Abstract
Regulation of gene expression in eukaryotes is an extremely complex process. In this review, we break down several critical steps, emphasizing new data and techniques that have expanded current gene regulatory models. We begin at the level of DNA sequence where cis-regulatory modules (CRMs) provide important regulatory information in the form of transcription factor (TF) binding sites. In this respect, CRMs function as instructional platforms for the assembly of gene regulatory complexes. We discuss multiple mechanisms controlling complex assembly, including cooperative DNA binding, combinatorial codes, and CRM architecture. The second section of this review places CRM assembly in the context of nucleosomes and condensed chromatin. We discuss how DNA accessibility and histone modifications contribute to TF function. Lastly, new advances in chromosomal mapping techniques have provided increased understanding of intra- and interchromosomal interactions. We discuss how these topological maps influence gene regulatory models.
Keywords: gene regulation, cis-regulatory module, cooperativity, DNA accessibility, chromosomal interactions
INTRODUCTION
Gene regulation is fundamental for every biological process. Reflective of its importance to cell survival and function, the regulatory mechanisms controlling gene expression are exquisitely sophisticated. Unraveling this complexity has broad implications, from understanding animal development to preventing and treating clinical pathologies. Innovative technologies and creative experimental design are rapidly expanding the current models of gene expression. From new insights into DNA-binding specificity to the contribution of nuclear architecture, this review aims to integrate recent discoveries into a more comprehensive picture of eukaryotic gene regulation.
Gene Regulation Stripped Down to the DNA
The first gene regulatory model was pioneered by François Jacob and Jacques Manod in the early 1960s. They postulated that gene products are able to feed back and regulate the expression of genes, a concept that laid the foundation for contemporary gene expression models. Currently, the most basic model dictates that regulatory proteins called transcription factors (TFs) act in trans to promote or inhibit expression from a locus by binding specific DNA sequences in cis-regulatory modules (CRMs) or enhancers (66). TFs are characterized by the sequence and structure of their DNA-binding domains. Throughout evolution, gene duplication events have expanded the number of TFs, resulting in groups or families of highly related TFs. As a consequence, evolutionarily related TFs often share similar DNA-binding domains and similar in vitro DNA-binding specificities. In some cases, related TFs display functional redundancy in vivo. However, there are many instances in which individual TFs with highly similar DNA-binding properties carry out distinct functions. Given that related TFs have both overlapping and unique functions, they must have the capacity to regulate both common and specific gene targets. Although it is easy to understand how TFs with similar binding properties recognize the same binding sites and regulate some of the same target genes, it is less obvious how binding to different CRMs is restricted to specific TFs (130). In the first section of this review, we focus on recent studies that provide new insights into how specificity is achieved at the level of DNA recognition. Throughout the review, many of the examples are taken from Drosophila melanogaster because of the many whole genome, genetic, and biochemical studies of it that have been carried out over the past several years.
CRMs usually harbor multiple TF-binding sites. In some cases, binding sites are stably occupied only by TFs that bind cooperatively. However, the mechanisms by which cooperative DNA binding increases TF specificity varies according to TF family and CRM (25, 51, 119). Focusing on recent examples with significant structural and functional data, we discuss different ways cooperative binding is achieved. Simply relying on the combinatorial binding and activity of multiple factors is another way CRMs coordinate gene expression, allowing integration of cell type and environmental inputs (Figure 1).
Figure 1.
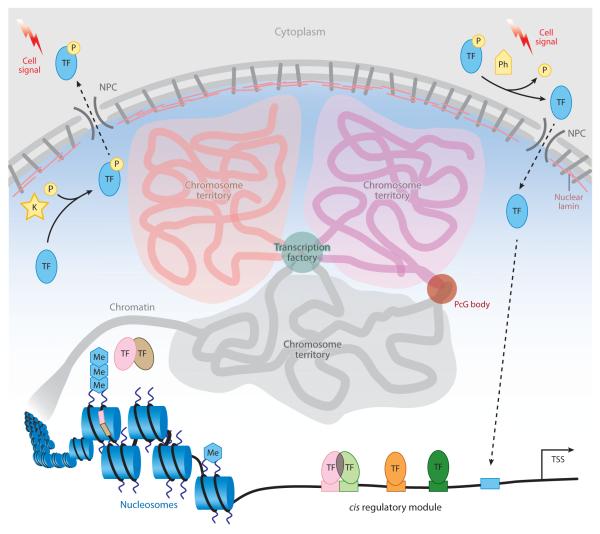
Overview of eukaryotic gene regulation. Within the nucleus of a cell, chromosomes occupy defined spatial regions called territories. Interactions between adjacent territories can correlate with transcriptionally active chromatin in transcription factories or silenced chromatin in Polycomb Group (PcG) bodies. Posttranslational modification of transcription factors (TFs), such as phosphorylation (P), can influence nuclear import (dashed arrows) through nuclear pore complexes (NPCs) in response to extracellular signals. Posttranslational modifications of histones, such as methylation (Me), can also correlate with the transcriptional state of associated genes. The position of nucleosomes can restrict access of TFs by occluding binding sites (colored rectangles). Lastly, TF recognition of specific binding sites, either as monomers or as a part of a complex with other proteins, also contributes to proper recruitment or release of RNA polymerase from the transcriptional start site (TSS). Abbreviations: K, kinase; Ph, phosphatase.
Dressing Up Gene Regulation with Chromatin
The biophysical realities of DNA packaged within a nucleus are in stark contrast to the naked DNA researchers typically think about when discussing protein-DNA binding. Although the stripped-down view is important for understanding the biophysical properties that govern TF-DNA interactions, it ignores all of the complexities of the nuclear environment. DNA in eukaryotic genomes is compacted into chromatin; the basic unit of chromatin, the nucleosome, consists of 147 base pairs of DNA wrapped around a histone octamer containing two copies of each of the core histones H2A, H2B, H3, and H4 (88, 98). DNA associated with histones is less accessible to TFs and RNA polymerase than is naked DNA, making chromatin transcriptionally more repressed compared with naked DNA. Additionally, chromatin structure is not homogenous along the entire genome and can adapt more complex local structures and higher-level three-dimensional arrangements. In the Assembling cis-Regulatory Module Complexes section below, we discuss how chromatin structure contributes to gene regulation (Figure 1).
Historically, chromatin has been described as existing in two distinct flavors: euchromatin and heterochromatin. Heterochromatin is condensed, transcriptionally inactive, and associated with repressive histone modifications, whereas euchromatin is relatively accessible and associated with actively transcribed genes and active histone modifications (42, 69). This low-resolution view is still accurate, but recent genomic studies have resulted in a much higher-resolution and more nuanced view of chromatin states. We discuss how nucleosome occupancy as well as histone modifications and variants correlate with gene regulation. Finally, with the help of several technological advances, our understanding of higher-order chromatin structure and the three-dimensional organization of chromosomes in nuclei has increased dramatically over the past decade. We discuss how nuclear architecture and chromosomal conformation have also been implicated in eukaryotic gene regulation (Figure 1).
ASSEMBLING CIS-REGULATORY MODULE COMPLEXES
In prokaryotes, single TFs are able to regulate gene expression. However, this type of gene regulation is usually insufficient for eukaryotic gene regulation. Instead, eukaryotes rely on combinatorial transcriptional inputs into CRMs to regulate gene expression in space and time (66). The specific recruitment of many individual factors refines expression on the basis of cellular context, timing of expression, and extracellular signals. For example, the same binding sites in the same CRM of the Drosophila nidogen gene have been shown to bind different forkhead domain TFs in different tissues, with distinct regulatory outputs (183). Alternatively, multiple homeobox (Hox) TFs, which have highly similar DNA-binding specificity as monomers, can target the same gene via distinct CRMs in different tissues (39). It also appears that TFs can bind non-canonical motifs in certain contexts, although the mechanism by which these motifs are distinguished from canonical motifs remains unclear (4, 20). Regulation can be further refined by posttranslational modifications (PTMs) of TFs, which can affect subcellular localization, DNA binding, and protein-protein interactions (Figure 1) (14, 16, 24, 27, 169). Some researchers propose a PTM code in which multisite PTM events provide an important regulatory mechanism for different signaling pathways to affect TF function and influence gene expression (14). In this section, we focus on two additional mechanisms, cooperative DNA binding and combinatorial codes that regulate the assembly and activities of CRM complexes.
Cooperativity
One mechanism cells use to increase the DNA-binding specificity of TFs is cooperative DNA binding. With an emphasis on structural data paired with in vitro DNA-binding assays, we distinguish three types of cooperative complex formation (Figure 2). The first type, which we refer to here as classical cooperativity, relies on direct protein-protein interactions between TFs and their cofactors to increase DNA-binding affinity (Figure 2). A variation on classical cooperativity, termed latent specificity, is when protein-protein interactions lead not only to increased DNA-binding affinity but also to a change in DNA-binding specificity (Figure 2) (161). We refer to a second form of cooperativity as enhanceosome or modular cooperativity (Figure 2). The distinguishing feature here is that, unlike classical cooperativity, which is typically defined for homo-and heterodimers of TFs, enhanceosome cooperativity is observed for large complexes of proteins and, at least in some cases, appears to not depend on protein-protein interactions (131). A third form of cooperativity is termed collaborative competition (Figure 2) (138). In this case, cooperative binding occurs only on a chromatin template because multiple TFs are more effective than are individual TFs at competing with nucleosomes for binding to target sequences (113). As the first two forms of cooperativity are measured on naked DNA, we discuss them here, whereas the third form, which is apparent only in the context of chromatin, is discussed in a later section.
Figure 2.
