Abstract
Streptococcus pyogenes is responsible for a variety of infectious diseases and immunological complications. In this study, 91 isolates of S. pyogenes recovered from oropharynx secretions were submitted to antimicrobial susceptibility testing, emm typing and pulsed-field gel electrophoresis (PFGE) analysis. All isolates were susceptible to ceftriaxone, levofloxacin, penicillin G and vancomycin. Resistance to erythromycin and clindamycin was 15.4%, which is higher than previous reports from this area, while 20.9% of the isolates were not susceptible to tetracycline. The macrolide resistance phenotypes were cMLSB (10) and iMLSB (4). The ermB gene was predominant, followed by the ermA gene. Thirty-two emm types and subtypes were found, but five (emm1, emm4, emm12, emm22, emm81) were detected in 48% of the isolates. Three new emm subtypes were identified (emm1.74, emm58.14, emm76.7). There was a strong association between emm type and PFGE clustering. A variety of PFGE profiles as well as emm types were found among tetracycline and erythromycin-resistant isolates, demonstrating that antimicrobial resistant strains do not result from the expansion of one or a few clones. This study provides epidemiological data that contribute to the development of suitable strategies for the prevention and treatment of such infections in a poorly studied area.
Keywords: Streptococcus pyogenes, antimicrobial resistance, epidemiological typing, genetic diversity
Streptococcus pyogenes [Group A Streptococcus (GAS)] is one of the most clinically relevant species of Streptococcus. It has been associated with human infections ranging from mild sore throat and impetigo to invasive, life-threatening diseases, such as necrotising fasciitis and toxic shock syndrome. Moreover, immunological complications such as acute rheumatic fever, rheumatic heart disease and post-streptococcal acute glomerulonephritis are significant streptococcal disease burdens, especially in the developing world (Steer et al. 2009, Dale et al. 2013).
Several efforts have been made over the past decades to understand the epidemiology of GAS infections. Contributing to these efforts, molecular techniques such pulsed-field gel electrophoresis (PFGE) and multilocus sequencing typing have been extensively applied in the evaluation of the genetic relationships among isolates (Dicuonzo et al. 2001, Ardanuy et al. 2010). Another approach is emm typing, a sequence-based typing of the M protein (Beall et al. 1996). M protein is a cell surface protein that inhibits phagocytosis and is considered the most important epidemiological marker of GAS infections. This approach has the potential to type GAS isolates from different areas of the world. However, most available data are from high-income countries, which has lead to several limitations, such as the lack of knowledge of emm type prevalence in developing countries where severe streptococcal infections and immunological complications occur frequently (Steer et al. 2009).
The development of an efficient vaccine for GAS infections is another important issue. Several candidates have been evaluated, including the variable N-terminal and the conserved C-terminal epitopes of M protein (McNeil et al. 2005, Dale et al. 2011, Postol et al. 2013). Knowledge of the global distribution of emm types is required to formulate an N-terminal M protein-based vaccine that covers the prevalent M types in different geographical areas (Steer et al. 2009).
Treatment of GAS infections is based on beta-lactams because this species remains susceptible to penicillin G; macrolides, lincosamides and fluoroquinolones are recommended for allergic individuals. However, GAS resistance to these later antimicrobials has been described worldwide (Smeesters et al. 2009, Friães et al. 2012). While fluoroquinolone resistance in GAS is due to point mutations in the gyr and par genes, macrolide resistance is due to active efflux (M phenotype) or modification of the target site (iMLSB and cMLSB phenotypes) and is mediated by the genetic determinants mefA/E, ermA and ermB (D‘Oliveira et al. 2003, Montes et al. 2010). It is worth noting that in some countries where macrolide resistance rates used to be high, such as Spain and China, unexpected reductions in the resistance rates have been recently observed (Huang et al. 2014, Montes et al. 2014).
Antimicrobial resistance rates and emm type distribution in a given population are essential to the development of suitable strategies for the prevention and treatment of GAS infections. The aim of this study was to investigate the antimicrobial susceptibility and emm type and to evaluate the genetic diversity of circulating GAS isolates in the metropolitan area of Rio de Janeiro, Brazil.
