Abstract
Community-associated (CA) infections with methicillin-resistant Staphylococcus aureus (MRSA) are on a global rise. However, analysis of virulence characteristics has been limited almost exclusively to the US endemic strain USA300. CA-MRSA strains that do not produce Panton-Valentine leukocidin (PVL) have not been investigated on a molecular level. Therefore, we analyzed virulence determinants in a PVL-negative CA-MRSA strain, ST72, from Korea. Genome-wide analysis identified 3 loci that are unique to that strain, but did not affect virulence. In contrast, phenol-soluble modulins (PSMs) and the global virulence regulator Agr strongly affected lysis of neutrophils and erythrocytes, while α-toxin and Agr had a major impact on in vivo virulence. Our findings substantiate the general key roles these factors play in CA-MRSA virulence. However, our analyses also showed noticeable differences to strain USA300, inasmuch as α-toxin emerged as a much more important factor than PSMs in experimental skin infection caused by ST72.
Keywords: alpha-toxin, community-associated infection, methicillin-resistant Staphylococcus aureus, Panton-Valentine leukocidin, phenol-soluble modulins
Infections with methicillin-resistant Staphylococcus aureus (MRSA) are the most frequent cause of death by an infectious agent in the United States and a leading source of morbidity and mortality in hospitals worldwide [1, 2]. While traditionally limited to hospitals and predisposed patients, community-associated (CA) MRSA infections emerged in the late 1990s, presenting largely as infections of the skin and soft tissues [3]. These occurred in otherwise healthy patients with no contact to the hospital setting. The strains causing CA-MRSA infections are different from those involved in hospital infections. In the United States, virtually all CA-MRSA cases are caused by a clone called pulsed-field type USA300. USA300 has become the leading cause of skin infections reporting to emergency departments in the United States [4]. Globally, endemic CA-MRSA clones have emerged at different geographical locations throughout the world [3], and many countries currently report a rise of CA-MRSA infections [5–7].
The reason for the extraordinary success of CA-MRSA strains as pathogens has been described as a situation where “resistance and virulence converge” [8]. All CA-MRSA strains harbor an SCCmec element, carrying methicillin resistance determinants, which is significantly smaller than those found in hospital-associated (HA)–MRSA clones. This is believed to cause less of a fitness cost, but experimental results have been inconsistent [9, 10]. Similarly, what underlies the increased virulence potential of CA-MRSA strains has remained controversial [11, 12]. Two hypotheses were developed. Both are based on the key role that cytolysis, in particular lysis of neutrophils, plays as immune evasion strategy of S. aureus during the establishment of infection [13], and the characteristically high cytolytic activity detected in CA-MRSA strains [14]. One hypothesis focuses on the acquisition of a mobile genetic element carrying the genes coding for Panton-Valentine leukocidin [15], a toxin that lyses leukocytes in a receptor-dependent fashion [16]. This hypothesis was mainly based on the finding that most initially found CA-MRSA clones harbor Panton-Valentine leukocidin (PVL)–encoding genes, while HA-MRSA commonly do not [15]. Another hypothesis explains the increased virulence of CA- as opposed to HA-MRSA strains by increased expression of core genome-encoded toxin genes, such as mainly phenol-soluble modulins (PSMs) and α-toxin [17–19].
While the molecular basis of CA-MRSA skin infections remains controversial in particular [20, 21], the considerable research efforts that have been made in this field now present a picture in which both expression of core genome-encoded toxins and acquisition of mobile genetic elements such as the prophage carrying the PVL-encoding genes played a role in the evolution of virulence of CA-MRSA [22]. However, virtually all investigations were performed using strain USA300, and a smaller number in the first isolated CA-MRSA strain USA400, which in the meantime was almost completely replaced by USA300 [3]. Both strains are prevalent mostly in the Americas. The factors contributing to virulence in other global CA-MRSA strains have received much less attention. Notably, this includes a series of CA-MRSA strains that do not have PVL-encoding genes and have emerged as causes of infections similarly severe as those caused by PVL-positive CA-MRSA [11]. For example, the PVL-negative sequence type (ST) 72 predominates among CA-MRSA strains in Korea and can cause severe skin infections [23]. Thus, to gain a more comprehensive understanding of virulence development in CA-MRSA clones, we investigated the basis of virulence in the CA-MRSA strain ST72 from Korea.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions
Bacteria were grown in tryptic soy broth with shaking at 180 rpm at 37°C. The bacterial strains used were ST72 (HL1), USA300 (LAC), USA400 (MW2), and COL (a standard HA-MRSA strain). Gene deletion mutants were constructed by a previously described allelic replacement procedure [24], after amplifying approximately 1 kb regions up- and downstream of the gene(s) of interest and cloning into plasmid pKOR1, with the resulting plasmids being first transformed into strain S. aureus RN4220 and then strain ST72. Oligonucleotides used are shown in Table 1.
Table 1.
