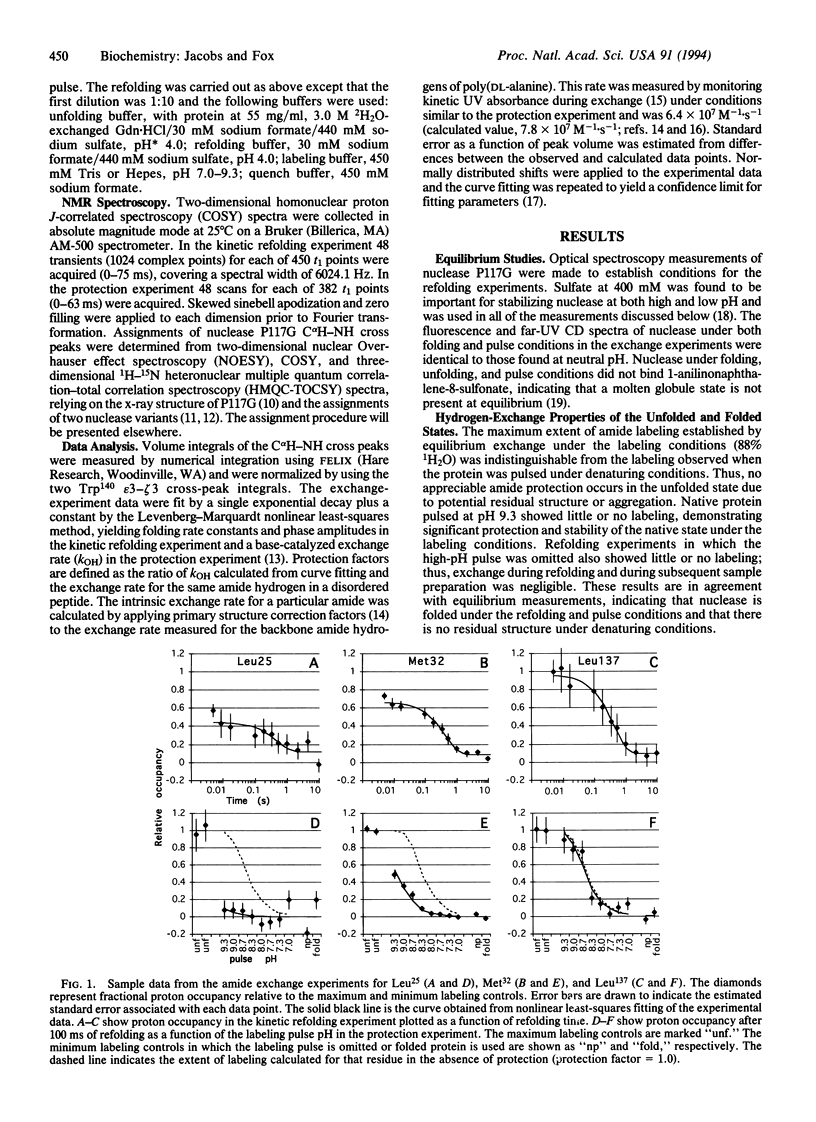
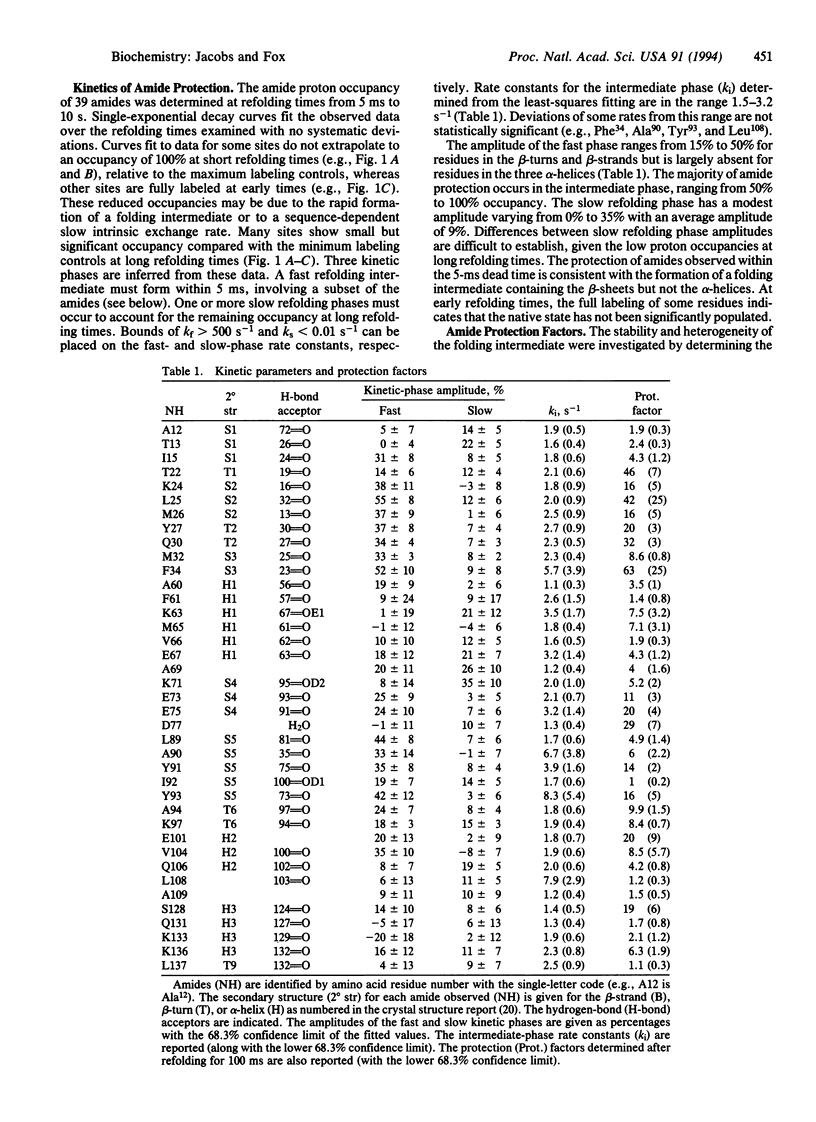
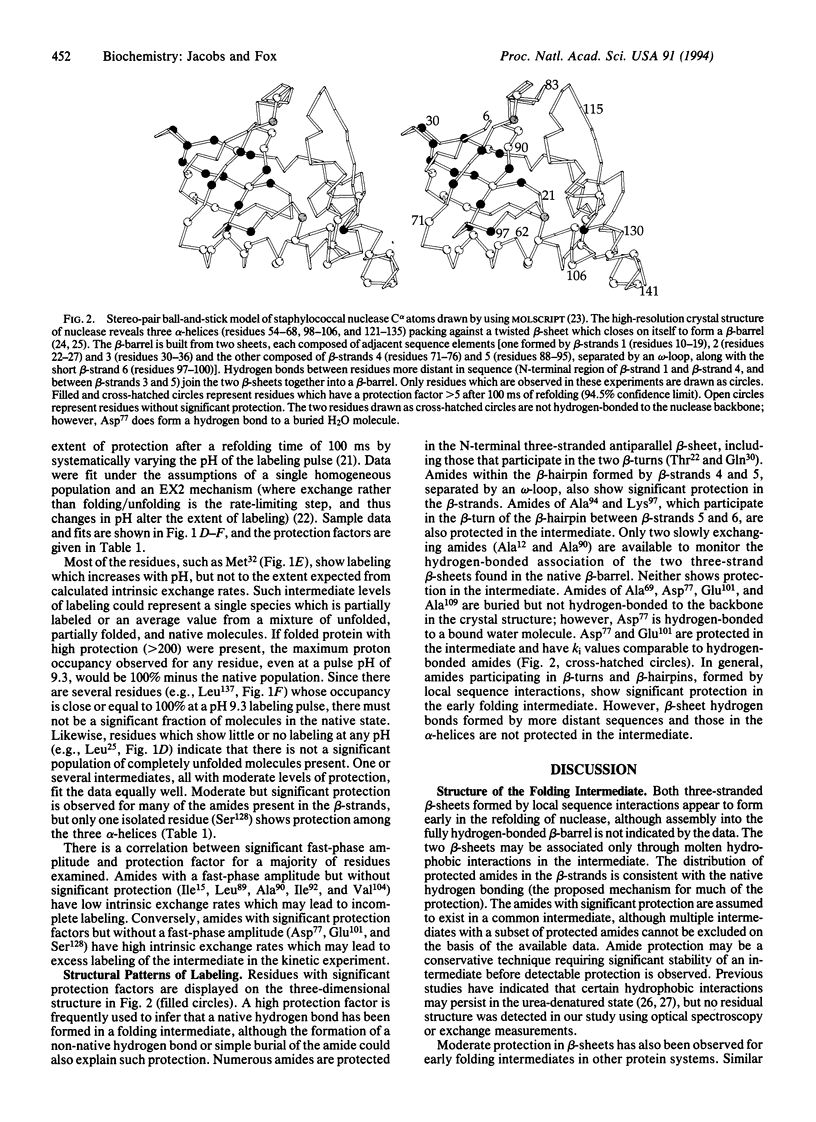
Abstract
Pulsed hydrogen-deuterium exchange during refolding was used to probe the protection of backbone amide hydrogens from solvent exchange of the staphylococcal nuclease Pro117-->Gly variant. The extent of exchange for 39 residues was determined by two-dimensional proton NMR after refolding for 5 ms to 10 s. Three kinetic phases are inferred. Modest protection of amides in the early refolding intermediate composed of two beta-sheets formed by local sequence interactions was observed after a 5-ms refolding period. Protection factors were determined by varying the high pH labeling pulse after refolding for 100 ms. The intermediate state has modest, yet significant, protection for residues in the beta-sheets (protection factors of 10-60) and almost no protection in the alpha-helices (protection factors of < 10). The pattern of labeling is consistent with a role for beta-turns and beta-hairpins in the formation of the early intermediate.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexandrescu A. T., Ulrich E. L., Markley J. L. Hydrogen-1 NMR evidence for three interconverting forms of staphylococcal nuclease: effects of mutations and solution conditions on their distribution. Biochemistry. 1989 Jan 10;28(1):204–211. doi: 10.1021/bi00427a028. [DOI] [PubMed] [Google Scholar]
- Alonso D. O., Dill K. A. Solvent denaturation and stabilization of globular proteins. Biochemistry. 1991 Jun 18;30(24):5974–5985. doi: 10.1021/bi00238a023. [DOI] [PubMed] [Google Scholar]
