Abstract
Purines are molecules essential for many cell processes, including RNA and DNA synthesis, regulation of enzyme activity, protein synthesis and function, energy metabolism and transfer, essential coenzyme function, and cell signaling. Purines are produced via the de novo purine biosynthesis pathway. Mutations in purine biosynthetic genes, for example phosphoribosylaminoimidazole carboxylase/phosphoribosylaminoimidazole succinocarboxamide synthetase (PAICS, E.C. 6.3.2.6/E.C. 4.1.1.21), can lead to developmental anomalies in lower vertebrates. Alterations in PAICS expression in humans have been associated with various types of cancer. Mutations in adenylosuccinate lyase (ADSL, E.C. 4.3.2.2) or 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC, E.C. 2.1.2.3/E.C. 3.5.4.10) lead to inborn errors of metabolism with a range of clinical symptoms, including developmental delay, severe neurological symptoms, renal stones, combined immunodeficiency, and autistic features. The pathogenetic mechanism is unknown for any of these conditions, and no effective treatments exist. The study of cells carrying mutations in the various de novo purine biosynthesis pathway genes provides one approach to analysis of purine disorders. Here we report the characterization of AdeD Chinese hamster ovary (CHO) cells, which carry genetic mutations encoding p.E177K and p.W363* variants of PAICS. Both mutations impact PAICS structure and completely abolish its biosynthesis. Additionally, we describe a sensitive and rapid analytical method for detection of purine de novo biosynthesis intermediates based on high performance liquid chromatography with electrochemical detection. Using this technique we detected accumulation of AIR in AdeD cells. In AdeI cells, mutant for the ADSL gene, we detected accumulation of SAICAR and SAMP and, somewhat unexpectedly, accumulation of AIR. This method has great potential for metabolite profiling of de novo purine biosynthesis pathway mutants, identification of novel genetic defects of purine metabolism in humans, and elucidating the regulation of this critical metabolic pathway.
Keywords: phosphoribosylaminoimidazole carboxylase/phosphoribosylaminoimidazole succinocarboxamide synthetase (PAICS), adenylosuccinate lyase (ADSL), de novo purine biosynthesis, electrochemical detection, Chinese Hamster Ovary (CHO) cells
1. Introduction
Purines are essential building blocks for RNA and DNA synthesis, and regulate energy metabolism and transfer, and protein synthesis, function, and enzyme activity. Purines are also vital components of many essential coenzymes (NAD, NADH, FAD, Coenzyme A), and signaling molecules (cAMP, guanine nucleotides). The enzymatic steps of de novo purine biosynthesis convert PRPP (5-phospho-α-D-ribosyl-1-pyrophosphate) to IMP (inosine monophosphate), and an additional four steps convert IMP to AMP (adenosine monophosphate) or GMP (guanosine monophosphate) (Figure 1). Recent evidence demonstrates that the proteins carrying out de novo purine biosynthesis form a multienzyme complex, the purinosome, which is induced by acute requirement for purines [1-3]. Formation of the purinosome can be disrupted by mutations in ADSL (E.C. 4.3.2.2) and ATIC (E.C. 2.1.2.3/E.C.3.5.4.10), with severe developmental consequences in humans [3]. In addition to ADSL and ATIC, 30 enzyme defects of purine and pyrimidine metabolism have been identified and 17 of these are known to cause human disease [4]. The clinical presentation of genetic disorders of purine metabolism includes a wide variety of symptoms, such as severe combined immunodeficiency, severe neurological defects, developmental delay, and abnormal brain development [4-6]. The consequences of inborn errors in purine metabolism are poorly understood, and misdiagnosis most likely results in underestimation of their incidence and prevalence [4].
