Abstract
Slow skeletal muscle troponin T (TNNT1) pre-messenger RNA alternative splicing (AS) provides transcript diversity and increases the variety of proteins the gene encodes. Here, we identified three major TNNT1 splicing patterns (AS1–3), quantified their expression in the vastus lateralis muscle of older adults, and demonstrated that resistance training modifies their relative abundance; specifically, upregulating AS1 and downregulating AS2 and AS3. In addition, abundance of TNNT1 AS2 correlated negatively with single muscle fiber–specific force after resistance training, while abundance of AS1 correlated negatively with V max. We propose that TNNT1 AS1, AS2 and the AS1/AS2 ratio are potential quantitative biomarkers of skeletal muscle adaptation to resistance training in older adults, and that their profile reflects enhanced single fiber muscle force in the absence of significant increases in fiber cross-sectional area.
Key Words: Troponin T, Alternative splicing, Skeletal muscle, Resistance exercise.
Skeletal muscle wasting, decreased strength, and increased frailty impair performance of daily living activities and lead to major medical and socioeconomic problems for the aging population. In rodents and humans, age-related decreases in muscle mass do not fully account for the decreases in strength as atrophy only partially explains muscular weakness (1–3). Resistance training (RT) may prevent the loss of strength in older adults, and its benefits are only partially explained by the prevention of loss of muscle mass (4–6). Publications from our laboratory and others support the concept that weakness in old age is the result of decreased muscle-specific force (force/cross-sectional area [CSA]) (2,7–11). Impairment of muscle’s intrinsic force-generating capacity with age may be partially due to alterations in excitation–contraction coupling (1,12) in which the troponin complex, consisting of the tropomyosin-binding (TnT), inhibitory, and Ca2+-binding subunits, plays a major role in Ca2+-dependent regulation of muscle contraction (13).
The fast-twitch skeletal TnT gene (troponin T [TNNT3]) undergoes extensive pre-messenger RNA (mRNA) alternative splicing (AS) in systems ranging from flies to rodents (14). TNNT3 pre-mRNA is encoded by 18 exons, including five AS exons near the 5′ end and a pair of mutually exclusive exons near the 3′ end, potentially generating 128 distinct splice forms. Changes in the relative frequency of TNNT3 AS forms would affect muscle contractile ability by altering the Ca2+ sensitivity of myofilaments as shorter forms shift the pCa–force relationship toward lower calcium concentrations (15–18). Recent evidence shows that normal body growth and experimentally increased body weight modify the relative frequency of TNNT3 AS forms. In addition to body weight, mechanical stretch stimulation on C2C12 skeletal muscle cells cultured in flexible-bottomed plates modifies TNNT3 AS (19). Therefore, variations in TNNT3 AS provide a quantitative molecular marker to track how an animal perceives and responds to changes in mechanical body loading (14).
Despite the reported modifications in TNNT3 AS in response to mechanical loading changes in various animal species, the effects of RT on TNNT3 and TNNT1 (the gene that encodes slow-twitch skeletal muscle TnT) pre-mRNA AS in human muscle are unknown. We hypothesized that TNNT1 AS is modified in aging human muscle in response to chronic loading via 5 months of progressive RT. Because a transition from fast-twitch to slow-twitch fiber type has been reported in aging rodents and humans (20,21), and type-I fibers largely outnumbered type-II fibers in our biopsies from the vastus lateralis of the quadriceps, we focused our research on the effects of RT on slow skeletal muscle TNNT1 AS.
Methods
Study Participants
Participants in this study included older men (n = 5) and women (n = 3) enrolled in the National Institutes of Health–funded Improving Muscle for Functional Independence Trial. All subjects were recruited from the Piedmont Triad area of North Carolina according to the following inclusion/exclusion criteria: (i) overweight/obese (body mass index [BMI] = 25–35kg/m2), (ii) older (age = 65–80 years), (iii) nonsmoking, (iv) not on hormone replacement therapy, (v) sedentary (<15 minutes of exercise, 2 times/wk) in the past 6 months, and (vi) weight stable (<5% weight change) for at least 6 months prior to enrollment. All had normal liver, kidney, pulmonary, and thyroid function; no history of excessive alcohol intake; and no major chronic illness, anemia, or orthopedic impairment. The study was approved by the Wake Forest Institutional Review Board for Human Research and all participants signed informed consent to participate in the study.
Exercise Intervention
The exercise intervention consisted of 5 months of progressive RT designed to elicit adaptations in skeletal muscle and increase strength and power. To optimize the resistance stimulus for maximal functional gain, the exercise prescription was based on a relative intensity level and progressed with each individual’s strength gain. Participants exercised 3 d/wk under the supervision of two exercise physiologists. Participants walked or cycled slowly for 5–10 minutes to warm up prior to RT. The resistance exercises were performed on eight Nautilus resistance machines (Nautilus, Vancouver, WA), on which the load could be adjusted in small increments. The program was based on American College of Sports Medicine guidelines for intensity, number of repetitions, number of sets, and number of days per week (ACSM position stand, 2009 (22)). The one repetition maximum, that is, the maximal weight that could be lifted with correct form in a single repetition was used to prescribe intensity. Intensity was progressively increased during the first month toward the training goal of three sets of 10 repetitions at 70% repetition maximum for a given exercise. The repetition maximum strength testing was repeated every 4 weeks, and the training loads adjusted to achieve the 70% repetition maximum goal.
Skeletal Muscle Biopsy
Needle biopsies of the vastus lateralis were collected under local anesthesia with 1% lidocaine for RNA and single muscle fiber contraction studies. All biopsies were performed in the early morning after an overnight fast. Subjects were asked to refrain from taking aspirin, prescription and over-the-counter nonsteroidal anti-inflammatory drugs, or other compounds that may affect bleeding, platelets, or bruising for the week prior to the biopsy, and to refrain from any strenuous activity (including RT) for at least 36 hours prior to the biopsy. Visible blood and connective tissue were removed from the muscle specimen before bundles of muscle fibers for contraction tests were carefully dissected and immediately placed in cold (4°C) 1× relaxing solution, where they were later divided into smaller bundles of approximately 30–50 fibers and tied with 10-0 silk surgical suture to glass capillary tubes at slightly stretched lengths. These bundles were chemically skinned for 24 hours in 50% 2× relaxing solution and 50% glycerol at 4°C, then stored at −20°C for up to 4 weeks as described (8). The muscle portion (10–20mg) used for RNA extraction was snap frozen in liquid nitrogen under RNAse-free conditions, and stored at −80°C until analysis.
Clinical Assessments
Participant height was assessed using a stadiometer and body mass was measured on a calibrated electronic scale. BMI was calculated as body mass in kilograms divided by height in meters squared.
Solutions
The composition of the relaxing and activating solutions used for single muscle fiber experiments were described previously (8).
