Abstract
Background
Woolly hair (WH) belongs to a family of disorders characterized by hair shaft anomalies that clinically presents with tightly curled hair, which can be divided into syndromic and non syndromic forms of WH. We have recently identified mutations in both LPAR6/P2RY5 and LIPH that are associated with autosomal recessive woolly hair (ARWH).
Objective
To study the underlying genetic causes of autosomal woolly hair in Pakistani population.
Methods
We studied ten Pakistani families with ARWH for mutations in LPAR6/P2RY5 and LIPH and then performed haplotype analysis to confirm their segregation in the families.
Results
We identified five mutations in LPAR6/P2RY5, among which three were recurrent and two were novel in eight Pakistani families. We then showed that two of the mutations in LPAR6/P2RY5 are founder mutations in Pakistani families. Moreover, we identified two recurrent mutations in the LIPH gene in two Pakistani families.
Conclusion
Our study extends the spectrum of mutations in LPAR6/P2RY5 gene and underscores that mutations in LPAR6/P2RY5 and LIPH result in similar phenotypes.
Keywords: Woolly hair, hypotrichosis, LIPH, P2RY5, LPAR6
Introduction
Woolly hair (WH) belongs to a group of disorders characterized by hair shaft anomalies that clinically presents with tightly curled hair.1 WH is distinct from the tightly curly hair in African populations in that WH shows hair shaft anomalies which can lead to hair loss and hair depigmentation.1 Woolly hair can be divided into two main categories. The first is syndromic WH, in which WH occurs in the setting of associated cutaneous and/or systemic anomalies. The second is non syndromic WH, that can be inherited in an autosomal dominant (ADWH [MIM 194300]) or autosomal recessive (ARWH [MIM 278150]) pattern.2 The distinction between the two categories is very critical because woolly hair can occur in the setting of syndromes that can be lethal at early ages due to cardiac disease. Naxos (OMIM 601214) and Carvajal syndromes (OMIM 605676) are two conditions that present with woolly hair, palmoplantar keratoderma and ventricular arrhythmias.3,4
Until recently, genes associated with non syndromic woolly hair were unknown. We and others have recently reported that mutations in the LIPH (MIM 607365) and LPAR6/P2RY5 (MIM 609239) genes underlie ARWH and/or localized autosomal recessive hypotrichosis (LAH [MIM 604379 and 611452]).5,6,7 Mutations in both genes, LPAR6 and LIPH act in the same signaling pathway and result in a clinically similar phenotype which can range from woolly hair to sparse hair and complete loss of hair.5,6,8 More recently, we have shown that mutations in keratin 74 are associated with ADWH.9 Here, we studied ten Pakistani families with ARWH/hypotrichosis and identified several mutations in LPAR6/P2RY5 and LIPH.
Materials and Methods
Patients
After obtaining informed consent, we collected peripheral blood samples from the family members and 100 unrelated healthy control individuals in EDTA-containing tubes (under institutional approval and in adherence to the Declaration of Helsinki Principles). Genomic DNA was isolated from these samples according to standard techniques.
Mutation Analysis
All exons and exon-intron boundaries of the LPAR6/P2RY5 and LIPH gene were amplified by PCR with primers and conditions described previously.5,10 The amplified PCR products were directly sequenced in an ABI Prism 310 Automated Sequencer, using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems).
Genotyping and haplotype analysis
To analyze whether the mutations c.69insCATGfsX29 (p.24insH52) and c.562A>T (p.I188F) are common founder mutations in Pakistani population, genomic DNA from members of families affected with either mutation were amplified by PCR using primers for four microsatellite markers, D13S168, D13S153, D13S1307 and D13S165 close to LPAR6 gene.5 PCR products were run on 8% polyacrylamide gels and genotypes were assigned by visual inspection.
Screening Assays
We performed screening assays for the novel mutations c.409T>C; c.410-426del17 and c.734A>G (p.Y245C) in the LPAR6 gene. For the mutation c.409T>C; c.410-426del17, we amplified DNA from affected individuals and 100 Pakistani controls using primers for exon 3 after which the products were run on 8% polyacrylamide gel and inspected visually. The wild type allele was 301bp while the mutant allele was 284bp. For the mutation p.Y245C we sequenced 100 Pakistani controls.