- Bai Y., Milne J. S., Mayne L., Englander S. W. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993 Sep;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs M. S., Roder H. Early hydrogen-bonding events in the folding reaction of ubiquitin. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2017–2021. doi: 10.1073/pnas.89.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. M., Markin V. S., Tsong T. Y. pH-induced folding/unfolding of staphylococcal nuclease: determination of kinetic parameters by the sequential-jump method. Biochemistry. 1992 Feb 11;31(5):1483–1491. doi: 10.1021/bi00120a027. [DOI] [PubMed] [Google Scholar]
- Chen H. M., You J. L., Markin V. S., Tsong T. Y. Kinetic analysis of the acid and the alkaline unfolded states of staphylococcal nuclease. J Mol Biol. 1991 Aug 5;220(3):771–778. doi: 10.1016/0022-2836(91)90116-n. [DOI] [PubMed] [Google Scholar]
- Chyan C. L., Wormald C., Dobson C. M., Evans P. A., Baum J. Structure and stability of the molten globule state of guinea-pig alpha-lactalbumin: a hydrogen exchange study. Biochemistry. 1993 Jun 1;32(21):5681–5691. doi: 10.1021/bi00072a025. [DOI] [PubMed] [Google Scholar]
- Collins K. D., Washabaugh M. W. The Hofmeister effect and the behaviour of water at interfaces. Q Rev Biophys. 1985 Nov;18(4):323–422. doi: 10.1017/s0033583500005369. [DOI] [PubMed] [Google Scholar]
- Connelly G. P., Bai Y., Jeng M. F., Englander S. W. Isotope effects in peptide group hydrogen exchange. Proteins. 1993 Sep;17(1):87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- Cotton F. A., Hazen E. E., Jr, Legg M. J. Staphylococcal nuclease: proposed mechanism of action based on structure of enzyme-thymidine 3',5'-bisphosphate-calcium ion complex at 1.5-A resolution. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2551–2555. doi: 10.1073/pnas.76.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A., Fiebig K. M., Chan H. S. Cooperativity in protein-folding kinetics. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1942–1946. doi: 10.1073/pnas.90.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A. Theory for the folding and stability of globular proteins. Biochemistry. 1985 Mar 12;24(6):1501–1509. doi: 10.1021/bi00327a032. [DOI] [PubMed] [Google Scholar]