Oligonucleotides Used for Allelic Replacement Plasmid Construction
| Name | Sequence |
|---|---|
| For deletion of SAKOR_00396 | |
| attB1-RAGM01350-P1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGCTACTCAAATACTCTGATAGAT |
| RAGM01350-P2 | AATTCCTCCTCCTTATATATAATATAAATATAAC |
| RAGM01350-P3 | TAAGGAGGAGGAATTAAAATTATACACTTAAAAACTTTATTCGAATAC |
| attB2-RAGM01350-P4 | GGGGACCACTTTGTACAAGAAAGCTGGGTTGCTTGCGACCATTTCATGATATTCAATTG |
| For deletion of SAKOR_02177 | |
| attB1-RAGM02170-P1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCAAGGATTACGAAAAGACCTAAG |
| RAGM02170-P2 | GATTTTAACACCTACATTTTAATTGAATTGGTAG |
| RAGM02170-P3 | GTAGGTGTTAAAATCAAAAACGTATAATTATTTTAACGAGCTATTATATC |
| attB2-RAGM02170-P4 | GGGGACCACTTTGTACAAGAAAGCTGGGTGGTTTGCCTGTTAATCTTCCAATAC |
| For deletion of SAKOR_00052 to SAKOR_00058 | |
| attB1-RAGM01691-01685-P1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTGTGTAGATAAACTGAGTTGTTTGAGAG |
| RAGM01691-01685-P2 | CCAATACTCTCCTTCTTGAAATTAAATATTCTATTTCGAAGTC |
| RAGM01691-01685-P3 | GAAGGAGAGTATTGGAAAAAATATAATAATATTACGTGTAAATGACTAG |
| attB2-RAGM01691-01685-P4 | GGGGACCACTTTGTACAAGAAAGCTGGGTCAGAACAGTTCCGTGAAACGAGTATTCAAC |
| For deletion of hla | |
| attB1-hla-P1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCTAAATCTAGCATCTTCTAGTTTGG |
| hla-P2 | TTTCATCATCCTTCTATTTTTTAAAACGATTTGAGGAAAC |
| hla-P3 | AGAAGGATGATGAAATGTAAATTATTTGTTCATGTACAAATAAATATAATTTATAAC |
| attB2-hla-P4 | GGGGACCACTTTGTACAAGAAAGCTGGGTGATGCGGATTTTGTATGGACTTCTTCATC |
| For deletion of psmα operon | |
| attB1-PSM-ABCD-P1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTGATAATTTCGCAGACTCACGTGGC |
| PSM-ABCD-P2 | TAAGATTACCTCCTTTGCTTATGAGTTAACTTC |
| PSM-ABCD-P3 | AAGGAGGTAATCTTATTTAAGCGAATTGAATACTTAAAATTCTCAGGCC |
| attB2-PSM-ABCD-P4 | GGGGACCACTTTGTACAAGAAAGCTGGGTTGCCAAATAATGTCGTTCGATCAAAAGG |
Analysis of the ST72 Genome Content
The comparison of the ST72 (HL1) genome content with those of other S. aureus strains was performed by IG Assets, Inc (Mount Prospect, IL), using a proprietary script. Every open reading frame (ORF) from the ST72 genome was compared against the ORFs from the 31 available S. aureus genomes. Each ORF was compared using 3 metrics: similarity score (10−10), functional annotation, and protein length. For ORFs to be considered for inclusion, the following criteria had to be satisfied. The considered ORFs had to have a P score similarity to an ORF from the ST72 genome 284 of 1 × 10−10 or less. In addition, the considered ORF either had to have the same functional annotation as the ST72 genome's ORF or have at least 80% of the protein length matching with the corresponding protein in the ST72 genome.
Lysis of Erythrocytes and Neutrophils by Culture Filtrates
Human blood was obtained from anonymized donors under approved protocols at the National Institutes of Health (NIH) Blood Bank. Supernatants were collected from bacterial cultures grown for 8 hours. Hemolytic activities were determined by incubating samples with human erythrocytes (2% v/v in Dulbecco's phosphate buffered saline [PBS]) for 1 hour at 37°C. Hemolysis was determined by measuring the optical density at 540 nm using an enzyme-linked immunosorbent assay reader. The assay was performed in triplicate.
Human neutrophils were isolated from heparinized venous blood with a standard method. Supernatants were collected from bacterial cultures grown for 8 hours and diluted to 1:1, 1:5, and 1:10 for the assay. Lysis was measured using a lactate dehydrogenase (LDH) cytotoxicity detection kit according to the manufacturer's protocol (Roche) as described elsewhere [14].
Neutrophil Lysis by Whole Bacteria
Human neutrophils were isolated as described above. Bacteria grown to mid-logarithmic growth phase and neutrophils were incubated for 30 minutes, 60 minutes and 90 minutes at a 10:1 ratio bacteria to neutrophils. Then, supernatants were collected and diluted 1:5 for the assays. Lysis after phagocytosis was measured using an LDH cytotoxicity detection kit according to the manufacturer's protocol (Roche) as described elsewhere [14].
Measurement of PSM Production
PSM concentrations were determined by reversed-phase high-pressure liquid chromatography/electrospray ionization mass spectrometry (RP-HPLC/ESI-MS) of S. aureus culture filtrates as described elsewhere [25]. Samples were taken from cultures inoculated to an optical density at 600 nm of 0.1 from precultures and grown for 8 hours.
Measurement of α-Toxin Production
Supernatants were taken from cultures inoculated to an optical density at 600 nm of 0.1 from precultures and grown for 8 hours. Equal amounts of proteins in culture supernatants were loaded on 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and run at 150 V for 1 hour. Proteins in the gels were blotted on nitrocellulose membranes by using an iBlot Western blotting system (Life Technologies, Grand Island, NY). Blotted membranes were incubated with Odyssey blocking buffer (LI-COR, Lincoln, NE) for 1 hour at room temperature. Anti-staphylococcal α-toxin rabbit serum (1:2000 dilution; Sigma-Aldrich, St. Louis, MO) was added to the blocking buffer and incubated for another hour at room temperature. Membranes were washed 5 times with washing buffer (Tris-buffered saline containing 0.1% Tween-20, pH 7.4) and incubated with 1:10 000 diluted Cy5-labeled goat antirabbit immunoglobin G (Life Technologies, Grand Island, NY) in Odyssey blocking buffer in the dark for 1 hour at room temperature. Membranes were washed 5 times with the washing buffer and scanned using a Typhoon TRIO+ Variable Mode Imager (GE Healthcare, Piscataway, NJ).