Experimental Setup
A single fiber segment about ~1.7±0.03mm long was carefully isolated from a muscle bundle using fine forceps. Both ends were securely tied with 10-0 sutures to titanium wires, one connected to an isometric force transducer (Model 403; Aurora Scientific, Aurora, Ontario, Canada) and the other to a high-speed servomotor (Model 315C; Aurora Scientific). The 3-mm wire connected to the motor was used to minimize the error in fiber length due to rotation of the lever arm (23).
The mounted fiber segment was suspended in one of several small glass-bottomed chambers in a stainless steel plate and activated by rapid motorized transfer from the chamber containing relaxing solution to the chamber containing activation solution. The experimental apparatus was mounted on the stage of an inverted microscope (Axiovert S100; Carl Zeiss, Inc., Oberkochen, Germany), enabling observation of the fiber at ×450 through the transparent bottom of each chamber. Images were obtained using a CCD camera and a scientific graphic acquisition board.
Fiber length and sarcomere length were measured using a calibrated eyepiece micrometer. Sarcomere length was set to ~2.5 µm by adjusting the overall segment length, and final sarcomere length confirmed by measuring 10 consecutive sarcomeres at a minimum of three points along the fiber. Fiber CSA was modeled as an ellipse and calculated from measurements of fiber width and depth using a calibrated eyepiece micrometer. Fiber width was measured at three points. At every measurement point, fiber depth was measured by recording the vertical displacement of the microscope nosepiece while focusing on the top and bottom surfaces of the fiber.
Single Muscle Fiber Physiology Tests and Experimental Protocols
All measurements were conducted at 15°C; temperature was continuously monitored by a thermocouple inserted into the experimental chamber (model 802B; Aurora Scientific). All functional data were collected and analyzed using a personal computer and a data acquisition board (Model 600A Digital controller; Aurora Scientific). A slack test was used to determine unloaded shortening velocity (V o) (8). In this procedure, fibers were transferred into activating solution (pCa 4.5), and once peak force was attained (monitored by real-time digital oscilloscope), subjected to a rapid slack step (≤20% of fiber length within 1ms). The procedure was repeated at different slack lengths, and the time required to redevelop tension was plotted against the corresponding slack distances. A straight line was fit by least squares linear regression, and the slope of the regression line, normalized to fiber length, defined V o.
After the slack test, we performed a series of isotonic contractions of the muscle fiber to determine the force/velocity relationship (8). Briefly, the fiber was placed in activating solution (pCa 4.5) and, after it reached peak force, subjected to three isotonic steps varying from 3% to 80% of P o. After the final step, the fiber was rapidly (<1 ms) slacked by 20% of its length. This zeroed the force transducer, providing a baseline for force measurement. Step duration was less than ~100ms. Velocity was calculated as the slope of the position record over the same time period and, like force, averaged over the final half of each step. A total of 5–6 separate fiber activations were applied to generate 15–18 pairs of velocity and relative force measurements.
The Hill equation (24) was fitted to the data using an iterative, nonlinear, curve-fitting procedure. The following parameters were used to describe the hyperbolic fit: V max (velocity extrapolated to a force of zero); P o (average force during the trial); and a/P o (the dimensionless curvature or shape of the force/velocity relationship) (25). We used five slack lengths normalized to fiber length to determine V max. Peak power was calculated from these three parameters and expressed as W/liter fiber (26). In all contractions, Ca2+-activated force was measured using zero transducer signal as a baseline. Forces were normalized to the fiber’s CSA to obtain specific force.
For quality control and assessment of fiber myosin heavy chain isoform, see reference (8). Briefly, fibers were excluded from analysis if force declined more than 5% or if they broke or showed partial myofibrillar tearing at any observation timepoint in the experimental protocol. Experiments were excluded from analysis if compliance, defined as displaced axis intercept of the slack test plot, exceeded 5% of fiber length and if the r 2 of the force–velocity regression was less than 0.98. Biopsies were performed on eight volunteers but two preparations failed quality control for specific force and three for power analysis.
Myosin Heavy Chain Isoforms Analysis in Single Muscle Fiber and Bundle of Fibers
For fiber typing, at the end of each recording, the single muscle fiber segment was saved in 20 µl of sodium dodecyl sulfate sample buffer (containing 62.5 mM Tris pH 6.8, 2% sodium dodecyl sulfate, 10% glycerol, 5% betamercaptoethanol, and 0.001% bromophenol blue) for sodium dodecyl sulfate–polyacrylamide gel electrophoresis to verify muscle fiber type by silver staining (8). In contrast to single fiber typing, bundles of fibers were sonicated with a F60 sonic dismembrator (Fisher, Waltham, MA) for 30 seconds.
RNA Isolation and Expression Analysis
Total RNA was extracted from human muscle tissues using the Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. RNA samples and the same volume of H2O (negative control) were reverse transcribed using SuperScript III Reverse Transcriptase (Invitrogen). TNNT1 complementary DNA was amplified by nested PCR, first using forward primer F1 (5′-AATATGAGGAGGAGCAGC-3′) and reverse primer R1 (5′-CCCTTACGGAACTTCTGG-3′); then using fluorescein (FAM)-labeled forward primer FAM-F2 (5′-FAM-GGAAGAGGAGGCTGCGGA-3′) and reverse primer R2 (5′-TGATGCGGTTGTACAGCA-3′). PCR was performed using GoTaq Green Master Mix (Promega, Madison, WI) under the following cycling conditions: 2 minutes at 94°C; 35 1-minute cycles at 94°C; 1 minute at 60°C; 1 minute at 72°C; ending with 7 minutes at 72°C.
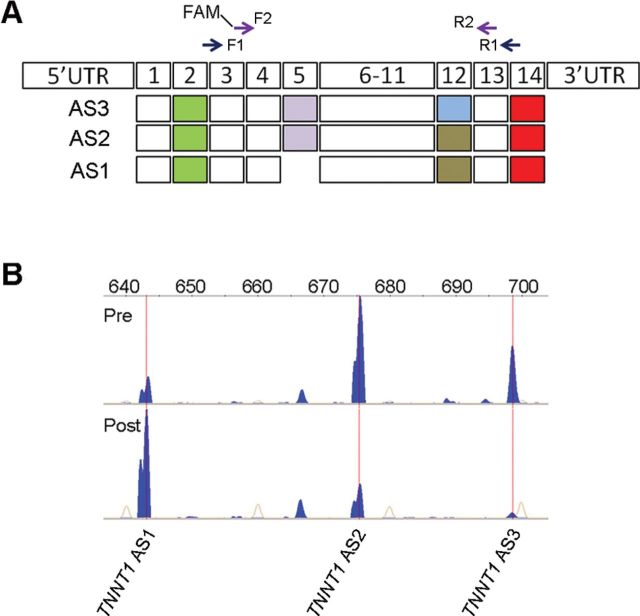
To quantify the splicing patterns and relative abundance of TNNT1, we used a protocol described recently (14,19). Briefly, we analyzed 1 μl of each FAM-labeled nested PCR product by capillary electrophoresis (ABI Hitachi 3730XL DNA Analyzer; Applied Biosystems, Carlsbad, CA), which allows precise determination of PCR fragment size and quantity in a PCR amplicon pool. The relative abundance of each TNNT1 amplicon in the PCR reaction was determined by dividing its peak height by the total of all TNNT1 amplicon peak heights (see also Figure 1). Amplicon fragment size was determined using the GS1200 LIZ internal size standard and Genemapper (Applied Biosystems) fragment analysis software. To examine the exon composition of the TNNT1 splicing patterns, amplicons were extracted from an agarose gel using a Gel and PCR Clean-up System (Promega), cloned using a TOPO-TA cloning kit (Invitrogen), and sequenced (Genscript, Piscataway, NJ). As the presence or absence of exon 5 together with the short or long exon 12 can generate four potential mRNAs, we prefer using pattern instead of the more classical denomination form, following previous publications (15,27,28) when we refer to our data.