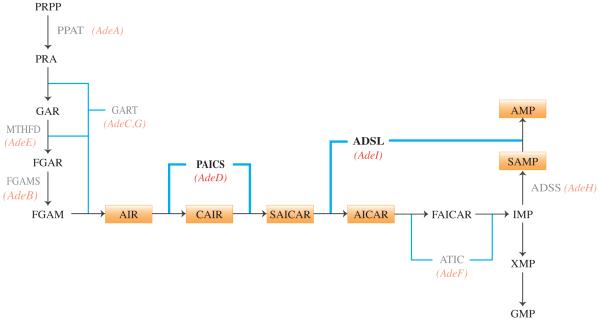
Figure 1.
The de novo purine biosynthesis pathway. Ten enzymatic steps produce IMP, and four additional steps produce either AMP or GMP. Orange boxes indicate pathway intermediates of interest. Red italics indicate CHO cell mutations, and affected enzymatic steps. Enzymes are indicated in gray text, and enzymatic steps are indicated (blue lines).
One approach utilized in the analysis of de novo purine biosynthesis and the role of the purinosome is the use of mammalian cells with mutations in the genes encoding enzymes of the pathway. These can be cells isolated from individuals with inborn errors of purine metabolism as reported previously [3], [7], or experimentally derived cells which carry mutations in de novo purine biosynthesis genes. We have previously reported the isolation and initial characterization of Chinese hamster ovary cell (CHO-K1) mutants presumably defective in each of the seven proteins required for de novo AMP synthesis [8], [9]. Here we report the further characterization of two of these, AdeD and AdeI. AdeD is deficient in bifunctional phosphoribosylaminoimidazole carboxylase/phosphoribosylaminoimidazole succinocarboxamide synthetase (PAICS, E.C. 6.3.2.6/E.C. 4.1.1.21) activity, responsible for conversion of AIR (5-aminoimidazole ribotide) to CAIR (5-amino-4-carboxyimidazole ribotide) and conversion of CAIR to SAICAR (5-amino-4-imidazole-N-succinocarboxamide ribotide). AdeI is deficient in bifunctional adenylosuccinate lyase (ADSL) activity, responsible for conversion of SAICAR to AICAR (5-amino-4-imidazolecarboxamide ribotide) and conversion of SAMP (adenylosuccinate) to AMP (adenosine monophosphate). We previously characterized AdeI, and presented evidence that AdeI is a useful model for the cellular consequences of ADSL deficiency [10].
The metabolic intermediates SAICAR (the product of PAICS and a substrate for ADSL) and AICAR (the product of ADSL) regulate the expression of purine pathway genes in yeast cells [11]. When purine levels are deficient, accumulated AICAR enhances binding of pho2p, pho4p, and bas1p transcription factors to purine and phosphate pathway gene promoters [11]. SAICAR enhances pho2p and bas1p binding, which specifically positively regulates purine regulon genes [11]. Whether these intermediates have similar effects in mammalian cells is not known. AICAr (the riboside form of AICAR) is a potential antitumor agent and promotes apoptosis in aneuploid cells [12], [13]. Administration of AICAr to sedentary mice mimicked the effects of exercise, resulting in increased oxidative biomarkers in cultured skeletal muscle cells and enhanced running endurance [14]. It has also been demonstrated that inhibition of adenylosuccinate lyase activity by SAICAR results in skeletal muscle dysfunction [15]. These findings show that altering the balance of SAICAR and AICAR disrupts cellular functions, including energy metabolism and the regulation of nucleotide synthesis and phosphate consumption.
In addition to its role in supplying the substrate for ADSL, PAICS is of interest because mutations in PAICS cause errors in vertebrate embryonic development [16], and because PAICS shows elevated expression in numerous cancers, and may be an important target for anticancer therapies [17], [18]. PAICS overexpression is likely due to the increased proliferation of tumor cells and consequent increased demand for purine nucleotides. Since the two steps catalyzed by PAICS in vertebrates are catalyzed by three enzymes (PurK, PurE, and PurC) in bacteria, and the mechanism of catalysis is different, PAICS is a target for development of antimicrobial drugs [19]. If the PAICS deficiency observed in AdeD is due to mutation in the PAICS gene, it could serve as a mammalian cell culture model for analysis of the mechanism of mammalian PAICS catalysis, which should be informative in the development of antimicrobials or vaccines that target SAICAR synthesis [20], the role of PAICS mutations in abnormal development, and cancer metabolism.
