SUMMARY
The second messenger molecule cyclic diguanylate (c-di-GMP) is essential for Y. pestis biofilm formation that is important for blockage-dependent plague transmission from fleas to mammals. Two diguanylate cyclases (DGCs) HmsT and Y3730 (HmsD) are responsible for biofilm formation in vitro and biofilm-dependent blockage in the oriental rat flea Xenopsylla cheopis, respectively. Here, we have identified a tripartite signaling system encoded by the y3729-y3731 operon that is responsible for regulation of biofilm formation in different environments. We present genetic evidence that a putative inner membrane-anchored protein with a large periplasmic domain Y3729 (HmsC) inhibits HmsD DGC activity in vitro while an outer membrane Pal-like putative lipoprotein Y3731 (HmsE) counteracts HmsC to activate HmsD in the gut of X. cheopis. We propose that HmsE is a critical element in transduction of environmental signal(s) required for HmsD-dependent biofilm formation.
INTRODUCTION
The gram-negative bacterium Yersinia pestis, infamous for causing plague pandemics, remains a serious problem with epidemic outbreaks occurring worldwide. Y. pestis is a successful zoonotic pathogen with large established sylvatic foci in Asia, Africa and the Americas. Y. pestis causes zoonotic disease that primarily involves rodents and associated fleas where fleas serve as a transmission vector to spread plague from animal to animal (Perry and Fetherston, 1997; Hinnebusch and Erickson, 2008; Stenseth et al., 2008). In the oriental rat flea Xenopsylla cheopis, a primary Y. pestis vector in many plague endemic areas throughout the world, Y. pestis grows as a biofilm in the midgut and eventually colonizes the proventriculus (a valve between the midgut and esophagus) causing partial or complete blockage. In completely blocked fleas, fresh blood cannot reach the midgut (stomach). Consequently blocked fleas increase their feeding attempts causing bacteria to dislodge from the biofilm; regurgitation of the now contaminated blood into the bite site infects the mammal. In partially blocked fleas, there is a small open channel in the proventriculus that allows some of the bloodmeal to enter the midgut of the flea. Partial blockage is also able to mediate efficient biofilm-dependent transmission of Y. pestis (Bacot and Martin, 1914; Bacot, 1915; Jarrett et al., 2004; Hinnebusch and Erickson, 2008; Hinnebusch, 2012). This blockage- or biofilm-dependent mechanism of transmission has been shown for many flea species that are established plague vectors in the majority of endemic plague areas (Bibikova and Klassovskii, 1974; Vatschenok, 1988; Anisimov et al., 2004; Krasnov et al., 2006).
Y. pestis biofilm formation requires a poly-β-1,6-N-acetyl-D-glucosamine exopolysaccharide (EPS) that is produced by the hmsHFRS gene products (Perry et al., 1990; Lillard et al., 1997; Bobrov et al., 2008; Erickson et al., 2008). The levels of Hms-dependent EPS in Y. pestis KIM6+ are regulated by enzymes involved in synthesis and degradation of the second messenger molecule cyclic di-GMP (c-di-GMP) (Kirillina et al., 2004; Romling et al., 2013). Although 10 genes encoding diguanylate cyclases (DGCs) and phosphodiesterases (PDEs) are present in the Y. pestis genome, only three produce enzymatically functional proteins. Under in vitro conditions, the DGC HmsT synthesizes c-di-GMP and activates production of the EPS polymer while the PDE HmsP degrades c-di-GMP and lowers EPS synthesis. The second functional DGC identified in Y. pestis KIM6+, Y3730 (HmsD), is responsible for a small fraction of cellular c-di-GMP and is not required for biofilm formation under in vitro conditions (Kirillina et al., 2004; Bobrov et al., 2005; Simm et al., 2005; Bobrov et al., 2008; Bobrov et al., 2011). In contrast, HmsD, not HmsT, is critical for the development of biofilm-dependent blockage in X. cheopis. The mechanism for differential regulation of biofilm formation by HmsT and HmsD in these two environments has not been identified. However, equivalent levels of transcription of hmsD and hmsT in vitro and in the flea have been demonstrated, suggesting a post-transcriptional mechanism for this regulation (Sun et al., 2011).
In this study, we show that the DGC HmsD is controlled by two linked genes, y3729 (hmsC) and y3731 (hmsE), which likely encode inner membrane (IM) –anchored and outer membrane (OM) proteins, respectively. A deletion of hmsC or overexpression of hmsE resulted in increased c-di-GMP production by HmsD and hyper-biofilm formation in vitro. In X. cheopis, an hmsE mutant had lower blockage rates – similar to an hmsD mutant. Although the hmsC mutant exhibits robust EPS production in vitro at 37°C similar to an hmsP mutant, unlike the hmsP mutant it does not have a virulence defect in mouse model of bubonic plague. We also show that the HmsCDE regulatory pathway is functional in Y. pestis strains representative of all biovars and subspecies.
While this manuscript was under revision, a manuscript by Ren et al showing regulation of HmsD by HmsC alone was accepted for publication (Ren et al., 2013). Consequently, we have adopted their terminology for the y3729-y3731 locus (hmsCDE). In our discussion, we include differences and similarities between the two studies.
RESULTS
HmsC and HmsE inversely regulate Hms-dependent biofilm formation in Y. pestis via control of c-di-GMP production by the DGC HmsD
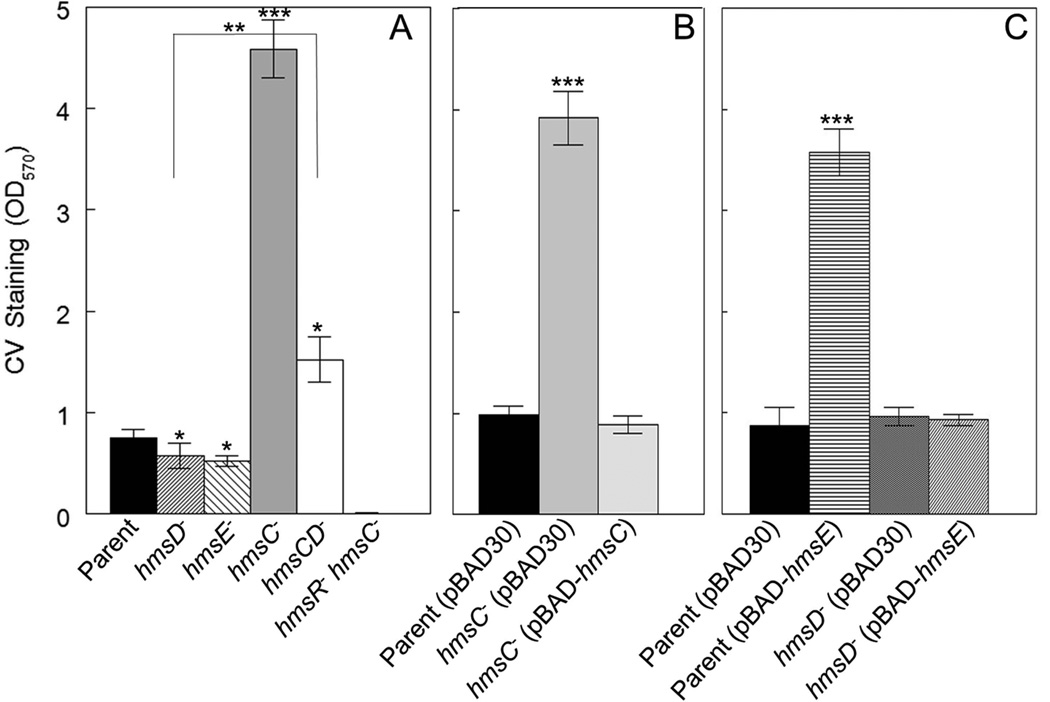
The hmsD gene is part of a three gene operon (hmsCDE), with the two flanking genes, hmsC and hmsE, predicted to encode an IM protein and a peptidoglycan associated lipoprotein (PAL) - like OM protein, respectively. All three gene products are orthologous to YfiRNB proteins from Escherichia coli and Pseudomonas species where YfiR (the HmsC orthologue) represses the function of the DGC YfiN (the HmsD orthologue) (Girgis et al., 2007; McDonald et al., 2009; Malone et al., 2010). We show here that deletion of hmsC in Y. pestis KIM6+ resulted in a drastic increase in biofilm formation in vitro while overexpression of this gene from an arabinose-inducible promoter complemented this mutant phenotype (Fig. 1A and 1B). Moreover, an hmsCD double mutant showed an ~ 3-fold reduction in biofilm formation compared to the hmsC mutant (Fig. 1A) indicating that the DGC HmsD is negatively regulated by HmsC. Curiously, biofilm formation by the hmsCD mutant was significantly higher than that of the parent strain or hmsD mutant suggesting that HmsC may regulate additional protein(s) that directly or indirectly influence biofilm development, at least under these growth conditions. These results independently confirm those of Ren et al (Ren et al., 2013) and suggest that HmsD does not contribute to in vitro biofilm formation because it is inhibited by HmsC. Therefore, HmsT is the dominant DGC controlling in vitro biofilm development, as previously shown (Kirillina et al., 2004).
