Abstract
The C-terminal domain of RNA polymerase II (CTD) modulates the process of transcription through sequential phosphorylation/dephosphorylation of its heptide repeats through which it recruits various transcription regulators. Ssu72 is the first characterized cis-specific CTD phosphatase that dephosphorylates Ser5 with a requirement for the adjacent Pro6 in a cis conformation. The recent discovery of Thr4 phosphorylation in the CTD calls into question whether such a modification can interfere with Ssu72 binding via the elimination of a conserved intra-molecular hydrogen bond in the CTD that is potentially essential for recognition. To test if Thr4 phosphorylation will abolish Ser5 dephosphorylation by Ssu72, we determined the kinetic and structural properties of Drosophila Ssu72-symplekin in complex with the CTD peptide with consecutive phosphorylated Thr4 and Ser5. Our mass spectrometric and kinetic data established that Ssu72 doesn’t dephosphorylate Thr4, but the existence of phosphoryl-Thr4 next to Ser5 reduces the activity of Ssu72 towards the CTD peptide by four fold. To our surprise, even though the intra-molecular hydrogen bond is eliminated due to the phosphorylation of Thr4, the CTD adopts an almost identical conformation to be recognized by Ssu72 with Ser5 phosphorylated alone or both Thr4/Ser5 phosphorylated. Our results indicate that Thr4 phosphorylation will not abolish the essential Ssu72 activity, which is needed for cell survival. Instead, the phosphatase activity of Ssu72 is fine-tuned by Thr4 phosphorylation and eventually may lead to changes in transcription. Overall, we report the first case of structural and kinetic effects of phosphorylated Thr4 on CTD modifying enzymes. Our results support a model in which a combinatorial cascade of CTD modification can modulate transcription.
Keywords: Ssu72 phosphatase, phosphorylation of RNA polymerase II, transcription regulation, CTD code, post-translational modification, intra-molecular hydrogen bond, x-ray crystallography
Introduction
The conformational states of the C-terminal domain (CTD) of eukaryotic RNA polymerase II represent a critical regulatory check point for transcription (1, 2), (3–6). The CTD, found only in eukaryotes, consists of 26–52 tandem hepta-peptide repeats with the general consensus sequence, 1TyrSerProThrSerProSer7. The CTD can spatially and temporally recruit various regulatory and processing factors to the transcriptional machinery (7). The precise recruitment of transcription factors is accomplished through the various conformations of the CTD, which are dominated by post-translational covalent modifications or prolyl isomerization (1). The sequential phosphorylation/dephosphorylation on Ser2 and Ser5 is used to achieve temporal control of the transcription process (8). The prolyl isomerization regulation of the CTD, which adds yet another layer to the complexity of the “CTD code”, was identified with the discovery that yeast prolyl isomerase Ess1 (Pin1 in human) can greatly affect the transcription by its enzymatic activity against the CTD (9–11). Indeed, the CTD is enriched with phosphorylated Ser-Pro motifs which are recognized by the WW domain of Pin1 (12). The Pin1-CTD peptide structure elucidates that even though both Ser2-Pro3 and Ser5-Pro6 are potentially recognized by Pin1, the prolyl peptide between Ser5-Pro6 is the preferred site of isomerization (13). This prompted the speculation that Pin1 affects transcription by isomerizing the Ser5-Pro6 prolyl peptide bond which affects the CTD conformation for downstream factors (14).
The importance of prolyl isomerization on the CTD was established by the characterization of the first cis-specific phosphatase Ssu72 (14–16). Ssu72 was first identified as a genetic suppressor for sua7 mutation in yeast (42) and later found to be a subunit of yeast cleavage and polyadenylation factor (CPF) (17). Highly conserved from yeast to human, the reduction of the phosphatase activity of Ssu72 causes the accumulation of phosphoryl-Ser5 of the CTD in the cell (18). Ssu72 forms a complex with an adapter protein Pta1, (symplekin in higher eukaryotes) which in turn associates with other CPF complex proteins involved in transcription processing and termination (17), (19). Upon binding to Ssu72, Pro6 of hepta-peptide repeats of the CTD adopts an unusual cis conformation which comprises a minority species in nature and this unusual conformation is stabilized by an intra-molecular hydrogen bond between Thr4 hydroxyl side chain and backbone carbonyl oxygen of Pro6 (16). Changing the isomerization state of Pro6 will affect the activity of Ssu72 (14). Indeed, with Pin1 to guarantee the supply of cis-Pro, which only comprises 15% of the overall proline species, Ssu72 exhibits an apparent enhancement of three-fold in phosphatase activity against the CTD (14–16). This activation effect only functions for cis-specific enzymes since human Scp, a phosphatase recognizing the same Ser5-Pro motif but with Pro6 in the trans-form, is not affected much by the Pin1 isomerase activity (14). This further emphasizes that the unique recognition of cis-proline by Ssu72 is used as a tunable regulatory mechanism by Pin1 for transcription.
Recently, novel phosphorylation modifications in vivo have been reported for the CTD residues at sites other than Ser2 and Ser5, namely at Tyr1, Thr4 and Ser7 (20). It has also been established that multiple phosphorylation in the CTD repeats are likely very frequent in stages of transcription process (21). The phosphorylation of Ser5 is most prominent at the initiation stage of transcription, but it can still be detected along the whole transcription. This has been detected with the newly improved antibodies, consistent with the detection of Ssu72 as Ser5 phosphatase at the end of transcription (22, 23). Studies also show that Thr4 phosphorylation occurs in vivo in late stage transcription (24, 25), suggesting the co-occurrence of Thr4/Ser5 phosphorylation even though direct observance has not yet been possible due to the lack of antibodies that recognize multiple phosphorylated CTD. Thr4 residues in the CTD heptad repeats were scrutinized recently due to the newfound discovery that Thr4 is indispensable for cell viability in mammalian and chicken cells (24, 25). This is in contrast to observation in lower eukaryotes, the fission and budding yeast, that Thr4 mutation is well tolerated (26, 27). Studies in chicken DT40 cells have shown that Thr4Val mutation in the CTD altered histone transcription without causing a defect in general transcription (24), whereas in human cells Thr4Ala mutation displayed a global defect in RNA elongation, implicating the importance of the hydroxyl side-chain of Thr4 (25). In this context, the recognition mechanism of the CTD peptide with multiple phosphorylation sites and the effect of flanking phosphorylation of Thr4 on the activity of CTD modifying enzymes are particularly interesting questions.
To address these questions, we focused on the case of cis-specific phosphatase Ssu72 to investigate how the phosphorylation of flanking residues affects its activity. The structure of Ssu72 binding to the CTD highlights an intra-molecular hydrogen bond between Thr4 and the peptide backbone, stabilizing the unique cis-conformation of proline. We investigated how the phosphorylation of Thr4 would affect this recognition of the CTD by Ssu72 whose phosphatase activity is essential for the termination of transcription. In this study, through the quantitative measurement of Ssu72-symplekin activity against various CTD peptides, we show that additional phosphorylation on Thr4 can affect the activity of Ssu72-symplekin complex but does not abolish it. Using crystallography, we show the molecular recognition state when Ssu72 binds to a CTD peptide with consecutive phosphorylated Thr4 and Ser5. Our results provide strong evidence for a model that combinatorial CTD modifications modulate the enzymatic activity of Ssu72 and possibly regulate transcription.