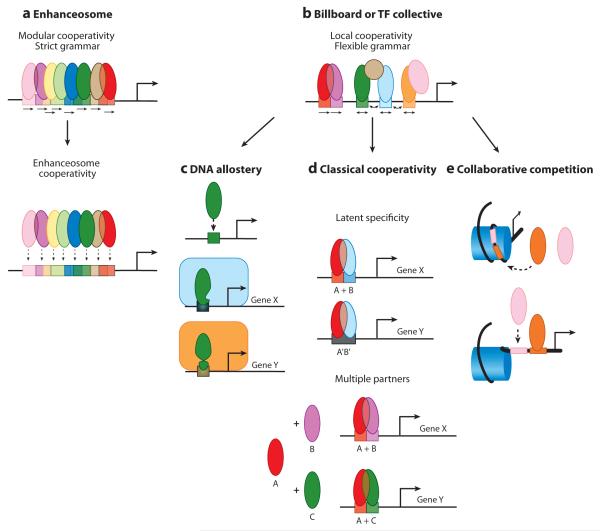
Cis-regulatory module (CRM) assembly and cooperative DNA-binding models. This image depicts three different models of CRM assembly and related cooperativity mechanisms. (a) The enhanceosome model requires strict modular cooperativity between all transcription factors (TFs) (129, 131). (b) In contrast, the flexibility of the Billboard and TF Collective models permits different cooperativity mechanisms to control CRM assembly. (c) In the case of DNA allostery, interactions between the DNA sequence and the TF can facilitate conformational changes in the TF that result in the recruitment of different regulatory complexes (rounded rectangles) in a sequence-specific manner (108). (e) Classical cooperativity uses protein-protein interactions between TFs to facilitate cooperative binding. These types of cooperative interactions help to increase TF DNA-binding specificity by restricting recruitment to dimeric sites (A+B). In the case of latent specificity, direct protein-protein interactions alter binding specificities so that TFs recognize novel composite sites (A’B’) (161). (f) Lastly, collaborative competition between TFs and nucleosomes can lead to cooperative binding when the binding of one TF provides access for another TF to bind a neighboring site (114, 120, 167).
Protein-protein interactions reveal latent specificities
Interactions between TFs not only increase DNA-binding affinity to their cognate binding sites but may also result in a modification of their DNA recognition properties (Figure 2). This phenomenon has recently been described for the Hox family of TFs, which provides a classic example of the TF specificity paradox. Characterized by a highly conserved DNA-binding domain called the homeodomain, all Hox proteins recognize similar AT-rich sequences in vitro (15, 127) but confer phenotypically distinct identities in vivo (65). One way they achieve functional specificity is through cooperative DNA binding with cofactors. In Drosophila, the best-characterized Hox cofactors are Extradenticle (Exd) and Homothorax (Hth), which have orthologs called Pbx and Meis, respectively, in vertebrates (87, 116). Exd/Pbx and Hth/Meis are members of the three amino acid loop extension (TALE) class of homeodomain-containing proteins (103, 116). Functional and structural studies have identified a conserved protein-protein interaction between Exd and Hox proteins that facilitates cooperative DNA binding (105, 112). Tryptophan-containing motifs (W motifs), most commonly YPWM, found N terminal to the Hox homeodomain, directly interact with the TALE motif in the Exd homeodomain (105, 112). Mutation of these W motifs can dramatically affect both cooperative DNA binding in vitro and in vivo function of some Hox proteins (87, 105, 110, 148). For example, for two Drosophila Hox proteins, Sex combs reduced (Scr) and Deformed (Dfd), mutation of the W motif is sufficient to abolish cooperative complex formation on a known specific binding site in vitro as well as several Exd-dependent functions measured in vivo (74, 87). In addition, minimal Hox proteins that contain only the homeodomains and W motifs retain many wild-type functions when assessed in vivo, suggesting that they are sufficient for many in vivo activities (134, 135).
In addition to increasing affinity, the binding of Exd with Hox proteins modifies their specificity, a phenomenon referred to as latent specificity (161). Using systematic evolution of ligands by exponential enrichment (SELEX)-seq, in which traditional SELEX (38, 170) is paired with deep sequencing, cooperative binding with Exd was shown to elicit changes in Hox DNA-binding preferences that are distinct from their monomeric DNA-binding preferences (161). Importantly, for at least one Hox protein (Ubx), the types of latent heterodimer specificities revealed by SELEX-seq were over-represented in DNA sequences bound by this factor in vivo, which suggests that the in vitro–measured specificities are biologically relevant (161). In another recent example, DNA-binding measurements from protein-binding microarrays for the Saccharomyces cerevisiae TFs Met4, Met28, and Cbf1 demonstrate that cooperative complex formation increases DNA-binding specificity (159). In this case, Cbf1, together with its non-DNA-binding cofactors Met28 and Met4, recognize additional DNA sequences that are adjacent to the traditional Cbf1 binding site (159). Importantly, these additional DNA sequences are essential for gene regulation in vivo. Together, these data suggest that interactions between TFs and their cofactors have the potential to reveal specificities that are not available in the absence of cofactors. Moreover, latent specificities can apparently be induced by both DNA-binding (e.g., Hox-Exd) and non-DNA-binding (e.g., Cbf1-Met4-Met28) cofactors.
The E twenty-six (ETS) family of TFs may provide an additional example of latent specificity, referred to in this case asacquired specificity (173). ETS proteins are characterized by a highly conserved winged helix-loop-helix DNA-binding domain and recognize 5′-GGA(A/T)-3′ sequence motifs. Similar to Hox proteins, ETS factors use direct interactions with cofactors to increase DNA-binding specificity (63). Cooperative DNA binding between PAX5 and ETS1 is critical for activation of mlb-1 during B cell development (46). Structural studies demonstrate that direct interactions between the paired domain of PAX5 and the ETS domain of ETS1 can rearrange protein-DNA contacts to increase DNA binding (50): A PAX5-induced rotation of a particular tyrosine residue in the recognition helix of ETS1 promotes binding to a low-affinity site (50). A similar mechanism has been proposed for other ETS/cofactor complexes. All members of the ternary complex factor subfamily of ETS factors, which includes ELK1 and SAP1, interact with serum response factor (SRF) to activate immediate-early genes (63). In the case of ELK1/SRF complexes, studies of related SAP1s/SRFs suggest that protein-protein interactions may induce conformational changes in an analogous tyrosine residue in the recognition helix of ELK1 to increase DNA-binding affinity (115). Altering the structure of the ETS domain may prove to be a general mechanism for increasing specificity, especially considering it mediates many of the functional interactions between ETS factors and several cofactors (173). An analogous cofactor-induced change in conformation was also proposed to underlie Hox-Exd latent specificity. In this case, X-ray crystal structures demonstrated that Exd positions a normally unstructured region of the Hox protein Scr so that it can interact with DNA, specifically, a narrow region of the minor groove (73, 105). The recognition of minor groove structure or, more generally, DNA shape is widespread among TFs, suggesting that it may be a common mode of DNA recognition (144, 145).
The similarities between ETS complexes and Hox complexes extend to another phenomenon called autoinhibition in which DNA binding of a TF is inhibited by domains in the TF itself. Mutual relief of autoinhibition has been shown to mediate cooperative complex formation between ETS1 and RUNX1 (63) as well as complex formation between Hox and Exd proteins, in which the Hox W motif apparently interferes with monomeric DNA binding (23). Therefore, protein-protein interactions can alter DNA-binding specificity and increase affinity by both rearranging protein-DNA contacts and suppressing autoinhibition.
It is noteworthy that some of the DNA-interacting residues in the Scr-Exd complex and ETS complexes are outside the traditionally defined DNA-binding domain. Although crystal structures are not yet available, the Drosophila Hox protein Dfd requires residues that are in an analogous position to Scr’s minor groove–interacting residues to bind and regulate some of its specific targets in vivo (74). These observations blur the traditional definition of a DNA-binding domain, in that they show that additional motifs can contribute directly to DNA binding when TFs interact with cofactors. Although it is currently not clear which residues in the Cbf1-Met28-Met4 complex contact its expanded binding site (159), it is plausible that, analogous to Hox-Exd, residues not normally considered part of the DNA-binding domain make some of these contacts. Additional nuclear magnetic resonance or X-ray crystal structures would be very helpful for resolving these questions.
Additional complexity and, perhaps, DNA-binding specificity also comes from the fact that some Hox proteins have additional ways to interact with Exd beyond their W motifs. For example, some Hox proteins, namely Ubx and AbdA, have multiple W and non-W motifs that are used to bind DNA cooperatively with Exd/Hth (22, 87, 109–111, 126, 148). Context-dependent interactions between motifs within the same Hox protein have also been proposed to contribute to functional diversity (110, 148). Unfortunately, no structural information is currently available to show how these additional motifs interact with each other, Exd, or Hth. In the case of AbdA, where up to four potential sequence motifs contribute to cooperative complex formation with Exd, some motifs were differentially required depending on the readout examined (87, 110, 126). Studies using vertebrate Hox proteins have also demonstrated differential requirements for W motifs (32, 45, 82, 142, 150, 157, 174, 178). For example, targeted mutagenesis of the murine Hoxa1 W motif parallels the Hoxa1 loss-of-function phenotype during hindbrain patterning (142). However, other functions, such as neural crest cell migration and cranial nerve development, are not as severely affected as in the Hoxa1 loss-of-function animals, suggesting that the W motif is required in a context-dependent manner, analogous to some of the Drosophila Hox proteins (142). These data suggest that, depending on the mode of Exd interaction, different target sites may be recognized, and the three-dimensional structure of the bound complex may vary. The presence of additional interaction modes also contributes to the phenomenon of posterior prevalence, also known as phenotypic suppression, in which more posterior Hox proteins dominate in a posttranslational manner over more anterior Hox proteins (34, 35, 52, 53, 104). A recent study comparing the abilities of Scr and AbdA to bind and regulate a shared target site found that the quality and quantity of AbdA’s Exd interaction modes both increased cooperative DNA-binding affinity in vitro and contributed to AbdA’s ability to outcompete Scr for target gene regulation in vivo (126).
