:183–188. [PubMed] [Google Scholar]
- 10.Borges HL, Linden R, Wang JY. DNA damage-induced cell death: lessons from the central nervous system. Cell research. 2008;18:17–26. doi: 10.1038/cr.2007.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lans H, Hoeijmakers JH. Cell biology: ageing nucleus gets out of shape. Nature. 2006;440:32–34. doi: 10.1038/440032a. [DOI] [PubMed] [Google Scholar]
- 12.Beyer DC, Ghoneim MK, Spies M. Structure and Mechanisms of SF2 DNA Helicases. Adv Exp Med Biol. 2013;767:47–73. doi: 10.1007/978-1-4614-5037-5_3. [DOI] [PubMed] [Google Scholar]
- 13.Raney KD, Byrd AK, Aarattuthodiyil S. Structure and Mechanisms of SF1 DNA Helicases. Adv Exp Med Biol. 2013;767:17–46. doi: 10.1007/978-1-4614-5037-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singleton MR, Dillingham MS, Wigley DB. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 15.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Op Struct Biol. 1993;3:419–429. [Google Scholar]
- 16.Morris PD, Tackett AJ, Raney KD. Biotin-streptavidin-labeled oligonucleotides as probes of helicase mechanisms. Methods. 2001;23:149–159. doi: 10.1006/meth.2000.1116. [DOI] [PubMed] [Google Scholar]
- 17.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 18.Sung P, Bailly V, Weber C, Thompson LH, Prakash L, Prakash S. Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature. 1993;365:852–855. doi: 10.1038/365852a0. [DOI] [PubMed] [Google Scholar]
- 19.Sung P, Prakash L, Matson SW, Prakash S. RAD3 protein of Saccharomyces cerevisiae is a DNA helicase. Proceedings of the National Academy of Sciences. 1987;84:8951–8955. doi: 10.1073/pnas.84.24.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathieu N, Kaczmarek N, Ruthemann P, Luch P, Naegeli H. DNA quality control by a lesion sensor pocket of the xeroderma pigmentosum group D helicase subunit of TFIIH. Current Biology. 2013 doi: 10.1016/j.cub.2012.12.032. [DOI] [PubMed] [Google Scholar]
- 21.Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 22.Sommers JA, Rawtani N, Gupta R, Bugreev DV, Mazin AV, Cantor SB, Brosh RM. FANCJ Uses Its Motor ATPase to Destabilize Protein-DNA Complexes, Unwind Triplexes, and Inhibit RAD51 Strand Exchange. Journal of Biological Chemistry. 2009;284:7505–7517. doi: 10.1074/jbc.M809019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, Tissenbaum H, West SC, Rose AM, Boulton SJ. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vannier J-B, Pavicic-Kaltenbrunner Mark V, Petalcorin IR, Ding H, Boulton Simon J. RTEL1 Dismantles T Loops and Counteracts Telomeric G4-DNA to Maintain Telomere Integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 26.Amann J, Kidd VJ, Lahti JM. Characterization of Putative Human Homologues of the Yeast Chromosome Transmission Fidelity Gene, CHL1. Journal of Biological Chemistry. 1997;272:3823–3832. doi: 10.1074/jbc.272.6.3823. [DOI] [PubMed] [Google Scholar]
- 27.Kuper J, Kisker C. DNA Helicases in NER, BER, and MMR. In: Spies M, editor. DNA Helicases and DNA Motor Proteins. Springer; New York: 2013. pp. 203–224. [DOI] [PubMed] [Google Scholar]
- 28.Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, Wiegant WW, Elghalbzouri-Maghrani E, Steltenpool J, Rooimans MA, Pals G, Arwert F, Mathew CG, Zdzienicka MZ, Hiom K, De Winter JP, Joenje H. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group. J Nature genetics. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 29.Levran O, Attwooll C, Henry RT, Milton KL, Neveling K, Rio P, Batish SD, Kalb R, Velleuer E, Barral S, Ott J, Petrini J, Schindler D, Hanenberg H, Auerbach AD. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nature genetics. 2005;37:931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- 30.Uringa E-J, Lisaingo K, Pickett HA, Brind’Amour J, Rohde J-H, Zelensky A, Essers J, Lansdorp PM. RTEL1 contributes to DNA replication and repair and telomere maintenance. Molecular Biology of the Cell. 2012;23:2782–2792. doi: 10.1091/mbc.E12-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, ONNJ, Rose AM, West SC, Meyer BJ, Boulton SJ. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327:1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parish JL, Rosa J, Wang X, Lahti JM, Doxsey SJ, Androphy EJ. The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells. Journal of Cell Science. 2006;119:4857–4865. doi: 10.1242/jcs.03262. [DOI] [PubMed] [Google Scholar]