Mouse Model of Skin Infection
The mouse skin infection model was performed essentially as described previously [17]. Briefly, female Crl:SKH1-hrBR mice were injected subcutaneously with approximately 2 × 107 bacterial cells in 50 µL of PBS in the back. The length and width of the abscess or lesion caused by the bacterial infection was measured with an electronic caliper daily for 14 days after infection and calculated using the formula length × width. All mouse experiments were performed at the animal care facility of the National Institute of Allergy and Infectious Diseases (NIAID), Building 33, and were in compliance with the guidelines of the NIAID/NIH Institutional Animal Care and Use Committee.
Statistics
Statistical analysis was performed using GraphPad Prism 6.02. One- or 2-way analysis of variance was used for comparison of multiple groups, as appropriate; t tests for 2 groups. Error bars show standard deviation.
RESULTS
The Three Gene Loci Unique to CA-MRSA Strain ST72 Do Not Contribute to Virulence
We previously sequenced the genome of the CA-MRSA clone ST72 (strain HL1) to completion [26], giving us the opportunity to compare its genome for the presence of virulence determinants, in particular such that could potentially substitute for the lack of the PVL-encoding genes. We already established previously that there is no obvious novel toxin gene in that strain that could have been attributed such a role [26]. Therefore, we here analyzed the ST72 genome in a genome-wide comparison with all other S. aureus strains, whose genomes have been sequenced to date, searching for genes that are unique to ST72 and which could potentially have a yet undetermined, ST72-specific role in virulence (see Supplementary data).
We found 19 annotated genes that were unique. However, 10 of those appeared to be pseudogenes, based on evaluation of length and absence of potential ribosome binding sites. These were not further considered. Two single genes (SAKOR_00396, SAKOR_02177) and 1 likely operon consisting of 7 genes (SAKOR_00052 to SAKOR_00058) were found to be unique to ST72 and likely to represent true protein-encoding genes. They had no annotated functions. We deleted the genes, or the SAKOR_00052 to SAKOR_00058 operon, respectively, by allelic replacement, and analyzed in vitro growth, impact on expression of PSMs and α-toxin, impact on erythrocyte and neutrophil lysis, and behavior in a mouse skin–infection model. Growth patterns were indistinguishable from that of the wild-type strain (Figure 1A) and no impact on the expression of PSMs (Figure 1B) or α-toxin (Figure 1C) was measurable. Furthermore, capacities to lyse neutrophils or erythrocytes were not different between wild-type and mutant strains (Figure 1D and 1E). Finally, no difference was detected between ST72 and the 3 isogenic deletion mutants when analyzed for in vivo virulence (Figure 1F). These findings indicate that the 3 gene loci unique to ST72 do not impact growth, virulence, or expression of key toxins presumed important for CA-MRSA skin infection.
Figure 1.
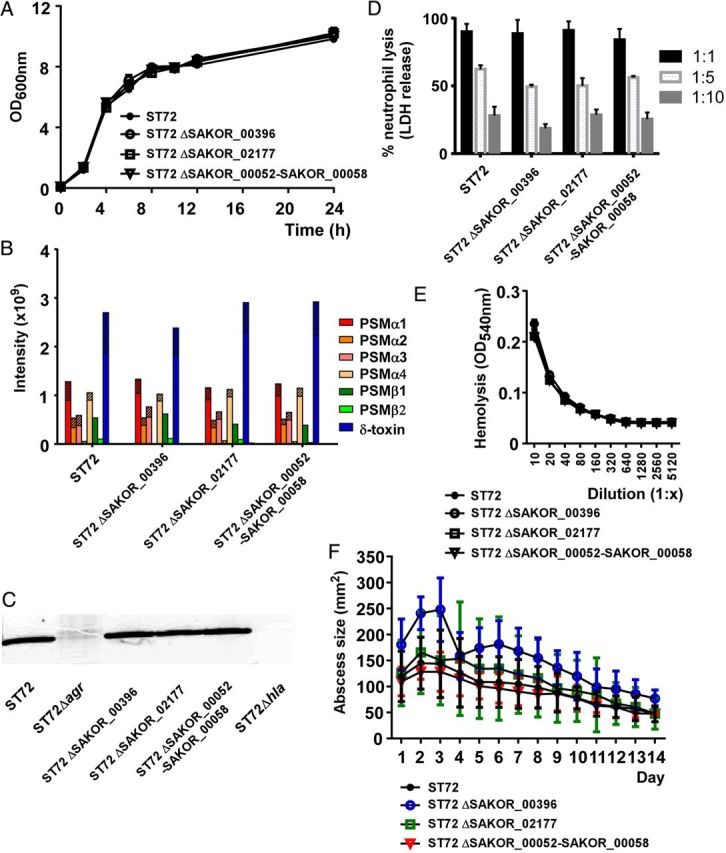
Impact of 3 unique loci in ST72 on virulence, virulence phenotypes, and virulence factor expression. A, In vitro growth in TSB of the used 3 isogenic deletion mutants in the respective unique gene loci compared to wild-type ST72. B, PSM production. PSM production was measured by HPLC/MS in 8-hour cultures grown in TSB. Striped parts of bars represent the N-deformylated portion of the respective PSM. C, Production of α-toxin during growth in TSB, Western blot with anti-α-toxin antiserum of 8-hour cultures. D, Neutrophil lysis by culture filtrates of 8-hour cultures, diluted by the given factor. E, Hemolysis by culture filtrates of 8-hour cultures, diluted by the given factor. F, Mouse model of skin infection. Crl:SKH1-hrBR mice were injected subcutaneously with approximately 2 × 107 bacterial cells and abscess sizes were measured daily. Number of mice per group, 5. A, D, E, Measurements were performed in triplicate. A, D, E, F, Error bars depict SD. Abbreviations: HPLC/MS, high-pressure liquid chromatography/mass spectrometry; LDH, lactate dehydrogenase; OD, optical density; PSM, phenol-soluble modulin; SD, standard deviation; TSB, tryptic soy broth.