Methods
Materials
CTD peptides were custom-ordered from Anaspec and CPC BioSciences. Drosophila symplekin cDNA was purchased from Bloomington Drosophila Stock Center at Indiana University. Malachite green reagent (BIOMOL) was purchased from Enzo Life Sciences. All other reagents and chemicals were purchased from Sigma-Aldrich unless specified otherwise.
Protein expression and purification
Drosophila melanogaster symplekin cDNA was amplified and cloned into a pET28b derivative vector. The final construct includes an N-terminal histidine tag followed by a PreScission protease recognition site connected to the N-terminal domain of symplekin (residues 19–351). The construct was transformed into Escherichia coli strain BL21 (DE3) and cells were grown at 37°C in Luria-Bertani medium until OD600 reached 0.5–0.8. The culture temperature was dropped to 16°C to induce protein production with the addition of 0.5 mM IPTG for 16 hours. Harvested cells were lysed by sonication in a buffer containing 50 mM Tris pH 8.0, 500 mM NaCl, 10mM Imidazole, 0.1% Triton X-100 and 10% glycerol. The recombinant protein was first purified with Ni-NTA column (Qiagen) and then incubated with PreScission protease (GE Healthcare) overnight at 4°C (at ratio 1:100) to remove the N terminal hexahistidine tag. Finally, the protein was further purified to homogeneity by gel filtration using a Superdex-75 column (GE Healthcare). The full length gene of Drosophila melanogaster Ssu72 (residue 1–195) was cloned into a pET28b derivative vector with an N-terminal 8xHis-SUMO tag to facilitate solubility and stability. The protein expression and purification followed a protocol described previously (28). Ssu72 mutants were generated by QuikChange site-directed mutagenesis methods (Stratagene, CA) and purified using a similar protocol as for the wild-type (28).
To obtain the Drosophila melanogaster Ssu72-symplekin complex, symplekin (residues 19–351) and full length Ssu72 proteins were mixed at a molar ratio of 1: 1.5 and incubated overnight at 4°C. The excess Ssu72 was separated from the Ssu72-symplekin complex by gel-filtration chromatography using a Superdex-200 column (GE Healthcare). Purified symplekin-Ssu72 complex was concentrated to ~10 mg/ml and stored in a buffer of 25 mM Tris-HCl at pH 8.0, 200 mM NaCl and 1 mM DTT before flash freezing to −80°C for future crystallization or quantitative kinetics analysis.
Phosphatase activity assay
The phosphatase activity of Ssu72-symplekin complex was measured by a malachite green assay using various 19-mer CTD peptides as substrates. The peptide frame consists of two full repeats (II and III) and two partial repeats (I and IV) which include residues Ser5I-Pro6I-Ser7I-Tyr1II-Ser2II-Pro3II-Thr4II-Ser5II-Pro6II-Ser7II-Tyr1III-Ser2III-Pro3III-Thr4III-Ser5III-Pro6III-Ser7III-Tyr1IV-Ser2IV. The peptides were phosphorylated at the Ser5 or both Thr4/Ser5 at the third repeat. The reactions were initiated by mixing ~85 ng wild type Drosophila melanogaster Ssu72-symplekin complex proteins with the assay buffer (20 mM MES (pH 6.0) 50 mM NaCl) containing various concentrations of CTD peptides (0.05 to 1 mM). The assay was performed in a 200 μL PCR tube with a total reaction volume of 20 μl. The reaction was held at 28° C in Mastercycler PCR machine (Eppendorf North America) for 6 or 12 minutes. The reaction was quenched with the addition of 40 μL malachite green reagent and incubated at room temperature for 20 minutes for the color development. The OD620 absorbance was then measured with a Tecan Infinite M200 microplate reader (Tecan, CA) and the liberated inorganic phosphate was calculated based on a phosphate standard curve.
In the kinetic analysis, reactions showed substrate inhibition when the peptide concentration exceeded 0.6 mM, therefore the specificity constant kcat/Km was used to quantify phosphatase activity. The kinetic data were analyzed using Origin 6.0 software (Origin Lab). The specificity constant value (kcat/Km) was calculated using the following equation (1) where kon = kcat/Km:
| (1) |
Mass Spectrometry and 193 nm Ultraviolet Photodissociation Analysis of the CTD peptide
CTD peptides (19 mer with single Ser5 phosphorylation and Thr4/Ser5 double phosphorylation and Ser5/Ser7 double phosphorylation, at a concentration of 0.2 mM) were treated with ~500 ng Drosophila Ssu72-symplekin complex protein in a reaction buffer (20 mM MES, pH6.0, 50 mM NaCl) for 3.5 hours or overnight. Following phosphatase exposure for 0 hrs, 3.5 hrs, and overnight at 28°C, the peptides were separated from the enzyme using 3 kDa molecular weight cutoff filters (Millipore, Billerica, MA). The filtrates containing the peptides were evaporated using a speedvac and the concentrated peptides were re-suspended in 500 μL of 50/49/1 acetonitrile/water/formic acid. Solutions were infused directly at 5 μL per minute by electrospray ionization into a Velos dual linear ion trap mass spectrometer (ThermoFisher, San Jose, CA) for molecular weight confirmation. Mass spectra were acquired in the positive mode in the range m/z 400–2000. The analyzed spectra were the result of fifty averaged scans. The Ser5 monophosphorylated and Thr/Ser5 diphosphorylated peptides were also analyzed using dual selected ion monitoring (SIM) on an Orbitrap Elite mass spectrometer (ThermoFisher, San Jose, CA) customized to accommodate 193 nm ultraviolet photodissociation (UVPD) via an excimer laser in the HCD cell as described previously (29). Dual selected ion monitoring (SIM) scans were performed encompassing the expected mass-to-charge ratios of both the unreacted and the dephosphorylated peptides. Negative mode 193 nm UVPD was performed throughout the acquisition in a data independent manner on the m/z corresponding to the singly phosphorylated peptide to confirm the site of modification. Triple SIM scans were performed in a similar fashion for analysis of the phosphoryl-Ser5 and phosphoryl-Ser7 peptides.
Crystal soaking and structure determination
All initial protein crystal screening and subsequent optimization processes were carried out using a sitting-drop vapor diffusion method at 20°C by mixing Ssu72-symplekin (C13D/D144N) variant with equal volume of well solution. Optimized Ssu72-symplekin crystals were obtained with reservoir solution consisting of 12% PEG3350 (w/v) and 100 mM HEPES pH 8.5. To obtain the tertiary complex structure of Drosophila Ssu72-symplekin-CTD, crystals of Ssu72-symplekin were soaked in a mother solution containing 2 mM 19-mer CTD peptides overnight at room temperature and then cryo-protected with 25% glycerol prior to x-ray data collection.