- 33.van Brabant AJ, Stan R, Ellis NA. DNA helicases, genomic instability, and human genetic disease. Annu Rev Genomics Hum Genet. 2000;1:409–459. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- 34.Suhasini AN, Brosh RM., Jr Disease-causing missense mutations in human DNA helicase disorders. Mutation Research/Reviews in Mutation Research. 2012 doi: 10.1016/j.mrrev.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Shete S, Etzel CJ, Scheurer M, Alexiou G, Armstrong G, Tsavachidis S, Liang FW, Gilbert M, Aldape K, Armstrong T, Houlston R, Hosking F, Robertson L, Xiao Y, Wiencke J, Wrensch M, Andersson U, Melin BS, Bondy M. Polymorphisms of LIG4, BTBD2, HMGA2, and RTEL1 genes involved in the double-strand break repair pathway predict glioblastoma survival. J Clin Oncol. 2010;28:2467–2474. doi: 10.1200/JCO.2009.26.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, Giannini C, Halder C, Kollmeyer TM, Kosel ML, LaChance DH, McCoy L, O’Neill BP, Patoka J, Pico AR, Prados M, Quesenberry C, Rice T, Rynearson AL, Smirnov I, Tihan T, Wiemels J, Yang P, Wiencke JK. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nature genetics. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ballew B, Yeager M, Jacobs K, Giri N, Boland J, Burdett L, Alter B, Savage S. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in Dyskeratosis congenita. Human Genetics. 2013:1–8. doi: 10.1007/s00439-013-1265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D’Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panes J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D’Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nature genetics. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mancini M, Petretto E, Kleinert C, Scavone A, De T, Cook S, Silhavy J, Zidek V, Pravenec Gd’Amati M, Camici PG. Mapping genetic determinants of coronary microvascular remodeling in the spontaneously hypertensive rat. Basic Res Cardiol. 2013;108:316. doi: 10.1007/s00395-012-0316-y. [DOI] [PubMed] [Google Scholar]
- 40.van der Lelij P, Chrzanowska KH, Godthelp BC, Rooimans MA, Oostra AB, Stumm M, Zdzienicka MgZ, Joenje H, de Winter JP. Warsaw Breakage Syndrome, a Cohesinopathy Associated with Mutations in the XPD Helicase Family Member DDX11/ChlR1. The American Journal of Human Genetics. 2010;86:262–266. doi: 10.1016/j.ajhg.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y, Suhasini AN, Brosh RM., Jr Welcome the Family of FANCJ-like Helicases to the Block of Genome Stability Maintenance Proteins. Cellular and Molecular Life Sciences. 2009;66:1209–1222. doi: 10.1007/s00018-008-8580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y, Brosh RM. DNA helicase and helicase/nuclease enzymes with a conserved iron-sulfur cluster. Nucleic acids research. 2012;40:4247–4260. doi: 10.1093/nar/gks039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudolf J, Makrantoni V, Ingledew WJ, Stark MJ, White MF. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol Cell. 2006;23:801–808. doi: 10.1016/j.molcel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 44.Voloshin ON, Vanevski F, Khil PP, Camerini-Otero RD. Characterization of the DNA damage-inducible helicase DinG from Escherichia coli. The Journal of biological chemistry. 2003;278:28284–28293. doi: 10.1074/jbc.M301188200. [DOI] [PubMed] [Google Scholar]