PSMs Have a Key Role in Neutrophil and Erythrocyte Lysis by ST72
Neutrophil lysis is considered a premier virulence mechanism by which S. aureus escapes elimination by innate host defense [27]. Notably, it was found that virulence of MRSA strains correlates with the potential to lyse neutrophils [14, 28]. PSMα peptides are premier determinants of that immune evasion mechanism in the PVL-positive CA-MRSA strains LAC (USA300) and MW2 (USA400), accounting for virtually the total difference compared to the low capacity exerted by HA-MRSA strains to lyse neutrophils [17].
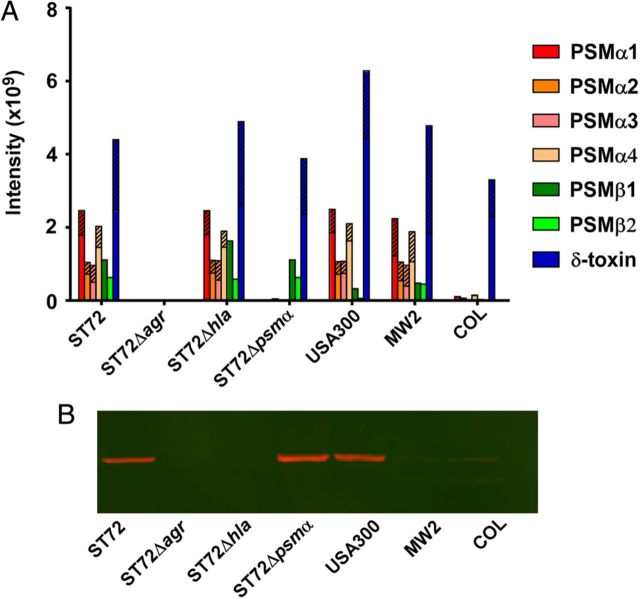
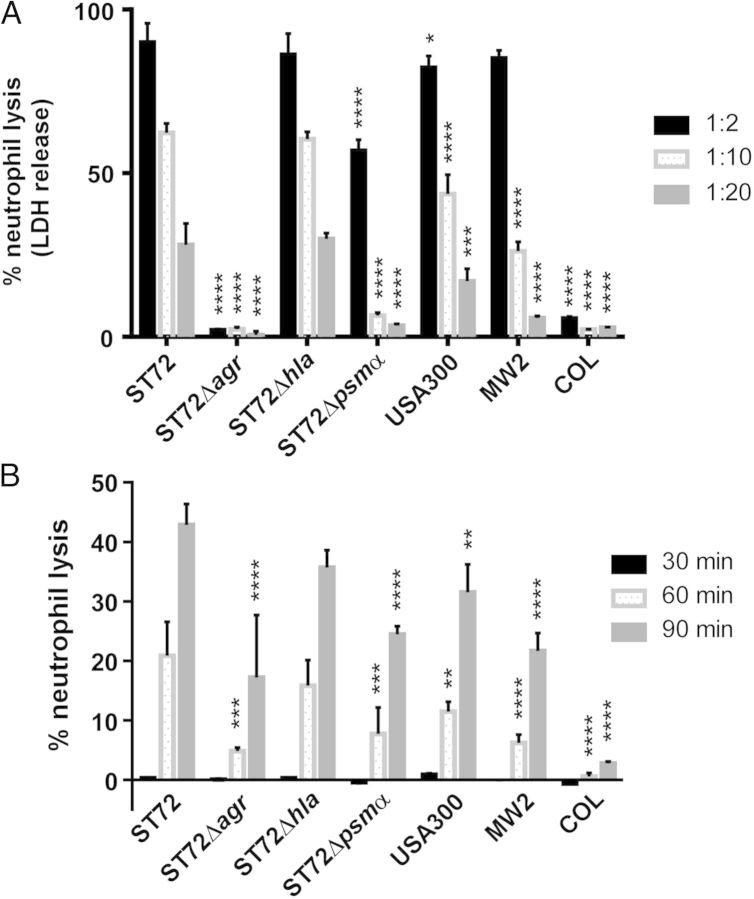
We therefore compared in vitro production levels of PSMs and their contribution to lyse human neutrophils in ST72 as compared to other CA- and HA-MRSA strains. To that end, we constructed isogenic deletion mutants in the psmα locus and the global virulence regulator Agr, which controls all leukotoxic proteins of CA-MRSA (classic leukotoxins such as PVL, and PSMs) [29] in strain ST72. We also constructed a mutant in the α-toxin gene hla as a further major virulence determinant described for USA300 [18, 20]. Growth of the mutant strains was comparable to that of the wild-type ST72 strain (Supplementary Figure 1). PSM production levels were very similar to those in USA300, except for a somewhat higher production of PSMβ peptides, which, however, are barely lytic to neutrophils [30] (Figure 2). Overall capacity of ST72 culture filtrates to lyse neutrophils was even slightly higher than that exerted by USA300 culture filtrates (Figure 3A). Notably, PSMα peptides had a strong and significant contribution to that phenotype. Leukotoxicity was completely absent from the agr mutant, as expected. No effect was found in an hla mutant. This is in accordance with previous findings showing absence of lytic potential of α-toxin toward human neutrophils [31]. Furthermore, we measured neutrophil lysis by whole live bacteria, because S. aureus survival inside neutrophils contributes to infection [32] and PSMα peptides were shown to contribute to neutrophil lysis after ingestion into the neutrophil phagosome [33]. In a fashion similar to the results obtained using culture filtrates, ST72 had higher lytic capacity than USA300. We also compared to strain MW2 (USA400), and an HA-MRSA strain, COL; ST72 exerted significantly higher capacity to lyse neutrophils compared to both. Notably, PSMα peptides contributed significantly to neutrophil lysis, almost reaching the level detected with the agr mutant (Figure 3B).
