Abstract
Bartonella species are blood-borne, re-emerging organisms, capable of causing prolonged infection with diverse disease manifestations, from asymptomatic bacteremia to chronic debilitating disease and death. This pathogen can survive for over a month in stored blood. However, its prevalence among blood donors is unknown, and screening of blood supplies for this pathogen is not routinely performed. We investigated Bartonella spp. prevalence in 500 blood donors from Campinas, Brazil, based on a cross-sectional design. Blood samples were inoculated into an enrichment liquid growth medium and sub-inoculated onto blood agar. Liquid culture samples and Gram-negative isolates were tested using a genus specific ITS PCR with amplicons sequenced for species identification. Bartonella henselae and Bartonella quintana antibodies were assayed by indirect immunofluorescence. B. henselae was isolated from six donors (1.2%). Sixteen donors (3.2%) were Bartonella-PCR positive after culture in liquid or on solid media, with 15 donors infected with B. henselae and one donor infected with Bartonella clarridgeiae. Antibodies against B. henselae or B. quintana were found in 16% and 32% of 500 blood donors, respectively. Serology was not associated with infection, with only three of 16 Bartonella-infected subjects seropositive for B. henselae or B. quintana. Bartonella DNA was present in the bloodstream of approximately one out of 30 donors from a major blood bank in South America. Negative serology does not rule out Bartonella spp. infection in healthy subjects. Using a combination of liquid and solid cultures, PCR, and DNA sequencing, this study documents for the first time that Bartonella spp. bacteremia occurs in asymptomatic blood donors. Our findings support further evaluation of Bartonella spp. transmission which can occur through blood transfusions.
Author Summary
Bartonella is a genus of small bacteria with worldwide distribution, transmitted by blood-sucking insects, and is capable of causing disease in humans and animals. Some of the clinical presentations of Bartonella spp., such as cat scratch disease, trench fever, and bacillary angiomatosis are well documented; however, novel presentations have been described in the last two decades, ranging from cyclic flu-like syndrome to neurologic disease and life-threatening endocarditis. Asymptomatic human infection is possible and accidental blood transmission has been reported. Bacterium isolation is very difficult because they grow slowly and require special culture media and procedures. Serology testing poorly predicts active Bartonella infection, except in infection of cardiac valves. Therefore, diagnosis is generally challenging. However, when molecular detection techniques are coupled with special culture protocols, enhanced sensitivity and specificity can be achieved. We investigated Bartonella spp. infection prevalence in a large blood donor population and confirmed bacteremia in 1.2% of the subjects. Bloodstream infection was detected with at least three different molecular methods in 3.2% of donors. These results indicate that Bartonella is a genus of importance for transfusion medicine.
Introduction
Bartonella, a genus of fastidious bacteria with worldwide distribution, is responsible for persistent infections in animals and humans [1]. Bartonella spp. are considered neglected zoonotic pathogens, presumed to be transmitted to humans by a variety of arthropod vectors including sandflies, body lice, fleas, ticks, and keds [1,2]. During the past several years, the spectrum of clinical manifestations associated with bartonellosis, a term that now encompasses infection with any Bartonella spp., has widened substantially [3]. In humans, Bartonella spp. are known causative agents of Peruvian bartonellosis, cat scratch disease, trench fever, and bacillary angiomatosis [1]. However, more recent studies have documented bloodstream infections in patients with cardiovascular, neurological, and rheumatologic disease manifestations [4,5]. With the exception of localized lymphadenopathy or blood-culture-negative endocarditis, physicians rarely consider Bartonella sp. infection among differential diagnoses [6].
Bartonella spp. are able to infect and survive inside erythrocytes [7], resulting in a long-lasting intraerythrocytic and presumably intraendothelial infection, which can be associated with a relapsing pattern of bacteremia [8]. In vitro, these bacteria have been shown to invade, multiply within, and persist for the lifetime of the infected host cell [9,10]. Prolonged bacteremia allows greater opportunity for arthropod vector and other modes of transmission to occur between hosts. Although at least fifteen Bartonella spp. have been associated with human infections, B. henselae is the most frequent species identified from humans, as well as from companion animals such as cats and dogs [1,9]. There is no single gold standard methodology to diagnose bartonellosis and multi-step platforms are necessary to decrease false-negative test results [1]. Culture in liquid and solid media, multiple PCR reactions and serology have been used together to improve the diagnostic sensitivity [8,11].
Previous studies by our group using transmission electron microscopy and culture isolation have documented the ability of B. henselae to survive in stored blood for 35 days, suggesting the potential for transfusion-associated transmission [12]. We also documented B. henselae adhered to human erythrocytes 10 hours after inoculation of the bacteria into blood and intraerythrocytic infection after 72 hours [13]. These results suggested a potentially important role for Bartonella sp. in transfusion medicine, particularly as blood transfusion infection has been documented in cats [14] and needle stick transmission of Bartonella sp. has also been reported in two veterinarians [15,16]. Since the presence of selected Bartonella spp. was previously documented in blood samples of asymptomatic subjects [17–20], we hypothesized that bloodstream infection with Bartonella sp. occurs in blood donors at the time of donation. The objective of this study was to determine the seroprevalence and frequency of bacteremia caused by Bartonella spp. in a large asymptomatic blood donor population in Campinas, São Paulo State, Brazil.