Results
Clinical features
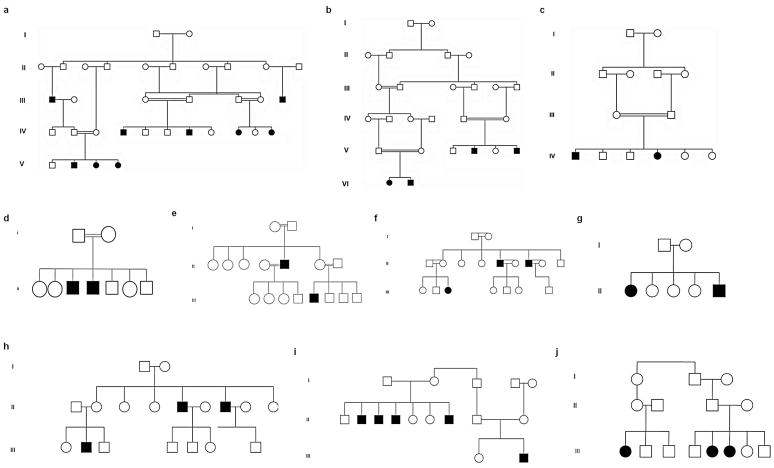
We studied 10 consanguineous Pakistani families (Family A, B, C, D, E, F, G, H, I and J) (Fig. 1) that had multiple affected individuals showing features consistent with recessively inherited woolly hair that were present since birth. All the families shared similar phenotypes that at times were variable within the same family. The hair over the entire scalp region was coarse, lusterless, dry and tightly curled, leading to a diffuse woolly hair phenotype with varying degrees of hypotrichosis or sparse hair. Additionally several patients showed hair depigmentation (Fig. 2). Eyebrow, eyelash and beard hairs appeared normal. Affected individuals in all families showed normal teeth, nails and sweating and did not show palmoplantar hyperkeratosis or keratosis pilaris. There was no familial history of cardiac disease.
Figure 1.
We studied ten consanguineous Pakistani families (families A to J) with autosomal recessive woolly hair and/or hypotrichosis.
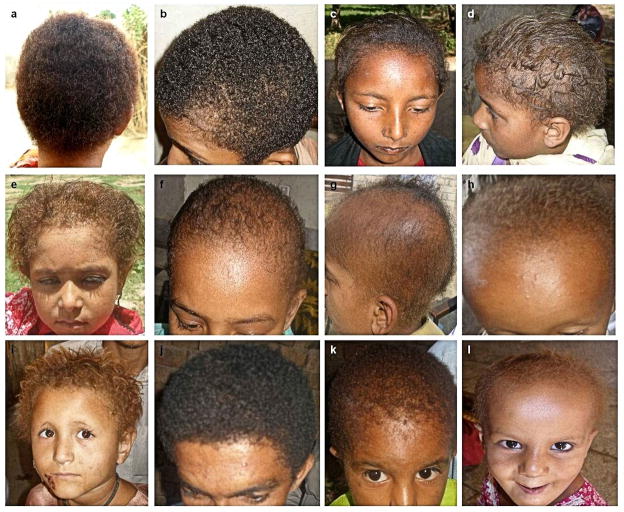
Figure 2.
Affected members from the ten families showed similar phenotypes that varied from persistent woolly hair, to variable degrees of hypotrichosis and hair depigmentation. (a and h) are affected individuals from family A, (b and F) are affected individuals from family B, (c) is an affected individual from family C, (d) is an affected individual from family D, (e) is an affected individual from family E, (g) is an affected individual from family F, (i) is an affected individual from family G, (j) is an affected individual from family H, (k) is an affected individual from family I, (l) is an affected individual from family J.
Mutation Analysis and Haplotype Analysis
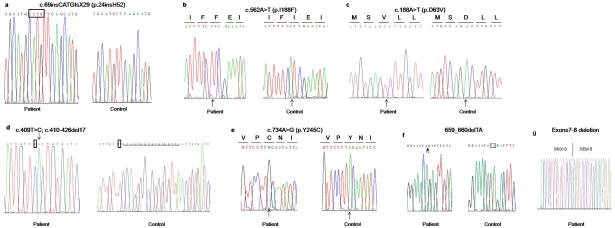
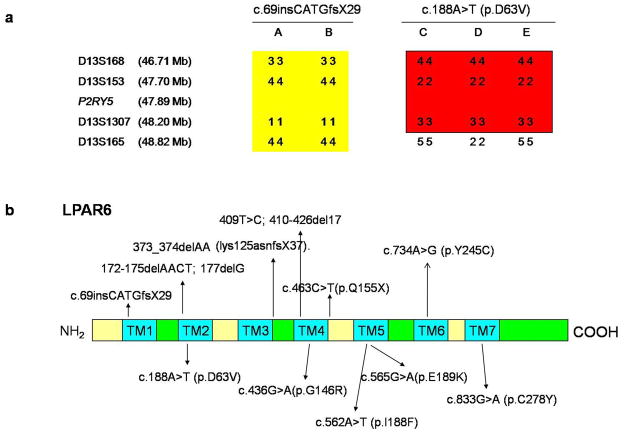
We identified five mutations in the LPAR6/P2RY5 gene among which three were recurrent and two novel mutations. Moreover, we identified two recurrent mutations in the LIPH gene. Families A and B had a recurrent mutation, designated c.69insCATGfsX29, in the LPAR6 gene (Fig. 3a). Families C, D and E had a recurrent mutation designated, p.I188F in the LPAR6 gene (Fig. 3b). Family F had a recurrent mutation, designated c.188A>T (p.D63V), in the LPAR6 gene (Fig. 3c). Family G had a novel mutation designated c.409T>C, c.410-426del17 in the LPAR6 gene (Fig. 3d). This mutation was not present in 100 Pakistani control individuals. Family H had a novel mutation, designated p.Y245C, in the LPAR6 gene (Fig. 3e). This mutation was not present in 100 Pakistani control individuals. Family I had a recurrent mutation designated c.659_660delTA in the LIPH gene (Fig. 3f). Family J had a recurrent mutation that consisted of deletion of exons 7 and 8 in the LIPH gene (Fig. 3g). Haplotype analysis showed that the mutations c.69insCATG and p.I188F are founder mutations in the Pakistani population (Fig. 4a).