Fig. 1. HmsC and HmsE inversely modulate Hms-dependent biofilm formation in vitro via HmsD.
A crystal violet (CV) staining assay was used to assess Hms biofilm formation in the following strains: KIM6+ (Parent), KIM6-2159.1+ (hmsD−), KIM6-2174+ (hmsE−), KIM6-2173.1+ (hmsC−), KIM6-2198+ (hmsCD−) and KIM6-2173.2 (hmsC− hmsR−) (panel A). Panel B shows the results of complementation with hmsC expressed from plasmid pBAD-hmsC. Panel C demonstrates the effect of hmsE expressed from pBAD-hmsE on biofilm formation in the parent and hmsD mutant strains. Results are from duplicate assays on two independent cultures (4 biological samples) (in A and B) and from triplicate assays on three independent cultures (9 biological samples) (in C). Error bars indicate standard deviations. The asterisks indicate statistically significant differences between the parent strains and mutants by the Student’s T-test (*P<0.05, **P<0.005 *** P<0.001). The asterisks with the bracket indicate a statistically significant difference between the two indicated strains.
An hmsE mutation showed a subtle reduction of biofilm production, similar to that observed in an hmsD mutant (Fig. 1A). Alternatively, overexpression of hmsE in the parent strain greatly increased biofilm formation (Fig. 1C). In contrast, overexpression of hmsE in the hmsD mutant did not cause increased biofilm formation (Fig. 1C), indicating that HmsE specifically activates HmsD. Furthermore, inactivation of hmsC or overexpression of hmsE in an hmsT− background caused robust biofilm formation (Fig. 2) further supporting our conclusion that HmsC and HmsE control Y. pestis biofilm formation via the DGC HmsD. The effects of HmsE and HmsC are specific to Hms-dependent biofilm formation, since deletion of hmsR, encoding a putative glycosyltransferase essential for Hms EPS biosynthesis, negates the phenotypes of the hmsC mutant and the hmsE overexpressing strain (Fig. 1A and data not shown).
Fig. 2. HmsE counteracts HmsC to activate HmsD function.
The following strains were compared in a CV staining assay: KIM6-2051.1+ (hmsT−), KIM6-2173.3+ (hmsT− hmsC−), KIM6-2173.4+ (hmsT− hmsC− hmsE−) as well as KIM6-2051.1+ and KIM6-2173.3+ carrying pBAD30 or pBAD30-hmsE. Results are averages of triplicate assays from three independent cultures. Error bars indicate standard deviations. The asterisks indicate a statistically significant difference between the indicated strains by the Student’s T-test (*** P<0.001).
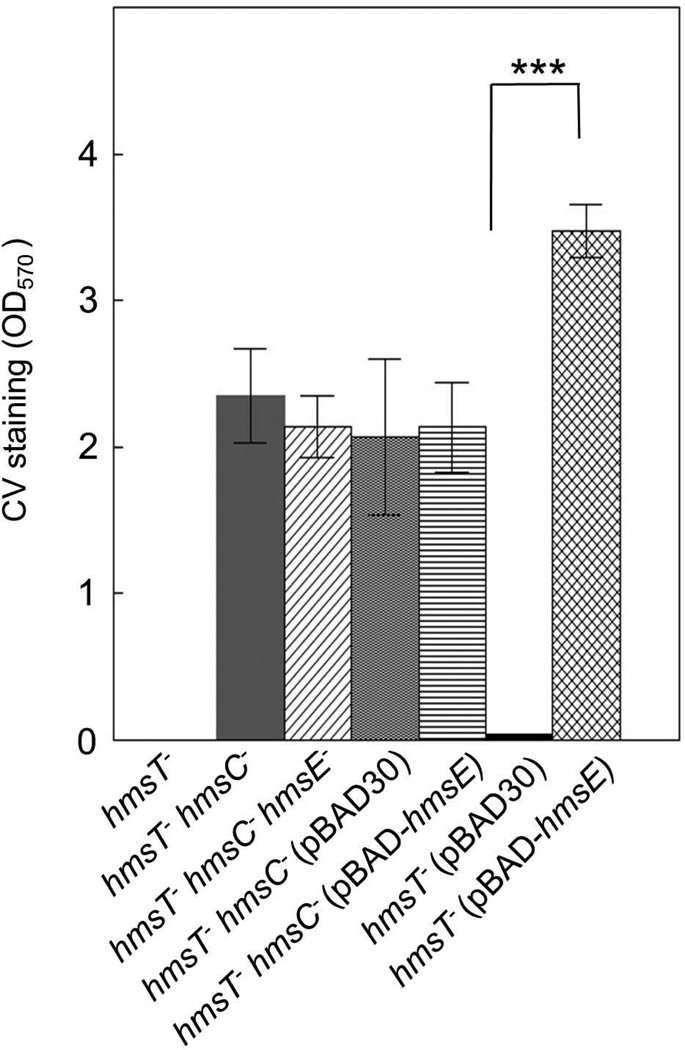
To demonstrate that HmsC and HmsE affect biofilm formation by modulating HmsD-dependent c-di-GMP levels in Y. pestis, the intracellular concentrations of c-di-GMP in an hmsT mutant were compared to those in a double hmsT hmsC mutant and an hmsT mutant overexpressing hmsE. Both deletion of hmsC or overexpression of hmsE resulted in increased levels of c-di-GMP compared to the hmsT mutant where c-di-GMP levels were undetectable (Fig. 3). Thus, HmsC inhibits and HmsE promotes the HmsD-dependent production of c-di-GMP in Y. pestis KIM6+.
Fig. 3. HmsC and HmsE respectively reduce and increase cellular levels of c-di-GMP through HmsD.
Relative cellular concentrations of c-di-GMP in KIM6-2051+ (hmsT−) and KIM6-2173.3+ (hmsT− hmsC−) carrying either pBAD30 or pBAD-hmsE are shown. Results are from duplicate assays from two independent cultures. Error bars indicate standard deviations. Statistical significance was not calculated because the c-di-GMP concentration in the parent hmsT− strain was below detection limits.
To provide genetic evidence that HmsE affects HmsD function via HmsC, we inactivated hmsE in the hyper-biofilm-forming double hmsT hmsC mutant. The level of biofilm formation in a triple hmsT hmsC hmsE mutant was not significantly different from the double hmsT hmsC mutant. Additionally, overexpression of hmsE in the hmsT hmsC double mutant did not significantly increase biofilm formation. In contrast, overexpression of hmsE in the hmsT mutant results in increased biofilm formation (Fig. 2). This suggests that HmsE acts indirectly to activate the DGC activity of HmsD by counteracting HmsC.
Topology of proteins encoded by the hmsCDE operon
We used bioinformatics to predict the cellular localizations and topologies of HmsCDE proteins. To confirm the bioinformatics predictions, we performed enzymatic assays with translational fusions of HmsD and HmsC to β-galactosidase (β-Gal) or alkaline phosphatase A (PhoA), which provides topological information about the subcellular location of these translational fusions. β-Gal exhibits enzymatic activity in the cytoplasm and PhoA becomes active after its translocation into the periplasm (van Geest and Lolkema, 2000).