- Englander J. J., Calhoun D. B., Englander S. W. Measurement and calibration of peptide group hydrogen-deuterium exchange by ultraviolet spectrophotometry. Anal Biochem. 1979 Jan 15;92(2):517–524. doi: 10.1016/0003-2697(79)90693-6. [DOI] [PubMed] [Google Scholar]
- Englander S. W., Downer N. W., Teitelbaum H. Hydrogen exchange. Annu Rev Biochem. 1972;41:903–924. doi: 10.1146/annurev.bi.41.070172.004351. [DOI] [PubMed] [Google Scholar]
- Englander S. W., Mayne L. Protein folding studied using hydrogen-exchange labeling and two-dimensional NMR. Annu Rev Biophys Biomol Struct. 1992;21:243–265. doi: 10.1146/annurev.bb.21.060192.001331. [DOI] [PubMed] [Google Scholar]
- Evans P. A., Kautz R. A., Fox R. O., Dobson C. M. A magnetization-transfer nuclear magnetic resonance study of the folding of staphylococcal nuclease. Biochemistry. 1989 Jan 10;28(1):362–370. doi: 10.1021/bi00427a050. [DOI] [PubMed] [Google Scholar]
- Evans P. A., Kautz R. A., Fox R. O., Dobson C. M. A magnetization-transfer nuclear magnetic resonance study of the folding of staphylococcal nuclease. Biochemistry. 1989 Jan 10;28(1):362–370. doi: 10.1021/bi00427a050. [DOI] [PubMed] [Google Scholar]
- Flanagan J. M., Kataoka M., Fujisawa T., Engelman D. M. Mutations can cause large changes in the conformation of a denatured protein. Biochemistry. 1993 Oct 5;32(39):10359–10370. doi: 10.1021/bi00090a011. [DOI] [PubMed] [Google Scholar]
- Hughson F. M., Wright P. E., Baldwin R. L. Structural characterization of a partly folded apomyoglobin intermediate. Science. 1990 Sep 28;249(4976):1544–1548. doi: 10.1126/science.2218495. [DOI] [PubMed] [Google Scholar]