SUBJECTS, MATERIALS AND METHODS
Isolates - Ninety-one GAS isolates recovered from oropharynx secretions were included in this study. Clinical specimens were processed during routine diagnoses by one clinical laboratory in Rio de Janeiro from January 2008-July 2012. The subjects’ ages varied from two-56 years, but 46% of the isolates were recovered from children two-11 years of age and 51.6% were from women. In our laboratory, these isolates were cultured on blood agar plates (Difco Laboratories, USA) and submitted to conventional tests (PYR test, bacitracin susceptibility and streptococcal serogrouping) to confirm species identification.
Antimicrobial susceptibility testing - All isolates were submitted to susceptibility tests to ceftriaxone (30 µg), clindamycin (2 µg), erythromycin (15 µg), levofloxacin (5 µg), penicillin (10 U), tetracycline (30 µg) and vancomycin (30 µg) (CECON, Brazil) using the disk diffusion method on Mueller-Hinton blood agar (Difco) according to CLSI guidelines (2013). Macrolide resistance phenotypes were determined by the double disk test using erythromycin (15 µg) and clindamycin (2 µg) disks placed 12 mm apart (CLSI 2013). The erythromycin minimum inhibitory concentration (MIC) was determined in all resistant and intermediate isolates by the agar dilution method (CLSI 2009).
Investigation of erythromycin resistance-associated genes - DNA preparation was performed as previously described (Dmitriev et al. 2002) with modifications. Briefly, suspensions with turbidity adjusted to McFarland Standard 3.0 were prepared in 300 µL of 10 mM Tris-EDTA buffer and boiled for 5 min. The presence of ermA, ermB and mefA/E genes was investigated in erythromycin-resistant isolates using specific polymerase chain reaction (PCR) protocols (Sutcliffe et al. 1996, Perez-Trallero et al. 2007). Cycling was carried in a GeneAmp 9700 Thermocycler (Applied Biosystems, USA). PCR products were resolved on 1% agarose gels.
Determination of emm types - emm types were determined by a sequence-based protocol (cdc.gov/ncidod/biotech/strep/protocol_emm-type.htm) using BigDye Terminator Cycle Sequencing Kit (Applied Biosystems). Sequencing was performed using a 3130 Genetic Analyzer (Applied Biosystems). Sequences were edited using Bioedit software v.7.0 and compared with reference sequences using the BLAST algorithm (blast.ncbi.nlm.nih.gov/Blast.cgi). Sequences that did not match with 100% similarity to any sequence deposited in GenBank were submitted to the Centers for Disease Control and Disease (CDC) emm sequence database (cdc.gov/ncidod/biotech/strep/strepblast.htm) for assignment to new emm subtypes.
Analysis of DNA restriction patterns by PFGE - All 44 isolates belonging to the five prevalent emm types (emm1, emm4, emm12, emm22 and emm81) were ana- lysed by PFGE after the DNA was digested with SmaI according to a previous protocol (Teixeira et al. 1997) with modifications as described below. Briefly, bacteria were grown on blood agar plates. A 300-µL aliquot of bacterial suspension in PIV buffer was mixed with an equal volume of low melting point agarose (Promega, USA) and distributed into plug moulds. Plugs were incubated in 2 mL of lysis solution containing 5 mg/mL lysozyme. SmaI digestion was performed according to the manufacturer’s recommendations (Invitrogen, USA). DNA fragments were separated by the CHEF-DRIII system (Bio-Rad Laboratories, USA). The dice coefficient was calculated by visual analysis and dendrograms based on Unweighted Pair Group Method with Arithmetic Mean were constructed using genomes.urv.cat/UPGMA.
Statistical analyses - The discriminatory power of emm typing, regarding the overall population as well as erythromycin and tetracycline resistance, was measured using Simpson’s index of diversity (SID) by calculating the 95% confidence intervals (CI) (Hunter & Gaston 1988, Grundmann et al. 2001). All calculations were performed using the Comparing Partitions Tool available from comparingpartitions.info.