Promiscuous cooperativity with multiple cofactors
As described above, some TFs, such as Hox proteins, increase their specificity by interacting with a small number of cofactors and using a variety of mechanisms to reveal latent specificities. Another strategy used by other TFs to expand their regulatory repertoire is to interact with a large number of protein partners, which allows TFs to gain cell- and tissue-specific control of gene expression depending on the cell-type availability of cofactors (Figure 2). For example, ETS factors utilize a variety of partners to bind DNA (63). With more than 40 different regulatory partners, the Sox [Sex-determining region Y (SRY)-related-high mobility group (HMG)-box] family of proteins provides another example of how multiple cofactors can contribute to specificity (16, 83). Classified by their HMG box DNA-binding domain, Sox family proteins control a variety of developmental processes and are key regulators of pluripotency (16, 83). During melanocyte development the SOX10/PAX3 pair activates expression of the TF MITF, which subsequently functions as another SOX10 partner to promote progression of melanocyte differentiation (16, 83). Additionally, recent data suggest that MEF2C can also function as a SOX10 partner to promote maintenance of the melanocyte fate (1). An analogous regulatory mechanism is observed during SOX10 regulation of Schwann cell development (16, 83).
Interactions with different partners can also affect DNA-binding specificity, perhaps using mechanisms that are analogous to the latent specificity model described above. Recently, two studies have demonstrated that single amino acid substitutions in SOX2 and SOX17 can disrupt or promote, respectively, cooperative binding with OCT4 in vitro. The ability to form cooperative complexes with OCT4 correlated with cell-reprogramming potential (68, 123). Because most Sox proteins recognize similar binding sites, restricting interactions to only a specific set of cofactors is important for regulating proper CRM binding.
Enhanceosome cooperativity
The CRM responsible for viral-inducible expression of interferon-β (IFN-β) is among the most-studied human transcriptional regulatory elements. Eight proteins cooperatively bind this 55–base pair (bp) enhancer in a structure termed the enhanceosome: one ATF2/c-Jun dimer, four interferon response factors (initially IRF-3, which is replaced with IRF-7 after IFN-β induction), and one NFκB dimer (p50/RELA) (131, 165). Activated by three different pathways, the specific expression of IFN-β is ultimately regulated by the coincidental activation and cooperative binding of all of these factors. Each factor is unable to individually activate IFN-β expression, and loss of any single protein abolishes IFN-β activation (165). Despite the binding sites within this 55-bp element being tightly packed, several crystal structures that capture subsets of the enhanceosome display a paucity of protein-protein interactions between pairs of dimers (131–133). Additionally, recent molecular dynamics simulations suggest that the DNA-bound complexes display an unusually high level of flexibility (129). From these observations, it seems unlikely that direct interactions between TFs are mediating the observed cooperative DNA binding. Instead, these studies suggest that sequence-dependent structural changes in the DNA may facilitate binding of TFs to overlapping sites (129, 131, 133). Use of a combination of higher- and lower-affinity sites can regulate the order in which complexes assemble (129); in this way, binding of one factor could facilitate the cooperative binding of another factor to an overlapping low-affinity site through complementary structural changes in shared nucleotides. Further, the architectural factor HMGA1a and other secondary factors may also enhance cooperative DNA binding on the IFN-β enhancer (131). Therefore, the IFN-β enhanceosome represents a type of modular cooperativity with strict requirements for binding-site arrangement and overlap (Figure 2).
Binding-site allostery
Lastly, we wish to emphasize that the binding site itself can be an active player in TF function and activity. Studies on the glucocorticoid receptor (GR) have shown that DNA can function as an allosteric regulator of TF activity (108). Structural studies of the GR demonstrate that a region called the lever arm within the DNA-binding domain adopts different conformations according to the DNA sequence bound (108). These structural changes parallel functional variations in cognate regulatory complexes for the different binding sites (108). Therefore, different conformations of the DNA-binding domain induced by the DNA-binding site can affect the transcriptional activity of GR in a site-specific manner (108). As with latent specificity, we speculate that a variety of influences on the three-dimensional structure of TF complexes, in part influenced by the DNA-binding site, can affect TF functions. Therefore, understanding the sequence and shape of CRMs and how they impact the structure of bound factors is critical to decoding the regulatory logic driving gene expression.
cis-Regulatory Module Architecture
A common feature of all CRMs is that they provide a scaffold for a combinatorial logic code in which the assembly of multiple factors provides cell type– and environment-dependent gene regulation (66). In the above section, we focused on several types of DNA-binding cooperativity used by TFs to bind CRMs. With the exception of enhanceosome cooperativity, in which all factors must bind to an inflexible CRM, the other types of cooperativity described above allow for much greater CRM flexibility: A single CRM can potentially integrate a large variety of inputs, some of which may be cooperative. For example, the Hox-targeted CRM from reaper integrates not only the Hox protein Deformed (Dfd) but also at least eight additional TFs (162). In contrast, a Dfd autoregulatory target requires multiple Dfd-Exd heterodimer inputs plus additional, as yet unidentified, inputs (74). The ability to interchange CRM inputs provides cells with the flexibility to maintain a tight regulatory control in a wide variety of distinct contexts. Given this requirement, how are CRMs organized, and can generalizations be gleaned from the data?
Three different models have been proposed to describe CRM architecture: the enhanceosome, the billboard, and the TF collective (Figure 2) (3, 75). As described above, the enhanceosome model posits that cooperative binding of a group of TFs using a strict arrangement or grammar of binding sites in the DNA is necessary for CRM activity (Figure 2). Evidence for this model comes primarily from the IFN-β enhanceosome, described above, as well as from the tumor necrosis factor-α enhanceosome (13, 131). Given the small number of identified enhanceosome-like CRMs in which the arrangement of the binding sites is inflexible, they may be more the exception than the rule. Enhanceosomes may be limited to regulatory events that must be controlled with exquisite precision, as they can be activated only when all factors are present. In addition, once formed, enhanceosomes are unusually stable, allowing them to activate transcription until subsequent mechanisms disassemble them (165).
A second model for CRM architecture is the billboard model (Figure 2) (3). At the opposite end of the spectrum from inflexible enhanceo-some CRMs, billboard CRMs are hypothesized to be very flexible; each binding site is critical, but their relative orientation and spacing do not contribute to CRM function (3, 85). According to this view, individual TF inputs might act independently to recruit different components of the basal transcription machinery, adapter complexes, or chromatin-modifying complexes to promote transcription (3).
A third view of CRM architecture refers to CRMs as TF collectives (Figure 2) (75). According to this view, TFs are cooperatively recruited to CRMs but without a precise motif grammar (75). Using whole-embryo chromatin immunoprecipitation (ChIP)-chip and ChIP-seq at different stages during Drosophila embryogenesis, binding events for a set of five factors involved in cardiac gene regulation were analyzed (75). Interestingly, the authors found that a large number of CRMs included all five factors and that some combinations were very rare (75). For example, when one was missing [Tinman (Tin)], the other four factors were rarely found together (75). Although such observations could represent cooperative binding or cooperative recruitment of these five factors, they could also represent an evolutionary selection for functional CRMs that contain all five inputs and selection against CRMs that have only four of the five inputs. Another potential explanation for these results is that Tin functions as a so-called pioneer factor that is required to initiate binding to these CRMs in a chromatin environment. Additional biochemical experiments are required to determine if cobinding of these factors is cooperative rather than a consequence of selection.
Another feature of the TF-collective model is that binding sites need not necessarily be present for every TF present at the CRM (75). Accordingly, some factors are indirectly bound at CRMs because of protein-protein interactions alone (Figure 2). Although this is certainly plausible, it is also possible that low-affinity binding sites or latent specificity mechanisms make it difficult for current computational methods to recognize all of the essential binding sites in CRMs. Consistent with this idea, low-affinity binding sites are critical for the accurate activities of some CRMs (136).
Detailed in vivo structure-function analysis of the sparkling CRM of Drosophila Pax2 in eye development provides another informative view of CRM logic (163). In this case, the authors asked how binding sites for the known TFs rearrange relative to each other during the course of evolution (163). Several interesting conclusions come from this work. For one, sparkling CRMs from different Drosophila species can have a very different arrangement of binding sites but still function to drive accurate expression in D. melanogaster, similar to conclusions obtained from evolutionary comparisons of the stripe 2 CRM from even-skipped (96, 97). In addition, despite rapid evolutionary turnover of binding sites at the sparkling CRM, the authors were able to recognize that the spacing between pairs of some binding sites was conserved (163). This conserved CRM grammar suggests that there may be interactions, either direct or mediated by additional factors, between the bound TFs. The idea that cooperative or interacting subelements could be among a set of otherwise independent inputs was also part of the original billboard model (3) and is consistent with the important role of TF DNA-binding specificity discussed in the previous section.
Regardless of whether biologists refer to CRMs as a billboard, a TF collective, or some other nom du jour, the emerging view from numerous studies is that many, perhaps the majority of, enhancer elements are not enhanceosome in nature but instead flexibly integrate multiple TF inputs in a surprisingly large number of arrangements. Consistent with a key role for multiple TF inputs, a recent comparative study found that in vivo binding sites for the mesodermal TF Twist (Twi) are highly conserved across several Drosophila species, and loss of Twi binding in one species is often associated not with loss of a Twi motif but with loss of a cofactor binding site (59). Some of these inputs may be singly bound TFs, others may be cooperatively bound pairs of TFs, and yet others may interact indirectly via a third factor (Figure 2). Such flexibility in CRM architecture makes the de novo identification of CRMs, based solely on DNA sequence, a big challenge for biologists because apparently the same set of inputs can be encoded in the DNA sequence in many ways.
CHROMATIN STRUCTURE
In the previous section, we discussed how TFs recognize and bind target sites in the context of naked DNA. However, to fit within the miniscule confines of the nucleus, DNA is wrapped around histones and condensed into chromatin. In addition to limiting TF access, histone-DNA complexes are subject to many PTMs that affect gene expression. Furthermore, chromatin is not uniformly distributed throughout the nucleus, so distant regions of the genome, based on linear DNA sequence, may actually be in close proximity. In this section, we describe how modifications of histone-DNA complexes and chromatin architecture contribute to gene regulation. It is not possible to catalog all of the many modifications known to occur on hi-stones; for this, the reader is referred to many recent reviews covering this topic (7, 12, 86, 139, 181). Instead, we focus on the small subset of modifications that are correlated with CRMs or CRM activity.
