Abstract
The human beta defensin 1 (hBD-1) antimicrobial peptide is a member of the innate immune system known to act in the first line of defence against microorganisms, including viruses such as human papillomavirus (HPV). In this study, five functional polymorphisms (namely g-52G>A, g-44C>G and g-20G>A in the 5’UTR and c.*5G>A and c.*87A>G in the 3’UTR) in the DEFB1 gene encoding for hBD-1 were analysed to investigate the possible involvement of these genetic variants in susceptibility to HPV infection and in the development of HPV-associated lesions in a population of Brazilian women. The DEFB1 g-52G>A and c.*5G>A single-nucleotide polymorphisms (SNPs) and the GCAAA haplotype showed associations with HPV-negative status; in particular, the c.*5G>A SNP was significantly associated after multiple test corrections. These findings suggest a possible role for the constitutively expressed beta defensin-1 peptide as a natural defence against HPV in the genital tract mucosa.
Keywords: antimicrobial peptides, human papillomavirus, genetic polymorphisms, cervical lesion
Antimicrobial peptides and proteins (AMPs) are abundant and widely distributed molecules that play important roles in host defence as part of the innate immune system. Several AMPs contribute to human skin and mucosae defence (such as in the lining of the female genital tract) (Selsted & Ouellette 2005, Schröder & Harder 2006).
While the effects of AMPs against bacterial infections have been studied in detail, there is little information regarding their antiviral effects.
Among the AMPs, defensins, important components of the mucosal innate immunity of the female genital tract, may be involved in the defence against human papillomavirus (HPV) infection. Human beta defensin (hBD)-1 is constitutively expressed, whereas hBD-2, hBD-3 and hBD-4 are inducible in vaginal mucosa. The expression of human alpha defensin (HNP-1)-1, HNP-3 and HD-5 is also inducible in the vaginal mucosa (Buck et al. 2006, Klotman & Chang 2006).
Few studies have been published concerning the relationships between defensins and HPV infection. Buck et al. (2006) used papillomaviral vectors to perform high-throughput screenings for compounds that could block the initial stages of HPV infection. The authors showed that HNP-1, HNP-2 and HNP-3 and HD-5 are antagonists of cutaneous and mucosal-tropic strain HPV infections, whereas hBD-1 and hBD-2 do not posses anti-HPV activity. This study was conducted on an in vitro model without in vivo confirmation.
Additionally, hBD-1 might possess anti-cancer activities. The loss of hBD-1 expression was detected at high frequencies in renal and prostate cancer (Donald et al. 2003). Sun et al. (2006) found that synthetic hBD-1 was able to inhibit bladder cancer cell proliferation and promote apoptosis in human renal carcinoma cells. Bullard et al. (2008) observed that hBD-1 was cytotoxic to late-stage prostate cancer cell lines and might be involved in the recognition and elimination of other cancer cells, as well as tumour progression control.
Regarding the relationships between the DEFB1 gene (also reported as DEFB101, encoding hBD-1 and localised at 8p22-23) and viral infections, DEFB1 polymorphisms have been associated with susceptibility to human immunodeficiency virus (HIV) infection in two independent studies performed in Italian and Brazilian populations (Braida et al. 2004, Milanese et al. 2006).
In our study, we evaluated whether polymorphisms in the DEFB1 gene were associated with susceptibility to HPV infection and/or the development of genital tract lesions.
Three functional polymorphisms in the DEFB1 5’ untranslated region (UTR), namely g-52G>A (rs1799946), g-44C>G (rs1800972) and g-20G>A (rs11362) and two regulatory 3’UTR DEFB1 single-nucleotide polymorphisms (SNPs), namely c.*5G>A (rs1047031) and c.*87A>G (rs1800971), located at potential microRNA-binding sites were analysed in Brazilian gynaecological patients.
SUBJECTS, MATERIALS AND METHODS
We enrolled 356 women at the Oswaldo Cruz University Hospital and Clinical Hospital in the state of Pernambuco, northeastern Brazil. The study included 154 HPV-infected women (mean age, 36.3 years; range, 16-70 years): 40 women with normal cytology (no lesions), 48 women with cervical intraepithelial neoplasia (CIN) grade I, 28 women with CIN II, 25 women with CIN III and 13 women with cervical cancer.