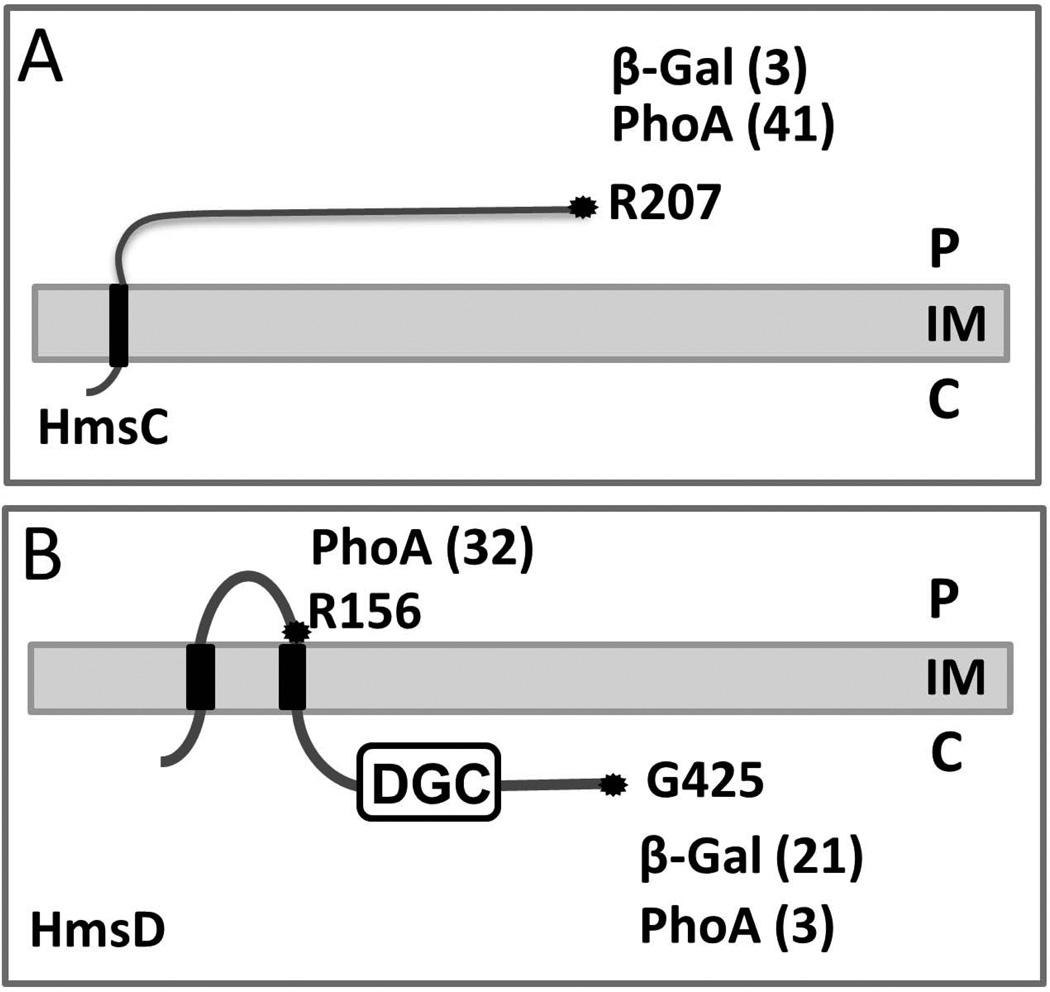
HmsCDE proteins are orthologous to components of the YfiBNR signal transduction system that modulates c-di-GMP levels in P. aeruginosa (Malone et al., 2010; Malone et al., 2012). However, HmsCDE have only modest similarities to the YfiRNB proteins - 42–56% identity over 79–99% of the Y. pestis ORFs. Bioinformatics analyses predict that a large periplasmic domain of HmsC, an orthologue of YfiR, is anchored to the IM via a single transmembrane domain while YfiR is a periplasmic protein. In contrast to YfiR, neither the Philius nor Phobius programs predicted the presence of a signal peptide in HmsC. Instead the Philius, Phobius and TMHMM programs predict that HmsC is an IM protein with one transmembrane domain with its N- and C-termini located in the cytoplasm and periplasm, respectively. Using β-Gal and PhoA translational fusions to the C-terminus of the hmsC ORF, PhoA but not β-Gal activity was detected in an hmsC mutant (Fig S1). This supports the bioinformatics analyses that the C-terminus of HmsC is located in the periplasm (Fig. 4A).
Fig. 4. Topology models of HmsC and HmsD.
Both topologies are computer predictions supported by analysis of strains expressing β-galactosidase (β-Gal) or alkaline phosphatase (PhoA) fusion proteins. HmsC and HmsD fusions were expressed in KIM6-2173.1+ (hmsC−), and KIM6-2159.1+ (hmsD−) strains, respectively. The positions of fusions are indicated by amino acid residue number; assay values are indicated in parentheses and statistical analyses are shown in Fig. S1. The periplasm (P) cytoplasm (C), inner and outer membrane (IM and OM) are labeled for orientation.
HmsD was consistently predicted as an IM protein with two transmembrane domains separating a central periplasmic region from cytoplasmic N- and C-termini. We confirmed these bioinformatics predictions using translational fusions to the C-terminus and the central putative periplasmic loop of HmsD (at position of R156). For unknown reasons, we were unable to obtain a β-Gal fusion at R156. C-terminal HmsD fusions showed high β-Gal and low PhoA activities in an hmsD mutant indicating a cytoplasmic location for its C-terminus. Conversely, the high PhoA activity of the R156 fusion supports a periplasmic location for this central region of HmsD (Fig. 4B and Fig. S1).
For HmsE, the Philius and Phobius programs predicted the presence of a signal peptide and the LipoP program predicted a lipoprotein signal sequence. Additionally, HmsE does not have an aspartate residue at the +2 position that has been determined to be necessary for localization of processed lipoproteins to the IM (Okuda and Tokuda, 2011). Moreover, HmsE is predicted to have a PAL-like peptidoglycan binding domain shown to be critical for the function of the orthologous protein YfiB in P. aeruginosa (Malone, 2012). Consequently, HmsE is expected to be sorted to the periplasmic face of the OM. The OM location of HmsE is supported by data on localization of protein YPTB0591 (>99% identity with HmsE), in the OM fraction of Yersinia pseudutoberculosis cells (Thein et al., 2010).
Thus, bioinformatics and our data suggest that HmsD is an IM protein with the DGC domain located in the cytoplasm and a central domain in the periplasm, that HmsC is an IM-anchored (or possibly a periplasmic) protein with an extensive C-terminal periplasmic domain and that HmsE is an OM lipoprotein with the C-terminus likely facing the periplasm.
HmsC and HmsE regulate biofilm formation in epidemic and endemic Y. pestis strains
To date, the c-di-GMP-dependent mechanism of Y. pestis biofilm regulation has been studied primarily in the KIM6+ and CO92 strains. Despite 99% identity among sequenced Y. pestis genomes, we identified significant variations in a number of the c-di-GMP metabolic enzymes among epidemic Y. pestis strains or “main subspecies” (phylogenetically divided into biovars: Antiqua, Medievalis, and Orientalis) and the more ancient endemic Pestoides strains (non-main subspecies). Pestoides strains are reported to be avirulent for humans and are thought to be intermediates between the Y. pestis ancestor, Y. pseudotuberculosis, and epidemic Y. pestis strains (Anisimov et al., 2004; Cui et al., 2008; Bearden et al., 2009; Bobrov et al., 2011). In addition, there is evidence in the literature of different degrees of biofilm-dependent blockage in a Caenorhabditis elegans model and some flea species by epidemic and endemic Y. pestis strains (Vatschenok, 1988; Anisimov, 2002; Eroshenko et al., 2010). To address whether the HmsCDE system may play a role in biofilm regulation inY. pestis strains other than KIM6+, we examined strains from our collection that represent all the main phylogenetic branches of Y. pestis – the 3 epidemic biovars and 2 clades of endemic strains. Biofilm formation by these strains varied greatly under the in vitro conditions tested, with Antiqua strains (particularly Kuma) exhibiting very robust biofilms and the endemic strains as well as KIM6+ forming the smallest biofilms (Fig. S2). However, all strains showed similar patterns in the regulation of in vitro biofilm formation by HmsC and HmsE. Like in KIM6+, deletion of hmsC or overexpression of hmsE resulted in hyper-biofilm formation in vitro indicating their roles as negative and positive regulators, respectively, of biofilm formation (Fig. 5A and 5B) in all strains tested. Although the KIM6+ hmsE region was overexpressed in all of these strains, the nucleotide sequence of the cloned region is 100% identical in all Y. pestis strains tested. Thus, HmsC and HmsE appear to be functional in epidemic and endemic strains of Y. pestis and likely act on HmsD as shown in KIM6+ (Fig. 1) indicating that this locus has a conserved role in biofilm formation.
Fig. 5. HmsC and HmsE respectively reduce and increase in vitro biofilm formation in epidemic and endemic strains of Y. pestis.
A crystal violet staining assay was used to assess Hms biofilm formation in: A) parent strains and their hmsC mutants (hmsC−); B) strains carrying pBAD30 or pBAD-hmsE; C) Kuma strains carrying pBAD30 or pBAD-hmsC. Results are from duplicate assays from two independent cultures. Error bars indicate standard deviations. The asterisks indicate statistically significant differences in biofilm formation between parent strains and their respective hmsC mutants (A) or between strains carrying pBAD-hmsE versus the pBAD30 vector (B) by Student’s T-test (*P<0.05, **P<0.005 *** P<0.001). The asterisks with brackets indicate statistically significant differences between the two indicated strains.
Strain Kuma had increased biofilm formation relative to the other Y. pestis strains we tested (Fig. S2). This could be due to a defect in HmsC in this strain. However, overexpression of the cloned KIM6+ hmsC gene caused a significant reduction in biofilm formation in the Kuma hmsC mutant but had no effect in the parent Kuma strain (Fig. 5C). This suggests that hyper-biofilm formation under in vitro conditions by Y. pestis Kuma is not due to an HmsC defect but occurs via other regulatory mechanisms.