DNA Accessibility
DNA accessibility is increasingly recognized as an important variable in gene regulation (79, 80, 91, 166). Although it has been difficult to mechanistically test the causal relationship between chromatin structure and gene transcription, many genome-wide studies have demonstrated significant correlations. Through modeling of TF binding and DNase1 sensitivity data, two recent studies revealed that chromatin accessibility has a significant impact on the genome-wide binding patterns of a number of developmental regulatory factors expressed in the Drosophila embryo (79, 91). Within regions of open chromatin, TF binding is primarily determined by sequence specificity (79). A similar correlation between accessibility and TF binding has been observed in mammalian cells (70) and yeast (182). Additionally, several genome-wide nucleosome-mapping studies reveal trends suggesting that nucleosome positioning may influence gene expression (5). First, promoter regions tend to contain nucleosome-depleted regions (NDRs) (5). Second, nucleosomes around transcription start sites are often well organized (5). The formation of NDRs is predicted to reveal binding sites and facilitate TF binding at regulatory sequences (106). Many mechanisms have been proposed to regulate nucleosome positioning and NDR formation (5, 67, 81, 152). In addition to the controversial role of the primary DNA sequence (70, 76–78), ATP-dependent chromatin remodelers, such as the Swi/Snf complex, and binding by specific TFs have also been implicated in NDR formation (6, 168, 176, 179). The formation of NDRs falls under the more general heading of establishing gene- and cell type–specific chromatin architectures, which can include the positioning of nucleosomes at regulatory elements and promoters (47, 95).
Similar to nucleosome-depleting factors, so-called pioneer factors have been proposed to prime chromatin environments to initiate subsequent TF binding. Unlike many TFs, pioneer factors such as the FOXA and GATA factors, PU.1, and AP1 have the ability to bind their target motifs in a closed chromatin environment (17, 61, 102, 180). Upon binding their target DNA, the pioneer factors can drive local chromatin remodeling and create accessible enhancers for additional TF binding (102, 180). This type of synergy has previously been termed nucleosome-mediated cooperativity or collaborative competition (Figure 2) (113, 114, 138). Thermodynamic and in vitro studies have demonstrated that TFs can compete with nucleosomes for DNA binding and, by unwrapping or evicting the overlapping nucleosome, induce the cooperative binding of another TF (114, 120, 167). Because protein-protein interactions are not required and the relative arrangement and orientation of binding sites are flexible, collaborative competition may be one explanation for why many CRMs have multiple flexible inputs without a strict grammar (e.g., the billboard and TF collective models; see above) (3, 75). Additionally, mathematical simulations suggest that by mediating chromatin reorganization, pioneer factors can increase the steady-state binding of other TFs, a process called assisted loading (175).
The Drosophila zinc-finger protein Zelda (Zld) may represent another type of pioneer factor. Zld is bound to its target motifs throughout the Drosophila genome during the maternal-to-zygotic transition, the point when zygotic transcription commences in the developing embryo (57, 92, 124, 149). Most Zld-targeted regions remain bound by Zld and highly accessible later in embryonic development and are also targeted by numerous developmental TFs at these later stages (57, 124). Because Zld is highly associated with the relatively open genome before zygotic transcription begins, it has been proposed that rather than reorganizing or opening chromatin, Zld binding prevents nucleosome occupancy and maintains DNA accessibility in certain regions of the genome (57).
Histone Modifications
Recent studies have greatly expanded the traditional view that chromatin exists in two states, heterochromatin and euchromatin. The emerging view is that there are many varieties of active and inactive chromatin present in a given cell type (44, 146). Moreover, active chromatin territories are not simply permissive for DNA binding. Two recent genome-wide studies making use of different techniques (171) in different cell types have provided higher-resolution views that reveal intricate patterns of histone modifications and DNA accessibility at enhancers and promoters (44, 80, 146). Despite experimental differences, and the fact that different chromatin factors were studied, the results of the two studies had a number of similarities. Of note, both studies found that TFs were more likely to bind their DNA motifs in the enhancer chromatin state, a subregion of active chromatin that is characterized in part by monomethylation of histone H3 on lysine 4 (H3K4me1). Methylation patterns on H3K4 appear to be closely linked to regulatory enhancers, promoters, and active transcription. Monomethylated H3K4 is associated with enhancers, dimethylated H3K4 (H3K4me2) is associated with both enhancers and promoters or transcription start sites (TSSs) of actively transcribed genes, and trimethylated H3K4 (H3K4me3) is associated only with the promoters/TSSs of actively transcribed genes (11, 60, 140). The presence of H3K4me1 is strongly enriched on developmental enhancers, but this chromatin modification does not correlate with enhancer activity (19). A similar finding—enhancer marking in the absence of transcriptional activation—has been reported for the H3K4me2 chromatin mark (58).
The list of chromatin modifications with the potential to affect TF DNA recognition extends well beyond methylation of histone H3. Enhancers marked with H3K4me1 are susceptible to both transcriptional repression and activation, and these repressed or activated states usually coincide with additional chromatin modifications. At one end of the spectrum, gene silencing and a repressive regulatory state are associated with trimethylation of H3 on lysine 27 (H3K27me3). H3K27me3-modified nucleosomes are generated by the Polycomb repressive complex 2 (PRC2) and recognized by another Polycomb complex (PRC1) (160). PRC1 silences gene expression, but the mechanism by which this occurs is unclear (107, 160). At the other end of the spectrum, active, H3K4me1-marked enhancers are often also associated with marks such as H3K79me3 and acetylation of H3K27 (19), but little is known about the role of these modifications in active chromatin.
Despite these links between histone modifications and TF binding, whether TFs can recognize a given modification when targeting the genome remains unclear. However, it is becoming increasingly evident that protein-DNA binding can be influenced by histone PTMs; the TF FOXA1 provides evidence for such a mechanism. FOXA1 can bind both DNA and histones and is selectively recruited to genomic regions with nucleosomes containing mono- or dimethylated H3K4 (72, 99, 153–155). Importantly, FOXA1 chromatin binding is attenuated upon loss of H3K4me1 and H3K4me2. This has been taken to suggest that FOXA1’s reading of these chromatin marks influences its genomic DNA targeting, although it is also possible that the loss of these chromatin marks indirectly prevents FOXA1 binding (99, 102). Additionally, inducible DNA binding of Drosophila heat shock factor (HSF) is also influenced by chromatin state (55). HSF binding to heat shock elements (HSEs) is dependent on the presence of a canonical HSE DNA motif. This motif is necessary for binding but not sufficient (i.e., only a fraction of HSEs are bound in vivo), so binding specificity is dependent on something other than DNA sequence. Additionally, the contribution of repressive chromatin to HSF-binding selectivity is minimal, as most of the unbound HSEs are not associated with the H3K27me3 repressive chromatin mark (55). Ultimately, it seems that, aside from the HSE motif, the most significant contributor to heat shock–inducible HSF binding is an active chromatin state; prior to heat shock, inducibly bound HSEs are not depleted of nucleosomes but do contain chromatin modifications that are viewed as active and thought to weaken histone-DNA interactions (H3K4me3 and acetylated histones) (55). Although it is possible that HSF is interacting with one of the active histone modifications, it is just as likely that these modifications have generated an environment where the weakened histone-DNA interactions are more permissive to HSF binding.
Many questions remain with regard to the functional role of these chromatin modifications and to whether something resembling a histone code influences genome-wide TF DNA binding (49, 139). Are certain posttranslational histone modifications simply indicators of repressive or permissive transcriptional environments? Or do histone modifications play a more active role in helping TFs recognize certain subsets of potential regulatory elements? Beyond the level of chromatin modifications, the presence of histone variants might also influence nucleosome stability and, therefore, DNA accessibility and TF-DNA interactions (88, 164). Although much is still to be understood, the combinatorial possibilities for both binding and regulatory specificity are daunting.
Chromatin Interactions in Three Dimensions
Until this point we have maintained a two-dimensional, linear view of DNA and chromatin. Although this is advantageous for understanding basic principles of protein-DNA interactions, it omits all of the structural complexities associated with regulating gene expression within the three-dimensional confines of the nucleus. Beginning with the initial observation that chromosomes occupy particular regions within the nucleus (184), many studies suggest that the nucleus is divided into distinct functional domains (Figure 1). However, these domains are dynamic, making them difficult to characterize (21). Nevertheless, new techniques are beginning to yield important insights into nuclear architecture that provide strong support for the idea that interactions between distant chromosomal regions contribute in critical ways to gene regulation.
Nuclear organization
Our understanding of chromosomal structure has increased dramatically over the past decade as a result of advances in chromosome conformation capture (3C) and 3C-based high-throughput technologies (30, 31) as well as microscopy-based techniques for studying subnuclear DNA or RNA localization (40, 93, 128). A general conclusion from these studies is that the interior of the nucleus is not a uniform compartment. This is not unexpected, considering the existence of subnuclear structures such as the nucleolus, but the level of organized structure associated with chromatin is striking. Termed chromosome territories, these domains are often further organized on the basis of gene density and activity (30, 33, 41, 156, 172). Gene-rich regions are found more toward the center of the nucleus, whereas gene-poor regions are located closer to the nuclear periphery. Although the mechanisms are unclear, rearrangements toward the periphery are proposed to be a consequence of interactions with the nuclear envelope (185). Additionally, active genes are generally found at the surface of a chromosomal territory, whereas inactive or repressed genes are buried in the interior (30, 33, 41, 172). Highly expressed genes have also been observed to reside in foci that have been termed transcription factories (141). These observations and others have led to the idea that colocalization may lead to coregulation (26). Recent Hi-C data in Drosophila further correlate intra- and interchromosmal interactions with transcriptionally active regions, whereas inactive domains remain confined within their respective chromosomal territories (Figure 3) (156). Additionally, physical domains of interaction correlate with specific sets of epigenetic marks and are demarcated by insulators (122, 156).