The control group consisted of 202 women (mean age, 36.5 years; range, 18-65 years) with normal cytology and HPV-negative status [healthy controls (HCs)]; no information was available concerning the possible exposure of the HCs to HPV. All of the women were from the same geographical area (Recife metropolitan region), were HIV-negative and were not being treated with immunosuppressive medication.
We evaluated the genomic ancestries of the patients and controls following the methodology of Kosoy et al. (2009), with an in-house modification consisting of the genotyping of 12 genetic ancestry markers (Coelho et al. 2014, unpublished observations). The cases and controls presented the following similar distributions of genomic contribution: approximately 60% contribution from European, 23% from African and 17% from Amerindian ancestral populations.
DNA isolation and HPV typing - Genomic DNA was extracted from cervical swabs using the DNeasy Blood and Tissue Kit (Qiagen Inc, USA) in accordance with the manufacturer’s manual.
HPV typing was performed by polymerase chain reaction (PCR)-based amplification of the viral L1 gene fragment using the MY09/11 degenerate primers (Manos et al. 1989, Karlsen et al. 1996).
DEFB1 genotyping - The three DEFB1 polymorphisms in the 5’UTR and the two in the 3’UTR were genotyped using Taqman (Life Technologies, USA) allele-specific fluorescent probes C__11636795_20 (g-52G>A), C__11636794_10 (g-44C>G), C__11636793_20 (g-20G>A), C___8845558_10 (c.*5G>A) and C___8845559_10 (c.*87A>G) on the ABI 7500 SDS real-time PCR platform (Life Technologies). The genotyping results were checked by direct sequencing of 50 randomly chosen amplicons and a 100% concordance rate was achieved.
Statistical analysis - The allele and genotype frequencies of the DEFB1 polymorphisms were calculated by direct gene counting; the haplotype frequencies and linkage disequilibrium (LD) were computed using Arlequin software v.3.5 (Excoffier & Lischer 2010) and Haploview (Barrett et al. 2005).
Fisher’s exact test was used to test the differences between the proportions of the study groups (comparing the allele, the genotype in general, the dominant models and the haplotype frequencies using 2 x 2 and 3 x 2 contingency tables, as appropriate) and the odds ratios (ORs) and 95% confidence intervals (CIs) for the comparisons were calculated. Adjustments for multiple tests were performed using several correction methods (Supplementary data). R software v.3.0.0 (R Development Core Team 2013) was employed for the statistical analyses.
Research ethics - The study procedures were performed in accordance with the ethical standards of the Declaration of Helsinki. The Federal University of Pernambuco (Recife, Brazil) and the University of Pernambuco Research Ethical committees approved this study (protocol CAAE 03606212.7.0000.5208, HUOC/PROCAPE 64/2010). Written informed consent was obtained from all of the patients and controls.
RESULTS
The five DEFB1 gene polymorphisms were in Hardy-Weinberg equilibrium (HWE) in the group of HPV patients and in the subgroups stratified according to the cervical lesions (HPV-positive patients without lesions, HPV-positive patients with CIN I, CIN II or CIN III lesions and HPV-positive patients with cervical cancer); among the HCs, the rs1800972 (g-44C>G) and rs1800971 (c.*87A>G) polymorphisms were in HWE, whereas the others were not (Table).
TABLE. DEFB1 polymorphisms allele, genotype and haplotype counts (and frequencies) in healthy controls (HC) and human papillomavirus (HPV) patients.