Methods
Study design and participants
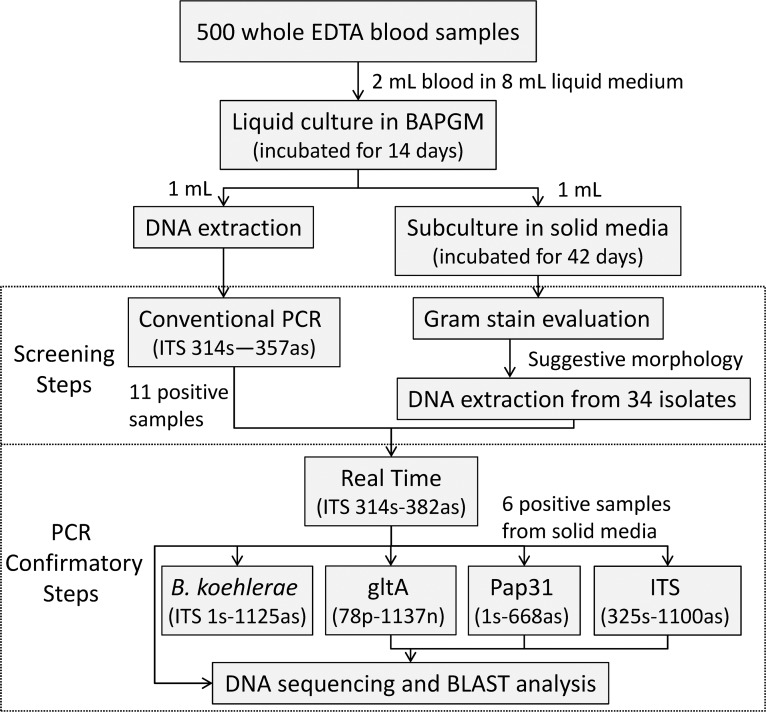
A cross-sectional study was conducted at the UNICAMP Blood Bank (HEMOCENTRO), which serves a geographic region with an estimated population of 6.4 million people in the São Paulo state, Brazil. Healthy blood donors were randomly recruited from November 19th to December 23rd 2009, at the time of their voluntary blood donation. Sample size was estimated in 473 subjects to allow for estimation of at least 5% prevalence of bloodstream infection with Bartonella spp. in blood donors, with a desired precision of 5% given a 95% confidence limit and a design effect of 1. Therefore, with possible attrition, we enrolled 500 donors. This study was approved by the Research Ethics Committee of the University of Campinas (UNICAMP), Brazil (CEP122/2005). An informed written consent, approved by the UNICAMP Research Ethics Committee, was obtained from each participant. Donor selection, blood collection, and infectious disease screening were performed in accordance with current international standards [21,22]. Following aseptic preparation of the venipuncture site and immediately after the collection of a blood unit, an additional 5 mL of whole blood was collected into a tube with ethylenediaminetetraacetic acid (EDTA) and another 5 mL was collected into a serum separator tube via the accessory port. Samples were stored at −20°C until analysis at the University of Campinas and subsequently at Western University of Health Sciences. An overview of the diagnostic procedures performed in this study is presented in the Fig. 1.
Figure 1. Flowchart of the culture and PCR-based procedures performed to determine Bartonella prevalence in 500 blood donors from Campinas, Brazil.
Blood culture
Culture procedures used in this study followed the previous description by Duncan et al. [11] and Maggi et al. [8], with modifications. Two milliliters of whole EDTA blood from each subject were thawed and added into 8 mL of liquid Bartonella alpha-Proteobacteria growth medium (BAPGM) [23], incubated at 37°C in 5% CO2, water-saturated atmosphere and maintained with a constant shaking motion for 14 days. Blood donor sample cultures were manipulated in batches of 75 flasks. A negative control flask containing only BAPGM medium was added to each batch of samples tested and subjected to the same laboratory procedures and culture conditions. Subsequently, 1 mL of the blood-inoculated liquid culture was sub-inoculated onto agar enriched slant tubes containing 30% Bartonella spp.-negative sheep blood (confirmed by PCR and culture methods) [24] for additional 42 days. BAPGM-negative controls were also subcultured onto blood-agar. The agar slant tubes were inspected weekly for evidence of bacterial growth. Because blood cultures in BAPGM may yield other species of alpha-Proteobacteria [25], an initial screening process was performed as demonstrated in Fig. 1. Colonies were Gram-stained and those isolates with suggestive morphology were suspended and frozen in Brain and Heart Infusion (BHI) for future identification by DNA amplification and sequencing. All Bartonella spp. culture methods were carried out in a class 2 biosafety cabinet in order to minimize the possibility of specimen contamination and to protect laboratory personnel.