Figure 2.
Production of PSMs and α-toxin in ST72; constructed isogenic agr, hla, and psmα mutants; and USA300. A, PSM production. PSM production was measured by HPLC/MS in 8-hour cultures grown in TSB. Striped parts of bars represent the N-deformylated portion of the respective PSM. B, Production of α-toxin during growth in TSB, Western blot with anti-α-toxin antiserum of 8-hour cultures. Abbreviations: HPLC/MS, high-pressure liquid chromatography/mass spectrometry; PSMs, phenol-soluble modulins; TSB, tryptic soy broth.
Figure 3.
Neutrophil lysis in ST72, constructed isogenic agr, hla, and psmα mutants, and USA300. A, Neutrophil lysis by culture filtrates of strains grown for 8 hours in TSB and diluted at the given factor. B, Neutrophil lysis by whole bacteria, measured at the given time points after addition of bacteria to neutrophils (ratio of bacteria to neutrophils, 10:1). A and B, Measurements were performed in triplicate. Error bars depict SD. **P < .01; ***P < .001; ****P < .0001. Statistical analysis is by 2-way analysis of variance with Dunnett post tests comparing to the data obtained with strain ST72. Abbreviations: LDH, lactate dehydrogenase; SD, standard deviation; TSB, tryptic soy broth.
Hemolysis (lysis of erythrocytes) is an important virulence determinant of S. aureus, representing a crucial way for the bacteria to acquire iron, whose concentration is growth limiting in human serum. Similar to the results obtained with neutrophils, hemolytic capacity of culture filtrate was higher in ST72 than USA300, with the data obtained for the isogenic deletion mutants indicating a premier role for PSMα peptides and Agr, while α-toxin had no detectable role (Figure 4). These findings are in accordance with data obtained for synthetic PSM peptides and the contribution of PSMα peptides and Agr to hemolysis by strain USA300 [34]. The toxicity of α-toxin toward erythrocytes varies greatly between species; those of humans are only moderately sensitive [35], explaining the results we obtained for the hla deletion mutant.
Figure 4.

Hemolysis in ST72, constructed isogenic agr, hla, and psmα mutants, and USA300. Hemolytic activity of culture filtrates grown for 8 hours in TSB was measured at the given dilutions. Measurements were performed in triplicate. Error bars depict SD. *P < .05; **P < .01; ****P < .0001. Statistical analysis is by 2-way analysis of variance with Dunnett post tests comparing to the data obtained with strain ST72. Abbreviations: OD, optical density; SD, standard deviation; TSB, tryptic soy broth.
α-Toxin Dominates the Impact of ST72 Virulence Determinants on Experimental Skin Infection in Mice
To determine the basis of virulence of ST72 in vivo, we used a mouse skin infection model, because skin infections predominate among CA-MRSA infections in general and with ST72 in particular [3, 36]. The USA300 strain was somewhat more virulent in that model, as measured by abscess sizes, than strain ST72, which is in accordance with results we previously obtained in a rabbit model [28]. Notably, both strains caused open necrotic lesions, a phenotype in that model that is limited to some apparently very virulent strains (Figure 5).
Figure 5.