| DEFB1 | HC | HPV | HPV vs. HCa |
|---|---|---|---|
|
| |||
| (n = 202) n (%) | (n = 154) n (%) | ||
| g-52G>A rs1799946 | |||
| G | 223 (0.55) | 160 (0.52) | ref |
| A | 181 (0.45) | 148 (0.48) | p = 0.40; CI = 0.83-1.55; OR = 1.13 |
| G/G | 69 (0.34) | 38 (0.25) | ref |
| G/A | 85 (0.42) | 84 (0.54) | p = 0.02; CI = 1.06-3.05; OR = 1.79 |
| A/A | 48 (0.24) | 32 (0.21) | p = 0.54; CI = 0.64-2.29; OR = 1.21 |
| G/A + A/A | 133 (0.66) | 116 (0.75) | p = 0.06; CI = 0.97-2.61; OR = 1.58 |
| HWE | χ2 = 4.40; p = 0.03 | χ2 = 1.32; p = 0.25 | - |
| g-44C>G rs1800972 | |||
| C | 334 (0.83) | 254 (0.82) | ref |
| G | 70 (0.17) | 54 (0.18) | p = 1.00; CI = 0.67-1.53; OR = 1.01 |
| C/C | 140 (0.69) | 106 (0.69) | ref |
| C/G | 54 (0.27) | 42 (0.27) | p = 1.00; CI = 0.62-1.70; OR = 1.03 |
| G/G | 8 (0.04) | 6 (0.04) | p = 1.00; CI = 0.27-3.37; OR = 0.99 |
| C/G + G/G | 62 (0.31) | 48 (0.31) | p = 1.00; CI = 0.63-1.65; OR = 1.02 |
| HWE | χ2 = 0.09; p = 0.34 | χ2 = 0.50; p = 0.48 | - |
| g-20G>A rs11362 | |||
| G | 253 (0.63) | 201 (0.65) | ref |
| A | 151 (0.37) | 107 (0.35) | p = 0.48; CI = 0.65-1.23; OR = 0.89 |
| G/G | 86 (0.43) | 64 (0.42) | ref |
| G/A | 81 (0.40) | 73 (0.47) | p = 0.42; CI = 0.75-1.95; OR = 1.21 |
| A/A | 35 (0.17) | 17 (0.11) | p = 0.25; CI = 0.31-1.32; OR = 0.65 |
| G/A + A/A | 116 (0.57) | 90 (0.58) | p = 0.91; CI = 0.67-1.63; OR = 1.04 |
| HWE | χ2 = 4.15; p = 0.04 | χ2 = 0.32; p = 0.57 | - |
| c.*5G>A rs1047031 | |||
| G | 346 (0.86) | 286 (0.93) | ref |
| A | 58 (0.14) | 22 (0.07) | p = 0.003; CI = 0.26-0.78; OR = 0.46 |
| G/G | 154 (0.76) | 134 (0.87) | ref |
| G/A | 38 (0.19) | 18 (0.12) | p = 0.05; CI = 0.28-1.03; OR = 0.54 |
| A/A | 10 (0.05) | 2 (0.01) | p = 0.07; CI = 0.02-1.11; OR = 0.23 |
| G/A + A/A | 48 (0.24) | 20 (0.13) | p = 0.01; CI = 0.26-0.87; OR = 0.48 |
| HWE | χ2 = 11.15; p = 0.001 | χ2 = 2.18; p = 0.14 | - |
| c.*87A>G rs1800971 | |||
| A | 363 (0.90) | 276 (0.90) | ref |
| G | 41 (0.10) | 32 (0.10) | p = 1.00; CI = 0.61-1.72; OR = 1.03 |
| A/A | 163 (0.81) | 124 (0.81) | ref |
| A/G | 37 (0.18) | 28 (0.18) | p = 1.00; CI = 0.55-1.77; OR = 0.99 |
| G/G | 2 (0.01) | 2 (0.01) | p = 1.00; CI = 0.09-18.35; OR = 1.31 |
| A/G + G/G | 39 (0.19) | 30 (0.19) | p = 1.00; CI = 0.57-1.77; OR = 1.01 |
| HWE | χ2 = 0.003; p = 0.95 | χ2 = 0.08; p = 0.77 | - |
| Haplotypes | |||
| ACGGA | 132 (0.33) | 113 (0.37) | ref |
| GCAGA | 90 (0.22) | 84 (0.27) | p = 0.69; CI = 0.72-1.64; OR = 1.09 |
| GGGGA | 68 (0.17) | 53 (0.17) | p = 0.74; CI = 0.57-1.44; OR = 0.91 |
| GCAAA | 55 (0.13) | 21 (0.07) | p = 0.005; CI = 0.24-0.80; OR = 0.45 |
| ACGGG | 39 (0.10) | 32 (0.10) | p = 0.89; CI = 0.54-1.68; OR = 0.96 |
| Others | 20 (0.05) | 5 (0.02) | p = 0.02; CI = 0.08-0.84; OR = 0.29 |
: uncorrected p-values; CI: confidence interval; HWE: Hardy Weinberg equilibrium; OR: odds ratio.