Bartonella genus PCR screening
Molecular techniques were performed in four separate rooms to avoid DNA contamination. A uni-directional workflow was strictly enforced between pre-PCR areas (sample handling, PCR set up, DNA extraction) and post-PCR areas (DNA amplification, gel analysis, and amplicon purification). Dedicated sets of equipment, pipettes, and supplies were used in each of these locations. Strict laboratory procedures were implemented in order to avoid potential contamination of reagents and samples with amplicons. In order to prevent PCR contamination, different negative controls testing the different stages in the PCR process was included in every experiment as described in the S1 Appendix.
After a 14-day incubation period, a 1 mL aliquot of liquid culture medium was centrifuged and the pellet was subjected to DNA extraction using the QIAamp® DNA Mini Kit (QIAGEN Inc., Valencia, CA). The average of DNA yield obtained was 6.19 ng/ul (SD: 32.8 ng/ul), and the average 260/280 ratio was 1.47 (SD: 2.03). Screening of the 500 liquid culture samples was performed using Bartonella genus-specific single tube PCR. This assay was manually designed to target a hypervariable region of the 16S-23S rRNA gene intergenic transcribed spacer (ITS) of Bartonella species using primers and conditions described in the S1 Appendix. For all PCR reactions three controls were incorporated: negative BAPGM control (BAPGM medium with no blood inoculate and incubated simultaneously with each batch of liquid cultures), Mastermix reagent control, and a positive control (DNA extracted from a Houston 1-ITS strain of B. henselae; ATCC 49882). These screening tests were performed at the Multidisciplinary Center of Biological Investigation (CEMIB), UNICAMP, Brazil.
Bartonella species identification
DNA obtained from Bartonella sp. positive liquid cultures and DNA from suspected subculture isolates were retested at Western University of Health Sciences, USA, for species identification. These DNA samples were tested by a Real-Time ITS PCR using primers manually designed to amplify a fragment of the 16S-23S rRNA ITS region of all Bartonella species. PCR primers and conditions are described in the S1 Appendix.
DNA samples from six isolates that were confirmed to contain Bartonella sp. DNA were further characterized using previously published conventional PCR assays for the ITS region [26], for the citrate synthase gene (gltA) [27], and the heme-binding phage-associated protein (pap31) gene [26]. In addition, each sample was also tested using a specific conventional PCR for Bartonella koehlerae [28]. DNA samples from liquid cultures were not tested by these assays due to insufficient genomic material. Amplicons generated from any one of the five PCR assays used during the confirmatory steps (Fig. 1) were sequenced for bacterial species identification (S1 Appendix).
Serology
Using B. henselae and B. quintana antigens supplied by the Centers for Disease Control and Prevention (CDC-Atlanta, USA), serum samples were analyzed for IgG antibodies to B. henselae and B. quintana antigens by an indirect immunofluorescence assay (IFA) as described in the S1 Appendix. Sera samples were tested at a 1:64 dilution. A positive test was warranted if brightly stained bacteria could be detected by fluorescence microscopy at 400× magnification. Previously IFA negative serum samples were used as negative controls.
Data analysis
Subjects with positive ITS PCR results from isolates were considered bacteremic. Subjects positive from liquid culture were considered with Bartonella sp. bloodstream infection. Molecular and serologic prevalence of Bartonella spp. were described as absolute frequencies, percentages, and 95% confidence intervals (computed using score method) using JMP Pro 10 for Windows (SAS Institute Inc., Cary, NC).
Results
From the 500 blood donors tested, 16 (3.2%, 95% Confidence Interval [CI]: 2.0%–5.1%) subjects were infected with a Bartonella spp. based on culture in liquid and/or isolation on solid media followed by ITS Real-Time and/or conventional PCR in two different laboratories. ITS amplicon sequence analysis revealed B. henselae in 15 of the 16 cultures (3%, 95% CI: 1.8%–4.9%), and B. clarridgeiae in one culture (0.2%, 95% CI: 0%–1.1%) (Table 1). Among the 16 Bartonella sp. bloodstream-infected donors, 11 were confirmed by liquid culture followed by PCR amplifications and DNA sequencing, whereas six bacteremic individuals were confirmed after subculture onto blood slant tubes followed by PCR and DNA sequence confirmation (Table 1). Only one subject was PCR positive in both liquid and solid subcultures.
Table 1. Serology and DNA sequencing results of 16 blood donors tested for exposure or infection with Bartonella species.
| Blood donor |
Serology (IFA)a
|
Bartonella species identified by DNA sequencing
|
|||||
|---|---|---|---|---|---|---|---|
| B. henselae | B. quintana | Sample tested | ITSb 314s-382as | ITSb 325s-1100as | Pap31c 1s-668as | gltAd 781p-1137n | |
| 1 | Negative | Negative | Liquid culture | B. henselae | NPe | NP | NP |
| 2 | Negative | Negative | Liquid culture/isolate | B. henselaef | B. henselae | Negative | Negative |
| 3 | Negative | Negative | Isolate | B. henselae | Negative | Positiveg | B. henselae |
| 4 | Positive | Negative | Liquid culture | B. henselae | NP | NP | NP |
| 5 | Negative | Negative | Isolate | B. henselae | B. henselae | B. henselae | B. henselae |
| 6 | Negative | Negative | Isolate | B. henselae | Negative | Negative | B. henselae |
| 7 | Negative | Negative | Isolate | B. henselae | B. henselae | B. henselae | B. henselae |
| 8 | Negative | Negative | Liquid culture | B. henselae | NP | NP | NP |
| 9 | Negative | Negative | Liquid culture | B. henselae | NP | NP | NP |
| 10 | Negative | Negative | Liquid culture | B. clarridgeiae | NP | NP | NP |
| 11 | Negative | Negative | Liquid culture | B. henselae | NP | NP | NP |
| 12 | Negative | Negative | Liquid culture | B. henselae | NP | NP | NP |
| 13 | Negative | Negative | Liquid culture | B. henselae | NP | NP | NP |
| 14 | Negative | Positive | Liquid culture | B. henselae | NP | NP | NP |
| 15 | Negative | Negative | Liquid culture | B. henselae | NP | NP | NP |
| 16 | Positive | Positive | Isolate | B. henselae | B. henselae | B. henselae | B. henselae |
a Indirect immunofluorescence assay with cut-off of 1:64.