An hmsC mutation does not affect virulence in a mouse model of bubonic plague
Previously we have shown that Y. pestis KIM strains unable to produce the Hms biofilm (hmsH and hmsR mutants) are fully virulent in mouse models of bubonic and pneumonic plague (Lillard et al., 1999; Abu Khweek et al., 2010; Bobrov et al., 2011). On the other hand, a hyper-biofilm producer (an hmsP mutant) that had increased Hms EPS production at 37°C compared to the parent strain, showed a >1,350-fold loss of virulence (LD50 > 1.35 × 104) in the bubonic plague model and a delay in time-to-death in the pneumonic plague model. Virulence was restored in an hmsR hmsP double mutant that was unable to express Hms EPS. (Bobrov et al., 2011).
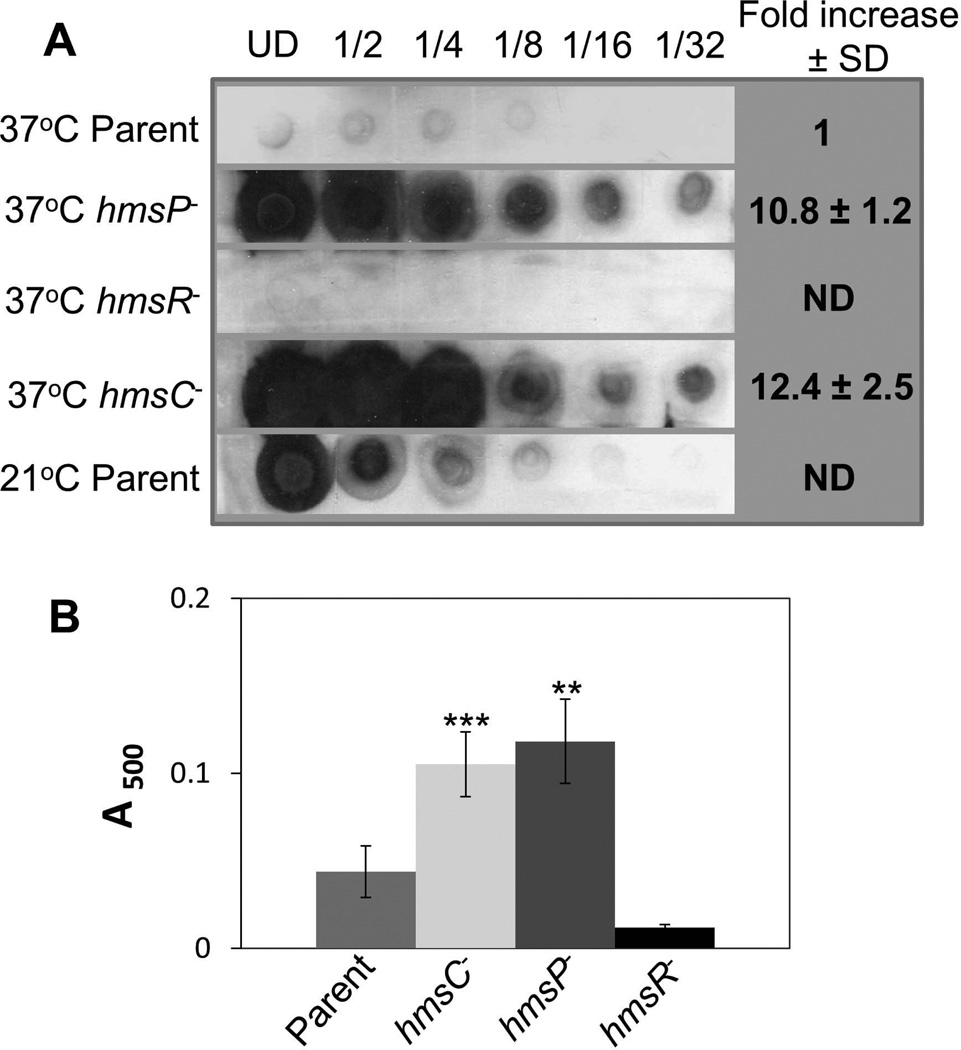
We found that the hmsC mutant had increased levels of Hms EPS production at 37°C similar to those of the hmsP mutant (Fig. 6) suggesting that this strain might have a defect in virulence similar to that of the hmsP mutant. However, in subcutaneous infections of mice, the hmsC mutant carrying the virulence plasmid encoding the Yersiniae type III secretion system was highly virulent (LD50 < 118 cells, the lowest dose tested). The parental hmsC+ strain had an LD50 of <12 cells. Thus, despite similar EPS overproduction in vitro at 37°C, the hmsC mutant was, at least, 100-fold more virulent than the hmsP mutant.
Fig. 6. The hmsP and hmsC mutants show similar increases in Hms exopolysaccharide (EPS) production at 37°C.
(A) Dot-blot analysis of the production of the poly-β-1,6-GlcNAc-like EPS by Y. pestis strains. Aliquots of undiluted (UD) samples and serial dilutions of crude cell extracts were spotted onto a nitrocellulose membrane and EPS was detected with antisera against purified PIA from S. epidermidis. (B) Congo red binding assays on Y. pestis strains. Results are from duplicate assays on two independent cultures. Strains: KIM6+ (Parent); KIM6-2118 (hmsR−); KIM6-2090.1+ (hmsP−), KIM6-2173.1+ (hmsC−). Error bars indicate standard deviations. The asterisks indicate statistically significant difference in Congo red binding by the parent strain and the mutants by Student’s T-test (**P<0.005 *** P<0.001). The values for the hmsP versus hmsC mutants were not statistically significant in both assays.
The HmsCDE system regulates blockage of Xenopsylla cheopis
The rat flea X. cheopis is an efficient plague vector and a paradigm for studying the interaction of Y. pestis with the flea. Y. pestis KIM6+ produced a biofilm in the proventriculus and the midgut of X. cheopis with the proventricular biofilm eventually causing complete or partial blockage of the flea. Classically flea blockage is defined as the inability of a fresh blood meal to reach the midgut (Jarrett et al., 2004). The DGC HmsD is critical for the proventricular blockage in X. cheopis (Sun et al., 2011). Our data indicate that c-di-GMP production by HmsD is inhibited by HmsC and activated by HmsE. If HmsD function is stimulated by HmsE in the flea environment then inactivation of hmsE should significantly reduce biofilm-dependent blockage of X. cheopis. Indeed, two independent experiments showed greatly decreased proventricular blockage of X. cheopis by an hmsE mutant compared to the parent KIM6+ strain (Table 1). In the first experiment, the hmsE mutant showed an ~ 3-fold reduction in the number of infected fleas that became blocked. Moreover, about 1/3 of these blocked fleas were only partially blocked. We observed in partially blocked hmsE-mutant-infected fleas that the biofilm in the flea midgut appeared fragmented unlike the sticky coherent biofilm extending in its entirety from the proventriculus into the midgut formed by the parent KIM6+ strain (Fig 7; Table 1). In the second experiment, no infected fleas were blocked by the hmsE mutant. An hmsD mutant showed a 6.5-fold lower blockage rate than the parent strain (Table 1), confirming the original observation of Sun et al. (2011) that blockage development in X. cheopis is primarily controlled by HmsD. An ~ 66% of fleas determined as blocked by the hmsD mutant were only partially blocked (Table 1). Thus, our results show that efficient HmsD-dependent proventricular blockage of X. cheopis requires HmsE activity. However, when HmsD is absent or not stimulated by HmsE, HmsT activity may be responsible for biofilm formation which appears to cause a high percentage of partial blockage of X. cheopis fleas.
Table 1.
The role of HmsE and HmsD in the blockage of X. cheopis.
|
Y. pestis strain genotype† |
Fleas (n) |
Excess mortality |
Day T-0 CFU/flea* (% fleas infected) |
Day T-28 CFU/flea* (% fleas infected) |
Blockage Rate |
|---|---|---|---|---|---|
| 1st Trial | |||||
| Parent | 94 | 23% | 1.0 × 105 ± 8.9 × 104 (100%) | 4.6 × 105 ± 4.2 × 104 (100%) | 21% |
| hmsE− | 106 | 0% | 3.1 × 105 ± 2.0 × 105 (60%) | 6.0 × 105 ± 4.2 × 105 (65%) | 8%# |
| 2nd Trial | |||||
| Parent | 122 | 30% | 4.5 × 105 ± 3.6 × 104 (100%) | 4.4 × 105 ± 2.4 × 105 (100%) | 13% |
| hmsE− | 105 | 16% | 1.2 × 105 ± 1.8 × 105 (100%) | 1.4 × 105 ± 2.0 × 105 (30%) | 0% |
| hmsD− | 132 | 18% | 1.9 × 104 ± 2.0 × 104 (100%) | 3.3 × 105 ± 1.2 × 105 (45%) | 2%§ |
Strains: KIM6+ (Parent; hmsCDE+), KIM6-2174.1+ (hmsE−), KIM6-2159.1+ (hmsD−)
CFU – colony forming units ± standard deviations; only fleas that had taken a blood meal were followed for 28 days. Number of fleas per infection ranged from 94 to 132.