Initially, the five frequency distributions of the SNPs in the HPV patients and the HCs were compared.
The 5’UTR rs1799946 (g-52G>A) G/G genotype was more frequent among the healthy subjects than in the HPV patients and was thus associated with protection against HPV infection when compared to the G/A genotype (uncorrected p = 0.02; CI = 0.33-0.94; OR = 0.56). The allele distribution was similar between the two groups.
The 3’UTR rs1047031 (c.*5G>A) A allele was more frequently observed in the HCs than in the HPV patients and was associated with the absence of HPV infection (uncorrected p = 0.003; CI = 0.26-0.78; OR = 0.46). The genotype distributions presented a similar trend, with the A/A and G/A genotypes being more frequent among the HCs and the G/G genotype being more represented among the patients, according to a dominant genetic model for the A allele (A/A plus A/G vs. G/G; uncorrected p = 0.01; OR = 0.48; CI = 0.26-0.87). The c.*5G>A association remained statistically significant (corrected p < 0.05) after correcting for the multiple tests using several correction methods (Supplementary data).
The 5’UTR g-44C>G, g-20G>A and 3’UTR rs1800971 alleles and genotype distributions showed no significant differences between the patients and controls.
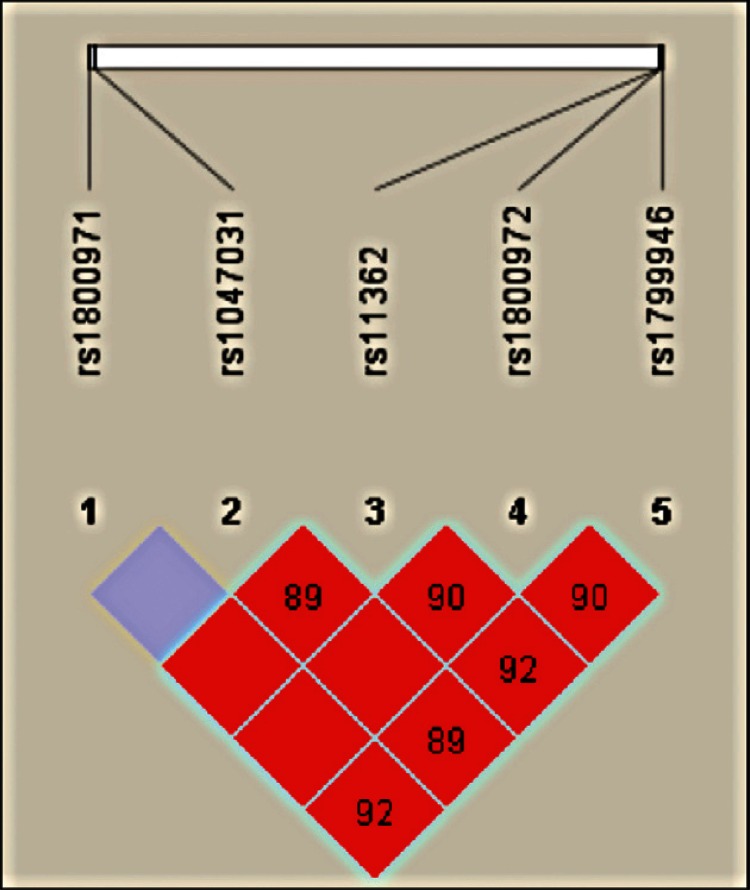
The five polymorphisms were in LD (p < 0.01, D’ > 0.91) and combined to form five major haplotypes (frequency > 0.01) and other minor haplotypes (frequency ≤ 0.01) (Figure, Table).
Haploview output of linkage disequilibrium (LD) across DEFB1 gene in Brazilian women. The figure illustrates on top the single-nucleotide polymorphisms (SNPs) that were genotyped in this study. On the bottom, each square represents a pair-wise LD relationship between two SNPs (with D’ values depicted within the box).