- 45.Pugh RA, Honda M, Leesley H, Thomas A, Lin Y, Nilges MJ, Cann IK, Spies M. The Iron-containing Domain Is Essential in Rad3 Helicases for Coupling of ATP Hydrolysis to DNA Translocation and for Targeting the Helicase to the Single-stranded DNA-Double-stranded DNA Junction. The Journal of biological chemistry. 2008;283:1732–1743. doi: 10.1074/jbc.M707064200. [DOI] [PubMed] [Google Scholar]
- 46.Cantor S, Drapkin R, Zhang F, Lin Y, Han J, Pamidi S, Livingston DM. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2357–2362. doi: 10.1073/pnas.0308717101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirota Y, Lahti JM. Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic acids research. 2000;28:917–924. doi: 10.1093/nar/28.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolski SC, Kuper J, Hanzelmann P, Truglio JJ, Croteau DL, Van Houten B, Kisker C. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 2008;6:e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, McRobbie AM, Brown SE, Naismith JH, White MF. Structure of the DNA repair helicase XPD. Cell. 2008;133:801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuper J, Wolski SC, Michels G, Kisker C. Functional and structural studies of the nucleotide excision repair helicase XPD suggest a polarity for DNA translocation. EMBO J. 2012;31:494–502. doi: 10.1038/emboj.2011.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pugh RA, Wu CG, Spies M. Regulation of translocation polarity by helicase domain 1 in SF2B helicases. EMBO J. 2012;31:503–514. doi: 10.1038/emboj.2011.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rudolf J, Rouillon C, Schwarz-Linek U, White MF. The helicase XPD unwinds bubble structures and is not stalled by DNA lesions removed by the nucleotide excision repair pathway. Nucleic acids research. 2010;38:931–941. doi: 10.1093/nar/gkp1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Honda M, Park J, Pugh RA, Ha T, Spies M. Single-molecule analysis reveals differential effect of ssDNA-binding proteins on DNA translocation by XPD helicase. Mol Cell. 2009;35:694–703. doi: 10.1016/j.molcel.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buechner CN, Heil K, Michels G, Carell T, Kisker C, Tessmer I. Strand specific recognition of DNA damages by XPD provides insights into Nucleotide Excision Repair substrate versatility. The Journal of biological chemistry. 2013 doi: 10.1074/jbc.M113.523001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pugh RA, Lin Y, Eller C, Leesley H, Cann IK, Spies M. Ferroplasma acidarmanus RPA2 facilitates efficient unwinding of forked DNA substrates by monomers of FacXPD helicase. J Mol Biol. 2008;383:982–998. doi: 10.1016/j.jmb.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 57.Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol. 2006;59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 58.Mui TP, Fuss JO, Ishida JP, Tainer JA, Barton JK. ATP-Stimulated, DNA-Mediated Redox Signaling by XPD, a DNA Repair and Transcription Helicase. J Am Chem Soc. 2011;133:16378–16381. doi: 10.1021/ja207222t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White MF, Dillingham MS. Iron-sulphur clusters in nucleic acid processing enzymes. Current Opinion in Structural Biology. 2012;22:94–100. doi: 10.1016/j.sbi.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 60.Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 2011;8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Wietmarschen N, Moradian A, Morin GB, Lansdorp PM, Uringa EJ. The mammalian proteins MMS19, MIP18, and ANT2 are involved in cytoplasmic iron-sulfur cluster protein assembly. J Biol Chem. 2012;287:43351–43358. doi: 10.1074/jbc.M112.431270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pugh RA, Honda M, Spies M. Ensemble and single-molecule fluorescence-based assays to monitor DNA binding, translocation, and unwinding by iron-sulfur cluster containing helicases. Methods. 2010 doi: 10.1016/j.ymeth.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clegg RM. FRET tells us about proximities, distances, orientations and dynamic properties. J Biotechnol. 2002;82:177–179. [PubMed] [Google Scholar]