Disruption of de novo purine biosynthesis can result in the accumulation of intermediary metabolites in cells and body fluids. The detection of these intermediates is difficult, which is problematic for studying de novo purine biosynthesis and identifying inborn errors of purine metabolism. This may be why mutations in PAICS have not yet been found in humans. A previous study used high performance liquid chromatography (HPLC) coupled with pulsed amperometry detection to detect phosphoribosylglycinamide (GAR), the product of the second step of de novo purine biosynthesis. This suggests that electrochemical detection may be feasible for detection of de novo purine biosynthesis pathway intermediates [21]. Since the system described requires gradient elution and has limited dynamic range, we used HPLC coupled coulometric detection (HPLC-EC) to analyze intermediate accumulation in AdeD and AdeI cells. Coulometric detection has a number of advantages over amperometric detection, including increased sensitivity, dynamic range, and simplicity of quantitation [22], [23].
Here we report the identification of the mutations in AdeD cells in the PAICS gene and their effect on PAICS mRNA and protein expression. Moreover, we report the comparative analysis of AdeD and AdeI intermediate accumulation using HPLC-EC. This method readily detects accumulation of AIR in AdeD cells and SAICAR and SAMP in AdeI cells and, somewhat unexpectedly, accumulation of AIR in AdeI cells.
2. Materials and Methods
2.1 Identification and analysis of PAICS mutations
2.1.1 Identification of PAICS mutations in AdeD by RT-PCR
Total RNA was isolated from CHO-K1 and AdeD cells using the RNAqueous4PCR kit (Ambion). AdeD and CHO-K1 cDNA was prepared from the isolated total RNA using the RETROscript kit (Ambion). The PAICS cDNA was amplified with appropriate PCR primers (Table 1) using the Expand Hi-Fidelity PCR kit (Roche) and thermocycler parameters: 95° C for 5 minutes, (95° C for 30 seconds, 60° C for 30 seconds, and 72° C for 30 seconds) for 35 cycles, and 72° C for 7 minutes. PCR products were analyzed by TAE agarose gel electrophoresis. Single bands were excised from the gel and the PCR product was purified using the Zymoclean Gel DNA Recovery kit (Zymo Research). DNA concentration was estimated using TAE agarose gel electrophoresis by comparison with Low Mass DNA or High Mass DNA ladders (Invitrogen). DNA samples containing ~60 ng/kb fragment length were sequenced by the University of Colorado Cancer Center DNA Sequencing and Analysis Core.
Table 1.
Primers used in this study
| PAICS Primers | ||
|---|---|---|
| Primers | 5′-3′ Sequence | Purpose |
| 245_PAICS_CHO_F | GGGAGACTGCTTTCATTGCACCCCA | RT-PCR & Sequencing |
| 1509_PAICS_CHO_R | TCAGCCTCTGGTGTGCTGAGGTTAC | |
| 406_PAICS_CHO_F | AATGACCCGCAGTGGTCTGAGGAGC | |
| 1510_PAICS_CHO_R | CTCAGCCTCTGGTGTGCTGAGGTT | |
| 1095_PAICS_CHO_R | GGATGACCACACATCCTGAGCACCC | |
| 807_PAICS_CHO_F | AGTGCTGATGGGTTCCACTTCTGAC | |
| 1265_PAICS_CHO_R | TCTCTGATTTTCTTGTCAGCTTGCTTT | |
| 5U-PAICS-1F | GCGCTGCTTCTCTTTCCCGCGCC | |
| T7 | TAATACGACTCACTATAGGG | pTarget Sequencing |
| pTarget.seq | TTACGCCAAGTTATTTAGGTGACA | |
| VP1.5 | GGACTTTCCAAAATGTCG | pCMV6-hPAICS Sequencing |
| XL39 | ATTAGGACAAGGCTGGTGGG | |
| 5U-IF-PAICS-4F | CGTGATATCTTTCCCTGCGTTGCGCCGCCCTTTCC | CHO PAICS Cloning |
| 3U-IF-PAICS-4R | GTCGACGGTACCCCCTTTCTGCCTCAGCCTCTGGTGTGC | |
| CHO_PAICS_F1 | CTGATGGGTTCCACTTCTGACCTTG | qPCR |