Figure 3.
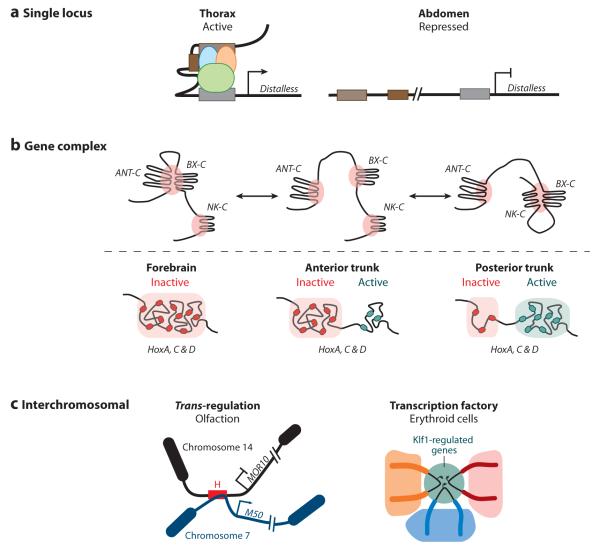
Different types of long-range interactions. (a) Interactions between elements in a single locus. Tissue-specific conformations of elements within the Distalless (Dll) locus correlate with gene activity. In the thorax, where Dll is expressed, the locus is more compact, with several regions interacting over long distances. However, in the abdomen, where Dll is repressed, the locus adapts a more extended conformation, and no interactions are observed between regulatory elements (2). (b) Interactions between elements in a gene complex. In Drosophila three gene complexes, antennapedia (ANT-C), bithorax (BX-C), and NK homeobox (NK-C), create foci of extensive intracomplex interactions when repressed by polycombs called Polycomb group (PcG) bodies (9). Additionally, intercomplex interactions suggest that PcG bodies can encompass more than one complex (9). However, PcG bodies containing all three complexes have not been observed (9). During vertebrate development, Hox genes cluster according to gene activity (125). In the forebrain, where none of the Hox genes are expressed, chromosome conformation capture-on-chip analysis indicates that all members of the complex group together (red). However, in trunk regions Hox genes adopt a bimodal distribution in which all the active genes (green) are found in one cluster and the inactive ones (red) interact within a different cluster (125). Furthermore, this three-dimensional organization correlates with collinear gene activation (125). (c) Interactions between chromosomes. During odorant receptor choice in mouse neurons, trans interactions can occur between the H enhancer and genes on different chromosomes (94). In this case, we have depicted interactions between the H enhancer on chromosome 14 and the M50 gene on chromosome 7 (94). Additionally, interactions between Klf1-regulated genes (green) on different chromosomes have been observed in erythroid cells (151). This colocalization of active genes constitutes a transcription factory.
Chromosomal interactions
Since the original finding that CRMs can be far from the promoters they regulate, scientists have strived to understand how different pieces of the genome communicate with each other. The identification of the β-globin locus control region and gypsy insulators in Drosophila established looping as the predominant paradigm for enhancer-promoter interactions, and there are now many examples of enhancer-promoter communication that occur as a result of looping (84). Recent experiments in Drosophila directly correlate enhancer-promoter interactions with cell type–specific gene expression using a new method called cgChIP (cell- and gene-specific ChIP) (2). Using cell type–specific expression of the bacterial DNA-binding protein LacI, enhancers of the gene Distalless (Dll), tagged with LacI binding sites (lacO), were observed to interact with the Dll promoter in limb primordia cells, where Dll is expressed (Figure 3) (2). In contrast, enhancer-promoter communication was not observed in homologous abdominal cells, where Dll is repressed by Hox proteins, implying a more extended DNA conformation in these cells (Figure 3) (2). These results imply that local chromatin structure, in this case, enhancer-promoter communication, varies in a cell type–specific manner. In addition to cis-looping models, enhancers can also interact with promoters in trans. In Drosophila, this process is called transvection and has been observed between Hox complexes on homologous chromosomes (36). In vertebrates, trans enhancer-promoter interactions have been observed during odorant receptor (OR) choice in olfactory neurons (94). Using 3C, the H enhancer on chromosome 14 was observed to interact with OR promoters on chromosomes 7, 9, and 14 (Figure 3) (94). The possibility for a single enhancer to govern gene expression through interchromosomal interactions provides one possible mechanism to address how olfactory neurons choose one out of 1,300 possible ORs (158).
Further evidence supporting a role for long-range interactions in gene regulation comes from studies on Hox clusters. Polycomb-dependent silencing of the antennapedia and bithorax complexes in Drosophila demonstrate intra- and intercomplex interactions (Figure 3) (9). These foci of corepressed genes by Poly-comb group (PcG) proteins are called PcG bodies (Figures 1 and 3) (8). Furthermore, removal of participating elements from one complex can weaken gene silencing in the other (9). These data suggest that PcG body formation is not just a consequence of corepression but may functionally contribute to gene regulation. Spatial clustering of Hox genes is conserved between vertebrates and invertebrates (43, 117, 125). Recent 3C and chromosome conformation capture-on-chip (4C) analysis of the mammalian HoxD gene cluster in developing limb buds found that functional regulatory regions dispersed within a gene desert upstream of the coding region interacted with the active Hoxd promoter (117) (Figure 3). These three-dimensional interactions were proposed to form a regulatory archipelago that through regulation of Hoxd gene expression could function to modulate digit morphology; this concept of partially redundant or shadow enhancers working together to regulate a single gene has been described for multiple Drosophila genes (2, 10, 37, 48, 64, 137). At the Hoxd regulatory locus, alterations in chromatin interactions correlated with collinear activation of the Hox genes during development (125). In tissues where the Hox clusters are silent, genes were observed to reside in a single three-dimensional domain marked by H3K27me3 (Figure 3) (125). However, once gene expression began, a bimodal organization was observed (Figure 3) (125). In samples from either anterior or posterior portions of the embryo, genes known to be actively transcribed in those regions occupied one domain that correlated with H3K4me3, whereas genes known to be silent occupied a separate domain that correlated with H3K27me3 (125).
Although these Hox complex studies provide strong correlations, there is some controversy as to the relationship between three-dimensional chromatin structure and gene regulation. In erythroid cells, specific intra- and interchromosomal interactions between coregulated genes were dependent on a single TF, Klf1 (Figure 3) (151). Additionally, transgenes carrying Klf1-regulated genes relocate to transcription factories when inserted into other genomic locations (151). These results suggest that active, coregulated genes can preferentially organize into transcriptional interactomes (151). However, other studies using glucocorticoid-inducible gene expression in cell lines did not observe significant chromosomal rearrangements upon activation (56). Instead, GR activates genes within preexisting loci that are enriched for DNase1-hypersensitive sites (56). These studies highlight the functional complexity that can be elucidated using 3C and related strategies. However, they also caution against making broad interpretations regarding the role of nuclear architecture in gene regulation, as observations can be highly specific to a particular gene or group of genes. Furthermore, interactions, or lack thereof, can be highly cell-type specific, as was recently shown in a chromosome conformation–based study of RNA polymerase II transcription (89) and by cgChIP in Drosophila (2). New advances in microscopy may help to sort out data from cross-linking-based studies by visualizing interactions in situ (71).
Regulatory factories and the hierarchically structured nature of chromatin in general suggest a regulatory environment in the nucleus in which the local concentration of TFs and accessory factors can vary significantly from region to region. Protein concentration is an important determinant of TF-DNA interaction and has been incorporated into recent models of genome-wide DNA binding (18, 79, 177), so the potential for regional variation in TF concentrations throughout the nucleus must be taken into account. Such foci might also lead to the identification of indirect protein-DNA interactions when using a cross-linking-based protocol such as ChIP; whether this is the case will become evident as more genome-wide 3C and cell type–specific data become available. Thus, although the study of chromosomal conformation is a relatively nascent field when compared with most of TF biology, these studies have the potential to impact both our interpretation of genome-wide ChIP data and our protein concentration–centric models of genome-wide TF binding.
CONCLUDING REMARKS
Much has been made of the finding that many TFs bind thousands of genomic regions in vivo, perhaps because these numbers easily exceed the number of expected target genes for most sequence-specific TFs (90, 100, 101). However, the number of binding events is still lower than the predicted number of sites throughout the genome on the basis of DNA sequence alone and close to what is expected when accounting for both DNA sequence and chromatin accessibility (177). TFs are most typically viewed as components of discrete regulatory networks, with separate target and nontarget genes for each TF (28, 29, 121). However, it has been proposed, on the basis of genome-wide binding data, that TF regulatory networks should instead be viewed as continuous networks (18). The continuous network model of TF function posits that, because of the high nuclear concentration of most expressed TFs, specificity mediated by protein-protein interactions is unnecessary, and essentially all genes are targeted by all TFs. According to this view, biological function is determined by quantitative differences in TF binding at accessible DNA rather than binary on/off TF binding (18). Aspects of this model are supported by data from a survey of Drosophila TFs expressed early in Drosophila embryogenesis (79, 91). However, the over-expression of many TFs, for example, by the Gal4-UAS method in Drosophila, generally do not lead to aberrant phenotypes; instead, higher than normal levels of TFs in cells typically result in wild-type readouts, which suggests that other factors besides concentration must be limiting for TF function. Moreover, there are many examples in which small differences in TF DNA-binding domains are important for their specific in vivo functions, arguing that DNA-binding specificity is critical. In addition, the continuous network model assumes that there are no cell type–specific differences in binding and that the signal generated by ChIP is indicative of direct DNA binding. But this may not always be the case, as there is ample evidence for ChIP signals resulting from indirect DNA binding via protein-protein interactions or interactions between regulatory elements (2, 54, 62, 118). On the basis of the current data, we suggest that chromatin accessibility is critical for limiting which TF-binding sites and which CRMs are available in specific cell types, but within accessible regions, DNA-binding specificities of TFs and TF complexes are essential for determining which binding sites are productively bound within these accessible regions.