X-ray diffraction data were obtained at Advanced Light Source (ALS) beamlines (5.0.2 and 5.0.3) and processed with HKL2000 (30). The crystals of Ssu72-symplekin-CTD peptides (19-mer peptides with Ser5III phosphorylated or Thr4III and Ser5III phosphorylated) were collected and scaled up to resolution at 2.58 Å and 2.35 Å, respectively. The initial phases were calculated using molecular replacement method with the structure of human Ssu72-symplekin (PDB ID: 3O2S) (15) as the search model. The fitted model was rebuilt with COOT (31) and refined by autoBUSTER (32) with iterative cycles of optimization. The quality of final refined structures was evaluated by MolProbity (33) and Procheck (34). In MolProbity validation, a global Molprobity score of 1.50 was assigned to Ssu72-symplekin in complexed CTD peptide with Ser5 phosphorylated, corresponding to the 100th percentile among the structures at similar resolution. Ssu72-symplekin with doubly phosphorylated CTD peptide obtained an overall score of 1.45, ranking in the 99th percentile among the structures of comparable resolution. The full statistic data of complex structures are summarized in Table 1.
Table 1.
Crystallographic data statistics for Ssu72-symplekin in complexed with CTD peptides
| Statistics | Ssu72-symplekin-CTD (phos.Ser5) | Ssu72-symplekin-CTD (phos.Thr4-phos.Ser5) |
|---|---|---|
| Data collection statistics | ||
| Advanced light source | Beamline 5.0.3 | Beamline 5.0.2 |
| Wavelength (Å) | 0.9765 | 0.9793 |
| Resolution (Å) | 50.00-2.58 (2.62-2.58) | 50.00-2.35 (2.39-2.35) |
| Space group | P4 | P4 |
| Unit cell (Å) a, b, c | 128.0, 128.0, 105.8 | 128.3, 128, 3, 106.1 |
| Number of unique reflections | 53955 (2693) * | 71330 (3531) * |
| Redundancy | 7.5 (7.3) * | 5.0 (4.8) * |
| Completeness (%) | 100 (99.8) | 100 (99.8) |
| I/σ | 24.2 (2.0) | 24.3 (1.4) |
| Rsym (%) | 9.6 (84.5) | 7.7 (88.9) |
| Refinement statics | ||
| Resolution limits (Å) | 41.60-2.58 (2.65-2.58) | 49.03-2.35 (2.41-2.35) |
| Number of reflections/test | 53782/2731 | 71311/3611 |
| Rwork/Rfree# (%) | 21.0/24.3 | 21.9/23.9 |
| Number of atoms protein | 8368 | 8157 |
| Number of atoms water | 207 | 199 |
| B-factors for protein chains (Å2) (Ssu72/symplekin) | 74.0/69.7 | 71.3/68.2 |
| B-factors for ligand chains: | 84.8 | 77.6 |
| B-factors for water (Å2) | 51.2 | 50.6 |
| Bond rmsd length (Å)/angles (°) | 0.010/1.05 | 0.010/1.02 |
| Most favored (%) | 94.5 | 94.3 |
| Additionally allowed (%) | 5.4 | 5.5 |
| Generously allowed (%) | 0.1 | 0.2 |
| Disallowed (%) | 0 | 0 |
Highest resolution shell is shown in parenthesis.
Rfree is calculated with 5% of the data randomly omitted from refinement.
Results and Discussion
To study the effect of additional phosphorylation on the activity of CTD modifying protein, we quantitatively characterized the phosphatase activity of Ssu72-symplekin system. As the first reported cis-specific CTD phosphatase (15), (14, 16), the protein recognizes substrate CTD with a tight β-turn conformation which seems to be maintained by the intra-molecular hydrogen bond between the Thr4 hydroxyl side chain and carbonyl group of cis-form Pro6, (16). Placing a phosphate group at the position of the hydroxyl group of Thr4 would introduce steric clashes to this tight turn unless some conformational changes occur. Therefore, phosphorylation modification at Thr4 might shift the ratio balance of cis:trans proline distribution and eventually influence the activity of downstream enzymes such as Ssu72. In addition, this will also answer the essential question for the CTD field: what determines the outcome of the CTD code; the transient secondary structure of CTD dominated by the combination of modification or the recruitment of CTD binding proteins whose local active site determine the substrate conformation.
However, the quantitative measurement of Ssu72 activity has been technically challenging for several reasons: (1) the CTD domain of eukaryotic RNA polymerase II is not homogenously phosphorylated in vivo, (2) the activity of Ssu72 towards synthetic CTD peptides is relatively low, and (3) Ssu72 is not thermally stable in vitro. To quantify the activity of Ssu72 towards the CTD, we successfully characterized the kinetic activity of Ssu72 from Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and human and found that Drosophila Ssu72 exhibited the highest activity in vitro (28). Furthermore, the formation of a complex with the template protein symplekin increased the apparent activity of Ssu72 (15). Thirdly, Drosophila Ssu72 is the only reported protein in this family that can crystallize in the absence of template protein. This enables the comparison of enzyme with and without template for structural variation. Therefore, we used the Drosophila Ssu72-symplekin complex to study the effect of flanking phosphorylation on the CTD in the present study.
Mass spectrometric analysis of the phosphorylation state of CTD peptide
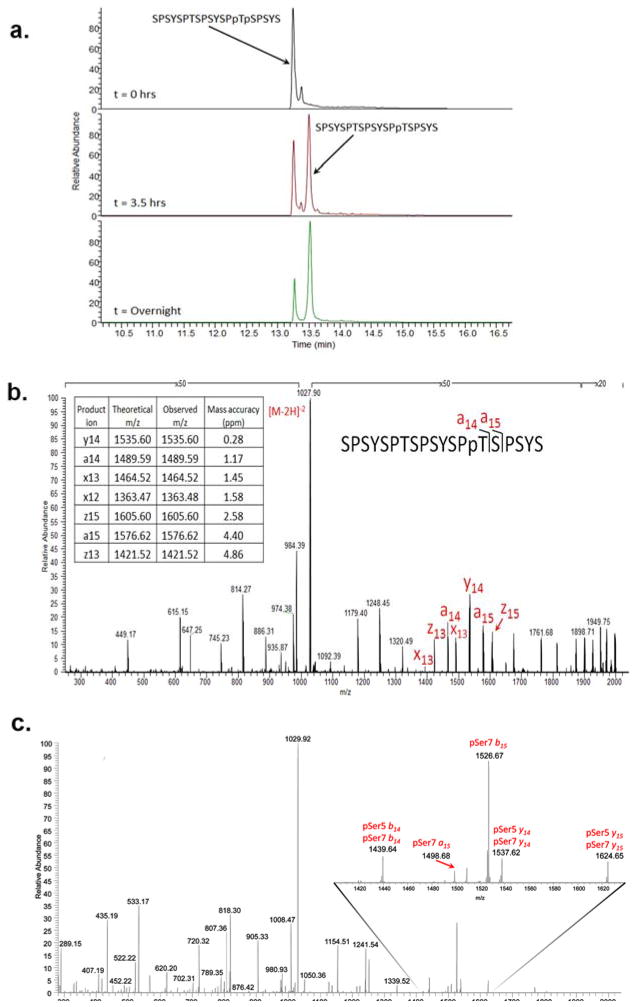
Phosphoryl-CTD is the only reported substrate for Ssu72 even though the reaction mechanism utilized for phosphoryl transfer is similar to low molecular weight tyrosine phosphatases (28). It has yet to be tested whether Ssu72 can dephosphorylate Thr4. In order to determine if Thr4 is also subject to dephosphorylation by Ssu72, we used mass spectrometry to qualitatively assess the extent of CTD peptide dephosphorylation mediated by Ssu72. Singly and doubly phosphorylated CTD peptides (i.e. Ser5, Thr4/Ser5 and Ser5/Ser7 phosphoryl-peptides) were incubated with Ssu72 individually for zero hours, 3.5 hours and overnight. The incubated samples were analyzed by LC-MS (SIM mode) by continuously monitoring the m/z values corresponding to the phosphoryl-peptides and their dephosphorylated counterparts. After the 3.5 hour incubation of the diphosphorylated Thr4/Ser5 peptide, a product with a mass 80 Da lower than the mass of initial dephosphorylated peptide is detected; corresponding to a singly dephosphorylated CTD peptide (Figure 1a). Following overnight incubation, the observed abundance of the original phosphoryl-peptide is greatly diminished compared to the product with one phosphate group removed. However, no additional dephosphorylation species appeared.