b 16S-23S rRNA gene intergenic transcribed spacer
c Heme-binding phage-associated protein
d Citrate synthase gene
e Not performed due to insufficient genomic material
f Both liquid culture and isolate yield the same Bartonella species. Other PCR assays were only performed on isolate due to insufficient genomic material from liquid culture.
g A DNA sequence was not obtained for this sample.
When subsequently amplified and sequenced, four of these six isolates contained a larger ITS region fragment (559 bp in size) that was 100% similar to B. henselae sequences deposited in GenBank (accession number NC_005956.1). From one liquid culture sample, a 190 bp ITS DNA sequence was obtained, being 100% similar to B. clarridgeiae (accession number NC_014932.1). Similarly, the presence of B. henselae DNA was confirmed in five isolates by also amplifying and sequencing the gltA gene (338 bp, 100% similarity to accession number BX897699.1), and from three by amplification and sequencing of the pap31 gene (501 bp, 100% similarity to accession number DQ529248.1). By testing the isolates, no blood donor was B. koehlerae PCR positive using species-specific ITS primers. The methodologies applied in our laboratory allowed the isolation of fastidious bacteria morphologically similar to Bartonella by Gram stain from 34 blood donors, with six isolates confirmed as Bartonella spp. by DNA sequencing. Some of the possible genera of the other isolates obtained in this study include Arthrobacter, Bacillus, Dermabacter, Methylobacterium, Propionibacterium, Pseudomonas, Sphingomonas, Staphylococcus and unknown “non-cultured” bacteria, as previously reported by Cadenas et al. using the same liquid enrichment culture method [25].
Of the 500 blood donors tested, antibodies against B. henselae or B. quintana were detected in 16.2% (81/500, 95% CI: 13.2%–19.7%) and 32% (160/500, 95% CI: 28.0%–36.2%), respectively. Seropositivity for both antigens was detected in 13.4% of blood donors (67/500, 95% CI: 10.6%–16.7%). B. quintana DNA was not detected in any blood donor in this study. Only two of the B. henselae seroreactive blood donors were confirmed by liquid culture/PCR as infected with B. henselae. However, two B. quintana seroreactive donors had confirmed B. henselae bloodstream infection, one of whom was also seroreactive to B. henselae. The B. clarridgeiae bacteremic subject was seronegative to both Bartonella spp. antigens (Table 1).
Discussion
Using a combination of liquid and solid cultures, PCR, DNA sequencing, and serology, this study documented the presence of Bartonella spp. bloodstream infections in 16 (3.2%) of 500 healthy blood donors presented to a major blood bank in Southeastern Brazil. Despite the fact that exposure of blood donors to Bartonella spp. has been previously documented by serology methods [29,30], no previous study has confirmed the presence of Bartonella spp. in blood donors using similar culture and molecular diagnostic methods. A total of six B. henselae isolates were obtained in this study, with other ten blood donors having blood infection with B. henselae or B. clarridgeiae identified by liquid culture enrichment coupled with DNA amplification by PCR at two different laboratories. These results indicate, for the first time, that asymptomatic blood donors can be infected with Bartonella spp. at the time of blood donation.
Bartonella spp. are re-emerging infectious agents that can induce asymptomatic and intraerythrocytic infection in preferred and accidental hosts. These bacteria have been isolated from both immunocompetent and immunodeficient human patients [8,9,31]. Any asymptomatic infection with a blood-borne pathogen has the potential for transfusion transmission. The survival of B. henselae in collected human blood units [12] associated with the bacterium’s ability to cause infection by the intravenous route as described in animal models [14,32], and in humans after needle stick accidents [15,16], supports a potential risk for transmission through blood transfusions. Although well-documented human cases of blood transfusion transmission have not yet been published, our data further support the hypothesis that this can occur.
Transmission via blood transfusion is of great relevance if the transfused agent subsequently causes short or long-term disease manifestation in the donor recipient. In humans, cat-scratch fever, bacillary angiomatosis and endocarditis are the most common historically recognized disease entities caused by Bartonella sp. infection [1]. However, non-specific symptoms have been described, including fever of unknown origin, local or generalized lymphadenopathy, severe or recurrent anemia, chronic uveitis, optic neuritis, frequent headaches, fatigue, intermittent paresthesias, and granulomatous or angioproliferative reactions of undefined etiologies [15,16,33,34]. Similar non-specific clinical signs have been reported in Bartonella sp. bacteremic cats and dogs [35–37].