RESULTS
Conventional tests identified all isolates as S. pyogenes. By the disk diffusion method, all isolates were susceptible to ceftriaxone, levofloxacin, penicillin G and vancomycin. Tetracycline-resistant and intermediate isolates comprised 18.7% and 2.2% of isolates, respectively. Clindamycin resistance was observed in 15.4% of isolates, while erythromycin-resistant and intermediate isolates comprised 14.3% and 1.1% of isolates, respectively. The erythromycin MIC varied from 8-256 µg/mL and therefore, the resistance rate was 15.4%. Ten and four isolates showed cMLSB and iMLSB resistance phenotypes, respectively. The genetic determinant ermB was predominant and was detected in 78.6% of the erythromycin-resistant isolates, alone or in association with ermA (64.3%). Neither the M phenotype nor the mefA/E gene was observed in this study. Erythromycin MIC values, the distribution of macrolide resistance phenotypes and genotypes and emm types of erythromycin-resistant isolates are shown in Table.
TABLE. Phenotypic and genotypic characteristics of macrolide resistant isolates.
| Isolate number | Year of recovery | emm type | MIC (µg/mL) | Phenotype | Macrolide resistance
genes |
||
|---|---|---|---|---|---|---|---|
| ermA | ermB | mefA/E | |||||
| 186 | 2008 | 68.1 | > 256 | cMLSB | - | + | - |
| 274 | 2008 | 58 | > 256 | iMLSB | + | - | - |
| 302 | 2008 | 58 | > 256 | iMLSB | + | - | - |
| 402 | 2009 | 1 | > 256 | cMLSB | - | + | - |
| 418 | 2009 | 58.14 | 8 | iMLSB | + | - | - |
| 425a | 2009 | 3.3 | 16 | cMLSB | - | + | - |
| 536 | 2010 | 22 | > 256 | cMLSB | - | + | - |
| 637A | 2011 | 1 | > 256 | cMLSB | + | + | - |
| 638 | 2011 | 76.7 | > 256 | cMLSB | + | + | - |
| 710 | 2011 | 28 | > 256 | cMLSB | - | + | - |
| 749 | 2011 | 73 | > 256 | cMLSB | + | + | - |
| 750 | 2011 | 6 | > 256 | cMLSB | + | + | - |
| 780 | 2012 | 11 | > 256 | cMLSB | + | + | - |
| 798 | 2012 | 11 | > 256 | iMLSB | + | + | - |
: this isolate was intermediate by disk diffusion; cMLSB: constitutive MLSB phenotype; iMLSB: inducible MLSB phenotype; MIC: minimal inhibitory concentration (susceptible < 0.25 µg/mL, intermediate 0.5 µg/mL, resistant > 1 µg/mL); +: presence of the gene; -: absence of the gene.
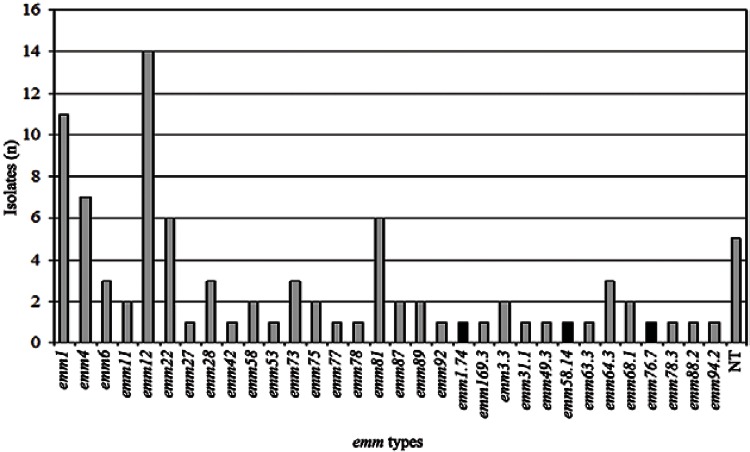
Thirty-two emm types or subtypes were identified among 86 of the 91 GAS isolates. Five isolates were non-typeable, even after three attempts. The most frequent types were emm1, emm4, emm12, emm22 and emm81, accounting for 48% of all isolates. Three new sequences were designated by the CDC Streptococcus Laboratory as new emm subtypes (emm1.74, emm58.14 and emm76.7) and deposited in the emm sequence database (cdc.gov/pub/infectious_diseases/biotech/tsemm) and in GenBank (accession KM364527-KM364529). These new subtypes differed from their parental types in 1-2% of the nucleotide sequence. The emm typing revealed a high level of diversity among the overall population (SID = 0.941; 95% CI, 0.917-0.965). The frequency of each emm type is shown in Fig. 1.
Fig. 1. : distribution of emm types and subtypes among isolates of Streptococcus pyogenes recovered from oropharynx secretion. NT: non-typeable; : new subtypes.