Figure 1.
Mass spectrometric analysis of CTD peptides treated with Ssu72. a. 19-mer CTD peptide doubly phosphorylated at Thr4 and Ser5 was treated with Ssu72 for zero, 3.5 hours and overnight. LC-MS selected ion monitoring (SIM) chromatograms of the dephosphorylated Thr4/Ser5 precursor (left-most peak, monitoring m/z 1068 ± 25) and the singly dephosphorylated product (right-most peak, monitoring m/z 1028 ± 25) after exposure of diphosphoryl-peptide SPSYSPTSPSYSPpTpSPSYS to Ssu72. b. UVPD characterization to determine the retained site of phosphorylation on the product CTD peptide in (a) after overnight incubation with Ssu72. c. Analysis of Ser5/Ser7 dephosphorylation by CTD. Shown is a 193nm negative mode UVPD spectrum of the mass representing doubly phosphorylated CTD (phosphorylated Ser5/Ser7) treated by Ssu72. Shown in the inset are specific ions of interest that confirm the presence of the phosphoryl group at position Ser7 of the peptide.
Negative mode UVPD was used to pinpoint the site of the single dephosphorylation event of the doubly phosphorylated Thr4/Ser5 CTD peptide. Our result confirmed that the singly dephosphorylated product was consistent with loss of the phosphate group solely from Ser5 and that the phosphate group on Thr4 remained in place (Figure 1b). Therefore, we established that Ssu72 dephosphorylates Ser5 but not Thr4 in vitro.
As a control experiment, the same analysis was conducted on 19mer CTD peptide with a single Ser5 phosphorylation site. A similar dephosphorylation process is observed for the mono-phosphorylated peptide upon incubation with Ssu72 with 80 Da removed from the peptide (data not shown). Another control experiment was conducted on doubly phosphorylated CTD peptide with both Ser5 and Ser7 phosphorylated. Ssu72 has been shown to dephosphorylate phosphoryl-Ser7 in vitro and in vivo, but exhibited a much higher activity against Ser5 over Ser7 in vitro (22, 23). Indeed, when doubly phosphorylated CTD at Ser5/Ser7 was treated with Ssu72, a singly phosphorylated CTD peptide species was detected first, which was later confirmed to be phosphorylated Ser7 (Figure 1c). Overnight incubation of doubly-phosphorylated CTD discloses that the completely dephosphorylated species was detected in high abundance (data not shown).
Kinetic characterization of Ssu72-symplekin towards phosphorylated CTD peptides
The mass spectrometric analysis established that phosphoryl-Thr4 is not subject to dephosphorylation by Ssu72. Next we want to evaluate the effect of phosphorylated Thr4 on the dephosphorylation of Ser5 by Ssu72 using steady state kinetics. We quantified the specific activity of Ssu72-symplekin towards CTD peptides when both Thr4 and Ser5 are phosphorylated or Ser5 alone is phosphorylated in the 19-mer CTD peptide. We found a decrease in the initial rate (s−1) when the substrate concentration exceeds 0.6 mM (data not shown), indicating substrate inhibition. Therefore, we determined the specificity constant (kcat/Km) for both CTD peptides instead of kcat or Km. Upon measurement of specificity constant, we found a reduction of 4.2 fold when the CTD peptide was doubly phosphorylated at Thr4/Ser5 (kcat/Km = 1.96 ± 0.20 mM−1 s−1) in comparison to Ser5 phosphorylation alone (kcat/Km = 8.22±1.24 mM−1 s−1) (Figure 2). This result shows that the phosphorylation of Thr4 preceding phosphoryl Ser5 doesn’t abolish Ssu72 activity, but reduces Ssu72 activity by more than four fold.
Figure 2.
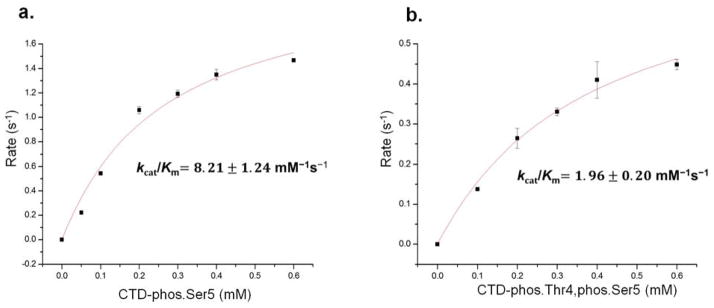
Steady-state kinetics of Drosophila Ssu72-symplekin towards 19mer CTD peptides: SPSYSPTSPSYSPT(phos.)SPSYS (a) and SPSYSPTSPSYSP(phos.)T(phos.)SPSYS (b).
Obtaining Drosophila Ssu72-Symplekin-CTD Ser5 complex structure
Even though the phosphorylation of flanking Thr4 reduced the Ssu72-mediated dephosphorylation of Ser5 by four fold, the remaining activity indicates that Ssu72 can still effectively bind to the CTD in the absence of intra-molecular hydrogen bond upon the phosphorylation of Thr4. To analyze the interaction between Ssu72 and phosphorylated CTD peptide, we seek to determine the complex structure of Drosophila Ssu72-symplekin with various CTD peptides. Furthermore, since Drosophila Ssu72 is the only member of the family that can be crystallized without the template symplekin, the complex structure of Drosophila Ssu72-symplekin opens the door to study the structural changes in Ssu72 upon template protein association.