Figure 1.
Characterization and quantification of troponin T (TNNT1) alternative splicing (AS) in the vastus lateralis muscle before and after resistance training (RT). (A) Three major human TNNT1 pre-messenger RNAs (mRNAs) are comprised of 14 exons (boxes 1–14) with a translation–initiation codon localized in exon 2 (green boxes) and a stop codon in exon 14 (red boxes). Exon 5 is excluded from the short TNNT1 AS1, and exon 12 is longer in TNNT1AS3 than in the other two splice forms. All three were detected by reverse transcription–PCR and complementary DNA sequencing. Two pairs of forward (F1 and FAM-F2) and reverse (R1 and R2) PCR primers were designed to perform nested PCR with increased sensitivity and specificity. (B) Representative electropherogram. Fluorescently labeled DNA fragment peaks showing the diversity of TNNT1 splice forms, denoted by position in the horizontal axis (expressed in base pairs). Relative abundance is indicated by the peak height (blue) of each form in the vastus lateralis muscle of the same subject before (pre) and after (post) RT. Internal size standards are represented by orange traces.
Statistical Analysis
Data analysis was performed with SigmaPlot 11.0 (Systat Software, San José, CA) or Prism 5.0 (Graphpad, La Jolla, CA). All data are presented as means ± SEM. The alpha level was set at p = .05. Linear regression analysis was used to identify the relationship between TNNT1 AS and specific functional variables, and the Pearson product moment correlation to measure the strength of the association between pairs of variables.
Results
RT Increases Abundance of TNNT1 AS1, But Decreases Abundance of AS2 and AS3, Splicing Patterns
We detected three major TNNT1 splicing patterns expressed in the human vastus lateralis muscle. Figure 1A shows the exon structure of each AS pattern amplified by reverse transcription–PCR and nested PCR screening of total RNA. Sequencing of complementary DNA clones confirmed that the shortest mRNA splice pattern (AS1) lacks exon 5, while AS2 has a short, and AS3, a long exon 12. Note that the 5-month RT intervention modified the relative abundance of all three TNNT1 splicing patterns; electropherograms show that the abundance of AS1 increases, while the abundance of AS2 and AS3 decreases (Figures 1B and 2A). Lower abundance splicing patterns were not analyzed further. The AS2 is the most abundant pattern in this sample of sedentary older adults.
Figure 2.
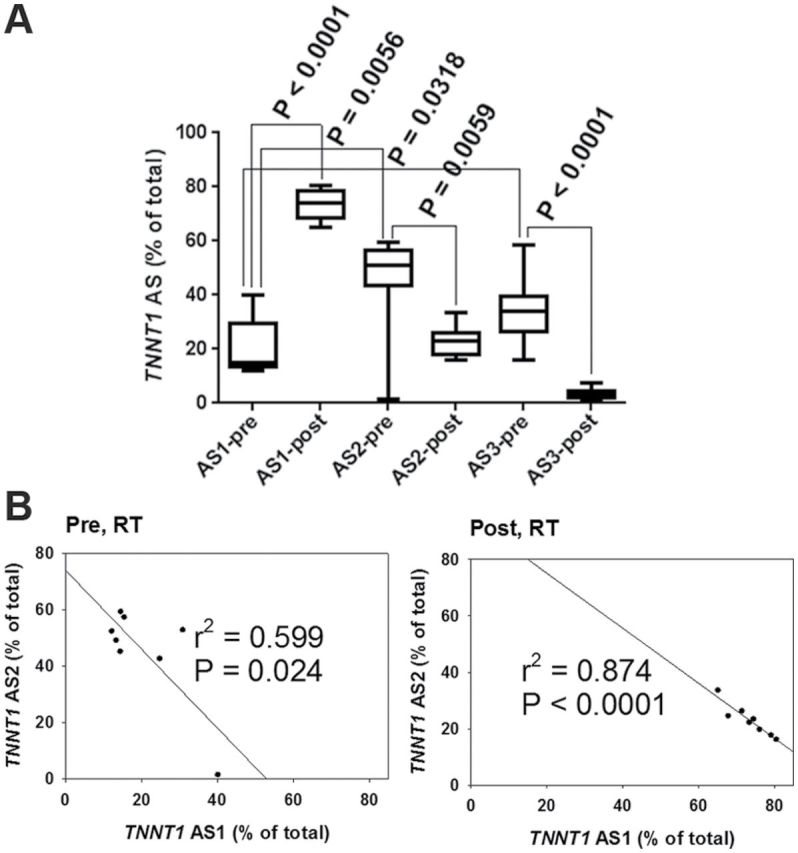
Effects of resistance training (RT) on troponin T (TNNT1) splice forms. (A) Relative abundance of TNNT1 splice forms before (pre) and after (post) RT. Significant changes were observed in all three: alternative splicing 1(AS1) increased, and AS2 and AS3 decreased. (B) Correlation analysis indicates a strong inverse correlation between AS1 and AS2 before and after RT (pre RT, r = −.774; post RT, r = −.935).
Abundance levels of AS1 and AS2 were negatively correlated both before and after the RT intervention, with a higher correlation seen after RT (Figure 2B). We reanalyzed our data after excluding the point at 40% AS1 in Figure 2B which seems to strongly influence the association before RT. With this data point removed, correlations between abundance of AS1 and AS2 before and after the RT intervention were r = −.217; r 2 = .047; p = .639, and r = −.993; r 2 = .912; p = .0022, respectively. There was no relationship between abundance of AS1 and AS3 patterns (pre, r = −.696; r 2 = .108, p = .082; post, r = −.092; r 2 = .03, p = .845).
Relationships Between TNNT1 AS Patterns and Participants’ Age, Body Mass, and BMI
To examine the association between age and TNNT1 AS patterns, we plotted the relative abundance of each splice pattern against the participants’ age, which ranged from 66 to 80 years. None of the three TNNT1 splice patterns showed any significant correlation with age at baseline (Figure 3A) or after RT (Figure 3B). Likewise, there was no relationship between abundance of any of the three TNNT1 AS patterns and body mass or BMI before RT (Figure 4C and E). After the 5-month RT intervention, body mass and BMI were not changed (Figure 4A and B); however, a positive correlation was observed between TNNT1 AS1 and body mass, while a negative correlation was found between abundance of TNNT1 AS2 and body mass (Figure 4D). No correlation between TNNT1 AS1 or AS2 and BMI was observed before and after RT (Figure 4E and F).