| CHO_PAICS_R1 | GCCATCGCCTTCATACTCTGCTT | |
| CHO_ACTB_F1 | CTTCGCGGGCGACGATGCT | |
| CHO_ACTB_R1 | GGCCCATGCCCACCATCACG | |
Sequence alignment with BLAST [24], [25] using the Mus musculus PAICS cDNA sequence (NCBI NM_025939.2) identified a CHO PAICS cDNA sequence, JP049561.1. Primers for RT-PCR were designed using the JP049561.1 sequence, however, nucleotides +1 to +217 were missing from the 5′-end. A reference CHO PAICS cDNA sequence was constructed by adding 431 nt from a predicted sequence for CHO PAICS (NCBI XM_003513528.1, 2168 bp) onto the 5′-end of JP049561.1. All PAICS RT-PCR primers (Table 1) were numbered relative to the +1 nucleotide in the XM_003513528.1 sequence. The PAICS amino acid sequence derived from JP049561.1 is identical to that from XM_003513528.1, except JP049561.1, like human, mouse, and rat PAICS sequence, has a K at p.418, while XM_003513528.1 has a Q at this position. The resulting CHO reference sequence was used to perform alignments with sequencing data from CHO-K1 and AdeD cDNA. The Needle program at EBI was used for pairwise alignments with our CHO reference sequence. After identification of polymorphisms in the CHO-K1 and AdeD RT-PCR data, sequencing reads were translated in all reading frames to determine whether any SNP resulted in an amino acid change.
2.1.2 Cloning of PAICS from CHO-K1 and AdeD cDNA
PAICS was amplified from CHO-K1 and AdeD cDNA using the same PCR protocol described above for RT-PCR. However, the Expand Hi-Fidelity Long Template PCR kit (Roche) was used instead of the Expand Hi-Fidelity Template PCR kit (Roche), following the manufacturer’s protocol. The In Fusion PCR primers for PAICS are 5U-IF-PAICS-4F and 3U-IF-PAICS-4R (Table 1). The PCR reactions were cleaned using Nucleospin Plasmid (Clontech).
The pTarget vector (Promega) was linearized overnight with Sma I restriction endonuclease and cleaned. The DNA concentration of linearized pTarget and PAICS AdeD PCR product was determined by gel electrophoresis as described earlier. The In Fusion HD Cloning Kit (Clontech) was used to ligate the PAICS AdeD In Fusion RT-PCR products into the pTarget vector.
2.1.3 Mini-preps and DNA sequencing of PAICS-AdeD and PAICS-CHO-K1-pTarget Clones
Mini-preps of plasmid DNA from ampicillin-resistant clones were performed using the Nucleospin Plasmid (Clontech) kit. TAE agarose gel electrophoresis was used to estimate the size of plasmids recovered from PAICS-AdeD-pTarget and PAICS-K1-pTarget clones, and to estimate DNA concentration. DNA sequencing was performed using the T7, pTarget seq, 807F, 1095R, and 1265R CHO PAICS primers (Table 1). Samples were sequenced by the University of Colorado Cancer Center DNA Sequencing and Analysis Core.
2.1.4 Maxi-prep of PAICS-K1-pTarget, E177K-PAICS-pTarget, and W363Stop-PAICS-pTarget plasmids
DNA maxi preps were performed using the Nucleobond Xtra Midi Plasmid DNA Purification kit (Clontech) according to the manufacturer’s protocol, with some modifications. The kit comes with a filter to clarify cell lysates simultaneous with column purification. We clarified cell lysates by centrifugation at 12,000 × g (8,500 RPM) for 50 minutes in a Sorvall RC-5B Superspeed Centrifuge, followed by filtration. The plasmid DNA was concentrated and desalinated using the NucleoBond Finalizer Plasmid DNA Concentration and Desalination kit (Clontech). The quality and quantity of plasmid DNA was quantified by UV spectroscopy at 260 and 280 nm.