The era of TF genomics has clearly changed our view of how TFs target the genome, but many of the methods routinely used are low resolution or are blind to cell type–specific differences. New, higher-resolution technologies will undoubtedly lead to refinement and restructuring of these models. For example, a new variation on ChIP, termed ChIP-exo, can generate genome-wide TF–DNA binding profiles down to single-base resolution (143). Not only does this high-resolution method provide a more precise view of a TF’s DNA-binding motifs, but it also eliminates a significant number of false positive binding events generated by traditional ChIP-chip or ChIP-seq (143). This method has the potential to refine the models of genome-wide TF binding that have been generated over the past five years.
Beyond advances in ChIP, increasingly sophisticated methods for monitoring nuclear organization will begin to generate a comprehensive picture of nuclear and chromosomal structure in vivo. This will require a combination of both 3C-based techniques and super-resolution microscopy techniques, such as stochastic optical reconstruction microscopy (147). Combining these approaches with high-resolution ChIP data will be essential for understanding TF-DNA interactions in the context of chromatin looping and transcription factories. Ultimately, integrating these multiple layers of data, in combination with the studies described in this review, will allow TF biologists to generate and test models regarding direct versus indirect, specific versus nonspecific, and functional versus nonfunctional binding.
SUMMARY POINTS.
Multiple mechanisms that provide site-specific TFs with increased specificity contribute to cooperative DNA binding.
For most CRMs, architecture is flexible, allowing the modular integration of multiple TF inputs, some of which may depend on cooperativity.
Multiple chromatin states have been described on the basis of histone PTMs and transcriptional activity.
Genetic material is structurally organized within nuclei in a manner that is relevant to gene regulation.
FUTURE ISSUES.
Structural studies are needed to address how different protein-protein and protein-DNA interactions affect CRM assembly and function at a molecular level.
Additional rigorous studies of CRM architecture would help build better models to identify CRMs and predict their functions.
Is there a causal relationship between histone modifications and transcriptional regulation?
High-resolution, cell type–specific studies of chromatin and nuclear architecture will be important for fully understanding gene regulation.
ACKNOWLEDGMENTS
We thank members of the Mann laboratory for helpful discussions. R.S.M. was supported by NIH grant GM54510 and K.L. by the 5T32DK07328 training grant.
Glossary
- TF
transcription factor
- cis-regulatory module (CRM)
collection of transcription factor binding sites that coordinate to regulate gene expression; also called enhancers
- PTM
posttranslational modification
- Cooperative DNA binding
occurs when the binding affinity of two or more factors is more than the sum of the individual affinities
- Latent specificity
novel DNA recognition properties that are revealed as a consequence of protein-protein interactions
- TALE
three amino acid loop extension
- SELEX
systematic evolution of ligands by exponential enrichment
- ChIP
chromatin immunoprecipitation
- NDR
nucleosome-depleted region
- Chromosome conformation capture (3C)
a cross-linking-based technique that can detect genomic regions in close proximity
- Chromosome conformation capture (3C)
a cross-linking-based technique that can detect genomic regions in close proximity
- Chromosome territories
defined spaces occupied by individual chromosomes
- Transcription factories
spatial clusters of genes being actively transcribed
- PcG bodies
spatial clusters of genes repressed by Polycomb group proteins
- 4C
chromosome conformation capture-on-chip
Footnotes
DISCLOSURE STATEMENT The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Agarwal P, Verzi MP, Nguyen T, Hu J, Ehlers ML, et al. The MADS box transcription factor MEF2C regulates melanocyte development and is a direct transcriptional target and partner of SOX10. Development. 2011;138:2555–65. doi: 10.1242/dev.056804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agelopoulos M, McKay DJ, Mann RS. Developmental regulation of chromatin conformation by Hox proteins in Drosophila. Cell Rep. 2012;1(4):350–59. doi: 10.1016/j.celrep.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnosti DN, Kulkarni MM. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J. Cell Biochem. 2005;94:890–98. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- 4.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–23. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai L, Morozov AV. Gene regulation by nucleosome positioning. Trends Genet. 2010;26:476–83. doi: 10.1016/j.tig.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Bai L, Ondracka A, Cross FR. Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter. Mol. Cell. 2011;42:465–76. doi: 10.1016/j.molcel.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bantignies F, Cavalli G. Polycomb group proteins: repression in 3D. Trends Genet. 2011;27:454–64. doi: 10.1016/j.tig.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–26. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 10.Barolo S. Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. BioEssays. 2012;34:135–41. doi: 10.1002/bies.201100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Barth TK, Imhof A. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem. Sci. 2010;35:618–26. doi: 10.1016/j.tibs.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Barthel R, Tsytsykova AV, Barczak AK, Tsai EY, Dascher CC, et al. Regulation of tumor necrosis factor alpha gene expression by mycobacteria involves the assembly of a unique enhanceosome dependent on the coactivator proteins CBP/p300. Mol. Cell. Biol. 2003;23:526–33. doi: 10.1128/MCB.23.2.526-533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benayoun BA, Veitia RA. A post-translational modification code for transcription factors: sorting through a sea of signals. Trends Cell Biol. 2009;19:189–97. doi: 10.1016/j.tcb.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–76. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernard P, Harley VR. Acquisition of SOX transcription factor specificity through protein-protein interaction, modulation of Wnt signalling and post-translational modification. Int. J. Biochem. Cell Biol. 2010;42:400–10. doi: 10.1016/j.biocel.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell. 2011;43:145–55. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biggin MD. Animal transcription networks as highly connected, quantitative continua. Dev. Cell. 2011;21:611–26. doi: 10.1016/j.devcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012;44:148–56. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 20.Busser BW, Shokri L, Jaeger SA, Gisselbrecht SS, Singhania A, et al. Molecular mechanism underlying the regulatory specificity of a Drosophila homeodomain protein that specifies myoblast identity. Development. 2012;139:1164–74. doi: 10.1242/dev.077362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavalli G. Chromosome kissing. Curr. Opin. Genet. Dev. 2007;17:443–50. doi: 10.1016/j.gde.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Chan SK, Mann RS. The segment identity functions of Ultrabithorax are contained within its homeodomain and carboxy-terminal sequences. Genes Dev. 1993;7:796–811. doi: 10.1101/gad.7.5.796. [DOI] [PubMed] [Google Scholar]
- 23.Chan SK, Popperl H, Krumlauf R, Mann RS. An extradenticle-induced conformational change in a HOX protein overcomes an inhibitory function of the conserved hexapeptide motif. EMBO J. 1996;15:2476–87. [PMC free article] [PubMed] [Google Scholar]
- 24.Charlot C, Dubois-Pot H, Serchov T, Tourrette Y, Wasylyk B. A review of post-translational modifications and subcellular localization of Ets transcription factors: possible connection with cancer and involvement in the hypoxic response. Methods Mol. Biol. 2010;647:3–30. doi: 10.1007/978-1-60761-738-9_1. [DOI] [PubMed] [Google Scholar]
- 25.Courey AJ. Cooperativity in transcriptional control. Curr. Biol. 2001;11:R250–52. doi: 10.1016/s0960-9822(01)00130-0. [DOI] [PubMed] [Google Scholar]
- 26.Dai Z, Dai X. Nuclear colocalization of transcription factor target genes strengthens coregulation in yeast. Nucleic Acids Res. 2012;40:27–36. doi: 10.1093/nar/gkr689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daitoku H, Sakamaki J, Fukamizu A. Regulation of FoxO transcription factors by acetylation and protein-protein interactions. Biochim. Biophys. Acta. 2011;1813:1954–60. doi: 10.1016/j.bbamcr.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–20. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidson EH, Levine MS. Properties of developmental gene regulatory networks. Proc. Natl. Acad. Sci. USA. 2008;105:20063–66. doi: 10.1073/pnas.0806007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 32.Delval S, Taminiau A, Lamy J, Lallemand C, Gilles C, et al. The Pbx interaction motif of Hoxa1 is essential for its oncogenic activity. PLoS ONE. 2011;6:e25247. doi: 10.1371/journal.pone.0025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dostie J, Bickmore WA. Chromosome organization in the nucleus: charting new territory across the Hi-Cs. Curr. Opin. Genet. Dev. 2012;22(2):125–31. doi: 10.1016/j.gde.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 34.Duboule D. Patterning in the vertebrate limb. Curr. Opin. Genet. Dev. 1991;1:211–16. doi: 10.1016/s0959-437x(05)80072-3. [DOI] [PubMed] [Google Scholar]
- 35.Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–64. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 36.Duncan IW. Transvection effects in Drosophila. Annu. Rev. Genet. 2002;36:521–56. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- 37.Dunipace L, Ozdemir A, Stathopoulos A. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development. 2011;138:4075–84. doi: 10.1242/dev.069146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 39.Enriquez J, Boukhatmi H, Dubois L, Philippakis AA, Bulyk ML, et al. Multi-step control of muscle diversity by Hox proteins in the Drosophila embryo. Development. 2010;137:457–66. doi: 10.1242/dev.045286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eskiw CH, Fraser P. Ultrastructural study of transcription factories in mouse erythroblasts. J. Cell Sci. 2011;124:3676–83. doi: 10.1242/jcs.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ethier SD, Miura H, Dostie J. Discovering genome regulation with 3C and 3C-related technologies. Biochim. Biophys. Acta. 2011;1819:401–10. doi: 10.1016/j.bbagrm.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 42.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–53. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 43.Ferraiuolo MA, Rousseau M, Miyamoto C, Shenker S, Wang XQ, et al. The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res. 2010;38:7472–84. doi: 10.1093/nar/gkq644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–24. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischback NA, Rozenfeld S, Shen W, Fong S, Chrobak D, et al. HOXB6 overexpression in murine bone marrow immortalizes a myelomonocytic precursor in vitro and causes hematopoietic stem cell expansion and acute myeloid leukemia in vivo. Blood. 2005;105:1456–66. doi: 10.1182/blood-2004-04-1583. [DOI] [PubMed] [Google Scholar]