Figure 3.
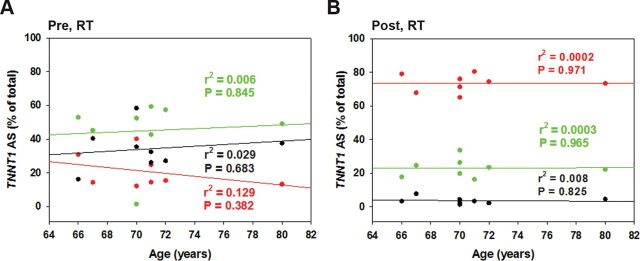
Troponin T (TNNT1) splice form profile and subjects’ age. No correlation between age and any of the three splice forms was observed pre-resistance training (RT; A) (r = −.360, .083, and .172 for AS1, AS2, and AS3, respectively) or post-RT (B; r = .016, .019, and −.094 for AS1, AS2, and AS3, respectively). AS1: red, AS2: green, AS3: black.
Figure 4.
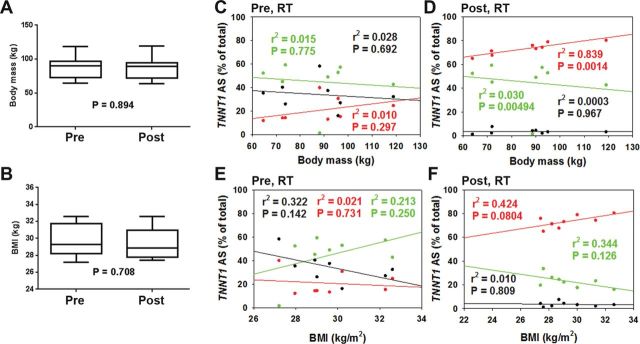
Relationships of troponin T (TNNT1) splice forms with body mass measures. Body mass (A) and body mass index (BMI; B) did not change significantly with resistance training (RT). TNNT1 alternative splicing 1 (AS1) correlated positively, and TNNT1 AS2 correlated negatively, with body mass (pre-RT, r = .422, −.121, and −.167; post-RT, r = .916, −.870, and −.0175 for AS1, AS2, and AS3, respectively) (C and D), and BMI (pre-RT, r = −.146, .462, and −.568; post-RT, r = .651, −.587, and −.102 for AS1, AS2, and AS3, respectively) (E and F). AS1: red, AS2: green, AS3: black.
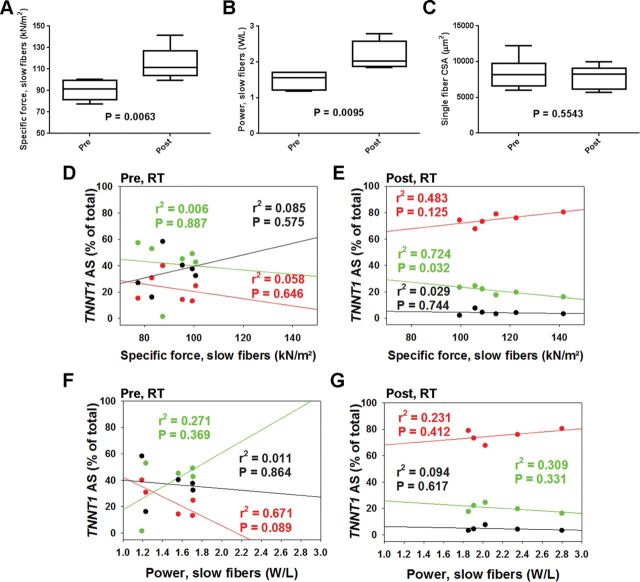
Changes in Single Muscle Fiber Force, Power, and V max With RT and Relationship With TNNT1 AS Patterns
Single muscle fiber–specific force and power, recorded on a subset of six and five participants, respectively, improved significantly with RT (Figure 5A and B). We next tested the relationship between single muscle fiber–specific force and power in slow fibers and abundance of TNNT1 AS patterns. Before RT, there were no relationships between TNNT1 AS patterns and single muscle fiber force or power (Figure 5D and F). On the other hand, single fiber force shows a negative correlation with abundance of AS2 after RT (Figure 5E). The relationship between single fiber power and TNNT1 AS1 or AS2 did not reach our conventional level of statistical significance after RT (Figure 5G). V max (pre: 0.698 ± 0.017; post: 0.806 ± 0.092) and a/P o (pre: 0.037 ± 0.008; post: 0.032 ± 0.001) did not differ significantly with RT. V max negatively correlated with TNNT1 AS1 while a trend to positively correlate with TNNT1 AS2 was apparent after RT (Figure 6). Additionally, negative correlations between V max and body mass, and power and BMI were observed after RT (Table 1). Interestingly, RT did not induce significant changes in fiber type composition in bundles of fibers from the subjects in which we examined single fiber contractile properties (pre-RT: type-1 = 44±3.1, type-2 = 56±3.1; post-RT: type-1 = 43±4.7, type-2 = 57± 4.6; p > .05).
Figure 5.
Changes in type-I single muscle fiber–specific force, power, and cross-sectional area (CSA), with resistance training (RT), and relationships with troponin T (TNNT1) alternative splicing (AS) forms. Both single muscle fiber–specific force (n = 6) (A) and power (n = 5) (B), but not CSA (C), increased with RT. Single muscle slow fiber force (pre-RT, r = −.240, .0754, and .291; post-RT, r = .695, −.851, and −.172 for AS1, AS2, and AS3, respectively) (D and E) and power (pre-RT, r = −.819, .520, and −.107; post-RT, r = .481, −.556, and −.306 for AS1, AS2, and AS3, respectively) (F and G) correlation with abundance of TNNT1 AS1-2 after RT (E and G). AS1: red, AS2: green, AS3: black.
Figure 6.
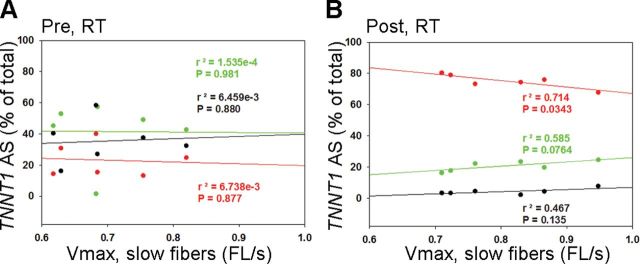
Relationships between troponin T (TNNT1) alternative splicing (AS) and single muscle fiber V max. (A) Single muscle fiber V max did not correlate with TNNT1 AS at baseline (r = −.0821, .0124, and .0804 for AS1, AS2, and AS3, respectively). (B) In contrast, single muscle slow fiber correlated negatively with TNNT1 AS1 and a trend to positively correlate with TNNT1 AS2 was observed after RT (r = −.845, .765, and .683 for AS1, AS2, and AS3, respectively). AS1: red, AS2: green, AS3: black.
Table 1.