1/3 of these were partially, not completely blocked
2/3 of these were partially, not completely blocked
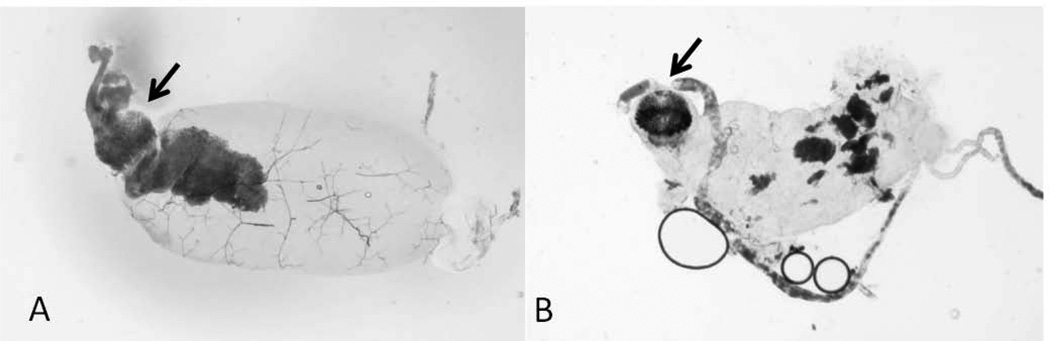
Fig. 7. Complete and partial blockage of the X. cheopis proventriculus by Y. pestis strains.
The digestive tracts dissected from X. cheopis infected with Y. pestis KIM6+ (parent strain) (A) and Y. pestis KIM6-2174.1+ (hmsE−) (B). Arrows indicate complete and partial blockage by the parent strain and the hmsE mutant, respectively.
DISCUSSION
The global second messenger signaling molecule c-di-GMP plays a critical role in controlling biofilm development in Y. pestis and life style switches during its obligate flea-rodent-flea cycle. At ambient temperatures, c-di-GMP is essential for Y. pestis Hms-dependent biofilm formation causing proventricular blockage that is critical for sustaining infections in fleas and for plague transmission to mammals by individual fleas such as X. cheopis. In contrast, high c-di-GMP levels and the resulting hyperbiofilm formation interferes with development of disease in mammals. However, most of the environmental signals and molecular mechanisms causing differential c-di-GMP fluctuations in Y. pestis cells remain unknown (Burroughs, 1947; Bibikova and Klassovskii, 1974; Perry and Bobrov, 2010; Wortham et al., 2010; Bobrov et al., 2011; Sun et al., 2011; Hinnebusch, 2012).
In this work, we identify that the HmsCDE c-di-GMP signaling system modulates Y. pestis biofilm formation in different environments. Our in vitro data indicate that the HmsCDE system is similar in function to the orthologous YfiBNR system of P. aeruginosa (Malone et al., 2010; Malone et al., 2012). We demonstrated that HmsC (a YfiR orthologue) inhibits HmsD (a YfiN orthologue)-dependent c-di-GMP synthesis and biofilm formation in vitro. (Fig. 1 and 3). This independently confirms the recently published findings of Ren et al. (Ren et al., 2013). However, we went one step further than Ren et al. to demonstrate that HmsE (a YfiB orthologue) counteracts HmsC to promote the activity of the DGC HmsD. Overexpression of HmsE on a low copy plasmid from an arabinose-inducible promoter increased biofilm formation and cellular c-di-GMP levels via HmsD (Fig. 1, 2, and 3). Using an IPTG-inducible promoter on a low-copy-number expression vector, Ren et al. failed to demonstrate a role for HmsE in vitro. Since no evidence of expression levels was provided to support this negative result (Ren et al., 2013), perhaps, little or no expression of HmsE accounts for their observation.
Similar to the YfiBNR system, bioinformatics and our data suggest that HmsE is a lipoprotein located in the OM and HmsD is an IM protein with its DGC domain located in the cytoplasm. In contrast to the periplasmic location of YfiR, HmsC may be an IM-anchored protein with a single transmembrane domain and a large periplasmic domain; all of the bioinformatics algorithms that we used showed the presence of a transmembrane domain typical for IM proteins and the absence of an N-terminal signal peptide in HmsC. Ren et al., using cell fractionation and an overexpressed HmsC-Flag3-His8 construct, concluded that the transmembrane domain of HmsC is cleaved with the remainder of HmsC in the periplasm (Ren et al., 2013). However, overexpression and the Flag3-His8 tag in the fusion protein may affect fractionation and cleavage/degradation. To determine an IM versus a periplasmic location, cell fractionation studies using the native HmsC protein expressed from the Y. pestis genome will need to be performed.
Based on the data presented in our manuscript, in Ren et al. (2013), and in Malone at al. (2010, 2012), the Pfam domain PF13689 (formerly Domain of unknown function DUF4154), which includes the YfiR/HmsC-like proteins from various bacteria, can be now annotated "Periplasmic domain, a negative regulator of diguanylate cyclase activity".
YfiR has been shown to directly interact with the DGC YfiN, while YfiB is strongly suggested to sequester YfiR to activate YfiN. Genetic analyses suggest that conserved hydrophobic patches in the YfiBNR proteins are needed for these protein-protein interactions (Malone et al., 2012). For the HmsCDE proteins, T-coffee analysis indicated the presence of identical or similar amino acid residues in analogous hydrophobic patches (data not shown) suggesting that HmsCDE proteins may be involved in protein-protein interactions. Biochemical data on the interaction of periplasmic domains of HmsC and HmsD supports this model (Ren at al., 2013). Thus, under in vitro conditions, the periplasmic domain of HmsC may interact with the periplasmic loop of HmsD disrupting its function. Therefore, the DGC HmsT is primarily responsible for in vitro biofilm formation (Fig. 8).
Fig. 8. A proposed model for regulation of the DGC HmsD in different environments.
In vitro HmsD function is inhibited by interaction with HmsC (right panel). However, in the oriential rat flea X. cheopis, the OM protein HmsE that is affected by an unknown signal interacts with HmsC which prevents inhibition of HmsD function causing increased production of c-di-GMP. OM and IM – outer and inner membranes, P – periplasm; the proposed interactions between proteins are highlighted by dotted ovals. HmsC is represented as an IM-anchored protein with a large periplasmic domain although Ren et al. (Ren et al., 2013) suggest it is a periplasmic protein.
A key finding of our study is that HmsE is required for blockage development in the flea X. cheopis. Similarly to the hmsD mutant, the hmsE mutant showed drastically reduced blockage compared to the parent KIM6+ strain (Table 1). We propose that, in the gut of X. cheopis, the OM protein HmsE is affected by an unknown environmental cue and then interacts with periplasmic domain of HmsC. This HmsE-HmsC interaction would effectively counteract HmsC and lead to increased c-di-GMP synthesis by HmsD, increased biofilm formation and proventricular blockage (Fig. 8).
We showed that HmsC-dependent inhibition and HmsE-dependent activation of HmsD is present in all biovars and subspecies of Y. pestis (Fig. 5) implying that all diverse Y. pestis strains could use the same mechanism for blockage development in fleas. While we demonstrated that this signaling mechanism is functional in X. cheopis, a common plague vector worldwide, it is possible that HmsE is critical for induction of biofilm formation in the other fleas that become blocked with high frequency. Such fleas include Xenopsylla skrjabini, Xenopsylla conformus, and Neopsylla setosa reported to be the major vectors for transmission of Y. pestis to rodents in multiple sylvatic foci (Bibikova and Klassovskii, 1974; Vatschenok, 1988; Anisimov et al., 2004). However, there are known or potential plague flea vectors with low rates of blockage including Oropsylla montana, Nosopsyllus laeviceps, and Cetanophilus tesquorum (Burroughs, 1947; Bibikova and Klassovskii, 1974; Vatschenok, 1988; Gage and Kosoy, 2005). The blockage rates for these fleas are comparable to the blockage rates in X. cheopis caused by the hmsD and hmsE mutants. (Sun et al., 2011; Table 1). The activity of HmsT may be responsible for biofilm formation that is sufficient for this low blockage rate especially since an hmsT mutation reduces blockage to an intermediate level (Sun et al., 2011) indicating that HmsT is still active in the flea. Perhaps, fleas with low blocking capability may lack the environmental cue(s) that activate HmsE, leaving HmsD inhibited by HmsC.