- Hynes T. R., Fox R. O. The crystal structure of staphylococcal nuclease refined at 1.7 A resolution. Proteins. 1991;10(2):92–105. doi: 10.1002/prot.340100203. [DOI] [PubMed] [Google Scholar]
- Jeng M. F., Englander S. W., Elöve G. A., Wand A. J., Roder H. Structural description of acid-denatured cytochrome c by hydrogen exchange and 2D NMR. Biochemistry. 1990 Nov 20;29(46):10433–10437. doi: 10.1021/bi00498a001. [DOI] [PubMed] [Google Scholar]
- Kuwajima K., Okayama N., Yamamoto K., Ishihara T., Sugai S. The Pro117 to glycine mutation of staphylococcal nuclease simplifies the unfolding-folding kinetics. FEBS Lett. 1991 Sep 23;290(1-2):135–138. doi: 10.1016/0014-5793(91)81243-2. [DOI] [PubMed] [Google Scholar]
- Lewis P. N., Momany F. A., Scheraga H. A. Folding of polypeptide chains in proteins: a proposed mechanism for folding. Proc Natl Acad Sci U S A. 1971 Sep;68(9):2293–2297. doi: 10.1073/pnas.68.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll P. J., Lattman E. E. The crystal structure of the ternary complex of staphylococcal nuclease, Ca2+, and the inhibitor pdTp, refined at 1.65 A. Proteins. 1989;5(3):183–201. doi: 10.1002/prot.340050302. [DOI] [PubMed] [Google Scholar]
- Lu J., Dahlquist F. W. Detection and characterization of an early folding intermediate of T4 lysozyme using pulsed hydrogen exchange and two-dimensional NMR. Biochemistry. 1992 May 26;31(20):4749–4756. doi: 10.1021/bi00135a002. [DOI] [PubMed] [Google Scholar]
- Nakano T., Antonino L. C., Fox R. O., Fink A. L. Effect of proline mutations on the stability and kinetics of folding of staphylococcal nuclease. Biochemistry. 1993 Mar 16;32(10):2534–2541. doi: 10.1021/bi00061a010. [DOI] [PubMed] [Google Scholar]
- Ptitsyn O. B., Pain R. H., Semisotnov G. V., Zerovnik E., Razgulyaev O. I. Evidence for a molten globule state as a general intermediate in protein folding. FEBS Lett. 1990 Mar 12;262(1):20–24. doi: 10.1016/0014-5793(90)80143-7. [DOI] [PubMed] [Google Scholar]