Correlation Between a/P o, and TNNT1 AS1–3, and Body Mass or BMI and Single Fiber Parameters
| Correlation | Pre-RT | Post-RT | |||||
|---|---|---|---|---|---|---|---|
| r | r 2 | p | r | r 2 | p | ||
| a/P o | TNNT1-AS1 | −.543 | .295 | .265 | .286 | .082 | .528 |
| TNNT1-AS2 | .520 | .269 | .291 | −.119 | .014 | .823 | |
| TNNT1-AS3 | −.330 | .109 | .523 | −.478 | .229 | .337 | |
| Body mass | Force | .138 | .019 | .794 | .508 | .258 | .304 |
| Power | .276 | .076 | .653 | −.487 | .237 | .405 | |
| V max | .785 | .617 | .064 | −.827 | .685 | .042 | |
| a/P o | −.315 | .099 | .543 | −.132 | .017 | .804 | |
| BMI | Force | −.049 | 2.395e−3 | .927 | −.092 | 8.497e−3 | .862 |
| Power | .635 | .404 | .175 | −.844 | .712 | .035 | |
| V max | .484 | .234 | .331 | −.596 | .355 | .212 | |
| a/P o | .319 | .102 | .538 | −.392 | .153 | .442 | |
Notes: AS = alternative splicing; BMI = body mass index; TNNT = troponin T.
Increased Force and Power Precede Any Detectable Increase in Single Muscle Fiber CSA In Vitro
As the gain in muscle force with RT is larger than gains in muscle mass in older adults (4–6), we examined whether improvements in single muscle fiber force and power are associated with increased CSA after RT. No significant mean changes in single muscle fiber CSA were detected with RT (Figure 5C). Also, no correlation between any of the three TNNT1 AS patterns and CSA was observed (data not shown).
TNNT1 AS1/AS2 Ratio Is a Sensitive Marker of Change in Single Muscle Fiber Force With RT
As the abundance of TNNT1 AS1 and AS2 patterns correlate with muscle function, we also examined whether changes in single muscle fiber force and power with RT are related to the ratio of these AS patterns. A significant positive correlation between the percent change in single muscle fiber force (Figure 7A) and AS1/AS2 ratio was observed. The correlation between percent change in single muscle fiber power or V max and AS1/AS2 ratio did not reach statistical significance (Figure 7B and C).
Figure 7.
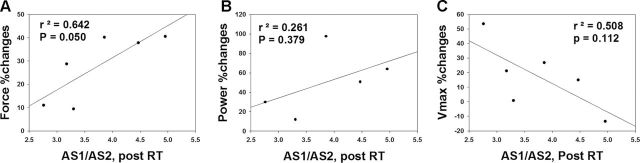
Troponin T (TNNT1) alternative splicing 1 (AS1)/AS2 ratio correlation with fiber force and power. (A) Single muscle fiber–specific force (% [post − pre]/pre) positively correlates with AS1/AS2 ratio after RT (r = .801). (B) Single muscle fiber power shows a trend toward a positive correlation with AS1/AS2 ratio after RT (r = .510). (C) Single muscle fiber V max shows a trend toward a negative correlation with AS1/AS2 ratio after RT (r = −.713).
Discussion
This work demonstrates that three major TNNT1 splice patterns are expressed in skeletal muscle from older adults and that RT modifies their relative abundance, with TNNT1 AS1 upregulated and TNNT1 AS2, AS3 downregulated. Strikingly, TNNT1 AS2 negatively correlates with single muscle fiber–specific force after RT. For the first time, we showed that the AS1/AS2 ratio positively correlates with the percent change in single muscle fiber force in response to RT. Our data suggest that TNNT1 AS2 relative abundance and the AS1/AS2 ratio can be promising biomarkers of skeletal muscle adaptation to overload (RT and/or body weight).
TNNT1 AS Patterns, a Quantitative Biomarker of Muscle Response to RT
Skeletal muscle undergoes dramatic changes in adapting to functional demands and stress (29). For instance, expression of slow-versus-fast TnT isoforms is highly sensitive to muscle unloading in simulated weightlessness (30). Nearly 95% of human multi-exon genes involve AS, which is a crucial mechanism of gene regulation and proteomic diversity (31,32). Altered relative abundance in mRNA isoforms in TNNT3 AS was reported in a rodent model of overweight and obesity, indicating that RNA AS is critical for muscle adaptation (14). Our current study further supports that RT alters the TNNT1 AS profile in older adults. It would be important to assess TNNT1 AS profile in young lean adults and determine whether age and/or body fat influences TNNT1 AS.
Aging is characterized by decreased lean muscle mass and increased body fat (33), both of which affect TNNT3 AS in rodents (14). As a transition from fast to slow fiber type has been reported and confirmed in aging humans and rodents (21,34), we examined the relationship between TNNT1 AS and age, body mass, and BMI in study participants before they engaged in the RT regimen. We did not observe any significant relationship with these factors, but these initially sedentary adults had substantially modified TNNT1 AS patterns after 5 months of RT, suggesting that RT may affect TNNT1 AS. One potential mechanism for this response could be through muscle mechanical stretch, as reported for mouse muscle C2C12 cells (19). The correlation between V max and AS1 (negative) or AS2 (trend to positive) indicates a change in contractile kinetics, which may be as important as enhanced specific force in response to RT.
TnT1 pre-mRNA AS is strongly correlated with muscle fiber contractile properties (30,35). Alternative splicing of exon 5 generates low molecular weight and high molecular weight isoforms of human slow TnT, and the low molecular weight isoform generates more specific force than the high molecular weight isoform (36). This result consistently supports our finding that TNNT1 AS2 is negatively correlated with muscle force. Additionally, that RT increased the relative abundance of TNNT1 AS1 and decreased the abundance of TNNT1 AS2 strongly supports the notion that TNNT1 AS2 and AS1/AS2 ratio are related to the beneficial effects of RT on muscle function.
TNNT1 AS, Aging, and Obesity
Obesity exacerbates age-associated decline in skeletal muscle strength and power, leading to physical disability and loss of independence (8,37,38). A recent study in older men and women associated obesity with lower muscle quality (39). However, decreased muscle mass in aging rodents and humans do not fully account for the diminished strength; atrophy only partially explains muscle weakness (1,2).
Decreased muscle force and power is determined, at least in part, by fast TNNT3 AS (14), a Ca2+-sensitive switch that turns muscle contraction on and off (15,17,18,40). TnT mediates the interaction between the troponin complex and tropomyosin, which is essential for Ca2+-activated striated muscle contraction. The thin filament regulatory complex (troponin–tropomyosin) acts as a switch in response to changes in intracellular Ca2+ concentration (41). Manipulating TnT genes and/or TnT protein isoforms in rodents (16,18,42–45) and single amino acid replacement mutations in humans (46) affect muscle Ca2+ sensitivity and maximal force development. Further, fibers containing fast troponin show higher co-operativity of Ca2+ activation than slow troponin fibers, which have higher Ca2+ sensitivity (16). Since mechanical stretch regulates the abundance of fast skeletal muscle TNNT3 splice forms (14,19), we propose that the changes in TNNT1 AS reported above may adjust single muscle fiber–specific force in response to chronic RT, probably through activation of the PI3K/Akt signaling pathway (19).