Fourteen different emm types or subtypes were found among 19 tetracycline non-susceptible isolates, while 11 distinct emm types or subtypes were found among 14 erythromycin and clindamycin-resistant isolates. Two new emm subtype isolates were resistant to these antimicrobials. The SID values calculated for erythromycin-resistant (0.967; 95% CI, 0.929-1.000) and tetracycline non-susceptible (0.959; 95% CI, 0.915-1.000) isolates were higher than those calculated for erythromycin (0.929; 95% CI, 0.898-0.960) and tetracycline (0.898; 95% CI, 0.855-0.940) susceptible isolates.
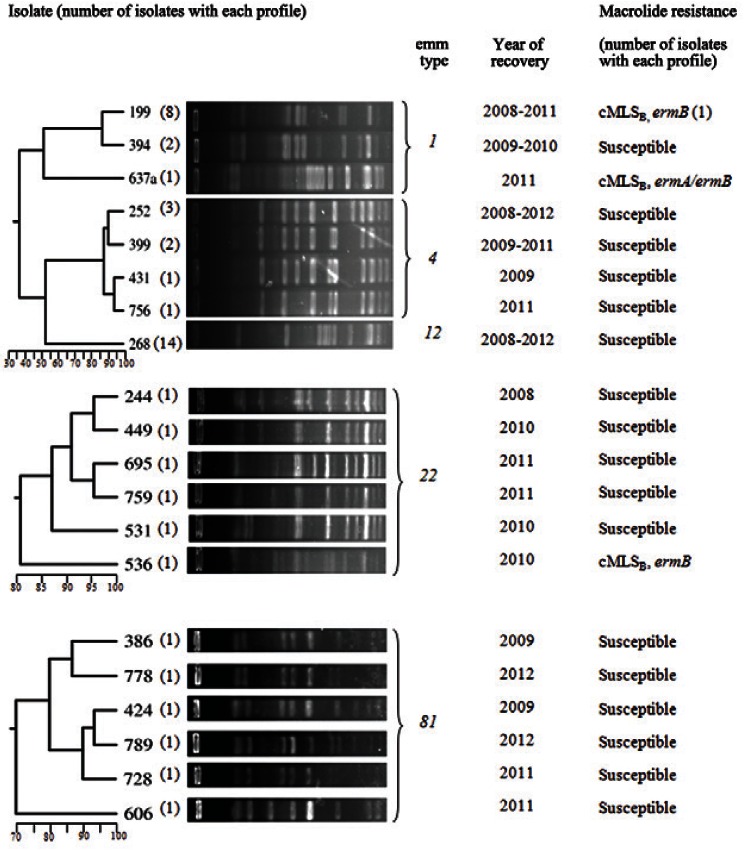
PFGE analysis of 44 isolates belonging to the five prevalent emm types generated 20 restriction profiles, which are shown in Fig. 2. One single PFGE profile was shared by all emm12 isolates. Three profiles, whose similarities varied from 50-85%, were observed among emm1 isolates. The most frequent profile was shared by eight isolates, including one resistant to erythromycin. Regarding emm4, four profiles were observed among seven isolates, ranging from 85-96% similarity. Among emm22 and emm81 isolates (6 isolates each), unique PFGE profiles were observed for each isolate, varying from 70-95% similarity.
Fig. 2. : pulsed-field gel electrophoresis profiles, dendrograms, year of isolation and macrolide resistance features of prevalent emm types isolates.
DISCUSSION
In this study, 91 GAS isolates recovered from oropharynx secretions of residents of Rio de Janeiro were submitted to susceptibility testing and typing methodologies. Despite the lack of data regarding the clinical conditions of the individuals, these results are relevant because both infected and colonised individuals can transmit this species to susceptible subjects.
Isolates were fully susceptible to beta-lactams and glycopeptides, as observed over decades of antibiotic usage. They were also susceptible to levofloxacin, despite previously detected resistance to this antimicrobial (Smeesters et al. 2009, Montes et al. 2010).