2.1.5 Transfection of cDNA expression plasmids PAICS-K1-pTarget, E177K-PAICS-pTarget, and W363Stop-PAICS-pTarget into AdeD
For each plate, 500 μl OptiMEM I reduced serum medium (Gibco), 20 μl Lipofectamine 2000 (Invitrogen), and 8 μg total DNA were combined according the manufacturer’s protocol (Invitrogen Lipofectamine 2000). The transfection mixture was added to confluent 60mm plates of AdeD cells and incubated overnight at 37° C, 5% CO2. After 24 hr, the medium was replaced with F12 medium supplemented with 10% FCS, Normocin (100 μg/ml), and 3 × 10−5 M adenine. After an additional 24 hr, the medium was changed to F12 medium containing Geneticin (400 μg/ml) for selection of pTarget-transfected cells and supplemented with 10% FCS, Normocin (100 μg/ml), and 3 × 10−5 M adenine. The purine requirement of AdeD cells stably transfected with wild type (WT) and mutant PAICS plasmid cDNAs was assessed by plating cells into α-MEMFCM10 with or without 1 × 10−4 M hypoxanthine.
2.2 Analysis of PAICS protein from CHO-K1 and AdeD cells
2.2.1 Protein Extraction
CHO-K1, AdeD and AdeI mutant cells were grown in F12 supplemented with 10% fetal calf serum (FCS), Normocin, and 3 × 10−5 M adenine when necessary. Confluent 60 mm plates of cells were rinsed with 1X PBS and the plates were scraped using cell scrapers. The total volume of extracted material was centrifuged for 10 minutes at 2,000 RPM at 4° C. The pelleted cells were resuspended in buffer (10 mM Tris-HCl pH 8.3, 10 mM KCl, 2 mM EDTA, 1 mM DTT, 4% glycerol) with Inhibitor Protease Cocktail Tablets (Roche) and lysed by sonication twice for 15 seconds at 40 W. The homogenate was centrifuged for 20 minutes at 17,000 × g at 4° C and protein in supernatant was measured by Bradford assay (Sigma).
2.2.2 Western Blot Analysis for PAICS protein
Total CHO-K1, AdeD, and AdeI protein (20 μg/lane) was separated by SDS-PAGE (10% gel) for 4 hours at 130 V. The gel was wet-blotted to PVDF membrane in blotting buffer (48 mM Tris, 39 mM glycine, 1.3 mM SDS, 20% methanol) at 100 mA constant current for 2 hours. The blot was blocked overnight at 4° C in block solution (5% BSA, 1X PBS, 0.05% Tween-20, pH 7.4) followed by a 3 hour incubation in anti-PAICS antibody (Sigma-Aldrich HPA035895) diluted 1:150 in block solution. This was followed by five washes (1, 3, 5, 10 and 15 minutes) in wash solution (1X PBS/0.05% Tween-20). The blot was then incubated in goat anti-rabbit IgG-HRP conjugate (Pierce-Thermo Scientific 31460) diluted 1:10,000 in block solution for 40 minutes followed by five washes in wash solution. Blots were treated with chemiluminescent SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) and visualized on a Syngene Genomic and Proteomic Gel Documentation System (Syngene, Cambridge, UK). The blot was rinsed in 1X PBS and incubated in mouse monoclonal anti-GAPDH antibody (Sigma-Aldrich G8795) diluted 1:10,000 in block solution for 90 minutes, followed by five washes. The blot was then incubated in goat anti-mouse IgM-HRP conjugate (Pierce-Thermo Scientific 31440) diluted at 1:20,000 in block solution for 40 minutes, followed by five washes and visualized as described above.