Diagnosis of bartonellosis remains a microbiological challenge because of the difficulty of culturing and isolating the bacterium from patient specimens. Bartonella sp. is highly fastidious and isolation by direct plating of blood or other clinical samples is not always successful. In order to improve the chances of isolation of Bartonella sp. from blood, samples should be cultured in liquid enrichment media to support growth [11], similar to culture techniques used for Haemophilus influenza and other bacteria species [38]. In our study, Bartonella spp. DNA was identified more frequently from BAPGM liquid culture samples than from subculture isolates, with only one subject PCR positive at both culture methods (Table 1). Similar discrepancies between results from liquid and solid culture methods were previously documented. Among 122 Bartonella-infected patients in the USA, 48 (39%) had BAPGM liquid culture samples positive for Bartonella DNA, but isolates were obtained only from five subjects (4%), including from two patients with PCR-negative liquid culture samples [4]. Causes for these differences are not completely elucidated and may be associated with low levels of bacteremia in asymptomatic humans [1]. In this study, the amount of viable Bartonella spp. in the bloodstream of these infected blood donors was not determined. Low bacterial burden in the bloodstream may limit the transmission of Bartonella spp. to a blood recipient or the development of infection. In addition, the discrepancy between the number of Bartonella sp. isolates obtained and the number of BAPGM samples positive by PCR could indicate that PCR-positive BAPGM samples contained non-viable bacteria. However, the two culture systems used in this study were designed to reproduce the vector environment (insect-based liquid culture medium BAPGM) and the host environment (blood-enriched agar medium), and each culture system may have facilitated the growth and detection of specific wild types of Bartonella. Consequently, the use of a combined diagnostic platform for testing the presence of Bartonella spp. DNA in BAPGM liquid culture and in subculture isolates in this study provided enhanced sensitivity, as previously demonstrated [39].
Evidence of Bartonella sp. infection may be confirmed by microbiological isolation, molecular techniques, and histopathologic visualization of Bartonella sp. antigens from tissue samples. Serology can be used to document exposure, but does not confirm infection. In this study, antibodies against B. quintana or B. henselae were detected in 32% (136/500) and 16.2% (78/500), respectively. A previous Brazilian study involving 437 healthy subjects from a rural area of Minas Gerais state documented B. quintana and B. henselae seroprevalences of 12.8% and 13.7%, respectively [40]. In Sweden, overall Bartonella spp. seroprevalence among 498 blood donors was 16.1%, but only 1.2% of those subjects were seropositive for B. henselae [29]. In New Zealand, 5% of 140 blood donors were seropositive for B. henselae [30]. In our study, antibodies against Bartonella sp. correlated poorly with infection as detected by PCR amplification followed by DNA sequencing or with the successful isolation of Bartonella organisms. Similar findings have been previously demonstrated in animals [14,41] and human patients [4,8], even in individuals with symptomatic disease [8,42]. Difficulties in Bartonella serodiagnosis are exemplified in the study by Vermeulen et al. [42]. It is suggested that Bartonella spp. manipulate the host immune system on a systemic scale to achieve a state of immunological attenuation, including stimulation of IL-10 secretion, which suppresses the capabilities of various immune cells, including T helper cells, monocytes/macrophages, and dendritic cells, thus interfering with both innate and adaptive immune responses [9,10]. Therefore, our results indicate that the predictive value of serology to detect Bartonella spp. infection in asymptomatic donors is low, supporting the recommendation that antibody status should not be used as a sole diagnostic modality to determine Bartonella spp. infection in blood donors. Moreover, negative results in one or more currently available diagnostic tests cannot exclude infection and, whenever possible, a combination of diagnostic tests is encouraged [42,43].
Another factor related to the apparent emergence of Bartonella spp. is the development of diagnostic techniques with improved sensitivity [4]. In recent years, the development of more sensitive and specific PCR methods, coupled with enrichment growth in specific culture media, has increased the detection of this pathogen in animal and human patient samples [4,8,11,26]. It is of clinical and epidemiological relevance that failure to amplify Bartonella sp. gene targets, following extraction of DNA from patient blood samples, does not rule out this bloodstream infection. It is estimated that Bartonella bacteremia in asymptomatic donors is approximately 10 CFU/mL of blood [1], which may be below the detection limit of most conventional or Real- Time PCR assays. Another reason for false-negative PCR or culture results is that Bartonella spp. typically cause a cyclic bacteremia [9]. It has been recently demonstrated that the detection of Bartonella sp. infection in humans is improved when three sequential blood samples are tested during a one-week period [39]. In that study, only 3 of 12 patients with Bartonella sp. bloodstream infection were documented as positive on more than one sample test date and no patient was positive on liquid culture/PCR for all three specimen test dates. Therefore, the number of bacteremic subjects may be underestimated in our study because only one blood sample from each blood donor was tested. We hypothesize that the actual number of blood donors infected with a Bartonella spp. may be higher in healthy humans than our current findings have documented. The low bacterial levels and the cyclic feature of Bartonella sp. bloodstream infection reinforce this hypothesis [9,10].