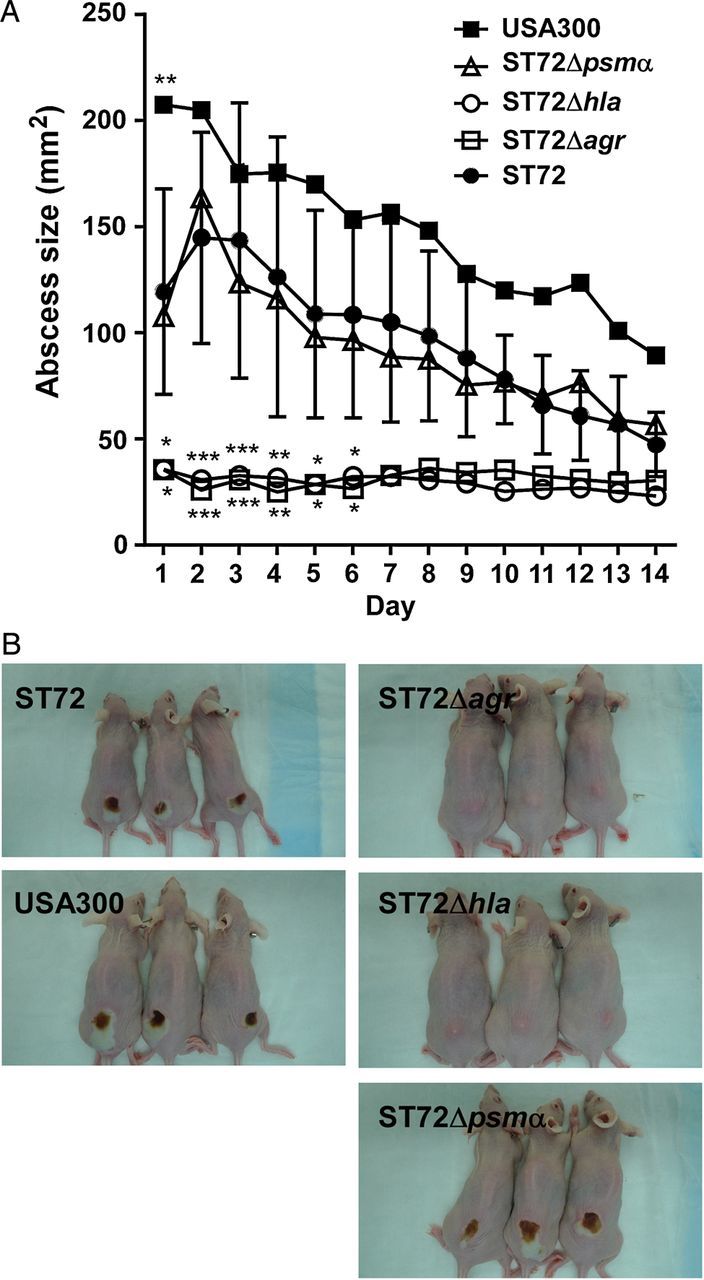
Mouse model of skin infection. A, Crl:SKH1-hrBR mice were injected subcutaneously with approximately 2 × 107 bacterial cells, and abscess sizes were measured daily. Number of mice per group, 5. Error bars depict SD. *P < .05; **P < .01; ***P < .001. Statistical analysis is by 2-way analysis of variance with Dunnett post tests comparing to the data obtained with strain ST72. B, Pictures of animals with representative abscesses. Abbreviation: SD, standard deviation.
The agr mutant of ST72 had the expected strong impact on virulence, with no necrotic lesions and almost no abscess formation visible. This is in accordance with findings we previously obtained with USA300 [37]. However, we detected strong differences as for the impact of PSMα peptides versus α-toxin on in vivo skin infection, despite equal in vitro production of PSMs and α-toxin in USA300 and ST72 (Figure 2). The hla mutant showed no detectable lesions, comparable to the agr mutant. In contrast, the psmα mutant showed no significantly different abscess formation compared to the wild-type strain. Notably, these findings establish an exceptionally great importance of α-toxin in skin infection by ST72.
DISCUSSION
In the present study, we analyzed the basis of virulence in a PVL-negative CA-MRSA strain, ST72, from Korea. We focused on the global virulence regulator Agr, PSMs, and α-toxin. These core genome-encoded factors have previously been shown to be strongly expressed and have a major impact on virulence in the PVL-positive strain USA300 [17–20, 28, 37]. Our study was thus aimed to analyze whether these features are generally conserved in CA-MRSA strains, independently of their PVL status.
The Agr global virulence regulator is a quorum-sensing system that controls expression of a large series of virulence determinants in a cell density–dependent fashion [29, 38]. Most toxins and degradative exoenzymes in S. aureus are under positive control by Agr. This includes α-toxin and PSMs, with the latter being under exceptionally strict and direct control via direct binding of the AgrA response regulator to the psm promoters [29]. We recently showed that Agr strongly impacts virulence of USA300 in skin infections [37], and our present result indicate the same important role in strain ST72.
PSMα peptides are the main factors responsible for neutrophil lysis in the CA-MRSA strains USA300 and USA400 [17]. They also have a key impact on hemolysis in these strains [34]. Our present results indicate a similarly important role in these phenotypes in strain ST72 when tested on human cells. However, despite the major contribution that neutrophil lysis is believed to have to the establishment of S. aureus infection, PSMα peptides showed no significant impact on the development of skin infection when tested in mice, in strong contrast to results obtained previously with strain USA300 [17]. Among several potential explanations for this discrepancy, it is possible that the in vivo environment leads to changes in gene expression that differ between the USA300 and ST72 strains, with other factors such as α-toxin possibly overshadowing the impact of PSMα peptides. In that regard, it is noteworthy that ST72 exerted higher rates of neutrophil and erythrocyte lysis than USA300, but was slightly inferior to USA300 regarding the capacity to cause lesions in the mouse skin infection model. Despite the previously reported correlation among MRSA strains between in vitro capacity to cause neutrophil and erythrocyte lysis and in vivo virulence [28], this indicates that in vivo virulence cannot always be predicted precisely from in vitro measurement of major virulence phenotypes.
Notably, we found that α-toxin has a dramatic impact on experimental skin infection by ST72, to an extent comparable to that exerted by the global virulence regulator Agr. α-toxin is not a significant contributor to lysis of human neutrophils or erythrocytes, which was reflected by our results, but has a considerable contribution to the establishment of invasive skin infection by stimulating regulatory cascades in epithelial cells that ultimately lead to a breach of the epithelial layer [39–41]. Our finding that α-toxin has a strong impact on virulence of ST72 during experimental skin infection, which is reminiscent of results achieved with USA300 [20, 42], further confirms the notion that α-toxin is a promising target for drug development aiming to interfere with CA-MRSA toxicity [40, 42–44].