Figure 3.
a) Affected individuals in families A and B had a common homozygous mutation designated, c.69insCATGfsX29 in the LPAR6 gene. b) Affected individuals in families C, D and E had a common homozygous mutation designated, p.I188F in the LPAR6 gene. c) Affected individuals in family F had a homozygous mutation designated, p.D63V in the LPAR6 gene. d) Affected individuals in family G had a homozygous mutation designated c.409T>C; c.410-426del17 in the LPAR6 gene. e) Affected individuals in family H had the homozygous mutation p.Y245C in the LPAR6 gene. f) Affected individuals in family I had the homozygous mutation c.659_660delTA in the LIPH gene. g) Affected individuals in family J had homozygous deletion of exons 7 and 8 in LIPH gene.
Figure 4.
a) Haplotype analysis showed that c.69insCATGfsX29 and p.I188F are founder mutations in the Pakistani population. b) LPAR6 encodes for a 7 transmembrane G protein coupled-receptor. Mutations occurring in any of the transmembrane (TM) regions are expected to be detrimental. The mutation c.69insCATGfsX29 occurs in TM1, p.D63V occurs in TM2, c.409T>C; c.410-426del17 occurs in TM4, p.I188F occurs in TM5 and p.Y245C occurs in TM6
Discussion
We and others have identified pathogenic mutations in the LPAR6/P2RY5 gene in several families with ARWH or hypotrichosis.5,6 Similarly, we have shown that mutations in LIPH gene lead to an identical phenotype.10 P2RY5 encodes for a seven transmembrane G protein coupled receptor (GPCR)1 (Fig. 4b) and is located within intron 17 of the retinoblastoma 1 (RB1) gene.5 LIPH encodes for a member of the phospholipase A1 family and is required for the synthesis of lysophosphatidic acid (LPA).11 LPA plays a critical role in promoting hair growth.12,13 LPA is a ligand for the receptor, P2Y5,6 which explains the similar phenotypes in patients with either LPAR6 or LIPH gene mutations. LPAR6/LIPH have overlapping expression in the inner root hair sheath of the hair follicle which arise from the hair matrix and differentiate before the keratinocytes of the central hair matrix thus forming a cylinder like structure providing a support for the normal development of the hair shaft14 which might explain why disruption in the LPA/P2Y5 signaling pathway results in a woolly hair.
We did not find evidence of phenotypic variability within the families we studied, which is in support of no genotype-phenotype correlations and the clinical variation can occur even within individuals of the same family.5,15 This suggests that other gene modifiers might play a role in phenotypic variability. There are no criteria to predict what patients will progress to develop hair loss and the severity of hair loss.
Here, we identified three recurrent and two novel mutations in the LPAR6 gene and two recurrent mutations in the LIPH gene. The mutation c.409T>C; c.410-426del17 occurs in the fourth transmembrane region (Fig. 4b) of LPAR6 resulting in premature termination codon. The mutation Y245C occurs in a highly conserved region in transmembrane 6 (Fig. 4b) and similarly to other mutations occurring in transmembrane regions is expected to destabilize the tertiary structure of the protein leading to its dysfunction. Moreover, we have shown that mutations c.60insCATGfsX29 and p.I188F are founder mutations in the Pakistani population.
In conclusion, our study increases the spectrum of mutations in LPAR6, provides more evidence for the lack of genotype-phenotype correlation and clinical variability in LPAR6 and LIPH and underscores the role of this G protein-coupled receptor, together with LIPH and lysophosphatidic acid (LPA), in determination of hair texture.