The tetracycline resistance rate was lower than that observed in a study conducted in Brazil one decade ago, in which the authors reported 43.1% resistance (D‘Oliveira et al. 2003). Instead, it was very similar to recent data from southern Brazil and Portugal (Torres et al. 2011, Friães et al. 2012). However, 73% resistance to this antimicrobial was reported in a recent study from India (Mathur et al. 2014). These findings may reflect that, while a consistent trend of decreasing tetracycline resistance has been recently observed in Brazil, circulating GAS isolates from other regions are highly resistant to this antimicrobial.
Regarding erythromycin and clindamycin resistance, the rates found here far exceeded those previously reported in the same geographical area (D‘Oliveira et al. 2003, Torres et al. 2011), but they are very similar to those described in some recent studies conducted in Europe (Friães et al. 2012, Syrogiannopoulos et al. 2013). While a trend of decreasing erythromycin resistance has been recently observed in Spain and Taiwan (Huang et al. 2014, Montes et al. 2014), the resistance rates in our study fluctuated over the years. Inducible and constitutive MLSB phenotypes, which are associated with ermA and/or ermB genes, were observed among isolates belonging to a variety of emm types, reflecting the polyclonal origin of such isolates. It is worth noting that the M phenotype and the mefA/E genotype, associated with emm12, were common in Brazil before the year 2000 (Torres et al. 2011), but they have not been detected since that time, either by those authors or in this study. In contrast, MLSB emerged after the 2000s and is linked to a variety of emm types, such as emm11, emm22, emm28 and emm73 (Torres et al. 2011). In contrast to this local replacement of the M phenotype with the cMLSB phenotype, the M phenotype has been prevalent over 12 years of survey (1998-2010) in Taiwan (Huang et al. 2014). However, the authors observed the replacement of emm12 with emm22 as the prevalent type associated with macrolide resistance, as well as a decreasing rate in macrolide resistance from 53.1% before 2000 to 10.7% from 2006-2010. These variations demonstrate how dynamic the bacterial population is worldwide and also highlight the need for changing therapeutic recommendations in geographical areas where MLSB phenotypes predominate.
A wide variety of emm types, including new alleles, were observed in our study, as demonstrated by the SID calculation. The diversity of emm types and the detection of new alleles from GAS isolates recovered from oropharynx secretions have been described in previous studies from Brazil (Teixeira et al. 2001, Tartof et al. 2010) and throughout the world (Shea et al. 2011). These findings reflect the genetic heterogeneity of such isolates and contribute to the expansion of the emm type database. The most frequent emm types found here are among the prevalent types in high-income countries, where the majority of studies have been performed (Steer et al. 2009, Shea et al. 2011). However, they differ significantly from those types observed in recent studies from India and the Pacific Region (Baroux et al. 2014, Mathur et al. 2014). Due to the limited amount of data from Latin America, our results contribute to a better understanding of emm type distribution in a poorly studied area. This issue is particularly important considering that M protein-based vaccines are under development (Dale et al. 2013). When comparing the two most promising 26 and 30-valent M vaccines, 49% and 64% of our isolates, respectively, have emm types included in these formulations. The potentially low impact of these vaccines can be explained by the fact that the M protein fragments for the 26-valent M vaccine were selected based on emm type distribution in North America (Steer et al. 2009).
There was strong agreement between emm type and PFGE clustering, as previously observed (Torres et al. 2011). No identical PFGE profile was observed among isolates belonging to distinct emm types, which could be related to the horizontal transfer of emm genes (Whatmore et al. 1994). Regarding erythromycin-resistant isolates, a great diversity of PFGE profiles and emm types were observed, illustrating that this characteristic is not due to the expansion of a specific clone. Moreover, both erythromycin-susceptible and erythromycin-resistant emm1 isolates shared a single PFGE profile. This finding suggests that the emm type, not erythromycin resistance, is more likely to be a determinant of clonality.
In conclusion, susceptibility testing and epidemiological typing techniques revealed an incidence of macrolide resistance not yet observed in this area and a great diversity of emm types, including new alleles, among GAS isolates circulating in Brazil. These data contribute to the improvement of prevention and treatment strategies of GAS infections.
ACKNOWLEDGEMENTS
To the Fleury Group, for strains donation, and to André V Barbosa, from the DNA Sequencing Platform, UFF.