- 64.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Masuda-Ozawa T, Hoang T, Seo YS, Chen LF, Spies M. Single-molecule sorting reveals how ubiquitylation affects substrate recognition and activities of FBH1 helicase. Nucleic Acids Res. 2013;41:3576–3587. doi: 10.1093/nar/gkt056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 67.Mackintosh SG, Raney KD. DNA unwinding and protein displacement by superfamily 1 and superfamily 2 helicases. Nucleic acids research. 2006;34:4154–4159. doi: 10.1093/nar/gkl501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moffitt JR, Chemla YR, Smith SB, Bustamante C. Recent Advances in Optical Tweezers. Annual Review of Biochemistry. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 69.Bustamante C, Chemla YR, Moffitt JR. High-resolution dual-trap optical tweezers with differential detection. Single-molecule techniques: a laboratory manual. 2008:297–324. [Google Scholar]
- 70.Qi Z, Pugh RA, Spies M, Chemla YR. Sequence-dependent base pair stepping dynamics in XPD helicase unwinding. eLife. 2013;2 doi: 10.7554/eLife.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ali JA, Lohman TM. Kinetic Measurement of the Step Size of DNA Unwinding by Escherichia coli UvrD Helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 72.Eoff RL, Raney KD. Intermediates revealed in the kinetic mechanism for DNA unwinding by a monomeric helicase. Nature structural & molecular biology. 2006;13:242–249. doi: 10.1038/nsmb1055. [DOI] [PubMed] [Google Scholar]
- 73.Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang XD, Dou SX, Xie P, Hu JS, Wang PY, Xi XG. Escherichia coli RecQ Is a Rapid, Efficient, and Monomeric Helicase. Journal of Biological Chemistry. 2006;281:12655–12663. doi: 10.1074/jbc.M513089200. [DOI] [PubMed] [Google Scholar]
- 75.Dillingham MS, Wigley DB, Webb MR. Demonstration of Unidirectional Single-Stranded DNA Translocation by PcrA Helicase: Measurement of Step Size and Translocation Speed. Biochemistry. 1999;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 76.Lee JY, Yang W. UvrD Helicase Unwinds DNA One Base Pair at a Time by a Two-Part Power Stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A Nonuniform Stepping Mechanism for E. coli UvrD Monomer Translocation along Single-Stranded DNA. Molecular Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheng W, Arunajadai SG, Moffitt JR, Tinoco I, Bustamante C. Single-Base Pair Unwinding and Asynchronous RNA Release by the Hepatitis C Virus NS3 Helicase. Science. 2011;333:1746–1749. doi: 10.1126/science.1206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Myong S, Bruno MM, Pyle AM, Ha T. Spring-Loaded Mechanism of DNA Unwinding by Hepatitis C Virus NS3 Helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramanagoudr-Bhojappa R, Chib S, Byrd AK, Aarattuthodiyil S, Pandey M, Patel SS, Raney KD. Yeast Pif1 Helicase Exhibits a One-base-pair Stepping Mechanism for Unwinding Duplex DNA. Journal of Biological Chemistry. 2013;288:16185–16195. doi: 10.1074/jbc.M113.470013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Betterton MD, Jülicher F. Opening of nucleic-acid double strands by helicases: Active versus passive opening. Physical Review E. 2005;71:011904. doi: 10.1103/PhysRevE.71.011904. [DOI] [PubMed] [Google Scholar]
- 83.Betterton MD, Julicher F. Velocity and processivity of helicase unwinding of double-stranded nucleic acids. J Phys Condens Matter. 2005;17:S3851–3869. doi: 10.1088/0953-8984/17/47/015. [DOI] [PubMed] [Google Scholar]
- 84.Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines Part I: Structures and properties of isolated helicases. Quarterly Reviews of Biophysics. 2002;35:431–478. doi: 10.1017/s0033583502003852. [DOI] [PubMed] [Google Scholar]