- Shortle D., Stites W. E., Meeker A. K. Contributions of the large hydrophobic amino acids to the stability of staphylococcal nuclease. Biochemistry. 1990 Sep 4;29(35):8033–8041. doi: 10.1021/bi00487a007. [DOI] [PubMed] [Google Scholar]
- Sugawara T., Kuwajima K., Sugai S. Folding of staphylococcal nuclease A studied by equilibrium and kinetic circular dichroism spectra. Biochemistry. 1991 Mar 12;30(10):2698–2706. doi: 10.1021/bi00224a018. [DOI] [PubMed] [Google Scholar]
- Torchia D. A., Sparks S. W., Bax A. Staphylococcal nuclease: sequential assignments and solution structure. Biochemistry. 1989 Jun 27;28(13):5509–5524. doi: 10.1021/bi00439a028. [DOI] [PubMed] [Google Scholar]
- Udgaonkar J. B., Baldwin R. L. Early folding intermediate of ribonuclease A. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8197–8201. doi: 10.1073/pnas.87.21.8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udgaonkar J. B., Baldwin R. L. NMR evidence for an early framework intermediate on the folding pathway of ribonuclease A. Nature. 1988 Oct 20;335(6192):694–699. doi: 10.1038/335694a0. [DOI] [PubMed] [Google Scholar]
- Wang J. F., Hinck A. P., Loh S. N., Markley J. L. Two-dimensional NMR studies of staphylococcal nuclease: evidence for conformational heterogeneity from hydrogen-1, carbon-13, and nitrogen-15 spin system assignments of the aromatic amino acids in the nuclease H124L-thymidine 3',5'-bisphosphate-Ca2+ ternary complex. Biochemistry. 1990 May 1;29(17):4242–4253. doi: 10.1021/bi00469a029. [DOI] [PubMed] [Google Scholar]
- Wright P. E., Dyson H. J., Lerner R. A. Conformation of peptide fragments of proteins in aqueous solution: implications for initiation of protein folding. Biochemistry. 1988 Sep 20;27(19):7167–7175. doi: 10.1021/bi00419a001. [DOI] [PubMed] [Google Scholar]