The role of PVL in the pathogenesis of CA-MRSA infections, in particular skin infection as the most common manifestation of CA-MRSA disease, is still controversial [12, 22]. Several observations have cast doubt on the initially assumed major role of PVL in those infections: first, PVL did not have a significant impact on virulence in several animal models comparing isogenic lukSF mutants to the respective wild-type strain [11], including in rabbits whose neutrophils are very sensitive to PVL [20]; second, there is no epidemiological connection, and no correlation when assayed in experimental infection, between the presence of the PVL-encoding genes and severity of skin infections [28, 45]; and third, PVL-negative CA-MRSA strains have emerged that are able to mount even very severe skin infections in otherwise healthy individuals in a fashion comparable to PVL-positive CA-MRSA strains [11, 23].
Another objective of our present study therefore was to further assess PVL's role in CA-MRSA virulence. We reasoned that if PVL plays a significant role in CA-MRSA virulence, then the comparable virulence of PVL-positive and PVL-negative strains must be caused by factors substituting for PVL's contribution in the latter. By whole-genome sequencing, we previously verified that there are no obvious toxin genes that would account for such substituting factors [26]. Here, we ascertained that there are no genes unique to ST72 with yet unknown function that could play such a role. Of the 3 loci that we identified to be unique to ST72, none had a measurable effect on premier virulence mechanisms or in vivo virulence. Furthermore, as far as can be implied from in vitro data, the other major factors found to contribute to CA-MRSA virulence, α-toxin and PSMs, did not show increased expression in ST72 compared to USA300, suggesting that differential expression of those factors does not compensate for a lack of PVL.
In conclusion, our findings indicate that virulence of the PVL-negative CA-MRSA strain ST72 in skin infection is mediated to a large extent by genome-encoded toxins, as shown previously for PVL-positive CA-MRSA strains. These results confirm the notion of a limited contribution of PVL to CA-MRSA skin infection and support the general idea that high expression of core genome-encoded toxins contributed to the evolution of CA-MRSA virulence. Furthermore, our study highlights strain-dependent differences in the contribution of α-toxin versus PSMs to CA-MRSA skin infection, whose mechanistic underpinnings warrant further investigation.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (ZIA AI000904-12 to M. O.) and the China Scholarship Council (to Y. C.). The authors thank Yuanjun Zhu for help with neutrophil isolation.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Klevens RM, Morrison MA, Nadle J, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 2.Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest. 2003;111:1265–73. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–68. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moran GJ, Krishnadasan A, Gorwitz RJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–74. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 5.Chuang YY, Huang YC. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Asia. Lancet Infect Dis. 2013;13:698–708. doi: 10.1016/S1473-3099(13)70136-1. [DOI] [PubMed] [Google Scholar]
- 6.Rolo J, Miragaia M, Turlej-Rogacka A, et al. High genetic diversity among community-associated Staphylococcus aureus in Europe: results from a multicenter study. PLOS One. 2012;7:e34768. doi: 10.1371/journal.pone.0034768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kock R, Becker K, Cookson B, et al. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill. 2010;15:19688. doi: 10.2807/ese.15.41.19688-en. [DOI] [PubMed] [Google Scholar]
- 8.Chambers HF. Community-associated MRSA—resistance and virulence converge. N Engl J Med. 2005;352:1485–7. doi: 10.1056/NEJMe058023. [DOI] [PubMed] [Google Scholar]
- 9.Diep BA, Stone GG, Basuino L, et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197:1523–30. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- 10.Lee SM, Ender M, Adhikari R, Smith JM, Berger-Bachi B, Cook GM. Fitness cost of staphylococcal cassette chromosome mec in methicillin-resistant Staphylococcus aureus by way of continuous culture. Antimicrob Agents Chemother. 2007;51:1497–9. doi: 10.1128/AAC.01239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otto M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu Rev Microbiol. 2010;64:143–62. doi: 10.1146/annurev.micro.112408.134309. [DOI] [PubMed] [Google Scholar]
- 12.Otto M. A MRSA-terious enemy among us: end of the PVL controversy? Nat Med. 2011;17:169–70. doi: 10.1038/nm0211-169. [DOI] [PubMed] [Google Scholar]
- 13.Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3:948–58. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 14.Voyich JM, Braughton KR, Sturdevant DE, et al. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175:3907–19. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 15.Tristan A, Bes M, Meugnier H, et al. Global distribution of Panton-Valentine leukocidin–positive methicillin-resistant Staphylococcus aureus, 2006. Emerg Infect Dis. 2007;13:594–600. doi: 10.3201/eid1304.061316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spaan AN, Henry T, van Rooijen WJ, et al. The staphylococcal toxin Panton-Valentine Leukocidin targets human C5a receptors. Cell Host Microbe. 2013;13:584–94. doi: 10.1016/j.chom.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Wang R, Braughton KR, Kretschmer D, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–4. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 18.Bubeck Wardenburg J, Bae T, Otto M, Deleo FR, Schneewind O. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat Med. 2007;13:1405–6. doi: 10.1038/nm1207-1405. [DOI] [PubMed] [Google Scholar]