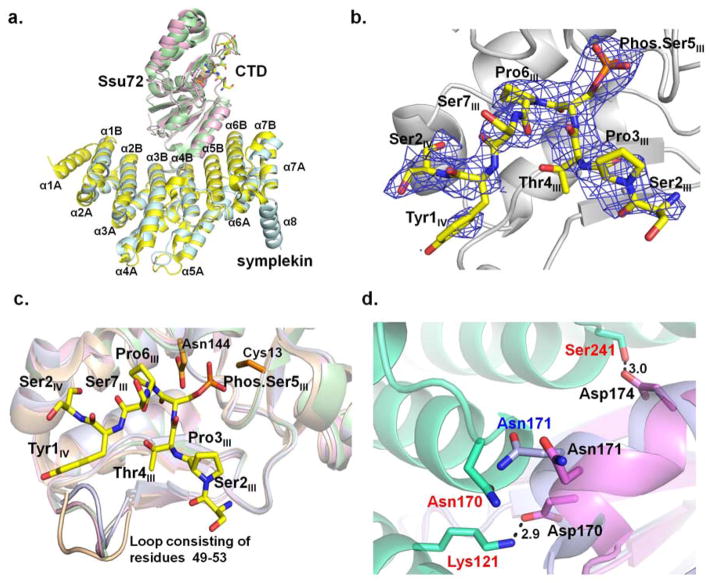
The primary sequences of Drosophila Ssu72 and symplekin are quite conserved with their human counterpart. Drosophila Ssu72 has a 61% sequence identity to human Ssu72 and the adapter protein symplekin exhibits a 38% identity between human and Drosophila in the region for Ssu72 interaction (N-terminal domain residues 1–350). To investigate if the binding of the CTD by Ssu72-symplekin is conserved in higher eukaryotes, we determined the tertiary structure of Drosophila Ssu72-symplekin in complex with CTD peptides with a single Ser5 phosphorylation sites (Figure 3a). In order to prevent the turnover of the substrate by Ssu72, mutations were made at the active site residue cysteine 13 to capture the mode of substrate binding. Crystals of C13S and C13D were both tested, but C13D crystals diffracted to higher resolution. In addition, another mutation D144N was reported to further improve diffraction quality (16) and therefore was used in the complex structural determination and analysis. During our crystallization trial, it was noticed that any CTD peptides with more than four residues at the C-terminal end of phosphoryl-Ser5 failed to be incorporated into the Ssu72-symplekin crystals. Analysis of the crystal lattice shows that longer peptides will interfere with the crystal packing, causing the crystals to lose diffraction upon soaking. Therefore, we used a 19-mer CTD peptide with one Ser5 phosphorylation site close to the C-terminus of the peptide, Ser5I-Pro6I-Ser7I-Tyr1II-Ser2II-Pro3II-Thr4II-Ser5II-Pro6II-Ser7II-Tyr1III-Ser2III-Pro3III-Thr4III-(phos.)Ser5III-Pro6III-Ser7III-Tyr1IV-Ser2IV (bold letters indicating residues modeled in the structure). Eight residues out of the 19 amino acids from the repeats were built into the electron density (Figure 3b). Even though the 19-mer substrate has 14 amino acids N-terminal to the phosphoryl-Ser5, only Ser2, Pro3 and Thr4 in the same repeat are visible in the complex structure with no additional secondary binding pocket identified. Both co-crystallization and soaking methods were successfully applied in the structural determination, resulting in identical models. Our description of the structures below is based on the model built from soaked crystals which diffracts to higher resolution.
Figure 3.
Structure of Drosophila melanogaster Ssu72-symplekin-CTD ternary complex in which the ligand is 19-mer CTD peptide with singly phosphorylated Ser5:
a. The superimposed structures of Drosophila and human Ssu72-symplekin-CTD complexes. Symplekin and Ssu72 of Drosophila melanogaster are colored pale cyan and light pink, respectively. Symplekin and Ssu72 of human are colored yellow and green. CTD peptides bound by Drosophila and human Ssu72s were shown in stick model with the carbon atom colored yellow and green, respectively. b. Composite omit annealing map of electron density of CTD peptide of singly phosphorylated Ser5 (yellow) in Drosophila melanogaster Ssu72-symplekin complex, contoured at 3 σ. c. Overlay of the structures of Drosophila apo Ssu72 (wheat), Ssu72 in complex with the CTD (light pink), Ssu72 in complex with symplekin (grey), and Ssu72 in complex with symplekin and the CTD (green): a loop consisting of Ssu72 49–53 residues display a significant conformational change upon the binding of the CTD or symplekin. d. Interaction formed between Drosophila Ssu72 (violet) upon its association with template protein symplekin (lime green). Superimposition with apo Ssu72 (light blue) structure reveals a small conformational adjustment to accommodate symplekin binding.
Recognition of the CTD by Drosophila Ssu72-symplekin
When we compared the Drosophila Ssu72 with and without the template protein (PDB code 3omw), no significant structural alternation is observed with a RMS deviation between the two structures is only 0.5 Å. The only significant conformational change upon symplekin binding occurs at the loop of Ssu72 residues 49–53. These residues comprise the tip of a highly flexible loop at the active site which encloses upon substrate incorporation (Figure 3c). Even though we cannot totally exclude the possibility that the change of conformation is induced by the remote interaction of symplekin, a more plausible explanation is the intrinsic flexibility of this loop. In the apo Ssu72 structure, this loop exhibits the highest thermal B factors, almost double the average B factors of the overall molecule (28). Upon the CTD association, the loop is locked into the similar conformation and provides a lid to cover the active site (Figure 3c). Therefore, the observed change of conformation at this loop is only due to the high flexibility of the loop rather than a direct result of template protein binding. Interestingly, a small change of conformation also occurs at two loops that interact with symplekin. A movement of 3 Å occurs for loop 168–174 in Ssu72 (Figure 3d). Upon the binding to symplekin, Asp170 forms a salt bridge with Lys121 from symplekin (Figure 3d). The side chain of Asn171 flips away to avoid clashing with Asn170 from symplekin, which in turn engages in hydrogen bonding to Asp168 from this loop in Ssu72. A hydrogen bond network between Asp174 and side chain of symplekin Ser241 further stabilizes the interaction (Figure 3d). The other loop that has a translational movement of the local backbone includes residues 127–130. The most significant change is the side chain of Asp128 which flips outside of the loop and forms a salt bridge with Arg198 in symplekin.