- 46.Fitzsimmons D, Lutz R, Wheat W, Chamberlin HM, Hagman J. Highly conserved amino acids in Pax and Ets proteins are required for DNA binding and ternary complex assembly. Nucleic Acids Res. 2001;29:4154–65. doi: 10.1093/nar/29.20.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–18. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–93. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gardner KE, Allis CD, Strahl BD. Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 2011;409:36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garvie CW, Hagman J, Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell. 2001;8:1267–76. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- 51.Georges AB, Benayoun BA, Caburet S, Veitia RA. Generic binding sites, generic DNA-binding domains: Where does specific promoter recognition come from? FASEB J. 2010;24:346–56. doi: 10.1096/fj.09-142117. [DOI] [PubMed] [Google Scholar]
- 52.Gonzalez-Reyes A, Morata G. The developmental effect of overexpressing a Ubx product in Drosophila embryos is dependent on its interactions with other homeotic products. Cell. 1990;61:515–22. doi: 10.1016/0092-8674(90)90533-k. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez-Reyes A, Urquia N, Gehring WJ, Struhl G, Morata G. Are cross-regulatory interactions between homoeotic genes functionally significant? Nature. 1990;344:78–80. doi: 10.1038/344078a0. [DOI] [PubMed] [Google Scholar]
- 54.Gordan R, Hartemink AJ, Bulyk ML. Distinguishing direct versus indirect transcription factor–DNA interactions. Genome Res. 2009;19:2090–100. doi: 10.1101/gr.094144.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guertin MJ, Lis JT. Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 2010;6:e1001114. doi: 10.1371/journal.pgen.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hakim O, Sung MH, Voss TC, Splinter E, John S, et al. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res. 2011;21:697–706. doi: 10.1101/gr.111153.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7:e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He HH, Meyer CA, Shin H, Bailey ST, Wei G, et al. Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 2010;42:343–47. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He Q, Bardet AF, Patton B, Purvis J, Johnston J, et al. High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat. Genet. 2011;43:414–20. doi: 10.1038/ng.808. [DOI] [PubMed] [Google Scholar]
- 60.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–18. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 61.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heldring N, Isaacs GD, Diehl AG, Sun M, Cheung E, et al. Multiple sequence-specific DNA-binding proteins mediate estrogen receptor signaling through a tethering pathway. Mol. Endocrinol. 2011;25:564–74. doi: 10.1210/me.2010-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 2011;80:437–71. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hughes CL, Kaufman TC. Hox genes and the evolution of the arthropod body plan. Evol. Dev. 2002;4:459–99. doi: 10.1046/j.1525-142x.2002.02034.x. [DOI] [PubMed] [Google Scholar]
- 66.Istrail S, Davidson EH. Logic functions of the genomic cis-regulatory code. Proc. Natl. Acad. Sci. USA. 2005;102:4954–59. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jansen A, van der Zande E, Meert W, Fink GR, Verstrepen KJ. Distal chromatin structure influences local nucleosome positions and gene expression. Nucleic Acids Res. 2012;40(9):3870–85. doi: 10.1093/nar/gkr1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jauch R, Aksoy I, Hutchins AP, Ng CK, Tian XF, et al. Conversion of Sox17 into a pluripotency reprogramming factor by reengineering its association with Oct4 on DNA. Stem Cells. 2011;29:940–51. doi: 10.1002/stem.639. [DOI] [PubMed] [Google Scholar]
- 69.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 70.John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–68. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones SA, Shim SH, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods. 2011;8:499–508. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joseph R, Orlov YL, Huss M, Sun W, Kong SL, et al. Integrative model of genomic factors for determining binding site selection by estrogen receptor-α. Mol. Syst. Biol. 2010;6:456. doi: 10.1038/msb.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, et al. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–43. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joshi R, Sun L, Mann R. Dissecting the functional specificities of two Hox proteins. Genes Dev. 2010;24:1533–45. doi: 10.1101/gad.1936910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, et al. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell. 2012;148:473–86. doi: 10.1016/j.cell.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 76.Kaplan N, Hughes TR, Lieb JD, Widom J, Segal E. Contribution of histone sequence preferences to nucleosome organization: proposed definitions and methodology. Genome Biol. 2010;11:140. doi: 10.1186/gb-2010-11-11-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaplan N, Moore I, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, et al. Nucleosome sequence preferences influence in vivo nucleosome organization. Nat. Struct. Mol. Biol. 2010;17(8):918–20. doi: 10.1038/nsmb0810-918. author reply, 920–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–66. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kaplan T, Li XY, Sabo PJ, Thomas S, Stamatoyannopoulos JA, et al. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS Genet. 2011;7:e1001290. doi: 10.1371/journal.pgen.1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–85. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khoueiry P, Rothbacher U, Ohtsuka Y, Daian F, Frangulian E, et al. A cis-regulatory signature in ascidians and flies, independent of transcription factor binding sites. Curr. Biol. 2010;20:792–802. doi: 10.1016/j.cub.2010.03.063. [DOI] [PubMed] [Google Scholar]
- 82.Knoepfler PS, Sykes DB, Pasillas M, Kamps MP. HoxB8 requires its Pbx-interaction motif to block differentiation of primary myeloid progenitors and of most cell line models of myeloid differentiation. Oncogene. 2001;20:5440–48. doi: 10.1038/sj.onc.1204710. [DOI] [PubMed] [Google Scholar]
- 83.Kondoh H, Kamachi Y. SOX-partner code for cell specification: regulatory target selection and underlying molecular mechanisms. Int. J. Biochem. Cell Biol. 2010;42:391–99. doi: 10.1016/j.biocel.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 84.Krivega I, Dean A. Enhancer and promoter interactions: long distance calls. Curr. Opin. Genet. Dev. 2011;22(2):79–85. doi: 10.1016/j.gde.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kulkarni MM, Arnosti DN. Information display by transcriptional enhancers. Development. 2003;130:6569–75. doi: 10.1242/dev.00890. [DOI] [PubMed] [Google Scholar]
- 86.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–85. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lelli KM, Noro B, Mann RS. Variable motif utilization in homeotic selector (Hox)-cofactor complex formation controls specificity. Proc. Natl. Acad. Sci. USA. 2011;108:21122–27. doi: 10.1073/pnas.1114118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr. Opin. Genet. Dev. 2011;21:175–86. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li XY, Thomas S, Sabo PJ, Eisen MB, Stamatoyannopoulos JA, Biggin MD. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol. 2011;12:R34. doi: 10.1186/gb-2011-12-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–3. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–13. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 95.Lomvardas S, Thanos D. Modifying gene expression programs by altering core promoter chromatin architecture. Cell. 2002;110:261–71. doi: 10.1016/s0092-8674(02)00822-x. [DOI] [PubMed] [Google Scholar]
- 96.Ludwig MZ, Manu, Kittler R, White KP, Kreitman M. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLoS Genet. 2011;7:e1002364. doi: 10.1371/journal.pgen.1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. Functional evolution of a cis-regulatory module. PLoS Biol. 2005;3:e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 99.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.MacArthur S, Li XY, Li J, Brown JB, Chu HC, et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.MacQuarrie KL, Fong AP, Morse RH, Tapscott SJ. Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet. 2011;27:141–48. doi: 10.1016/j.tig.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Magnani L, Eeckhoute J, Lupien M. Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet. 2011;27:465–74. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 103.Mann RS, Chan SK. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–62. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 104.Mann RS, Hogness DS. Functional dissection of Ultrabithorax proteins in D. melanogaster. Cell. 1990;60:597–610. doi: 10.1016/0092-8674(90)90663-y. [DOI] [PubMed] [Google Scholar]
- 105.Mann RS, Lelli KM, Joshi R. Hox specificity: unique roles for cofactors and collaborators. Curr. Top. Dev. Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mao C, Brown CR, Griesenbeck J, Boeger H. Occlusion of regulatory sequences by promoter nucleosomes in vivo. PLoS ONE. 2011;6:e17521. doi: 10.1371/journal.pone.0017521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–49. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–10. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Merabet S, Kambris Z, Capovilla M, Berenger H, Pradel J, Graba Y. The hexapeptide and linker regions of the AbdA Hox protein regulate its activating and repressive functions. Dev. Cell. 2003;4:761–68. doi: 10.1016/s1534-5807(03)00126-6. [DOI] [PubMed] [Google Scholar]
- 110.Merabet S, Litim-Mecheri I, Karlsson D, Dixit R, Saadaoui M, et al. Insights into Hox protein function from a large scale combinatorial analysis of protein domains. PLoS Genet. 2011;7:e1002302. doi: 10.1371/journal.pgen.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Merabet S, Saadaoui M, Sambrani N, Hudry B, Pradel J, et al. A unique extradenticle recruitment mode in the Drosophila Hox protein Ultrabithorax. Proc. Natl. Acad. Sci. USA. 2007;104:16946–51. doi: 10.1073/pnas.0705832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Merabet S, Sambrani N, Pradel J, Graba Y. Regulation of Hox activity: insights from protein motifs. Adv. Exp. Med. Biol. 2010;689:3–16. doi: 10.1007/978-1-4419-6673-5_1. [DOI] [PubMed] [Google Scholar]
- 113.Miller JA, Widom J. Collaborative competition mechanism for gene activation in vivo. Mol. Cell. Biol. 2003;23:1623–32. doi: 10.1128/MCB.23.5.1623-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mirny LA. Nucleosome-mediated cooperativity between transcription factors. Proc. Natl. Acad. Sci. USA. 2010;107:22534–39. doi: 10.1073/pnas.0913805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mo Y, Ho W, Johnston K, Marmorstein R. Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex. J. Mol. Biol. 2001;314:495–506. doi: 10.1006/jmbi.2001.5138. [DOI] [PubMed] [Google Scholar]