The GCAAA haplotype was more frequent in the HCs than in the HPV patient group and was associated with HPV-negative status when compared to the ACGGA haplotype (p = 0.005; CI = 0.24-0.80; OR = 0.45). These haplotypes differed for the g-52G>A, g-20G>A and c.*5G>A polymorphisms (Table).
No differences were observed for the DEFB1 allele, genotype and haplotype frequencies between the patients and controls when the patients were stratified according to cervical lesion grade (data not shown).
DISCUSSION
The possible involvement of hBD-1 in HPV-related diseases has been described. Elevated expression levels of this defensin (with hBD-2 and 3) have been reported in papillomavirus-induced lesions (Chong et al. 2006) and, more recently, in condylomata acuminata (Erhart et al. 2011).
Hubert et al. (2007) observed diminished expression levels of hBD-2 in high-grade squamous intraepithelial lesions and in squamous cell carcinomas compared with that in normal keratinocytes. The authors demonstrated that defensins are able to recruit dendritic cells (DCs) in organotypic cultures of HPV-transformed keratinocytes maintained in vitro or grafted in vivo, suggesting that these molecules might restore some immune functions that are altered during cervical carcinogenesis. In the Hubert et al. (2007) study, DEFB1 gene expression and the capacities of hBD-1 to recruit DCs and to restore the immune functions in HPV-infected cells were not evaluated.
In our study, we demonstrated an association between two DEFB1 polymorphisms, g-52G>A and c.*5G>A and one haplotype, GCAAA, with susceptibility to HPV infection in women from Northeast Brazil.
The g-52G>A G/G genotype, c.5*G>A A allele and GCAAA haplotype (presenting the g-52G>A G and c.5*G>A A alleles) were associated with the absence of HPV infection; they were observed in the HCs more than in the HPV patients. The g-52G>A and c.*5G>A polymorphisms, as well as the g-20G>A polymorphism, were not in HWE among the HCs. The deviation from HWE was not due to genotyping errors because the genotyping was performed using Taqman assays and the quality of the method was confirmed by direct sequencing of 50 randomly chosen amplicons, with a 100% rate of concordance being achieved. If genotyping errors occurred for a specific SNP, we would expect to find such errors in all of the groups, not only in the controls. Additionally, inbreeding, non-random mating, ethnicity, different genetic backgrounds and genetic drifting could be reasonably excluded in our case. Considering the reduced sample size and the small deviation from HWE only in the controls, we could assume that these deviations from HWE were random.
The associations reported in our study were quite weak and the statistical significance of the differences was lost for the g-52G>A SNP and the GCAAA haplotype after correcting for multiple testing. These differences could be indicative of the involvement of the DEFB1 gene in HPV susceptibility, considering that applying the standard methods of correction for multiple testing might be overly stringent and might fail to recognise single small effects, such as those expected by a single gene in a multifactorial complex disease, such as HPV infection, in which several factors (i.e., genetic and environmental) are involved.
We are confident of our results because, regardless of the relatively low numbers of individuals analysed and the deviation from the HWE in the controls, the c.5*G>A SNP was significantly associated with HPV infection after applying several methods of correction.
To the best of our knowledge, no previous studies have investigated the possible association between DEFB1 polymorphisms and HPV infection. Casalicchio et al. (2014) recently reported an association between the DEFB1 g-52G>A A/A genotype and protection against the development of atypical squamous cells of undetermined significance lesions in Italian gynaecological patients.
In our study, the DEFB1 g-52G>A polymorphism was not associated with the development or grade of cervical lesions/cancer, nor were any of the other investigated SNPs; however, the DEFB1 g-52G>A polymorphism was associated with the absence of HPV infection.
The lack of association between DEFB1 polymorphisms and the grade of cervical lesions/cancer that we reported could be a result of the small number of subjects analysed for each category and further studies in larger groups are needed to exclude the possible role of DEFB1 in HPV-related disease progression.
Our results suggest the involvement of DEFB1 polymorphisms, in particular that of the c.5*G>A SNP, in the susceptibility of women from Northeast Brazil to HPV infection.
The functional effect of the c.5*A>G polymorphisms has not been determined and in our study, the unavailability of other biological materials, such as vaginal washes, did not allow us to perform this analysis. Additionally, we are aware of the following limitations of our study: first, the small number of subjects analysed and second, the inability to verify whether the control subjects had been exposed to HPV.