- 85.Wu CG, Bradford C, Lohman TM. Escherichia coli RecBC helicase has two translocase activities controlled by a single ATPase motor. Nature structural & molecular biology. 2010;17:1210–1217. doi: 10.1038/nsmb.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Manosas M, Xi XG, Bensimon D, Croquette V. Active and passive mechanisms of helicases. Nucleic acids research. 2010;38:5518–5526. doi: 10.1093/nar/gkq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Winkler GS, Araujo SJ, Fiedler U, Vermeulen W, Coin F, Egly JM, Hoeijmakers JH, Wood RD, Timmers HT, Weeda G. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J Biol Chem. 2000;275:4258–4266. doi: 10.1074/jbc.275.6.4258. [DOI] [PubMed] [Google Scholar]
- 88.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 89.He Y, Fang J, Taatjes DJ, Nogales E. Structural visualization of key steps in human transcription initiation. Nature. 2013;495:481–486. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Savolainen L, Cassel T, Helleday T. The XPD subunit of TFIIH is required for transcription-associated but not DNA double-strand break-induced recombination in mammalian cells. Mutagenesis. 2010;25:623–629. doi: 10.1093/mutage/geq054. [DOI] [PubMed] [Google Scholar]
- 91.Yoder K, Sarasin A, Kraemer K, McIlhatton M, Bushman F, Fishel R. The DNA repair genes XPB and XPD defend cells from retroviral infection. Proc Natl Acad Sci U S A. 2006;103:4622–4627. doi: 10.1073/pnas.0509828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ito S, Tan LJ, Andoh D, Narita T, Seki M, Hirano Y, Narita K, Kuraoka I, Hiraoka Y, Tanaka K. MMXD, a TFIIH-independent XPD-MMS19 protein complex involved in chromosome segregation. Mol Cell. 2010;39:632–640. doi: 10.1016/j.molcel.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 93.Chen J, Larochelle S, Li X, Suter B. Xpd/Ercc2 regulates CAK activity and mitotic progression. Nature. 2003;424:228–232. doi: 10.1038/nature01746. [DOI] [PubMed] [Google Scholar]
- 94.Rajagopal V, Patel SS. Single Strand Binding Proteins Increase the Processivity of DNA Unwinding by the Hepatitis C Virus Helicase. Journal of Molecular Biology. 2008;376:69–79. doi: 10.1016/j.jmb.2007.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Spies M, Ha T. Inching over hurdles: How DNA helicases move on crowded lattices. Cell Cycle. 2010;9:1742–1749. doi: 10.4161/cc.9.9.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kerr ID, Wadsworth RI, Cubeddu L, Blankenfeldt W, Naismith JH, White MF. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of Sulfolobus SSB protein. Embo J. 2003;22:2561–2570. doi: 10.1093/emboj/cdg272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bochkarev A, Bochkareva E, Frappier L, Edwards AM. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. Embo J. 1999;18:4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rudolf J, Rouillon C, Schwarz-Linek U, White MF. The helicase XPD unwinds bubble structures and is not stalled by DNA lesions removed by the nucleotide excision repair pathway. Nucl Acids Res. 2009:gkp1058. doi: 10.1093/nar/gkp1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boulikas T. Xeroderma pigmentosum and molecular cloning of DNA repair genes. Anticancer Res. 1996;16:693–708. [PubMed] [Google Scholar]
- 100.Richard DJ, Bolderson E, Khanna KK. Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Critical Reviews in Biochemistry and Molecular Biology. 2009;44:98–116. doi: 10.1080/10409230902849180. [DOI] [PubMed] [Google Scholar]
- 101.Wolski SC, Kuper J, Kisker C. The XPD helicase: XPanDing archaeal XPD structures to get a grip on human DNA repair. Biol Chem. 2010;391:761–765. doi: 10.1515/BC.2010.076. [DOI] [PubMed] [Google Scholar]
- 102.Comstock MJ, Ha T, Chemla YR. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat Meth. 2011;8:335–340. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]