Acknowledgments
We gratefully acknowledge the families for having participated in this study. This study was supported by USPHS NIH grant from NIH/NIAMS RO1 AR44924 (to A.M.C.) and NIH Institutional Research Training Grant T32AR007605 (P.I. David Bickers), Postdoctoral Fellow, Department of Dermatology, Columbia University.
Footnotes
Institute where the work was performed: Columbia University
Conflict of interest: None.
References
- 1.Chien AJ, Valentine MC, Sybert VP. Hereditary woolly hair and keratosis pilaris. J Am Acad Dermatol. 2006;54:S35–S39. doi: 10.1016/j.jaad.2005.01.092. [DOI] [PubMed] [Google Scholar]
- 2.Hutchinson PE, Cairns RJ, Wells RS. Woolly hair. Clinical and general aspects. Trans St Johns Hosp Dermatol Soc. 1974;60:160–177. [PubMed] [Google Scholar]
- 3.McKoy G, Protonotarios N, Crosby A, et al. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease) Lancet. 2000;355:2119–2124. doi: 10.1016/S0140-6736(00)02379-5. [DOI] [PubMed] [Google Scholar]
- 4.Norgett EE, Hatsell SJ, Carvajal-Huerta L, et al. Recessive mutation in desmoplakin disrupts desmoplakinintermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum Mol Genet. 2000;9:2761–2766. doi: 10.1093/hmg/9.18.2761. [DOI] [PubMed] [Google Scholar]
- 5.Shimomura Y, Wajid M, Ishii Y, et al. Disruption of P2RY5, an orphan G protein-coupled receptor, underlies autosomal recessive woolly hair. Nat Genet. 2008;40:335–339. doi: 10.1038/ng.100. [DOI] [PubMed] [Google Scholar]
- 6.Pasternack SM, von Kugelgen I, Al Aboud K, et al. G protein-coupled receptor P2Y5 and its ligand LPA are involved in maintenance of human hair growth. Nat Genet. 2008;40:329–334. doi: 10.1038/ng.84. [DOI] [PubMed] [Google Scholar]
- 7.Kazantseva A, Goltsov A, Zinchenko R, et al. Human hair growth deficiency is linked to a genetic defect in the phospholipase gene LIPH. Science. 2006;314(5801):982–985. doi: 10.1126/science.1133276. [DOI] [PubMed] [Google Scholar]
- 8.Pasternack SM, von Kügelgen I, Müller M, et al. In vitro analysis of LIPH mutations causing hypotrichosis simplex: evidence confirming the role of lipase H and lysophosphatidic acid in hair growth. J Invest Dermatol. 2008;129(12):2772–2776. doi: 10.1038/jid.2009.154. [DOI] [PubMed] [Google Scholar]
- 9.Shimomura Y, Wajid M, Petukhova L, Kurban M, Christiano AM. Autosomal-dominant woolly hair resulting from disruption of keratin 74 (KRT74), a potential determinant of human hair texture. Am J Hum Genet. 2010;86:632–638. doi: 10.1016/j.ajhg.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimomura Y, Wajid M, Petukhova L, Shapiro L, Christiano AM. Mutations in the Lipase H (LIPH) gene underlie autosomal recessive woolly hair/hypotrichosis. J Invest Dermatol. 2009;129:622–628. doi: 10.1038/jid.2008.290. [DOI] [PubMed] [Google Scholar]
- 11.Sonoda H, Aoki J, Hiramatsu T, et al. A novel phosphatidic acid-selective phospholipase A1 that produces lysophosphatidic acid. J Biol Chem. 2002;277:34254–34263. doi: 10.1074/jbc.M201659200. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi T, Kamimura A, Hamazono-Matsuoka T, Honda S. Phosphatidic acid has a potential to promote hair growth in vitro and in vivo, and activates mitogen-activated protein kinase/extracellular signal-regulated kinase kinase in hair epithelial cells. J Invest Dermatol. 2003;121:448–456. doi: 10.1046/j.1523-1747.2003.12426.x. [DOI] [PubMed] [Google Scholar]
- 13.Inoue A, Arima N, Ishiguro J, Prestwich GD, Arai H, Aoki J. LPA-producing enzyme PA-PLA(1)α regulates hair follicle development by modulating EGFR signalling. EMBO J. 2011;30(20):4248–42460. doi: 10.1038/emboj.2011.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sundberg JP, Boggess D, Sundberg B, et al. Asebia-2J (Scd1(ab2J)): a new allele and a model for scarring alopecia. Am J Pathol. 2000;156:2067–2075. doi: 10.1016/S0002-9440(10)65078-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tariq M, Ayub M, Jelani M, et al. Mutations in the P2RY5 gene underlie autosomal recessive hypotrichosis in 13 Pakistani families. Br J Dermatol. 2009;160:1006–1010. doi: 10.1111/j.1365-2133.2009.09046.x. [DOI] [PubMed] [Google Scholar]