Molecular-signaling pathways that mediate the effects of mechanical load on muscle TNNT AS are poorly understood. When skeletal muscles are overloaded via RT, there is signaling through the Akt/mammalian target of rapamycin pathway (47), which may lead to changes in TNNT AS. The fast skeletal muscle TNNT3 AS controls the amplitude of muscle force by controlling Ca2+ sensitivity (48). Also, Akt and mammalian target of rapamycin phosphorylation (47,49,50) and its splicing response (51) are impaired in obese rodents (52). The chronic inflammatory state associated with obesity is mediated by adipose tissue macrophage infiltration (53), which impairs splice factor expression (54) and attenuates Akt (55) and mammalian target of rapamycin signaling (56). Although the sample examined was overweight or obese and their baseline TNNT1 AS poorly correlated with muscle function, this relationship significantly improved after exercise, which indicates that most of older adults retain the capacity to respond to a 5-month RT intervention.
This article provides the first evidence associating TNNT1 AS with muscle fiber force in older adults. Although single muscle fiber power increased significantly with RT, it did not show significant correlation with TNNT1 AS1–3. Body weight and RT may also play key roles in regulating TNNT1 AS, probably by a pathway similar to that reported for TNNT3 AS (14,19). As obesity interferes with normal TNNT3 AS regulation in the rat (14) and inhibits expression of certain splicing factors in humans (57), whole body function (body mass and obesity) may regulate TNNT1 AS by altering splicing factor function.
We did not detect any significant relationship among age and TNNT1 AS, which may be explained by the small number of participants examined. In addition, while the effect of aging and development on TNNT AS has been examined by cross-sectional studies in young, middle-aged, and old rodents (18,48,58), we could only examine two timepoints, 5 months apart. To clarify the effects of aging and obesity on slow TNNT AS, younger and leaner subjects must be compared in future studies.
Limitations of This Study
Analyzing TNNT1 AS patterns but not their proteins is a limitation because encoding RNAs may not reflect the relative abundance of their corresponding proteins. Exploring the diversity of TnT1 proteins instead of TNNT1 pre-mRNA AS may provide a more direct link with muscle fiber physiology; however, it should be noticed that as a first approach, human TNNT1 pre-mRNA AS must be characterized and the most abundant and reproducible splice variants determined. Our research also allowed us to accurately measure the size of the splice variants, which is possible but less reliable at the protein level by immunoblot. The small differences in TnT1 variants molecular weight, the lack of reliable size markers to define them, the lack of specific antibodies for each AS, and the possibility that a band includes more than one variant, with erroneous impact on its relative abundance, prevented us from performing this analysis at the protein level as our initial approach. The identification of TnT1 AS at the protein level is of our interest but will require technical developments. It is also worth noting that the three AS patterns studied involve two exons and could potentially have four different combinations in a given protein product, which may differ functionally. A more prolonged longitudinal study on a larger cohort may provide the power to better understand physical function, body composition, and TNNT1AS profile. Alternatively, a cross-sectional study of a broader age range might elucidate these variables. Analysis of TNNT3 and TNNT1 AS may clarify their relative contributions to RT-induced muscle fiber function improvement. We have not explored the mechanisms by which RT alters TNNT1 AS, which is required for targeting future interventions.
Clinical Implications and Future Directions
RT affects the TNNT1 splicing patterns associated with single muscle fiber force. TNNT3 shows an acidic-to-basic charge transition and sequence shortening with development due to the exclusion of exon 4 (shortened N-terminus), which correlates with increased muscle fiber Ca2+ sensitivity (15,59). Similarly, shorter TNNT1 may increase Ca2+ sensitivity to the myofilaments in slow-twitch fibers, but this hypothesis requires confirmation. Furthermore, RT may modify TNNT1 AS splicing in a relatively shorter time than fiber CSA, which indicates that RT-evoked increased specific force does not primarily depend on accruing protein.
The diversity of mRNA transcripts creates new opportunities for pharmacological interventions. For example, splice site–directed oligonucleotides that correct aberrant splicing are already being used in a clinical trial of a therapy for Duchenne muscular dystrophy (60). Future work should focus on identifying the splicing factors involved in RT-evoked TNNT AS modifications; muscleblind-like proteins or SFRS10, should be among the first examined (57,61). Future studies should also compare the effects of calorie restriction and regular diet on TNNT1 AS because the Akt/mammalian target of rapamycin pathway is also involved in a nutrient-sensing pathway (62).
Conclusions
We demonstrated that the TNNT1 AS2 pattern is negatively while the AS1/AS2 ratio is positively correlated with single muscle fiber–specific force. This finding identifies TNNT1 AS patterns as quantitative biomarkers of human muscle function and suggests that RT can influence TNNT AS patterns that may improve muscle function.
Funding
This work was supported by the National Institutes of Health grants, AG13934 and AG15820 to O.D., and AG020583 to B.N.; the Wake Forest Claude D. Pepper Older Americans Independence Center (P30-AG21332); and by a Glenn/AFAR Scholarship for Research in the Biology of Aging to A.B.