Intriguingly, 33% to 66% of the relatively few X. cheopis fleas that become blocked by the hmsD and hmsE mutants were largely partially blocked (Table 1). Partially blocked fleas are thought to be better transmitters of Y. pestis than completely blocked fleas since the bacterial biofilm prevents the proventricular valve from closing allowing the bacterial masses not only from the proventriculus but also from the midgut to be regurgitated, infecting the mammal. Additionally, partially blocked fleas can ingest blood and therefore likely survive longer than blocked fleas (Bacot, 1915; Hinnebusch, 2012). It has been reported that some fleas that demonstrate low or intermediate blockage, including Nosopsyllus fasciatus, Megabothris abantis, O. montana and Malaraeus telchinum, showed 25–100% transmission by fleas with partial blockage (Burroughs, 1947). It would be interesting to investigate if HmsT-dependent biofilm formation results in increases in partial blockage rates in fleas to properly understand the mechanism of transmission by some flea species with low blockage rates.
While c-di-GMP production is critical for biofilm formation and blockage in X. cheopis and likely other fleas, c-di-GMP-dependent biofilm formation in mammals appears to be detrimental for the development of bubonic plague. The phosphodiesterase HmsP has been shown to be essential for the progression of lethal infection in mice by down-regulating c-di-GMP-dependent Hms EPS production (Bobrov et al., 2011). Degradation of the DGC HmsT and other Hms proteins at mammalian temperatures may also contribute to this process (Perry et al., 2004). We show here that despite similar increased EPS production by the hmsC and hmsP mutants in vitro during growth at 37°C (Fig. 6) the hmsC mutant was 100-fold more virulent than the hmsP mutant and is likely fully virulent in the bubonic plague mouse model. The reason for this difference is unknown. Perhaps in the mouse, HmsP is able to counteract the increased c-di-GMP synthesis caused by the hmsC mutation. Alternatively, HmsD might not be an active DGC during mammalian infection regardless of the presence HmsC. Nevertheless, HmsC-dependent inhibiton of HmsD may still contribute to decreased c-di-GMP production in Y. pestis after transmission from X. cheopis to the mammalian host.
In this study, we identified a putative signal transduction system HmsCDE that modulates cellular c-di-GMP and Y. pestis biofilm levels under different environmental conditions. In vitro, in a variety of growth media, the DGC HmsD is inhibited by HmsC, leaving biofilm development under the control of the only other DGC in Y. pestis KIM6+, HmsT. In X. cheopis, HmsE promotes HmsD function leading to hyper-biofilm formation and efficient blockage of the flea proventriculus. Our findings provide new insights into the regulation of Y. pestis c-di-GMP modulation and biofilm formation in different environments. Future studies are needed to identify the environmental cue(s) that allow c-di-GMP synthesis by HmsD in the oriental rat flea.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids, primers and growth conditions
Bacterial strains and plasmids used and constructed in this study are listed in Table S1. The primers used are listed in Table S2. E. coli DH5α or DH5α (λ-pir) was used for construction and maintenance of recombinant plasmids and were grown in Luria broth (LB) or on LB agar at 28–37°C. Y. pestis avirulent strains lacking the pCD1 virulence plasmid were used for construction of all mutants. The Y. pestis strains were grown in Heart Infusion Broth (HIB) or Tryptose Blood Agar (TBA) (Difco) at 30–33°C. To test the Hms phenotype, Congo red (CR) agar was used (Surgalla and Beesley, 1969). To induce expression of genes encoded on pBAD vectors, arabinose was added to a final concentration 0.05% (w/v). Where necessary, ampicillin (Ap), chloramphenicol (Cm) and kanamycin (Km) were used at final concentrations of 100, 30 and 50 mg ml−1, respectively. For lambda Red recombinase mutant selection, Km and Cm were used at final concentrations of 25 mg ml−1 and 8 mg ml−1, respectively. TBA medium supplemented with 5% sucrose (TBAS) was used to cure suicide vectors.
Bioinformatics
The NCBI BLAST server was used to search for nucleotide and protein sequences in bacterial genomes. To predict protein transmembrane domains, signal peptides and subcellular localization, Philius (based on dynamic Bayesian networks), Phobius (based on Hidden Markov models [HMMs]) and TMHMM 2.0 (based on HMMs) programs were used (Krogh et al., 2001; Kall et al., 2004; Reynolds et al., 2008). The LipoP 1.0 program was used to predict lipoprotein signal peptides (Juncker et al., 2003). T-coffee alignment program was used to compare amino acid sequences of HmsCDE and YfiNBR (Notredame et al., 2000)
Construction of mutations
Deletions in hmsC, hmsE, hmsCE and hmsCDE were constructed in Y. pestis strains carrying pKD46 or pWL204 by gene inactivation using the lambda Red recombinase system (Datsenko and Wanner, 2000; Lathem et al., 2007). PCR products for replacement of hmsC with a kan cassette in KIM6+ and a cat cassette in the other four Y. pestis strains tested (CO92, Kuma, PestoidesF and PestoidesD) were amplified using primer pairs: Y3729 RED-1 and Y3729 RED-2, and Y3729 RED-1 and Y3729 RED-2-pKD3, respectively (Table S2). To generate hmsE deletions, primers Y3731 RED-1 and Y3731 RED-2 were used. For hmsCD deletion, pimers Y3729 RED-1 and Y3730 RED-2 were used. To eliminate kan or cat cassettes, flippase expressing plasmids pCP20 (Cherepanov and Wackernagel, 1995) or pSkippy (Price et al., 2012) were transformed into relevant mutants. The suicide plasmids pKNG-ΔhmsR2118 and pKNG-ΔhmsT-Δscar were used as previously described to generate in-frame deletions (Forman et al., 2006; Bobrov et al., 2011). All mutations and loss of antibiotic resistance cassettes were confirmed by PCR.
Construction of plasmids
The DNA fragments including the hmsC and hmsE ORFs and their upstream regions with putative Shine-Dalgarno sequence were amplified from Y. pestis KIM10+ genomic DNA using primers Y3729-1 and Y3729-XbaI-2, and Y3731pBAD-EcoRI and Y3731-down, respectively. The DNA fragment corresponding to hmsC was cloned into the XbaI and SmaI sites of pBAD30 generating pBAD-hmsC. The hmsE fragment was cloned using the EcoRI and SmaI sites of pBAD30 generating pBAD-hmsE. Both genes were cloned behind the arabinose inducible promoter.
To generate PhoA and β-Gal fusions with full-length HmsC and HmsD proteins for topology analyses, the entire DNA coding regions were amplified with primers Y3729-SD-fus-XbaI and Y3729-R207-PstI (for HmsC) and Y3730-SD-fus-XbaI and Y3730-G425-PstI (for HmsD) and cloned into vectors pRMCD28-T5 and pRMCD70-T5 (Bobrov et al., 2008), respectively, using XbaI and PstI sites. To construct C-end truncated fusions of HmsD with PhoA and β-Gal at position R156, Y3730-SD-fus-XbaI and Y3730-R156-PstI primers were used.
Sequencing of hmsE regions from Y. pestis Kuma and Pestoides D
For PCR amplification of an ~ 1.2 kb region that included the hmsE ORF from genomic DNA of Y. pestis Kuma and Pestoides F, high fidelity Phusion DNA polymerase and primers hmsA_region_F and hmsA_region_R were used (y3729-y3731 operon was originally designated hmsNDA but was changed to hmsCDE as in Ren et al.). For sequencing of the ~ 0.8 kb DNA fragment of the PCR amplicons by ACGT Inc, primers hmsA_region_Seq_F and hmsA_region_Seq_R were used. 611 bp Kuma and Pestoides D sequences were determined to be 100% identical to the KIM6+ hmsE region cloned in pBAD-hmsE.
Crystal violet assays
Cells attached to polystyrene 24 well plates were detected with crystal violet (CV) staining essentially as described by Bobrov et al. (2011). Briefly, cells were grown at 30–33°C overnight on TBA slants and used to inoculate HIB to an OD620 of 0.3. Cultures were grown for 16–18 hours with shaking at 21–23°C, washed once with water and then exposed to 0.01% CV for 15–20 min. The wells were washed three times with distilled water. The CV bound to the cells was solubilized with 33% acetic acid and the absorbance was measured at 570 nm using a Spectronics Genesys 5 spectrophotometer.
Determination of intracellular c-di-GMP levels
Overnight HIB cultures of Y. pestis, KIM6-2051+ carrying either pBAD30 or pBAD-hmsE and KIM6-2173.3+ carrying pBAD30 were diluted to an OD620 of ~ 0.1 and grown in TMH (Straley and Bowmer, 1986) at 30°C to an OD620 of ~ 0.8. The cultures were centrifuged and the samples were processed as previously described (Bobrov et al., 2011). Cyclic di-GMP was quantified using an Acquity Ultra performance liquid chromatography system (Waters) coupled with a Quattro Premier XE tandem mass spectrometer (Waters) as previously described (Massie et al., 2012). The concentration of c-di-GMP was determined by quantifying an 8-point standard curve of chemically synthesized c-di-GMP (Biolog) ranging from 1.9 nM to 250 nM.