Footnotes
Financial support: PROPPI/UFF, FAPERJ
REFERENCES
- Ardanuy C, Domenech A, Rolo D, Calatayud L, Tubau F, Ayats J, Martin R, Liñares J. Molecular characterization of macrolide- and multidrug-resistant Streptococcus pyogenes isolated from adult patients in Barcelona, Spain (1993-2008) J Antimicrob Chemother. 2010;65:634–643. doi: 10.1093/jac/dkq006. [DOI] [PubMed] [Google Scholar]
- Baroux N, D‘Ortenzio E, Amédéo N, Baker C, Alsuwayyid BA, Dupont-Rouzeyrol M, O‘Connor O, Steer A, Smeesters PR. The emm-cluster typing system for group A Streptococcus identifies epidemiologic similarities across the Pacific Region. Clin Infect Dis. 2014;59:e84–e92. doi: 10.1093/cid/ciu490. [DOI] [PubMed] [Google Scholar]
- Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–958. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI - Clinical Laboratory Standard Institute . Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, M07-A8. 8th ed. CLSI; Wayne: 2009. 65 [Google Scholar]
- CLSI - Clinical Laboratory Standard Institute . Performance standards for antimicrobial susceptibility testing, M100-S23. CLSI; Wayne: 2013. 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D‘Oliveira REC, Barros RR, Mendonça CRV, Teixeira LM, Castro ACD. Antimicrobial susceptibility and survey of macrolide resistance mechanisms among Streptococcus pyogenes isolated in Rio de Janeiro, Brazil. Microb Drug Resist. 2003;9:87–91. doi: 10.1089/107662903764736382. [DOI] [PubMed] [Google Scholar]
- Dale JB, Fischetti VA, Carapetis JR, Steer AC, Sow S, Kumar R, Mayosi BM, Rubin FA, Mulholland K, Hombach JM, Schödel F, Henao-Restrepo AM. Group A streptococcal vaccines: paving a path for accelerated development. Vaccine. 2013;31(Suppl. 2):B216–B222. doi: 10.1016/j.vaccine.2012.09.045. [DOI] [PubMed] [Google Scholar]
- Dale JB, Penfound TA, Chiang EY, Walton WJ. New 30-valent M protein-based vaccine evokes cross-opsonic antibodies against non-vaccine serotypes of group A streptococci. Vaccine. 2011;29:8175–8178. doi: 10.1016/j.vaccine.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicuonzo G, Gherardi G, Lorino G, Angeletti S, Cesaris M, Fiscarelli E, Bessen D, Beall B. Group A streptococcal genotypes from pediatric throat isolates in Rome, Italy. J Clin Microbiol. 2001;39:1687–1690. doi: 10.1128/JCM.39.5.1687-1690.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev A, Shakleina E, Tkáciková L, Mikula I, Totolian A. Genetic heterogeneity of the pathogenic potentials of human and bovine group B streptococci. Folia Microbiol (Praha) 2002;47:291–295. doi: 10.1007/BF02817655. [DOI] [PubMed] [Google Scholar]
- Friães A, Pinto FR, Silva-Costa C, Ramirez M, Melo-Cristino J, The Portuguese Group for the Study of Streptococcal Infections Group A streptococci clones associated with invasive infections and pharyngitis in Portugal present differences in emm types, superantigen gene content and antimicrobial resistance. 280BMC Microbiol. 2012;12 doi: 10.1186/1471-2180-12-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann H, Hori S, Tanner G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J Clin Microbiol. 2001;39:4190–4192. doi: 10.1128/JCM.39.11.4190-4192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Lai JF, Huang IW, Chen PC, Wang HY, Shlau IR, Cheng YW, Hsleh LY, Chang SC, Lauderdale TSY. Epidemiology and molecular characterization of macrolide-resistant Streptococcus pyogenes in Taiwan. J Clin Microbiol. 2014;52:508–516. doi: 10.1128/JCM.02383-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur P, Bhardwaj N, Mathur K, Behera B, Gupta G, Kapil A, Singh S, Misra MC. Clinical and molecular epidemiology of beta-hemolytic streptococcal infections in India. J Infect Dev Ctries. 2014;8:297–303. doi: 10.3855/jidc.3216. [DOI] [PubMed] [Google Scholar]