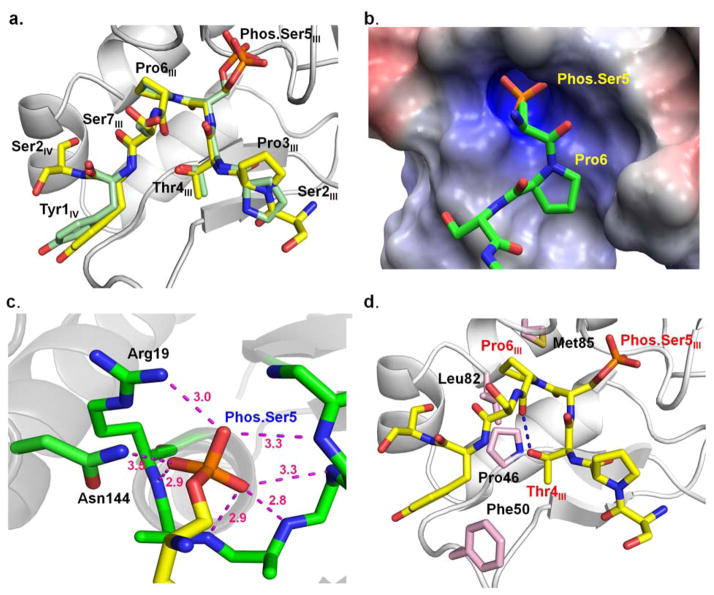
The structure at a resolution 2.6 Å shows that the recognition of the substrate is conserved between Drosophila and human Ssu72-symplekin complex (Figure 4a), making it a good surrogate for kinetic study to replace the human complex which is substantially less active and stable. Even though the nucleophile cysteine was replaced by an aspartic acid for crystallographic purposes, the negatively charged phosphate group still can be stably bound at the active site, as observed by the strong electron density indicating a tetrahedral-shaped phosphate group (Figure 3b). The rest of the substrate binding pocket is dominated by positively charged residues, along with an amide group in the backbone. These residues comprise an oxyanion hole to stabilize the binding of the phosphate group (Figure 4c), Similar oxyanion holes also exist in tyrosine phosphatases to promote the deprotonation state of the nucleophile and stabilize the phosphoryl-transfer intermediate (35). The phosphorylated Ser5 of CTD peptide protrudes into the shallow groove near the active site nucleophile located (Figure 4b). The positioning of Ser5 is mediated through a hair-pin shaped β-turn with Ser5 at the tip of the loop and fortified by an intra-molecular hydrogen bond through the side chain of Thr4 and the backbone carbonyl of Pro6 (Figure 4d).
Figure 4.
Recognition of CTD by Drosophila melanogaster symplekin-Ssu72.
a. Superimposition of singly phosphorylated Ser5 CTD peptide bound at the active site of Drosophila melanogaster Ssu72 (peptide carbon shown in yellow) and human Ssu72 (peptide carbon shown in green). b. The electrostatic surface potential of active site of Drosophila Ssu72, where blue indicates positive charge, red indicates negative charge, and white indicates hydrophobic regions. The surfaces were generated with the Adaptive Poisson–Boltzmann Solver (APBS) using the AMBER force field (APBS Tools 2.1 PyMoL plugin, M. G. Lerner). c. A hydrogen bond network formed around phosphate group of phos.Ser5 CTD peptide in active site of Ssu72. d. Key residues of Drosophila Ssu72 for CTD recognition. The amino acids of Ssu72 are colored pink while the carbon atoms of CTD peptide are shown in yellow.
Even though eight residues are modeled into the complex tertiary structure, the essential residues contributing to recognition are three hotspot residues: the phosphate group of Ser5, Pro6, and Tyr1 in subsequent repeat. The importance of the phosphate group is obvious for a phosphatase like Ssu72. In general, phosphate recognition is essential for CTD binding proteins and consistent with the theory that phosphorylation of the CTD recruits proteins to the transcribing RNA pol II. Extensive electrostatic interactions are formed between CTD binding enzymes and phosphorylated serine residues with the only exception being the CID domain of Pcf11, a weak binder for the CTD where the phosphate group of Ser2 only forms an intra-molecular hydrogen bond to stabilize the β-turn recognized by the protein. The second element proline residue is critical for the differentiation of Ser2 and Ser5. Pro3 and Pro6 are the only residues in the CTD motif that are not subject to phosphorylation regulation. However, they play an essential role in the CTD recognition by placing hydrophobic breaks at CTD sequence whose intervals can be used to distinguish the substrate. Indeed, all reported CTD complex structures showed hydrophobic interactions with proline residues. The position and conformation of proline determines how the enzymes distinguish Ser2 or Ser5. For example, Scp highly prefers Ser5 over Ser2 through its recognition of Pro3 by a unique hydrophobic pocket (36). Reducing the hydrophobicity of the binding pocket of Pro3 by mutation can eliminate the Ser5 over Ser2 preference of Scp (36). In Ssu72, Pro6 bound at the hydrophobic pocket is essential since mutation at this pocket abolishes CTD binding (28). Interestingly, when Ser7 is phosphorylated in the absence of Ser5 phosphorylation, a similar CTD binding mode is identified with Pro6 occupying the same binding site with the phosphate group from Ser7 at the active site for phosphate binding, resulting in a conformation with reversed orientation of CTD recognition in order to maintain the binding (37). Therefore, the electrostatic recognition at the phosphate group and the hydrophobic interaction with proline are dominating factors for the occupancy of the active site of Ssu72. The third element of recognition varies among individual CTD binding proteins. In Ssu72, Tyr1 at the subsequent repeat also plays a role in CTD binding. The CTD with a mutation at this site shows great reduction of dephosphorylation by the CTD (38) and the CTD synthetic peptide missing the last two residues of the 19-mer, Tyr and Ser, behaved as a poor substrate for Ssu72 without detectable activity (28).
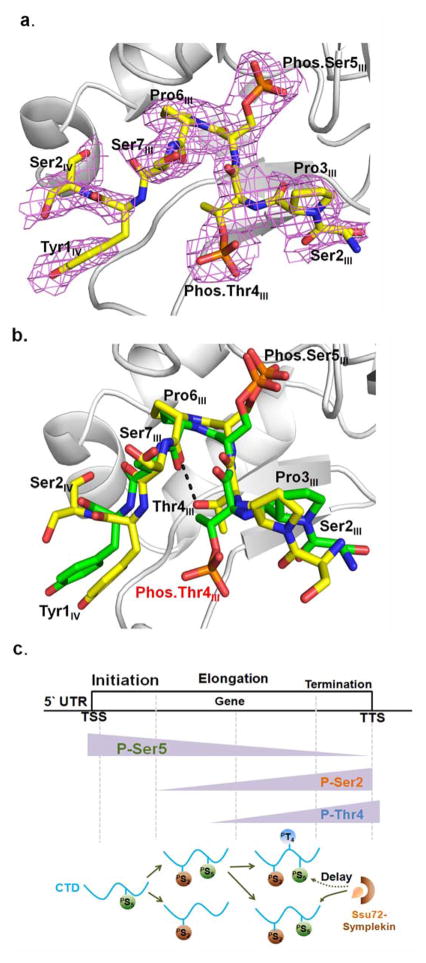
Structure determination of Ssu72-symplekin with CTD peptide when doubly phosphorylated at Thr4 and Ser5
Even though the phosphatase activity of Ssu72 is compromised when the Thr4 next to Ser5 is phosphorylated, the remaining activity indicates that Ssu72 can still effectively recognize the CTD. We therefore determined the structure of Drosophila Ssu72-symplekin binding to the same 19-mer CTD peptide when both Thr4 and Ser5 are phosphorylated (Figure 5a). The structure of Drosophila Ssu72-symplekin in complex with a 19-mer CTD peptide with both consecutive Thr4 and Ser5 phosphorylated was determined at 2.35 Å (Figure 5a). Surprisingly, the mode of recognition is similar to that of the CTD with non-phosphorylated Thr4 even though the phosphorylation of Thr4 prohibits the formation of an intra-molecular hydrogen bond between Thr4 and Pro6 (Figure 5b). Pro6 still exhibits the cis-conformation, but the Thr4 side-chain adapts an alternative isomer with the hydroxyl group facing the solvent (Figure 5b). This change of isomer conformation places the methyl group of Thr4 close to the carboxyl group of Pro6 where the intra-molecular hydrogen bond was. The loss of the intra-molecular hydrogen bond plus the placement of hydrophobic methyl group unfavorably adjacent to hydrophilic carboxyl group potentially reduce the stability of the conformation of the CTD being recognized by Ssu72.