PhoA and β-Gal assays
PhoA and β-Gal activities were quantified as previously described (Manoil, 1991; Miller, 1992). Y. pestis strains carrying reporter plasmids were cultivated in HIB medium at 30°C until an OD620 of ~ 0.3–0.4 was reached. Cells were permeabilized with chloroform and SDS and assayed for their PhoA and β-Gal activities using p-nitrophenyl phosphate (PNPP) and o-nitrophenyl galactopyranoside (ONPG) as substrates, respectively.
Immunodetection of the poly-β-1,6-GlcNAc-like polysaccharide in Y. pestis
Crude polysaccharide extracts were prepared essentially as previously described with the following modifications: Y. pestis strains were cultivated on solidified HIB medium supplemented with 0.2% galactose at 37°C or 21°C. Cells were collected with a plastic loop and resuspended in 18.9 µl of 0.5 M EDTA (pH 8.0) per 1 mg of wet cell weight. Sample preparation and detection proceeded as previously described (Bobrov et al., 2008). Densitometry analysis of the spots was performed using ImageJ software.
Congo red binding assay
The original CR-binding assay at 37°C (Kirillina et al., 2004) was modified slightly. Briefly, Y. pestis cells grown overnight in HIB at 37°C were pelleted and wet weights determined. The cells were resuspended in HIB/CR medium [1% (w/v) HIB containing 0.2% galactose and 15 µg CR ml−1] at concentration of 20 mg wet weight ml−1 and incubated for 3 hours on a rocking platform at 37°C. CR bound by the cells was determined by measuring the absorbance of cell-free supernatants at 500 nm with a Spectronic Genesys5 spectrophotometer and subtracting the values from the reading obtained with uninoculated HIB/CR medium.
Virulence testing
For virulence testing in mice, pCD1Ap was electroporated into various Y. pestis strains in the CDC-approved University of Kentucky BSL3/ABSL3 facility. The Lcr+ phenotype was confirmed by growth restriction at 37°C on TBA plates supplemented with 20 mM sodium oxalate and 20 mM MgCl2 and by a Western blot analysis with antisera against LcrV. For subcutaneous infections, the strains were grown overnight at 33°C, in HIB supplemented with Ap (50 µg/ml), CaCl2 (2.5 mM) and xylose (0.2%) and then inoculated to an OD620 of ~ 0.1 in the same medium and grown to OD620 of ~ 0.35–0.5. Six- to eight-week-old female Swiss Webster mice (Hsd::ND4) were injected subcutaneously with 100 µl of 10-fold serial dilutions of the bacterial suspensions in mouse isotonic phosphate-buffered saline (149 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4 [pH 7.0]). For the hmsC mutant [KIM5-2371.2(pCD1Ap)], infectious doses of ~102,103,104 and 105 were used in two independent trials, while infectious doses of ~10 and 102 were used for the HmsC+ parent (KIM5(pCD1Ap+) The number of cells injected was determined by plating serial dilutions on TBA-Ap plates. Four mice were used for each infectious dose. Mice were monitored daily for a period of 2 weeks and LD50 values were calculated.
All animal care and experimental procedures were conducted in accordance with the Animal Welfare Act, Guide for the Care and Use of Laboratory Animals, PHS Policy and the U.S. Government Principals for the Utilization of and Care for Vertebrate Animals in Teaching, Research, and Training and approved by the University of Kentucky Institutional Animal Care and Use Committee and University of Kentucky Institutional Biosafety Committee. The University of Kentucky Animal Care Program is accreditated by the Association for the Assessment and Accreditation of Laboratory Animal Care, Inc.
Infection and assessment of fleas
X. cheopis fleas were reared on CD-1 mouse neonates. Cohorts of X. cheopis fleas were infected with Y. pestis KIM6+ or the mutant strain using a previously described artificial feeding system (Hinnebusch et al., 1996; Hinnebusch et al., 2002). The infectious blood meal was prepared by growing the bacteria at 37°C in BHI medium, without aeration. A cell pellet containing 108–109 bacterial cells was resuspended in 1 ml PBS and added to 5 ml of heparinized CD-1 mouse blood (Bioreclamation, NY). The infected blood was added to the water-jacketed feeding chamber which was maintained at 37°C. The fleas were allowed to feed for 60–90 min through the mouse skin secured over the chamber. Fleas that took a blood meal were maintained at 21°C and 75% relative humidity, fed twice weekly on uninfected CD-1 neonate mice, and monitored as previously described (Hinnebusch et al., 1996). To determine the percent of bacterial survival and percent of infected fleas, 20 infected females were collected and processed as previously described (Erickson et al., 2006) to determine bacterial colony forming units (cfu) per flea at 1 hour, and 28 days post-infection. At the indicated times, biofilm blockage of the proventriculus was determined immediately following flea feeding on a neonatal mouse. Fleas were scored as blocked when fresh red blood was observed in the esophagus only and not in the midgut of the flea. A separate cohort of uninfected fleas was handled identically as a control for normal feeding and viability. Experiments were repeated to independently confirm phenotypes. The infection with the Y. pestis hmsD mutant (KIM6-2159.1+) was however performed only once to confirm previously reported findings (Sun et al., 2011). Dissected flea midguts were in PBS and visualized on an AMG EVOS xl core light microscope at 10× magnification. The use of mouse neonates for experiments and flea breeding and maintenance was approved by the Institutional Animal Care and Use Committee at Washington State University, USA, in accordance with institutional guidelines based on the U.S. National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
Supplementary Material
ACKNOWLEDGEMENTS
A.G.B, O.K., and R.D. P. were supported by Public Health Services grant AI25098 from the US National Institutes of Health. V.V. and A.K.H were supported by Public Health Services grant AI097974 from the US National Institutes of Health. C.M.W. was supported by grant U54AI057163. We thank Jackie Fetherston for her assistance with some animal experiments, thoughtful suggestions, and manuscript editing and Lauren O’Conner for critical reading of the manuscript.
Footnotes
The authors have no conflicts of interest to declare.