Although our findings are preliminary, they allow us to hypothesise a role for hBD-1 in the first-line defence of the genital mucosa because hBD-1 could inhibit or at least hamper virus entrance into the first epithelial layers of the mucosa. If the virus escapes the innate immune control exerted by hBD-1, it is then able to colonise the epithelium and hBD-1 is no longer able to control viral persistence, leading to lesion development and progression to cervical cancer. Because the functional effects of the polymorphisms reported in our study are not completely understood, our hypothesis requires confirmation in future studies.
The relationships between the innate immunity peptide hBD-1 and HPV should be more thoroughly investigated in terms of susceptibility to or protection against infection, considering the possible role of hBD-1 in the persistence of viral infections leading to lesions and cervical cancer.
Supplementary Data
TABLE. p-values of DEFB1 polymorphisms association with human papillomavirus according to different correction methods.
| DEFB1 SNPs | UNADJ | BONF | HOLM | SIDAK_SS | SIDAK_SD | FDR_BH | FDR_BY |
|---|---|---|---|---|---|---|---|
| Alleles | |||||||
| g-52G>A rs1799946 | 0.405 | 1 | 1 | 0.925 | 0.874 | 0.800 | 1 |
| g-44C>G rs1800972 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| g-20G>A rs11362 | 0.480 | 1 | 1 | 0.962 | 0.874 | 0.800 | 1 |
| c.*5G>A rs1047031 | 0.003a | 0.013a | 0.013a | 0.013a | 0.013a | 0.013a | 0.030a |
| c.*87A>G rs1800971 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Genotypes - dominant model | |||||||
| g-52G>A rs1799946 | 0.389 | 1 | 1 | 0.915 | 0.860 | 0.781 | 1 |
| g-44C>G rs1800972 | 0.942 | 1 | 1 | 1 | 0.993 | 0.943 | 1 |
| g-20G>A rs11362 | 0.468 | 1 | 1 | 0.958 | 0.860 | 0.781 | 1 |
| c.*5G>A rs1047031 | 0.002a | 0.013a | 0.012a | 0.012a | 0.012a | 0.013a | 0.029a |
| c.*87A>G rs1800971 | 0.916 | 1 | 1 | 1 | 0.993 | 0.943 | 1 |
: statistically significant p-values (< 0.05); BONF: Bonferroni single-step adjusted p-values; FDR_BH: adjusted p-values for Benjamini and Hochberg step-up false discovery rate controlling procedure; FDR_BY: adjusted p-values for Benjamini and Yekutieli step-up false discovery rate controlling procedure; HOLM: Holm step-down adjusted p-values; SIDAK_SD: Sidak step-down adjusted p-values; SIDAK_SS: Sidak single-step adjusted p-values; SNPs: single-nucleotide polymorphisms; UNADJ: unadjusted p-value.
Footnotes
Financial support: FACEPE (Brazil), APQ-1530-2.02/12, the Italian Ministry of Health (RC06/11) BSC is a recipient of a postdoctoral fellowship from CNPq (Brazil) and LZ is a recipient of a fellowship from IRCCS Burlo Garofolo, Trieste, Italy.