- 116.Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev. Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 117.Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–45. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 118.Moorman C, Sun LV, Wang J, de Wit E, Talhout W, et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2006;103:12027–32. doi: 10.1073/pnas.0605003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moretti R, Ansari AZ. Expanding the specificity of DNA targeting by harnessing cooperative assembly. Biochimie. 2008;90:1015–25. doi: 10.1016/j.biochi.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 120.Moyle-Heyrman G, Tims HS, Widom J. Structural constraints in collaborative competition of transcription factors against the nucleosome. J. Mol. Biol. 2011;412:634–46. doi: 10.1016/j.jmb.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Negre N, Brown CD, Ma L, Bristow CA, Miller SW, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–31. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ng CK, Li NX, Chee S, Prabhakar S, Kolatkar PR, Jauch R. Deciphering the Sox-Oct partner code by quantitative cooperativity measurements. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks153. doi: 10.1093/nar/gks153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nien CY, Liang HL, Butcher S, Sun Y, Fu S, et al. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011;7:e1002339. doi: 10.1371/journal.pgen.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. The dynamic architecture of Hox gene clusters. Science. 2011;334:222–25. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 126.Noro B, Lelli K, Sun L, Mann RS. Competition for cofactor-dependent DNA binding underlies Hox phenotypic suppression. Genes Dev. 2011;25:2327–32. doi: 10.1101/gad.175539.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–89. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–71. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 129.Pan Y, Nussinov R. The role of response elements organization in transcription factor selectivity: the IFN-β enhanceosome example. PLoS Comput. Biol. 2011;7:e1002077. doi: 10.1371/journal.pcbi.1002077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pan Y, Tsai CJ, Ma B, Nussinov R. Mechanisms of transcription factor selectivity. Trends Genet. 2010;26:75–83. doi: 10.1016/j.tig.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Panne D. The enhanceosome. Curr. Opin. Struct. Biol. 2008;18:236–42. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 132.Panne D, Maniatis T, Harrison SC. Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-β enhancer. EMBO J. 2004;23:4384–93. doi: 10.1038/sj.emboj.7600453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-β enhanceosome. Cell. 2007;129:1111–23. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Papadopoulos DK, Resendez-Perez D, Cardenas-Chavez DL, Villanueva-Segura K, Canales-del-Castillo R, et al. Functional synthetic Antennapedia genes and the dual roles of YPWM motif and linker size in transcriptional activation and repression. Proc. Natl. Acad. Sci. USA. 2011;108:11959–64. doi: 10.1073/pnas.1108686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Papadopoulos DK, Vukojevic V, Adachi Y, Terenius L, Rigler R, Gehring WJ. Function and specificity of synthetic Hox transcription factors in vivo. Proc. Natl. Acad. Sci. USA. 2010;107:4087–92. doi: 10.1073/pnas.0914595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Parker DS, White MA, Ramos AI, Cohen BA, Barolo S. The cis-regulatory logic of Hedgehog gradient responses: key roles for Gli binding affinity, competition, and cooperativity. Sci. Signal. 2011;4:ra38. doi: 10.1126/scisignal.2002077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 2010;20:1562–67. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Polach KJ, Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J. Mol. Biol. 1996;258:800–12. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- 139.Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr. Opin. Genet. Dev. 2012;22:148–55. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu. Rev. Biochem. 2009;78:245–71. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Razin SV, Gavrilov AA, Pichugin A, Lipinski M, Iarovaia OV, Vassetzky YS. Transcription factories in the context of the nuclear and genome organization. Nucleic Acids Res. 2011;39:9085–92. doi: 10.1093/nar/gkr683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Remacle S, Abbas L, De Backer O, Pacico N, Gavalas A, et al. Loss of function but no gain of function caused by amino acid substitutions in the hexapeptide Hoxa1 in vivo. Mol. Cell. Biol. 2004;24:8567–75. doi: 10.1128/MCB.24.19.8567-8575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–19. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010;79:233–69. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–53. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–97. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–95. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Saadaoui M, Merabet S, Litim-Mecheri I, Arbeille E, Sambrani N, et al. Selection of distinct Hox-Extradenticle interaction modes fine-tunes Hox protein activity. Proc. Natl. Acad. Sci. USA. 2011;108:2276–81. doi: 10.1073/pnas.1006964108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Satija R, Bradley RK. The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome Res. 2012;22:656–65. doi: 10.1101/gr.130682.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schnabel CA, Jacobs Y, Cleary ML. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene. 2000;19:608–16. doi: 10.1038/sj.onc.1203371. [DOI] [PubMed] [Google Scholar]
- 151.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–43. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–9. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: Genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol. Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Serandour AA, Avner S, Percevault F, Demay F, Bizot M, et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011;21:555–65. doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 157.Shen W, Chrobak D, Krishnan K, Lawrence HJ, Largman C. HOXB6 protein is bound to CREB-binding protein and represses globin expression in a DNA binding–dependent, PBX interaction–independent process. J. Biol. Chem. 2004;279:39895–904. doi: 10.1074/jbc.M404132200. [DOI] [PubMed] [Google Scholar]
- 158.Shykind BM. Regulation of odorant receptors: one allele at a time. Hum. Mol. Genet. 2005;14(Spec. No. 1):R33–39. doi: 10.1093/hmg/ddi105. [DOI] [PubMed] [Google Scholar]
- 159.Siggers T, Duyzend MH, Reddy J, Khan S, Bulyk ML. Non-DNA-binding cofactors enhance DNA-binding specificity of a transcriptional regulatory complex. Mol. Syst. Biol. 2011;7:555. doi: 10.1038/msb.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Simon JA, Kingston RE. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 161.Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011;147:1270–82. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stobe P, Stein MA, Habring-Muller A, Bezdan D, Fuchs AL, et al. Multifactorial regulation of a hox target gene. PLoS Genet. 2009;5:e1000412. doi: 10.1371/journal.pgen.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Swanson CI, Evans NC, Barolo S. Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev. Cell. 2010;18:359–70. doi: 10.1016/j.devcel.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Talbert PB, Henikoff S. Histone variants: ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 2010;11:264–75. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 165.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 166.Thomas S, Li XY, Sabo PJ, Sandstrom R, Thurman RE, et al. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 2011;12:R43. doi: 10.1186/gb-2011-12-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tims HS, Gurunathan K, Levitus M, Widom J. Dynamics of nucleosome invasion by DNA binding proteins. J. Mol. Biol. 2011;411:430–48. doi: 10.1016/j.jmb.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tolkunov D, Zawadzki KA, Singer C, Elfving N, Morozov AV, Broach JR. Chromatin remodelers clear nucleosomes from intrinsically unfavorable sites to establish nucleosome-depleted regions at promoters. Mol. Biol. Cell. 2011;22:2106–18. doi: 10.1091/mbc.E10-10-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tootle TL, Rebay I. Post-translational modifications influence transcription factor activity: a view from the ETS superfamily. BioEssays. 2005;27:285–98. doi: 10.1002/bies.20198. [DOI] [PubMed] [Google Scholar]
- 170.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 171.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000;18:424–28. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 172.Vaquerizas JM, Akhtar A, Luscombe NM. Large-scale nuclear architecture and transcriptional control. Subcell. Biochem. 2011;52:279–95. doi: 10.1007/978-90-481-9069-0_13. [DOI] [PubMed] [Google Scholar]
- 173.Verger A, Duterque-Coquillaud M. When Ets transcription factors meet their partners. BioEssays. 2002;24:362–70. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- 174.Vitobello A, Ferretti E, Lampe X, Vilain N, Ducret S, et al. Hox and Pbx factors control retinoic acid synthesis during hindbrain segmentation. Dev. Cell. 2011;20:469–82. doi: 10.1016/j.devcel.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–54. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang X, Bai L, Bryant GO, Ptashne M. Nucleosomes and the accessibility problem. Trends Genet. 2011;27:487–92. doi: 10.1016/j.tig.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 177.Wunderlich Z, Mirny LA. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 2009;25:434–40. doi: 10.1016/j.tig.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yaron Y, McAdara JK, Lynch M, Hughes E, Gasson JC. Identification of novel functional regions important for the activity of HOXB7 in mammalian cells. J. Immunol. 2001;166:5058–67. doi: 10.4049/jimmunol.166.8.5058. [DOI] [PubMed] [Google Scholar]
- 179.You JS, Kelly TK, De Carvalho DD, Taberlay PC, Liang G, Jones PA. OCT4 establishes and maintains nucleosome-depleted regions that provide additional layers of epigenetic regulation of its target genes. Proc. Natl. Acad. Sci. USA. 2011;108:14497–502. doi: 10.1073/pnas.1111309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–41. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 182.Zhou X, O’Shea EK. Integrated approaches reveal determinants of genome-wide binding and function of the transcription factor Pho4. Mol. Cell. 2011;42:826–36. doi: 10.1016/j.molcel.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhu X, Ahmad SM, Aboukhalil A, Busser BW, Kim Y, et al. Differential regulation of mesodermal gene expression by Drosophila cell type–specific Forkhead transcription factors. Development. 2012;139:1457–66. doi: 10.1242/dev.069005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zorn C, Cremer C, Cremer T, Zimmer J. Unscheduled DNA synthesis after partial UV irradiation of the cell nucleus: Distribution in interphase and metaphase. Exp. Cell Res. 1979;124:111–19. doi: 10.1016/0014-4827(79)90261-1. [DOI] [PubMed] [Google Scholar]
- 185.Zuleger N, Robson MI, Schirmer EC. The nuclear envelope as a chromatin organizer. Nucleus. 2011;2:339–49. doi: 10.4161/nucl.2.5.17846. [DOI] [PMC free article] [PubMed] [Google Scholar]