- McNeil SA, Halperin SA, Langley JM, Smith B, Warren A, Sharratt GP, Baxendale DM, Reddish MA, Hu MC, Stroop SD, Linden J, Fries LF, Vink PE, Dale JB. Safety and immunogenicity of 26-valent group A Streptococcus vaccine in healthy adult volunteers. Clin Infect Dis. 2005;41:1114–1122. doi: 10.1086/444458. [DOI] [PubMed] [Google Scholar]
- Montes M, Tamayo E, Mojica C, García-Arenzana JM, Esnal O, Perez-Trallero E. What causes decreased erythromycin resistance in Streptococcus pyogenes? Dynamics of four clones in a southern European region from 2005 to 2012. J Antimicrob Chemother. 2014;69:1474–1482. doi: 10.1093/jac/dku039. [DOI] [PubMed] [Google Scholar]
- Montes M, Tamayo E, Orden B, Larruskain J, Perez-Trallero E. Prevalence and clonal characterization of Streptococcus pyogenes clinical isolates with reduced fluoroquinolone susceptibility in Spain. Antimicrob Agents Chemother. 2010;54:93–97. doi: 10.1128/AAC.00780-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Trallero E, Montes M, Orden B, Tamayo E, García-Arenzana JM, Marimón JM. Phenotypic and genotypic characterization of Streptococcus pyogenes isolates displaying the MLSB phenotype of macrolide resistance in Spain, 1999 to 2005. Antimicrob Agents Chemother. 2007;51:1228–1233. doi: 10.1128/AAC.01054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postol E, Alencar R, Higa FT, Barros SF, Demarchi LMF, Kalil J, Guilherme L. StreptInCor: a candidate vaccine epitope against S. pyogenes infections induces protection in outbred mice. PLoS ONE. 2013;8: doi: 10.1371/journal.pone.0060969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea PR, Ewbank AL, Gonzalez-Lugo JH, Martagon-Rosado AJ, Martinez-Gutierrez JC, Rehman HA, Serrano-Gonzalez M, Fittipaldi N, Beres SB, Flores AR, Low DE, Willey BM, Musser JM. Group A Streptococcus emm gene types in pharyngeal isolates, Ontario, Canada, 2002-2010. Emerg Infect Dis. 2011;17:2010–2017. doi: 10.3201/eid1711.110159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeesters PR, Vergison A, Campos D, Jr, Melderen LV. Emerging fluoroquinolone-non-susceptible group A streptococci in two different paediatric populations. Int J Antimicrob Agents. 2009;34:44–49. doi: 10.1016/j.ijantimicag.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis. 2009;9:611–616. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrogiannopoulos GA, Grivea IN, Al-Lahham A, Panagiotou M, Tsantouli AG, Reinert ANMRR, van der Linden M. Seven-year surveillance of emm types of pediatric group A streptococcal pharyngitis isolates in Western Greece. PLoS ONE. 2013;8: doi: 10.1371/journal.pone.0071558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartof SY, Reis JN, Andrade AN, Ramos RT, Reis MG, Riley LW. Factors associated with Group A Streptococcus emm type diversification in a large urban setting in Brazil: a cross-sectional study. 327BMC Infect Dis. 2010;10 doi: 10.1186/1471-2334-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira LM, Barros RR, Castro ACD, Peralta JM, Carvalho MGS, Talkington DF, Vivoni AM, Facklam RR, Beall B. Genetic and phenotypic features of Streptococcus pyogenes strains isolated in Brazil that harbor new emm sequences. J Clin Microbiol. 2001;39:3290–3295. doi: 10.1128/JCM.39.9.3290-3295.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira LM, Carvalho MG, Merquior VL, Steirgerwalt AG, Brenner DJ, Facklam RR. Phenotypic and genotypic characterization of Vagococcus fluvialis, including strains isolated from human sources. J Clin Microbiol. 1997;35:2778–2781. doi: 10.1128/jcm.35.11.2778-2781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres RSLA, Torres RPA, Smeesters PR, Palmeiro JK, Messias-Reason IJ, Dalla-Costa LM. Group A Streptococcus antibiotic resistance in southern Brazil: a 17-year surveillance study. Microb Drug Resist. 2011;17:313–319. doi: 10.1089/mdr.2010.0162. [DOI] [PubMed] [Google Scholar]
- Whatmore AM, Kapur V, Sullivan DJ, Musser JM, Kehoe MA. Non-congruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol Microbiol. 1994;14:619–631. doi: 10.1111/j.1365-2958.1994.tb01301.x. [DOI] [PubMed] [Google Scholar]