2.2.3 Expression levels of PAICS transcript in CHO cells
PAICS expression levels in CHO cells (AdeD and CHO-K1) were determined by quantitative PCR (qPCR) and ΔΔCt analysis. Total RNA isolation and subsequent cDNA synthesis was carried out as described above (methods 2.1). The cDNA was used as a template in 25 ul reactions using the iQ SYBER Green Supermix kit (BioRad) with 500 nM CHO specific PAICS primers (Table 1). The qPCR cycling conditions were as follows: 95° C for 5 min followed by 35 cycles of denaturation at 95° C for 10 seconds, annealing at 60° C for 30 seconds followed by SYBER Green data collection, and extension at 72° C for 30 seconds. CHO beta-actin was amplified as a reference control using specific primers (Table 1).
2.3 HPLC-EC Analysis of CHO mutant intermediate accumulations
2.3.1 Accumulation of intermediates
CHO-K1 and the various adenine-requiring mutants were grown in 60 mm dishes in αMEM supplemented with 10% fetal calf serum (FCS), 3 × 10−5 M adenine, and 100 μg/mL Normocin. Accumulation of intermediates was induced by growth in purine-free medium (purine starvation) as follows. Medium in confluent plates was replaced with either fresh αMEM containing 10% FCM (twice dialyzed FCS), Normocin, and 10−4 M adenine (which we have previously shown drastically reduces de novo purine biosynthesis in CHO cells) or with fresh αMEM containing FCM and Normocin but lacking adenine. The cells were then incubated for 4-6 hours. After incubation plates were washed twice with 1 ml cold 1X PBS then incubated for 30 minutes in 500 μl of 80% ethanol at 4° C. The plates were scraped using cell scraper and the ethanol/extract mixture was transferred to 1.5 ml microcentrifuge tube and centrifuged for 10 minutes at 14,000 × g at 4 °C. The supernatant was transferred to a microcentrifuge tube and immediately frozen at −80°C. The frozen extracts were dried under a vacuum in a SpeedVac. The concentrated samples were resuspended in 200 μl HPLC grade water [ultrapure (18.2 MΩ cm) water, which was filtered through 0.2 μm filter and polished through a C18 Sep-Pak column]. Resuspended samples were centrifuged for 15 minutes at 14,000 × g at 4°C to remove any remaining cellular debris prior to analysis.
2.3.2 HPLC-EC Analysis
Separation and measurement of CHO mutant intermediate accumulation was performed using reverse phase HPLC-EC with a TSKgel ODS-80Tm C-18 column (250 mm × 4.6 mm ID, 5 μm) protected by Tosoh Bioscience TSKgel guard cartridge. A column temperature of 35° C was maintained throughout the analysis. A mobile phase consisting of 50 mM lithium acetate, 2% acetonitrile, 5 mM tetrabutyl ammonium phosphate (TBAP), pH 4.8 was delivered isocratically at a flow rate of 0.7 ml/min. Sample extracts and standards were kept at 10° C until a 20 μl aliquot of each sample was injected using an ESA autosampler (model 542). After injection and separation, analytes were detected using a CoulArray HPLC system (model 5600A, ESA) with three electrochemical detector modules. Each module contains four flow-through coulometric detectors in series set to a range of potentials from 0–900 mV in 100 mV increments. Also in series with the coulometric detectors was a UV detector set at 240 nm. ESA CoulArray software was used for baseline correction and analysis of all samples.
3. Results
3.1 Identification of the PAICS mutations in AdeD cells
We have shown previously that AdeD cells apparently accumulate AIR, due presumably to mutation of the PAICS gene [9]. The cDNA coding region for CHO-K1 and mouse PAICS is 1278 nucleotides long with 426 codons including the TAA stop codon. Sequencing of PAICS RT-PCR products from CHO-K1 and AdeD cells provided approximately 80% of the PAICS cDNA sequence. Comparison to our reference PAICS cDNA sequence revealed two sequence variants unique to AdeD, and one sequence variant common to both AdeD and CHO-K1 (Table 2). This latter variant is p.I237V (c.A709G), located in the loop between β14 and α5. A BLAST search reveals that most organisms (including mouse and human) have a V at this position, and we tentatively hypothesize that V is the correct residue in CHO cells. Interestingly, p.237 is missing in the structure of human PAICS (2H31.pdb); “Asp221-Thr238 were not visible in the electron density map and assumed being disordered in the crystal” [26]. This suggests that the region containing p.237 is quite flexible, and that replacement of valine with isoleucine (a conservative change) would have little effect on the monomer structure.