Based on recommendations by the Ethics Committee from UNICAMP Medical School, Bartonella sp. positive blood donors in this study were considered inappropriate for further blood donations. To the authors’ knowledge, there are no specific guidelines in the USA or other countries designed to prevent transfusion of human blood when donors are suspected to be infected with a Bartonella species. In Veterinary Medicine, at least two major medical boards have issued recommendations regarding blood donors. In 2005, the American College of Veterinary Internal Medicine (ACVIM) conditionally recommended the screening of canine and feline blood donors in order to obtain a Bartonella-free donor pool, especially for cats due to the high frequency of bacteremia in this host [44]. This recommendation for cats has been recently ratified by the European Advisory Board on Cat Diseases (ABCD) [45]. The results of our study indicate that guidelines for human blood transfusions should be designed, with special attention to the selection of Bartonella-free blood products for transfusion to immune suppressed subjects, which would include pediatric and geriatric patients.
The results of this study indicate that human exposure to Bartonella spp. frequently occurs in the Southeast region of Brazil, and that Bartonella sp. bacteremia occurs in asymptomatic blood donors. There is a risk of blood supply contamination with these pathogens from asymptomatic bacteremic donors. The impact of transmission of Bartonella spp. through blood transfusions recipients should be evaluated, as well as the use of advanced diagnostic techniques for the screening of Bartonella sp. infection among blood donors.
Supporting Information
(DOCX)
(DOC)
Acknowledgments
We thank the study patients, Dr. Steven Henriksen and Dr. Dominique Griffon for providing the research infrastructure and support needed at Western University of Health Sciences and Dr. Hannah Mirrashed, John Greenwood, Marizete Salvadego and Fabiana Cristina dos Santos for technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was the basis for the PhD thesis of LHUP and was partially funded by the Faculty Grant in Global Health to DGS from the Johns Hopkins Center for Global Health; a grant from FAEPEX—UNICAMP (Fundo de Apoio ao Ensino, à Pesquisa e à Extensão—UNICAMP) under protocols number 234/10 and 292/10; and a summer research scholarship to SS from the Office of the Vice President for Research and Biotechnology, Western University, Western University of Health Sciences, Pomona, CA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Breitschwerdt EB, Maggi RG, Chomel BB, Lappin MR (2010) Bartonellosis: an emerging infectious disease of zoonotic importance to animals and human beings. J Vet Emerg Crit Care 20: 8–30. 10.1111/j.1476-4431.2009.00496.x [DOI] [PubMed] [Google Scholar]
- 2. Chomel BB, Boulouis HJ, Breitschwerdt EB, Kasten RW, Vayssier-Taussat M, et al. (2009) Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet Res 40: 29 10.1051/vetres/2009011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boulouis HJ, Chang CC, Henn JB, Kasten RW, Chomel BB (2005) Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet Res 36: 383–410. 10.1051/vetres:2005009 [DOI] [PubMed] [Google Scholar]
- 4. Maggi RG, Mozayeni BR, Pultorak EL, Hegarty BC, Bradley JM, et al. (2012) Bartonella spp. bacteremia and rheumatic symptoms in patients from Lyme disease-endemic region. Emerg Infect Dis 18: 783–791. 10.3201/eid1805.111366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breitschwerdt EB, Maggi RG, Lantos PM, Woods CW, Hegarty BC, et al. (2010) Bartonella vinsonii subsp. berkhoffii and Bartonella henselae bacteremia in a father and daughter with neurological disease. Parasit Vectors 3: 29 10.1186/1756-3305-3-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garcia-Caceres U, Garcia FU (1991) Bartonellosis. An immunodepressive disease and the life of Daniel Alcides Carrion. Am J Clin Pathol 95: S58–66. [PubMed] [Google Scholar]
- 7. Pitassi LH, Cintra ML, Ferreira MR, Magalhaes RF, Velho PE (2010) Blood cell findings resembling Bartonella spp. Ultrastruct Pathol 34: 2–6. 10.3109/01913120903372761 [DOI] [PubMed] [Google Scholar]
- 8. Maggi RG, Mascarelli PE, Pultorak EL, Hegarty BC, Bradley JM, et al. (2011) Bartonella spp. bacteremia in high-risk immunocompetent patients. Diagn Microbiol Infect Dis 71: 430–437. 10.1016/j.diagmicrobio.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 9. Harms A, Dehio C (2012) Intruders below the radar: molecular pathogenesis of Bartonella spp. Clin Microbiol Rev 25: 42–78. 10.1128/CMR.05009-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pulliainen AT, Dehio C (2012) Persistence of Bartonella spp. stealth pathogens: from subclinical infections to vasoproliferative tumor formation. FEMS Microbiol Rev 36: 563–599. 10.1111/j.1574-6976.2012.00324.x [DOI] [PubMed] [Google Scholar]