Figure 5.
Complex structure of Drosophila Ssu72-symplekin bound to doubly phosphorylated CTD peptide at Thr4/Ser5.
a. Composite annealing omit map of CTD peptide with doubly phosphorylated Thr4 and Ser5, contoured at 3 σ.
b. Overlay of structures of Drosophila Ssu72-symplekin in complex with 19-mer CTD peptides, singly phosphorylated Ser5 with carbon atoms colored yellow and doubly phosphorylated Thr4 and Ser5 with carbon shown in green.
c. Model of combinatorial code of phosphorylation and isomerization. Thr4 phosphorylation occurring after the Ser2 phosphorylation will delay the dephosphorylation of Ser5.
The understanding of events occurring when phosphoryl-Thr4 coexists with phosphoryl-serine(s) on the CTD is limited due to the lack of kinetic and structural studies of such CTD peptides with CTD-binding proteins. Previously, it has been speculated that Thr4 phosphorylation would impair the recognition of the CTD by CTD binding proteins because the hydroxyl sidechain of Thr4 is often used in intramolecular hydrogen bonding. In the Pcf11-CTD interaction, phosphoryl-Ser2 enhances this interaction and its electrophilic interactions with Thr4 stabilized the β-turn (39). When SCAF8 was bound to phosphoryl-Ser5 or unphosphorylated CTD, the γ-oxygen atom of Thr4 is also within hydrogen bonding distance to the hydroxyl group of the unphosphorylated Ser2 residue, potentially stabilizing the tight β-turn (40). Our result on Ssu72 seems to suggest that the CTD structure is highly plastic and modification can be easily tolerated with local conformational changes. The disruption of the intra-molecular hydrogen bond between the Thr4 side-chain hydroxyl group and the Pro backbone carbonyl of CTD doesn’t necessarily prohibit CTD binding.
Induced fit of the CTD recognized by CTD-binding proteins
Our kinetic and structural analysis on the phosphorylation of Thr4 undoubtedly showed that the pre-existing intra-molecular hydrogen bond is not required for the recognition of Ssu72 towards substrate CTD. Instead, the CTD will form specific conformations upon enzyme binding (Figure 5b). No CTD structure was observed in the free RNA polymerase II crystal structure (41). Speculation on the importance of pre-formed secondary structure has resulted in a stable and compact beta-spiral model that is only temporally open by phosphorylation and subject to modification by CTD-binding enzymes (39). However, current structural studies on various CTD binding proteins seem to contradict such a model. First, so far, CTD conformations recognized by various CTD-binding enzymes are all distinct from each other. Second, the CTD doubly phosphorylated at Thr4 and Ser5 bound by Ssu72 in our structures showed that intra-molecular hydrogen bonds formed by the CTD are not essential for the recognition by Ssu72. Indeed, Ssu72 shows very little conformational change upon binding to the CTD peptide with or without the intra-molecular hydrogen bond to stabilize its tight beta-turn for Ssu72 association. Third, since the CTD is primarily composed of small hydrophilic residues such as serine and threonine, their phosphorylation might be well tolerated due to the size and alternative isomer conformation. In the case of Ssu72, Ser5 phosphorylation seems to be the prerequisite for the binding state while flanking residue phosphorylation can be tolerated, even though Ssu72 displays reduced activity. Overall, the flexibility and plasticity of the CTD tail allows it to be molded into the appropriate conformations in the active sites of various CTD binding proteins. The specific secondary structural elements of the CTD upon binding to various CTD binding proteins are governed by the local active site pocket of the protein.
Cascade of CTD coding
Our kinetic and structural studies have shown for the first time that the CTD code can fine-tune the activity of CTD modifying enzymes through the combination of various phosphorylation modifications at different sites in CTD heptide repeats. Ssu72 is an essential phosphatase that is involved in the transcript elongation, mRNA processing and transcription termination; whose inactivation leads to cell death in yeast (42). Experiments in yeast strains with Ssu72 variants with defective phosphatase activity compromised transcription, indicating that the level of phosphatase activity of Ssu72 important for the transcription (43). A mutant of Ssu72 that retains 40% of phosphatase activity exhibits reduced elongation efficiency in transcription (43). This genetic defect results in a temperature sensitive phenotype that causes cell death at non-permissive temperature. By using a compound 6-azauracil to reduce the overall transcription speed, we can alleviate the genetic defect of Ssu72 mutant with defective phosphatase activity. This indicates that the level of phosphatase activity of this enzyme essential for the coordination of transcription process (44). In vivo, any kinetic effect might get magnified under the context of 26–52 repeats of the heptide. The four fold reduction in Ssu72 caused by phosphorylation of Thr4 can be much more significant when Thr4/Ser5 are tandemly phosphorylated in many repeats of a CTD. Therefore, the reduction of Ssu72 phosphatase activity, even a moderate reduction, can cause changes in CTD phosphorylation states under abnormal conditions and therefore, different transcription outcomes.
The phosphorylation of Thr4 eliminates an intra-molecular hydrogen bond formed by the substrate and reduces the activity of Ssu72 against Ser5. These results point to a model where phosphorylation at Thr4 modulates the isomerization state of Pro6 which in turn fine-tunes the phosphorylation state of Ser5. As previously established that the phosphorylation of Thr4 is dependent on the phosphorylation of Ser2 (25), a regulatory cascade can be derived from Ser2 phosphorylation leading to Thr4 phosphorylation which in turn modulates the isomerization state and eventually affects the efficiency of Ser5 dephosphorylation mediated by Ssu72 that leads to changes in transcription results (Figure 5c). This highlights the sequential events that involve phosphorylation (Ser2 followed by Thr4), isomerization (Pro6), and dephosphorylation (Ser5) (Figure 5c). The combinatorial modification at all these sites can lead to divergent phosphorylation states of the CTD which will generate differentiated outcomes of transcription. In conclusion, our structure and kinetic analysis of Ssu72-symplekin complex with the CTD with consecutively phosphorylated Thr4 and Ser5 opens the door to understanding the role of coexisting phosphorylation marks on the CTD and the effect of their cooperative interaction on various stages of transcription.
Supplementary Material
Acknowledgments
Instrumentation and technical assistance for this work were provided by the Macromolecular Crystallography Facility, with financial support from the College of Natural Sciences, the Office of the Executive Vice President and Provost, and the Institute for Cellular and Molecular Biology at the University of Texas at Austin. We acknowledge the support of the Advanced Light Source (ALS), Berkeley, California for x-ray crystallographic data collection. The funding is provided by National Institute for Health (R03 DA030556, PI Zhang and R21 GM099028, PI Brodbelt) and Welch Foundation (F-1778, PI Zhang and F1155, PI Brodbelt). We thank Thermo Fisher Scientific with helping on the modifications to the Orbitrap Elite mass spectrometer to allow UVPD.