REFERENCES
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Braida L, Boniotto M, Pontillo A, Tovo PA, Amoroso A, Crovella S. A single-nucleotide polymorphism in the human beta-defensin 1 gene is associated with HIV-1 infection in Italian children. AIDS. 2004;18:1598–1600. doi: 10.1097/01.aids.0000131363.82951.fb. [DOI] [PubMed] [Google Scholar]
- Buck CB, Day PM, Thompson CD, Lubkowski J, Lu W, Lowy DR, Schiller JT. Human alpha-defensins block papillomavirus infection. Proc Natl Acad Sci USA. 2006;103:1516–1521. doi: 10.1073/pnas.0508033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard RS, Gibson W, Bose SK, Belgrave KJ, Eaddy AC, Wright CJ, Hazen-Martin DJ, Lage JM, Keane TE, Ganz TA, Donald CD. Functional analysis of the host defense peptide human beta defensin-1: new insight into its potential role in cancer. Mol Immunol. 2008;45:839–848. doi: 10.1016/j.molimm.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalicchio G, Freato N, Maestri I, Comar M, Crovella S, Segat L. Beta defensin-1 gene polymorphisms and susceptibility to atypical squamous cells of undetermined significance lesions in Italian gynecological patients. 10.1002/jmv.23878.J Med Virol. 2014 doi: 10.1002/jmv.23878. [DOI] [PubMed] [Google Scholar]
- Chong KT, Xiang L, Wang X, Jun EL, Xi LF, Schweinfurth JM. High level expression of human epithelial beta-defensins (hBD-1, 2 and 3) in papillomavirus induced lesions. 75Virol J. 2006;3 doi: 10.1186/1743-422X-3-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho AVC, Moura RR, Addobbati CJC, Guimarães RL, Sandrin-Garcia P, Crovella S, Brandão LAC. A rapid screening of ancestry for genetic association studies in an admixed population from Pernambuco, Brazil. Genet Mol Res. 2014 doi: 10.4238/2015.March.31.18. in press. [DOI] [PubMed] [Google Scholar]
- Donald CD, Sun CQ, Lim SD, Macoska J, Cohen C, Amin MB, Young AN, Ganz TA, Marshall FF, Petros JA. Cancer-specific loss of beta-defensin 1 in renal and prostatic carcinomas. Lab Invest. 2003;83:501–505. doi: 10.1097/01.lab.0000063929.61760.f6. [DOI] [PubMed] [Google Scholar]
- Erhart W, Alkasi Ö, Brunke G, Wegener F, Maass N, Arnold N, Arlt A, Meinhold-Heerlein I. Induction of human β-defensins and psoriasin in vulvovaginal human papillomavirus-associated lesions. J Infect Dis. 2011;204:391–399. doi: 10.1093/infdis/jir079. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. Arlequin suite version 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Hubert P, Herman L, Maillard C, Caberg JH, Nikkels A, Pierard G, Foidart JM, Noel A, Boniver J, Delvenne P. Defensins induce the recruitment of dendritic cells in cervical human papillomavirus-associated (pre)neoplastic lesions formed in vitro and transplanted in vivo. FASEB J. 2007;21:2765–2775. doi: 10.1096/fj.06-7646com. [DOI] [PubMed] [Google Scholar]
- Karlsen F, Kalantari M, Jenkins A, Pettersen E, Kristensen G, Holm R, Johansson B, Hagmar B. Use of multiple PCR primer sets for optimal detection of human papillomavirus. J Clinical Microbiol. 1996;34:2095–2100. doi: 10.1128/jcm.34.9.2095-2100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotman ME, Chang TL. Defensins in innate antiviral immunity. Nat Rev Immunol. 2006;6:447–456. doi: 10.1038/nri1860. [DOI] [PubMed] [Google Scholar]
- Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, Kittles R, Alarcon-Riquelme ME, Gregersen PK, Belmont JW, La Vega FM, Seldin MF. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manos MM, Ting Y, Wright DK, Lewis AJ, Broker TR, Wolinsky SM. The use of polymerase chain reaction amplification for the detection of genital human papillomavirus. Cancer Cells. 1989;7:209–214. [Google Scholar]
- Milanese M, Segat L, Pontillo A, Arraes LC, Lima JL, Filho, Crovella S. DEFB1 gene polymorphisms and increased risk of HIV-1 infection in Brazilian children. AIDS. 2006;1:1673–1675. doi: 10.1097/01.aids.0000238417.05819.40. [DOI] [PubMed] [Google Scholar]
- R Development Core Team R: a language and environment for statistical computing. 2013 r-project.org Available from.
- Schröder JM, Harder J. Antimicrobial skin peptides and proteins. Cell Mol Life Sci. 2006;63:469–486. doi: 10.1007/s00018-005-5364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- Sun CQ, Arnold R, Fernandez-Golarz C, Parrish AB, Almekinder T, He J, Ho SM, Svoboda P, Pohl J, Marshall FF, Petros JA. Human beta-defensin-1, a potential chromosome 8p tumor suppressor: control of transcription and induction of apoptosis in renal cell carcinoma. Cancer Res. 2006;66:8542–8549. doi: 10.1158/0008-5472.CAN-06-0294. [DOI] [PubMed] [Google Scholar]