References
- 1. Delbono O. Molecular mechanisms and therapeutics of the deficit in specific force in ageing skeletal muscle. Biogerontology. 2002;3:265–270. :5090822 [DOI] [PubMed] [Google Scholar]
- 2. Payne AM, Delbono O. Neurogenesis of excitation-contraction uncoupling in aging skeletal muscle. Exerc Sport Sci Rev. 2004;32:36–40. :00003677-200401000-00008 [DOI] [PubMed] [Google Scholar]
- 3. Goodpaster BH, Park SW, Harris TB, et al. The loss of skeletal muscle strength, mass, and quality in older adults: the health, aging and body composition study. J Gerontol A Biol Sci Med Sci. 2006;61:1059–1064. :10.1093/gerona/61.10.1059 [DOI] [PubMed] [Google Scholar]
- 4. Fiatarone MA, Marks EC, Ryan ND, Meredith CN, Lipsitz LA, Evans WJ. High-intensity strength training in nonagenarians. Effects on skeletal muscle. JAMA. 1990;263:3029–3034. :10.1001/jama.1990.03440220053029 [PubMed] [Google Scholar]
- 5. Leenders M, Verdijk LB, van der Hoeven L, van Kranenburg J, Nilwik R, van Loon LJ. Elderly men and women benefit equally from prolonged resistance-type exercise training. J Gerontol A Biol Sci Med Sci. 2013;68:769–779. :10.1093/gerona/gls241 [DOI] [PubMed] [Google Scholar]
- 6. Tracy BL, Ivey FM, Hurlbut D, et al. Muscle quality. II. Effects Of strength training in 65- to 75-yr-old men and women. J Appl Physiol (1985). 1999;86:195–201. [DOI] [PubMed] [Google Scholar]
- 7. González E, Messi ML, Delbono O. The specific force of single intact extensor digitorum longus and soleus mouse muscle fibers declines with aging. J Membr Biol. 2000;178:175–183. :10.1007/s002320010025 [DOI] [PubMed] [Google Scholar]
- 8. Choi SJ, Shively CA, Register TC, et al. Force-generation capacity of single vastus lateralis muscle fibers and physical function decline with age in African green vervet monkeys. J Gerontol A Biol Sci Med Sci. 2013;68:258–267. :10.1093/gerona/gls143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hairi NN, Cumming RG, Naganathan V, et al. Loss of muscle strength, mass (sarcopenia), and quality (specific force) and its relationship with functional limitation and physical disability: the Concord Health and Ageing in Men Project. J Am Geriatr Soc. 2010;58:2055–2062. :10.1111/j.1532-5415.2010.03145.x [DOI] [PubMed] [Google Scholar]
- 10. Delbono O. Expression and regulation of excitation-contraction coupling proteins in aging skeletal muscle. Curr Aging Sci. 2011;4:248–259. :BSP/CAS/E-Pub/000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiménez-Moreno R, Wang ZM, Gerring RC, Delbono O. Sarcoplasmic reticulum Ca2+ release declines in muscle fibers from aging mice. Biophys J. 2008;94:3178–3188. :10.1529/biophysj.107.118786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang S, Wong HC, Wang ZM, et al. Design and application of a class of sensors to monitor Ca2+ dynamics in high Ca2+ concentration cellular compartments. Proc Natl Acad Sci U S A. 2011;108:16265–16270. :10.1073/pnas.1103015108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wei B, Jin JP. Troponin T isoforms and posttranscriptional modifications: Evolution, regulation and function. Arch Biochem Biophys. 2011;505:144–154. :10.1016/j.abb.2010.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schilder RJ, Kimball SR, Marden JH, Jefferson LS. Body weight-dependent troponin T alternative splicing is evolutionarily conserved from insects to mammals and is partially impaired in skeletal muscle of obese rats. J Exp Biol. 2011;214(Pt 9):1523–1532. :10.1242/jeb.051763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Briggs MM, Schachat F. Physiologically regulated alternative splicing patterns of fast troponin T RNA are conserved in mammals. Am J Physiol. 1996;270(1 Pt 1):C298–C305. [DOI] [PubMed] [Google Scholar]
- 16. Brotto MA, Biesiadecki BJ, Brotto LS, Nosek TM, Jin JP. Coupled expression of troponin T and troponin I isoforms in single skeletal muscle fibers correlates with contractility. Am J Physiol Cell Physiol. 2006;290:C567–C576. :10.1152/ajpcell.00422.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gomes AV, Venkatraman G, Davis JP, et al. Cardiac troponin T isoforms affect the Ca(2+) sensitivity of force development in the presence of slow skeletal troponin I: insights into the role of troponin T isoforms in the fetal heart. J Biol Chem. 2004;279:49579–49587. :10.1074/jbc.M407340200 [DOI] [PubMed] [Google Scholar]
- 18. Ogut O, Granzier H, Jin JP. Acidic and basic troponin T isoforms in mature fast-twitch skeletal muscle and effect on contractility. Am J Physiol. 1999;276(5 Pt 1):C1162–C1170. [DOI] [PubMed] [Google Scholar]
- 19. Schilder RJ, Kimball SR, Jefferson LS. Cell-autonomous regulation of fast troponin T pre-mRNA alternative splicing in response to mechanical stretch. Am J Physiol Cell Physiol. 2012;303:C298–C307. :10.1152/ajpcell.00400.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andersen JL. Muscle fibre type adaptation in the elderly human muscle. Scand J Med Sci Sports. 2003;13:40–47. :10.1034/j.1600-0838.2003.00299.x [DOI] [PubMed] [Google Scholar]
- 21. Lexell J. Human aging, muscle mass, and fiber type composition. J Gerontol A Biol Sci Med Sci. 1995;50:11–16. [DOI] [PubMed] [Google Scholar]
- 22. American College of Sports Medicine position stand. Progression models in resistance training for healthy adults. Med Sci Sports Exerc. 2009;41:687–708. :10.1249/MSS.0b013e3181915670 [DOI] [PubMed] [Google Scholar]
- 23. Choi SJ, Kim S, Lim JY. Potential errors in fiber length measurements resulting from lever arm rotation during mechanical testing of muscle cells. J Biomech. 2011;44:1797–1800. :10.1016/j.jbiomech.2011.03.027 [DOI] [PubMed] [Google Scholar]
- 24. Hill AV. The heat of shortening and the dynamic constants of muscle. Proc R Soc Lond Ser B, Biol Sci. 1938;126:136–195. :10.1098/rspb.1938.0050 [DOI] [PubMed] [Google Scholar]
- 25. Woledge RC, Curtin NA, Homsher E. Energetic aspects of muscle contraction. Monogr Physiol Soc. 1985;41:1–357. [PubMed] [Google Scholar]
- 26. Widrick JJ, Trappe SW, Costill DL, Fitts RH. Force-velocity and force-power properties of single muscle fibers from elite master runners and sedentary men. Am J Physiol. 1996;271(2 Pt 1):C676–C683. [DOI] [PubMed] [Google Scholar]
- 27. Chen FC, Chuang TJ. Different alternative splicing patterns are subject to opposite selection pressure for protein reading frame preservation. BMC Evol Biol. 2007;7:179. :10.1186/1471-2148-7-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wong TK, Lam TW, Yang W, Yiu SM. Finding alternative splicing patterns with strong support from expressed sequences on individual exons/introns. J Bioinform Comput Biol. 2008;6:1021–1033. :10.1142/S0219720008003825 [DOI] [PubMed] [Google Scholar]
- 29. Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev. 1996;76:371–423. [DOI] [PubMed] [Google Scholar]
- 30. Yu ZB, Gao F, Feng HZ, Jin JP. Differential regulation of myofilament protein isoforms underlying the contractility changes in skeletal muscle unloading. Am J Physiol Cell Physiol. 2007;292:C1192–C1203. :10.1152/ajpcell.00462.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. :10.1038/nature08909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. :10.1038/ 418236a [DOI] [PubMed] [Google Scholar]