- 11. Duncan AW, Maggi RG, Breitschwerdt EB (2007) A combined approach for the enhanced detection and isolation of Bartonella species in dog blood samples: pre-enrichment liquid culture followed by PCR and subculture onto agar plates. J Microbiol Methods 69: 273–281. 10.1016/j.mimet.2007.01.010 [DOI] [PubMed] [Google Scholar]
- 12. Magalhaes RF, Pitassi LH, Salvadego M, de Moraes AM, Barjas-Castro ML, et al. (2008) Bartonella henselae survives after the storage period of red blood cell units: is it transmissible by transfusion? Transfus Med 18: 287–291. 10.1111/j.1365-3148.2008.00871.x [DOI] [PubMed] [Google Scholar]
- 13. Pitassi LH, Magalhaes RF, Barjas-Castro ML, de Paula EV, Ferreira MR, et al. (2007) Bartonella henselae infects human erythrocytes. Ultrastruct Pathol 31: 369–372. 10.1080/01913120701696510 [DOI] [PubMed] [Google Scholar]
- 14. Kordick DL, Brown TT, Shin K, Breitschwerdt EB (1999) Clinical and pathologic evaluation of chronic Bartonella henselae or Bartonella clarridgeiae infection in cats. J Clin Microbiol 37: 1536–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oliveira AM, Maggi RG, Woods CW, Breitschwerdt EB (2010) Suspected needle stick transmission of Bartonella vinsonii subspecies berkhoffii to a veterinarian. J Vet Intern Med 24: 1229–1232. 10.1111/j.1939-1676.2010.0563.x [DOI] [PubMed] [Google Scholar]
- 16. Lin JW, Chen CM, Chang CC (2011) Unknown fever and back pain caused by Bartonella henselae in a veterinarian after a needle puncture: a case report and literature review. Vector Borne Zoonotic Dis 11: 589–591. 10.1089/vbz.2009.0217 [DOI] [PubMed] [Google Scholar]
- 17. Arvand M, Schad SG (2006) Isolation of Bartonella henselae DNA from the peripheral blood of a patient with cat scratch disease up to 4 months after the cat scratch injury. J Clin Microbiol 44: 2288–2290. 10.1128/JCM.00239-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lantos PM, Maggi RG, Ferguson B, Varkey J, Park LP, et al. (2014) Detection of Bartonella species in the blood of veterinarians and veterinary technicians: a newly recognized occupational hazard? Vector Borne Zoonotic Dis 14: 563–570. 10.1089/vbz.2013.1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chamberlin J, Laughlin LW, Romero S, Solorzano N, Gordon S, et al. (2002) Epidemiology of endemic Bartonella bacilliformis: a prospective cohort study in a Peruvian mountain valley community. J Infect Dis 186: 983–990. 10.1086/344054 [DOI] [PubMed] [Google Scholar]
- 20. Foucault C, Barrau K, Brouqui P, Raoult D (2002) Bartonella quintana bacteremia among homeless people. Clin Infect Dis 35: 684–689. 10.1086/342065 [DOI] [PubMed] [Google Scholar]
- 21. AABB (2012) Standards for blood banks and transfusion services. Bethesda, MD: American Association of Blood Bank; [Google Scholar]
- 22. EDQMH (2010) Guide to the preparation, use and quality assurance of blood components. Strasbourg, France: European Directorate for the Quality of Medicines and HealthCare; [Google Scholar]
- 23. Maggi RG, Duncan AW, Breitschwerdt EB (2005) Novel chemically modified liquid medium that will support the growth of seven Bartonella species. J Clin Microbiol 43: 2651–2655. 10.1128/JCM.43.6.2651-2655.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Drummond MR, Pitassi LH, Lania BG, Dos Santos SR, Gilioli R, et al. (2011) Detection of Bartonella henselae in defibrinated sheep blood used for culture media supplementation. Braz J Microbiol 42: 430–432. 10.1590/S1517-83822011000200003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cadenas MB, Maggi RG, Diniz PP, Breitschwerdt KT, Sontakke S, et al. (2007) Identification of bacteria from clinical samples using Bartonella alpha-Proteobacteria growth medium. J Microbiol Methods 71: 147–155. 10.1016/j.mimet.2007.08.006 [DOI] [PubMed] [Google Scholar]
- 26. Diniz PP, Maggi RG, Schwartz DS, Cadenas MB, Bradley JM, et al. (2007) Canine bartonellosis: serological and molecular prevalence in Brazil and evidence of co-infection with Bartonella henselae and Bartonella vinsonii subsp. berkhoffii . Vet Res 38: 697–710. 10.1051/vetres:2007023 [DOI] [PubMed] [Google Scholar]
- 27. Norman AF, Regnery R, Jameson P, Greene C, Krause DC (1995) Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol 33: 1797–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Varanat M, Maggi RG, Linder KE, Breitschwerdt EB (2011) Molecular prevalence of Bartonella, Babesia, and hemotropic Mycoplasma sp. in dogs with splenic disease. J Vet Intern Med 25: 1284–1291. 10.1111/j.1939-1676.2011.00811.x [DOI] [PubMed] [Google Scholar]