Footnotes
Accession code
Coordinates of the Ssu72-symplekin in complex with 19-mer CTD peptides (singly phosphorylated at Ser5 and doubly phosphorylated at Thr4 and Ser5) have been deposited in the Protein Data Bank with the accession numbers 4imj and 4imi, respectively.
References
- 1.Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 2.Meinhart A, Silberzahn T, Cramer P. The mRNA Transcription/Processing Factor Ssu72 Is a Potential Tyrosine Phosphatase. Journal of Biological Chemistry. 2003;278:15917–15921. doi: 10.1074/jbc.M301643200. [DOI] [PubMed] [Google Scholar]
- 3.Palancade B, Bensaude O. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur J Biochem. 2003;270:3859–3870. doi: 10.1046/j.1432-1033.2003.03794.x. [DOI] [PubMed] [Google Scholar]
- 4.Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet. 2012;28:333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Zhang DW, Rodriguez-Molina JB, Tietjen JR, Nemec CM, Ansari AZ. Emerging Views on the CTD Code. Genet Res Int. 2012;2012:347214. doi: 10.1155/2012/347214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jasnovidova O, Stefl R. The CTD code of RNA polymerase II: a structural view. Wiley Interdiscip Rev RNA. 2013;4:1–16. doi: 10.1002/wrna.1138. [DOI] [PubMed] [Google Scholar]
- 7.Corden JL. Tails of RNA polymerase II. Trends Biochem Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 8.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu YX, Hirose Y, Zhou XZ, Lu KP, Manley JL. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 2003;17:2765–2776. doi: 10.1101/gad.1135503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu YX, Manley JL. Pinning down transcription: regulation of RNA polymerase II activity during the cell cycle. Cell Cycle. 2004;3:432–435. [PubMed] [Google Scholar]
- 11.Palancade B, Marshall NF, Tremeau-Bravard A, Bensaude O, Dahmus ME, Dubois MF. Dephosphorylation of RNA polymerase II by CTD-phosphatase FCP1 is inhibited by phospho-CTD associating proteins. J Mol Biol. 2004;335:415–424. doi: 10.1016/j.jmb.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 12.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 13.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 14.Zhang M, Wang XJ, Chen X, Bowman ME, Luo Y, Noel JP, Ellington AD, Etzkorn FA, Zhang Y. Structural and kinetic analysis of prolyl-isomerization/phosphorylation cross-talk in the CTD code. ACS Chem Biol. 2012;7:1462–1470. doi: 10.1021/cb3000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467:729–733. doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem. 2011;286:5717–5726. doi: 10.1074/jbc.M110.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He X, Khan AU, Cheng H, Pappas DL, Jr, Hampsey M, Moore CL. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 2003;17:1030–1042. doi: 10.1101/gad.1075203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 Is an RNA Polymerase II CTD Phosphatase. Molecular Cell. 2004;14:387–394. doi: 10.1016/s1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy SA, Frazier ML, Steiniger M, Mast AM, Marzluff WF, Redinbo MR. Crystal structure of the HEAT domain from the Pre-mRNA processing factor Symplekin. J Mol Biol. 2009;392:115–128. doi: 10.1016/j.jmb.2009.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidemann M, Eick D. Tyrosine-1 and threonine-4 phosphorylation marks complete the RNA polymerase II CTD phospho-code. RNA Biol. 2012;9:1144–1146. doi: 10.4161/rna.21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, Eick D, Ansari AZ. Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol. 2010;17:1154–1161. doi: 10.1038/nsmb.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell. 2012;45:158–170. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 23.Zhang DW, Mosley AL, Ramisetty SR, Rodriguez-Molina JB, Washburn MP, Ansari AZ. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J Biol Chem. 2012;287:8541–8551. doi: 10.1074/jbc.M111.335687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, Fenouil R, Ferrier P, Flatley A, Kremmer E, Chapman RD, Andrau JC, Eick D. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 2012;31:2784–2797. doi: 10.1038/emboj.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stiller JW, McConaughy BL, Hall BD. Evolutionary complementation for polymerase II CTD function. Yeast. 2000;16:57–64. doi: 10.1002/(SICI)1097-0061(20000115)16:1<57::AID-YEA509>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 27.Schwer B, Lehman K, Saha N, Shuman S. Characterization of the mRNA capping apparatus of Candida albicans. J Biol Chem. 2001;276:1857–1864. doi: 10.1074/jbc.M006072200. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Zhang M, Zhang Y. Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue. Biochem J. 2011;434:435–444. doi: 10.1042/BJ20101471. [DOI] [PubMed] [Google Scholar]
- 29.Han SW, Lee SW, Bahar O, Schwessinger B, Robinson MR, Shaw JB, Madsen JA, Brodbelt JS, Ronald PC. Tyrosine sulfation in a Gram-negative bacterium. Nat Commun. 2012;3:1153. doi: 10.1038/ncomms2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 35.Zhang YL, Yao ZJ, Sarmiento M, Wu L, Burke TR, Jr, Zhang ZY. Thermodynamic study of ligand binding to protein-tyrosine phosphatase 1B and its substrate-trapping mutants. J Biol Chem. 2000;275:34205–34212. doi: 10.1074/jbc.M004490200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Kim Y, Genoud N, Gao J, Kelly JW, Pfaff SL, Gill GN, Dixon JE, Noel JP. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol Cell. 2006;24:759–770. doi: 10.1016/j.molcel.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang K, Manley JL, Tong L. An unexpected binding mode for a Pol II CTD peptide phosphorylated at Ser7 in the active site of the CTD phosphatase Ssu72. Genes Dev. 2012;26:2265–2270. doi: 10.1101/gad.198853.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hausmann S, Koiwa H, Krishnamurthy S, Hampsey M, Shuman S. Different strategies for carboxyl-terminal domain (CTD) recognition by serine 5-specific CTD phosphatases. J Biol Chem. 2005;280:37681–37688. doi: 10.1074/jbc.M505292200. [DOI] [PubMed] [Google Scholar]
- 39.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 40.Becker R, Loll B, Meinhart A. Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2008;283:22659–22669. doi: 10.1074/jbc.M803540200. [DOI] [PubMed] [Google Scholar]
- 41.Bushnell DA, Kornberg RD. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc Natl Acad Sci U S A. 2003;100:6969–6973. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun ZW, Hampsey M. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol Cell Biol. 1996;16:1557–1566. doi: 10.1128/mcb.16.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reyes-Reyes M, Hampsey M. Role for the Ssu72 C-terminal domain phosphatase in RNA polymerase II transcription elongation. Mol Cell Biol. 2007;27:926–936. doi: 10.1128/MCB.01361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dichtl B, Blank D, Ohnacker M, Friedlein A, Roeder D, Langen H, Keller W. A Role for SSU72 in Balancing RNA Polymerase II Transcription Elongation and Termination. Molecular Cell. 2002;10:1139–1150. doi: 10.1016/s1097-2765(02)00707-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.