- 33. Visser M, Goodpaster BH, Kritchevsky SB, et al. Muscle mass, muscle strength, and muscle fat infiltration as predictors of incident mobility limitations in well-functioning older persons. J Gerontol A Biol Sci Med Sci. 2005;60:324–333. :10.1093/gerona/60.3.324 [DOI] [PubMed] [Google Scholar]
- 34. Larsson L. Motor units: remodeling in aged animals. J Gerontol A Biol Sci Med Sci. 1995;50 Spec No:91–95. [DOI] [PubMed] [Google Scholar]
- 35. Kischel P, Bastide B, Muller M, et al. Expression and functional properties of four slow skeletal troponin T isoforms in rat muscles. Am J Physiol Cell Physiol. 2005;289:C437–C443. 10.1152/ajpcell.00365.2004 [DOI] [PubMed] [Google Scholar]
- 36. Larsson L, Wang X, Yu F, et al. Adaptation by alternative RNA splicing of slow troponin T isoforms in type 1 but not type 2 Charcot-Marie-Tooth disease. Am J Physiol Cell Physiol. 2008;295:C722–C731. :10.1152/ajpcell.00110.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jensen GL, Hsiao PY. Obesity in older adults: relationship to functional limitation. Curr Opin Clin Nutr Metab Care. 2010;13:46–51. :10.1097/MCO.0b013e32833309cf [DOI] [PubMed] [Google Scholar]
- 38. Waters DL, Baumgartner RN. Sarcopenia and obesity. Clin Geriatr Med. 2011;27:401–421. :10.1016/j.cger.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 39. Koster A, Stenholm S, Alley DE, et al. ; Health ABC Study. Body fat distribution and inflammation among obese older adults with and without metabolic syndrome. Obesity (Silver Spring). 2010;18:2354–2361. :10.1038/oby.2010.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gahlmann R, Troutt AB, Wade RP, Gunning P, Kedes L. Alternative splicing generates variants in important functional domains of human slow skeletal troponin T. J Biol Chem. 1987;262:16122–16126. [PubMed] [Google Scholar]
- 41. Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. [DOI] [PubMed] [Google Scholar]
- 42. Chandra M, Tschirgi ML, Rajapakse I, Campbell KB. Troponin T modulates sarcomere length-dependent recruitment of cross-bridges in cardiac muscle. Biophys J. 2006;90:2867–2876. :10.1529/biophysj.105.076950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gomes AV, Guzman G, Zhao J, Potter JD. Cardiac troponin T isoforms affect the Ca2+ sensitivity and inhibition of force development. Insights into the role of troponin T isoforms in the heart. J Biol Chem. 2002;277:35341–35349. :10.1074/jbc.M204118200 [DOI] [PubMed] [Google Scholar]
- 44. MacFarland SM, Jin JP, Brozovich FV. Troponin T isoforms modulate calcium dependence of the kinetics of the cross-bridge cycle: studies using a transgenic mouse line. Arch Biochem Biophys. 2002;405:241–246. :10.1016/S0003-9861(02)00370-3 [DOI] [PubMed] [Google Scholar]
- 45. Nassar R, Malouf NN, Mao L, et al. cTnT1, a cardiac troponin T isoform, decreases myofilament tension and affects the left ventricular pressure waveform. Am J Physiol Heart Circ Physiol. 2005;288:H1147–H1156. :10.1152/ajpheart.00140.2004 [DOI] [PubMed] [Google Scholar]
- 46. Hernandez OM, Szczesna-Cordary D, Knollmann BC, et al. F110I and R278C troponin T mutations that cause familial hypertrophic cardiomyopathy affect muscle contraction in transgenic mice and reconstituted human cardiac fibers. J Biol Chem. 2005;280:37183–37194. :10.1074/jbc.M508114200 [DOI] [PubMed] [Google Scholar]
- 47. Katta A, Kundla S, Kakarla SK, et al. Impaired overload-induced hypertrophy is associated with diminished mTOR signaling in insulin-resistant skeletal muscle of the obese Zucker rat. Am J Physiol Regul Integr Comp Physiol. 2010;299:R1666–R1675. :10.1152/ajpregu.00229.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marden JH, Fescemyer HW, Saastamoinen M, et al. Weight and nutrition affect pre-mRNA splicing of a muscle gene associated with performance, energetics and life history. J Exp Biol. 2008;211(Pt 23):3653–3660. :10.1242/jeb.023903 [DOI] [PubMed] [Google Scholar]
- 49. Katta A, Kakarla S, Wu M, et al. Altered regulation of contraction-induced Akt/mTOR/p70S6k pathway signaling in skeletal muscle of the obese Zucker rat. Exp Diabetes Res. 2009;2009:384683. :10.1155/2009/384683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kim YB, Peroni OD, Franke TF, Kahn BB. Divergent regulation of Akt1 and Akt2 isoforms in insulin target tissues of obese Zucker rats. Diabetes. 2000;49:847–856. :10.2337/diabetes.49.5.847 [DOI] [PubMed] [Google Scholar]
- 51. Pan BS, Potter JD. Two genetically expressed troponin T fragments representing alpha and beta isoforms exhibit functional differences. J Biol Chem. 1992;267:23052–23056. [PubMed] [Google Scholar]
- 52. Funai K, Parkington JD, Carambula S, Fielding RA. Age-associated decrease in contraction-induced activation of downstream targets of Akt/mTor signaling in skeletal muscle. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1080–R1086. :10.1152/ajpregu.00277.2005 [DOI] [PubMed] [Google Scholar]
- 53. Wisse BE. The inflammatory syndrome: the role of adipose tissue cytokines in metabolic disorders linked to obesity. J Am Soc Nephrol. 2004;15:2792–2800. :10.1097/01.ASN.0000141966.69934.21 [DOI] [PubMed] [Google Scholar]
- 54. Xiong Z, Shaibani A, Li YP, et al. Alternative splicing factor ASF/SF2 is down regulated in inflamed muscle. J Clin Pathol. 2006;59:855–861. :10.1136/jcp.2005.032961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Varma V, Yao-Borengasser A, Rasouli N, et al. Muscle inflammatory response and insulin resistance: synergistic interaction between macrophages and fatty acids leads to impaired insulin action. Am J Physiol Endocrinol Metab. 2009;296:E1300–E1310. :10.1152/ajpendo.90885.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lang CH, Frost RA, Vary TC. Regulation of muscle protein synthesis during sepsis and inflammation. Am J Physiol Endocrinol Metab. 2007;293:E453–E459. :10.1152/ajpendo.00204.2007 [DOI] [PubMed] [Google Scholar]
- 57. Pihlajamäki J, Lerin C, Itkonen P, et al. Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell Metab. 2011;14:208–218. :10.1016/j.cmet.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bucher EA, Dhoot GK, Emerson MM, Ober M, Emerson CP., Jr Structure and evolution of the alternatively spliced fast troponin T isoform gene. J Biol Chem. 1999;274:17661–17670. :10.1074/jbc.274.25.17661 [DOI] [PubMed] [Google Scholar]
- 59. Schachat FH, Diamond MS, Brandt PW. Effect of different troponin T-tropomyosin combinations on thin filament activation. J Mol Biol. 1987;198:551–554. :10.1016/0022-2836(87)90300-7 [DOI] [PubMed] [Google Scholar]
- 60. Bauman J, Jearawiriyapaisarn N, Kole R. Therapeutic potential of splice-switching oligonucleotides. Oligonucleotides. 2009;19:1–13. :10.1089/oli.2008.0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ho TH, Charlet-B N, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. :10.1038/sj.emboj.7600300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gulati P, Thomas G. Nutrient sensing in the mTOR/S6K1 signalling pathway. Biochem Soc Trans. 2007;35(Pt 2):236–238. :10.1042/BST0350236 [DOI] [PubMed] [Google Scholar]