- 29. McGill S, Wesslen L, Hjelm E, Holmberg M, Auvinen MK, et al. (2005) Bartonella spp. seroprevalence in healthy Swedish blood donors. Scand J Infect Dis 37: 723–730. 10.1080/00365540510012152 [DOI] [PubMed] [Google Scholar]
- 30. Zarkovic A, McMurray C, Deva N, Ghosh S, Whitley D, et al. (2007) Seropositivity rates for Bartonella henselae, Toxocara canis and Toxoplasma gondii in New Zealand blood donors. Clin Experiment Ophthalmol 35: 131–134. 10.1111/j.1442-9071.2006.01406.x [DOI] [PubMed] [Google Scholar]
- 31. Mosepele M, Mazo D, Cohn J (2012) Bartonella infection in immunocompromised hosts: immunology of vascular infection and vasoproliferation. Clin Dev Immunol 2012: 612809 10.1155/2012/612809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guptill L, Slater L, Wu CC, Lin TL, Glickman LT, et al. (1997) Experimental infection of young specific pathogen-free cats with Bartonella henselae . J Infect Dis 176: 206–216. 10.1086/514026 [DOI] [PubMed] [Google Scholar]
- 33. Velho PE, Pimentel V, Del Negro GM, Okay TS, Diniz PP, et al. (2007) Severe anemia, panserositis, and cryptogenic hepatitis in an HIV patient infected with Bartonella henselae. Ultrastruct Pathol 31: 373–377. 10.1080/01913120701696601 [DOI] [PubMed] [Google Scholar]
- 34. Lamas C, Curi A, Boia M, Lemos E (2008) Human bartonellosis: seroepidemiological and clinical features with an emphasis on data from Brazil—a review. Mem Inst Oswaldo Cruz 103: 221–235. 10.1590/S0074-02762008000300001 [DOI] [PubMed] [Google Scholar]
- 35. Perez Vera C, Diniz PP, Pultorak EL, Maggi RG, Breitschwerdt EB (2013) An unmatched case controlled study of clinicopathologic abnormalities in dogs with Bartonella infection. Comp Immunol Microbiol Infect Dis; 10.1016/j.cimid.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 36. Perez C, Hummel JB, Keene BW, Maggi RG, Diniz PP, et al. (2010) Successful treatment of Bartonella henselae endocarditis in a cat. J Feline Med Surg 12: 483–486. 10.1016/j.jfms.2009.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Diniz PP, Wood M, Maggi RG, Sontakke S, Stepnik M, et al. (2009) Co-isolation of Bartonella henselae and Bartonella vinsonii subsp. berkhoffii from blood, joint and subcutaneous seroma fluids from two naturally infected dogs. Vet Microbiol 138: 368–372. 10.1016/j.vetmic.2009.01.038 [DOI] [PubMed] [Google Scholar]
- 38. Ajello GW, Feeley JC, Hayes PS, Reingold AL, Bolan G, et al. (1984) Trans-Isolate medium: A new medium for primary culturing and transport of Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenzae . Journal of Clinical Microbiology 20: 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pultorak EL, Maggi RG, Mascarelli PE, Breitschwerdt EB (2013) Serial testing from a 3-day collection period by use of the Bartonella Alphaproteobacteria growth medium platform may enhance the sensitivity of Bartonella species detection in bacteremic human patients. J Clin Microbiol 51: 1673–1677. 10.1128/JCM.00123-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. da Costa PS, Brigatte ME, Greco DB (2005) Antibodies to Rickettsia rickettsii, Rickettsia typhi, Coxiella burnetii, Bartonella henselae, Bartonella quintana, and Ehrlichia chaffeensis among healthy population in Minas Gerais, Brazil. Mem Inst Oswaldo Cruz 100: 853–859. 10.1590/S0074-02762005000800006 [DOI] [PubMed] [Google Scholar]
- 41. Perez C, Maggi RG, Diniz PP, Breitschwerdt EB (2011) Molecular and serological diagnosis of Bartonella infection in 61 dogs from the United States. J Vet Intern Med 25: 805–810. 10.1111/j.1939-1676.2011.0736.x [DOI] [PubMed] [Google Scholar]
- 42. Vermeulen MJ, Verbakel H, Notermans DW, Reimerink JHJ, Peeters MF (2010) Evaluation of sensitivity, specificity and cross-reactivity in Bartonella henselae serology Journal of Medical Microbiology 59: 743–745. 10.1099/jmm.0.015248-0 [DOI] [PubMed] [Google Scholar]
- 43. Wolf LA, Cherry NA, Maggi RG, Breitschwerdt EB (2014) In Pursuit of a Stealth Pathogen: Laboratory Diagnosis of Bartonellosis. Clinical Microbiology Newsletter 36: 33–39. [Google Scholar]
- 44. Wardrop KJ, Reine N, Birkenheuer A, Hale A, Hohenhaus A, et al. (2005) Canine and feline blood donor screening for infectious disease. J Vet Intern Med 19: 135–142. 10.1111/j.1939-1676.2005.tb02672.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pennisi MG, Marsilio F, Hartmann K, Lloret A, Addie D, et al. (2013) Bartonella Species Infection in Cats: ABCD guidelines on prevention and management. Journal of Feline Medicine and Surgery 15: 563–569. 10.1177/1098612X13489214 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.