References
- Abu Khweek A, Fetherston JD, Perry RD. Analysis of HmsH and its role in plague biofilm formation. Microbiology. 2010;156:1424–1438. doi: 10.1099/mic.0.036640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov AP. Factors of Yersinia pestis providing circulation and persistence of plague pathogen in ecosystems of natural foci. Communication 2 (in Russian) Mol Gen Mikrobiol Virusol. 2002:3–11. [PubMed] [Google Scholar]
- Anisimov AP, Lindler LE, Pier GB. Intraspecific diversity of Yersinia pestis. Clin Microbiol Rev. 2004;17:434–464. doi: 10.1128/CMR.17.2.434-464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacot AW. LXXXI. Further notes on the mechanism of the transmission of plague by fleas. J Hyg (Lond) 1915;14:774–776. 773. [PMC free article] [PubMed] [Google Scholar]
- Bacot AW, Martin CJ. LXVII. Observations on the mechanism of the transmission of plague by fleas. J Hyg (Lond) 1914;13:423–439. [PMC free article] [PubMed] [Google Scholar]
- Bearden SW, Sexton C, Pare J, Fowler JM, Arvidson CG, Yerman L, et al. Attenuated enzootic (pestoides) isolates of Yersinia pestis express active aspartase. Microbiology. 2009;155:198–209. doi: 10.1099/mic.0.021170-0. [DOI] [PubMed] [Google Scholar]
- Bibikova VA, Klassovskii LN. The Transmission of Plague by Fleas (in Russian) Moscow: Meditsina; 1974. [Google Scholar]
- Bobrov AG, Kirillina O, Perry RD. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol Lett. 2005;247:123–130. doi: 10.1016/j.femsle.2005.04.036. [DOI] [PubMed] [Google Scholar]
- Bobrov AG, Kirillina O, Forman S, Mack D, Perry RD. Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol. 2008;10:1419–1432. doi: 10.1111/j.1462-2920.2007.01554.x. [DOI] [PubMed] [Google Scholar]
- Bobrov AG, Kirillina O, Ryjenkov DA, Waters CM, Price PA, Fetherston JD, et al. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol Microbiol. 2011;79:533–551. doi: 10.1111/j.1365-2958.2010.07470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AL. Sylvatic plague studies: The vector efficiency of nine species of fleas compared with Xenopsylla cheopis. J Hyg (Lond) 1947;45:371–396. doi: 10.1017/s0022172400014042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Cui Y, Li Y, Gorgé O, Platonov ME, Yan Y, Guo Z, et al. Insight into microevolution of Yersinia pestis by clustered regularly interspaced short palindromic repeats. PLoS ONE: Public Library of Science. 2008:e2652. doi: 10.1371/journal.pone.0002652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson DL, Jarrett CO, Wren BW, Hinnebusch BJ. Serotype differences and lack of biofilm formation characterize Yersinia pseudotuberculosis infection of the Xenopsylla cheopis flea vector of Yersinia pestis. J Bacteriol. 2006;188:1113–1119. doi: 10.1128/JB.188.3.1113-1119.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson DL, Jarrett CO, Callison JA, Fischer ER, Hinnebusch BJ. Loss of a biofilm-inhibiting glycosyl hydrolase during the emergence of Yersinia pestis. J Bacteriol. 2008;190:8163–8170. doi: 10.1128/JB.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroshenko GA, Vidyaeva NA, Kutyrev VV. Comparative analysis of biofilm formation by main and nonmain subspecies Yersinia pestis strains. FEMS Immunol Med Microbiol. 2010;59:513–520. doi: 10.1111/j.1574-695X.2010.00719.x. [DOI] [PubMed] [Google Scholar]
- Forman S, Bobrov AG, Kirillina O, Craig SK, Abney J, Fetherston JD, Perry RD. Identification of critical amino acid residues in the plague biofilm Hms proteins. Microbiology. 2006;152:3399–3410. doi: 10.1099/mic.0.29224-0. [DOI] [PubMed] [Google Scholar]
- Gage KL, Kosoy MY. Natural history of plague: perspectives from more than a century of research. Annu Rev Entomol. 2005;50:505–528. doi: 10.1146/annurev.ento.50.071803.130337. [DOI] [PubMed] [Google Scholar]
- Girgis HS, Liu Y, Ryu WS, Tavazoie S. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 2007;3:1644–1660. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch BJ. Biofilm-dependent and biofilm-independent mechanisms of transmission of Yersinia pestis by fleas. Advances in Experimental Medicine and Biology. 2012;954:237–243. doi: 10.1007/978-1-4614-3561-7_30. [DOI] [PubMed] [Google Scholar]
- Hinnebusch BJ, Erickson DL. Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr Top Microbiol Immunol. 2008;322:229–248. doi: 10.1007/978-3-540-75418-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch BJ, Perry RD, Schwan TG. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, Forsberg A. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science. 2002;296:733–735. doi: 10.1126/science.1069972. [DOI] [PubMed] [Google Scholar]
- Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, Whitney AR, et al. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis. 2004;190:783–792. doi: 10.1086/422695. [DOI] [PubMed] [Google Scholar]
- Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol. 2004;54:75–88. doi: 10.1111/j.1365-2958.2004.04253.x. [DOI] [PubMed] [Google Scholar]
- Krasnov BR, Shenbrot GI, Mouillot D, Khokhlova IS, Poulin R. Ecological characteristics of flea species relate to their suitability as plague vectors. Oecologia. 2006;149:474–481. doi: 10.1007/s00442-006-0455-7. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lathem WW, Price PA, Miller VL, Goldman WE. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science. 2007;315:509–513. doi: 10.1126/science.1137195. [DOI] [PubMed] [Google Scholar]
- Lillard JW, Jr, Bearden SW, Fetherston JD, Perry RD. The haemin storage (Hms+) phenotype of Yersinia pestis is not essential for the pathogenesis of bubonic plague in mammals. Microbiology. 1999;145(Pt 1):197–209. doi: 10.1099/13500872-145-1-197. [DOI] [PubMed] [Google Scholar]
- Lillard JW, Jr, Fetherston JD, Pedersen L, Pendrak ML, Perry RD. Sequence and genetic analysis of the hemin storage (hms) system of Yersinia pestis. Gene. 1997;193:13–21. doi: 10.1016/s0378-1119(97)00071-1. [DOI] [PubMed] [Google Scholar]
- Malone JG, Jaeger T, Spangler C, Ritz D, Spang A, Arrieumerlou C, et al. YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog. 2010;6:e1000804. doi: 10.1371/journal.ppat.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone JG, Jaeger T, Manfredi P, Dotsch A, Blanka A, Bos R, et al. The YfiBNR signal transduction mechanism reveals novel targets for the evolution of persistent Pseudomonas aeruginosa in cystic fibrosis airways. PLoS Pathog. 2012;8:e1002760. doi: 10.1371/journal.ppat.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 1991;34:61–75. doi: 10.1016/s0091-679x(08)61676-3. [DOI] [PubMed] [Google Scholar]
- Massie JP, Reynolds EL, Koestler BJ, Cong JP, Agostoni M, Waters CM. Quantification of high-specificity cyclic diguanylate signaling. Proc Natl Acad Sci U S A. 2012;109:12746–12751. doi: 10.1073/pnas.1115663109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Gehrig SM, Meintjes PL, Zhang XX, Rainey PB. Adaptive divergence in experimental populations of Pseudomonas fluorescens. IV. Genetic constraints guide evolutionary trajectories in a parallel adaptive radiation. Genetics. 2009;183:1041–1053. doi: 10.1534/genetics.109.107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A Short Course in Bacterial Genetics. A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor, N. Y.: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Okuda S, Tokuda H. Lipoprotein Sorting in Bacteria. Annual Review of Microbiology, Vol 65. 2011;65:239–259. doi: 10.1146/annurev-micro-090110-102859. [DOI] [PubMed] [Google Scholar]
- Perry RD, Fetherston JD. Yersinia pestis-etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RD, Bobrov AG. Role of cyclic di-GMP in biofilm development and signaling in Yersinia pestis. In: Wolfe AJ, Visick KL, editors. The second messenger cyclic di-GMP. Washington, DC: ASM press; 2010. pp. 270–281. [Google Scholar]
- Perry RD, Pendrak ML, Schuetze P. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J Bacteriol. 1990;172:5929–5937. doi: 10.1128/jb.172.10.5929-5937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RD, Bobrov AG, Kirillina O, Jones HA, Pedersen L, Abney J, Fetherston JD. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J Bacteriol. 2004;186:1638–1647. doi: 10.1128/JB.186.6.1638-1647.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price PA, Jin J, Goldman WE. Pulmonary infection by Yersinia pestis rapidly establishes a permissive environment for microbial proliferation. Proc Natl Acad Sci U S A. 2012;109:3083–3088. doi: 10.1073/pnas.1112729109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren GX, Yan HQ, Zhu H, Guo XP, Sun YC. HmsC, a periplasmic protein, controls biofilm formation via repression of HmsD, a diguanylate cyclase in Yersinia pestis. Environ Microbiol. 2013 doi: 10.1111/1462-2920.12323. [DOI] [PubMed] [Google Scholar]
- Reynolds SM, Kall L, Riffle ME, Bilmes JA, Noble WS. Transmembrane topology and signal peptide prediction using dynamic bayesian networks. PLoS Comput Biol. 2008;4:e1000213. doi: 10.1371/journal.pcbi.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R, Fetherston JD, Kader A, Romling U, Perry RD. Phenotypic convergence mediated by GGDEF-domain-containing proteins. J Bacteriol. 2005;187:6816–6823. doi: 10.1128/JB.187.19.6816-6823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenseth NC, Atshabar BB, Begon M, Belmain SR, Bertherat E, Carniel E, et al. Plague: past, present, and future. PLoS Med. 2008;5:e3. doi: 10.1371/journal.pmed.0050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straley SC, Bowmer WS. Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect Immun. 1986;51:445–454. doi: 10.1128/iai.51.2.445-454.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YC, Koumoutsi A, Jarrett C, Lawrence K, Gherardini FC, Darby C, Hinnebusch BJ. Differential control of Yersinia pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLoS One. 2011;6:e19267. doi: 10.1371/journal.pone.0019267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surgalla MJ, Beesley ED. Congo red-agar plating medium for detecting pigmentation in Pasteurella pestis. Appl Microbiol. 1969;18:834–837. doi: 10.1128/am.18.5.834-837.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein M, Sauer G, Paramasivam N, Grin I, Linke D. Efficient subfractionation of gram-negative bacteria for proteomics studies. J Proteome Res. 2010;9:6135–6147. doi: 10.1021/pr1002438. [DOI] [PubMed] [Google Scholar]
- van Geest M, Lolkema JS. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol Mol Biol Rev. 2000;64:13–33. doi: 10.1128/mmbr.64.1.13-33.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatschenok VS. Fleas—vectors of pathogens causing diseases in humans and animals (in Russian) Leningrad: Nauka; 1988. [Google Scholar]
- Wortham BW, Oliveira MA, Fetherston JD, Perry RD. Polyamines are required for the expression of key Hms proteins important for Yersinia pestis biofilm formation. Environ Microbiol. 2010;12:2034–2047. doi: 10.1111/j.1462-2920.2010.02219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.