- 19.Li M, Diep BA, Villaruz AE, et al. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA. 2009;106:5883–8. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi SD, Malachowa N, Whitney AR, et al. Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. J Infect Dis. 2011;204:937–41. doi: 10.1093/infdis/jir441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipinska U, Hermans K, Meulemans L, et al. Panton-Valentine leukocidin does play a role in the early stage of Staphylococcus aureus skin infections: a rabbit model. PLOS One. 2011;6:e22864. doi: 10.1371/journal.pone.0022864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otto M. Community-associated MRSA: what makes them special? Int J Med Microbiol. 2013;303:324–30. doi: 10.1016/j.ijmm.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SH, Park C, Yoo JH, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus strains as a cause of healthcare-associated bloodstream infections in Korea. Infect Control Hosp Epidemiol. 2009;30:146–55. doi: 10.1086/593953. [DOI] [PubMed] [Google Scholar]
- 24.Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Joo HS, Otto M. The isolation and analysis of phenol-soluble modulins of Staphylococcus epidermidis. Methods Mol Biol. 2014;1106:93–100. doi: 10.1007/978-1-62703-736-5_7. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Chatterjee SS, Porcella SF, Yu YS, Otto M. Complete genome sequence of a Panton-Valentine leukocidin-negative community-associated methicillin-resistant Staphylococcus aureus strain of sequence type 72 from Korea. PLOS One. 2013;8:e72803. doi: 10.1371/journal.pone.0072803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rigby KM, DeLeo FR. Neutrophils in innate host defense against Staphylococcus aureus infections. Semin Immunopathol. 2012;34:237–59. doi: 10.1007/s00281-011-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M, Cheung GY, Hu J, et al. Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. J Infect Dis. 2010;202:1866–76. doi: 10.1086/657419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Queck SY, Jameson-Lee M, Villaruz AE, et al. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell. 2008;32:150–8. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung GY, Joo HS, Chatterjee SS, Otto M. Phenol-soluble modulins—critical determinants of staphylococcal virulence. FEMS Microbiol Rev. 2013;38:698–719. doi: 10.1111/1574-6976.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valeva A, Walev I, Pinkernell M, et al. Transmembrane beta-barrel of staphylococcal alpha-toxin forms in sensitive but not in resistant cells. Proc Natl Acad Sci USA. 1997;94:11607–11. doi: 10.1073/pnas.94.21.11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gresham HD, Lowrance JH, Caver TE, Wilson BS, Cheung AL, Lindberg FP. Survival of Staphylococcus aureus inside neutrophils contributes to infection. J Immunol. 2000;164:3713–22. doi: 10.4049/jimmunol.164.7.3713. [DOI] [PubMed] [Google Scholar]
- 33.Surewaard B, de Haas C, Vervoort F, et al. Staphylococcal alpha-phenol soluble modulins contribute to neutrophil lysis after phagocytosis. Cell Microbiol. 2013;15:1427–37. doi: 10.1111/cmi.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheung GY, Duong AC, Otto M. Direct and synergistic hemolysis caused by Staphylococcus phenol-soluble modulins: implications for diagnosis and pathogenesis. Microbes Infect. 2012;14:380–6. doi: 10.1016/j.micinf.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper LZ, Madoff MA, Weinstein L. Heat stability and species range of purified staphylococcal alpha-toxin. J Bacteriol. 1966;91:1686–92. doi: 10.1128/jb.91.5.1686-1692.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim ES, Song JS, Lee HJ, et al. A survey of community-associated methicillin-resistant Staphylococcus aureus in Korea. J Antimicrob Chemother. 2007;60:1108–14. doi: 10.1093/jac/dkm309. [DOI] [PubMed] [Google Scholar]
- 37.Cheung GY, Wang R, Khan BA, Sturdevant DE, Otto M. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun. 2011;79:1927–35. doi: 10.1128/IAI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Recsei P, Kreiswirth B, O'Reilly M, Schlievert P, Gruss A, Novick RP. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 39.Soong G, Chun J, Parker D, Prince A. Staphylococcus aureus activation of caspase 1/calpain signaling mediates invasion through human keratinocytes. J Infect Dis. 2012;205:1571–9. doi: 10.1093/infdis/jis244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berube BJ, Bubeck Wardenburg J. Staphylococcus aureus alpha-toxin: nearly a century of intrigue. Toxins. 2013;5:1140–66. doi: 10.3390/toxins5061140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inoshima I, Inoshima N, Wilke GA, et al. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat Med. 2011;17:1310–4. doi: 10.1038/nm.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kennedy AD, Bubeck Wardenburg J, Gardner DJ, et al. Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J Infect Dis. 2010;202:1050–8. doi: 10.1086/656043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hua L, Hilliard JJ, Shi Y, et al. Assessment of an anti-alpha-toxin monoclonal antibody for prevention and treatment of Staphylococcus aureus–induced pneumonia. Antimicrob Agents Chemother. 2014;58:1108–17. doi: 10.1128/AAC.02190-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tkaczyk C, Hua L, Varkey R, et al. Identification of anti-alpha toxin monoclonal antibodies that reduce the severity of Staphylococcus aureus dermonecrosis and exhibit a correlation between affinity and potency. Clin Vaccine Immunol. 2012;19:377–85. doi: 10.1128/CVI.05589-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bae IG, Tonthat GT, Stryjewski ME, et al. Presence of genes encoding the Panton-Valentine leukocidin exotoxin is not the primary determinant of outcome in patients with complicated skin and skin structure infections due to methicillin-resistant Staphylococcus aureus: results of a multinational trial. J Clin Microbiol. 2009;47:3952–7. doi: 10.1128/JCM.01643-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.