Table 2.
Identification of polymorphisms in CHO-K1 and AdeD PAICS cDNA by RT-PCR. The polymorphisms are p.E177K (c.G529A), p.W363* (c.G1088A), and p.I237V (c.A709G). ND indicates no polymorphisms detected.
| PAICS cDNA Sequence Polymorphisms | ||||
|---|---|---|---|---|
| Primers |
||||
| Cell Line | RT-PCR | Sequencing | Verified Sequence | Polymorphisms |
| K1 | 245F/1265R | 1095R | 244-1015 | A709G |
| 245F/1265R | 807F | 863-1263 | ND | |
| 5U-1F/1095R | 1095R | 433-883 | A709G | |
| 807F/1509R | 807F | 926-1278 | ND | |
| AdeD | 245F/1509R | 1095R | 277-1016 | G529A, A709G |
| 245F/1265R | 1265R | 716-1166 | G1088A | |
| 5U-1F/1095R | 1095R | 483-883 | G529A, A709G | |
| 807F/1509R | 807F | 883-1278 | G1088A | |
The two variants found only in AdeD PAICS are p.E177K (c.G529A), located in α4, and p.W363* (c. G1088A), located in α9. The AdeD and CHO-K1 cDNAs were ligated into pTarget and sequenced. The sequences were compared to the PAICS reference sequence described earlier (431 nt from a predicted CHO-K1-PAICS sequence stitched to the 5′-end of JP049561.1). Our CHO-K1 PAICS sequence is presented (see Figure 2) and differs from our reference sequence at c.G-5A (upstream of the coding region), c.A153G (a silent mutation), and c.A709G (p.I237V). In CHO-K1, it is possible that one PAICS allele has isoleucine at p.237 and the other has valine. The PAICS sequence in our pTarget clone has p.237V. Both JP049561.1 and XM_003513528.1 have c.A709 (p.I237), however valine is evolutionarily conserved at p.237 (Li et al. 2007).
Figure 2.
DNA sequence of PAICS-K1 cloned into pTarget. The adenine in the start (ATG) codon is designated +1. Mutations identified in AdeD are underlined: c.G1088A (p.W363*) and c.G529A (p.E177K). PAICS DNA sequence in capital letters represents the truncated cDNA sequence from JP049561.1. PAICS DNA sequence from the predicted sequence, NCBI XM_003513528.1, is in lowercase letters. Our K1 PAICS sequence is presented and differs from our XM_003513528.1: JP049561.1 reference sequence at c.G-5A and c.A153G (a silent mutation). The valine (V) at position p.237 (c.A709G) differs from the published K1 PAICS, JP049561.1, which when translated has isoleucine (I) at this position.
We hypothesized that the two PAICS variants in AdeD represent two different PAICS alleles. To test this, we ligated AdeD PAICS cDNA into pTarget and isolated subclones for sequencing. Some subclones contained the c.G529A (p.E177K) variant, and some contained the c.G1088A (p.W363*) variant. The c.A709G (p.I237V) variant was present only in c.G529A (p.E177K) subclones (Figure 3 A). None of the PAICS-AdeD-pTarget clones had the wild type sequence. The PAICS cDNA sequence of K1, and both AdeD alleles in our pTarget clones have been deposited in GenBank (NCBI accession numbers KC176530, KC176531, and KC176532). The AdeD p.E177K allele (KC176531) is referred to as “allele 1” and the p.W363* allele (KC176532) is referred to as “allele 2”.
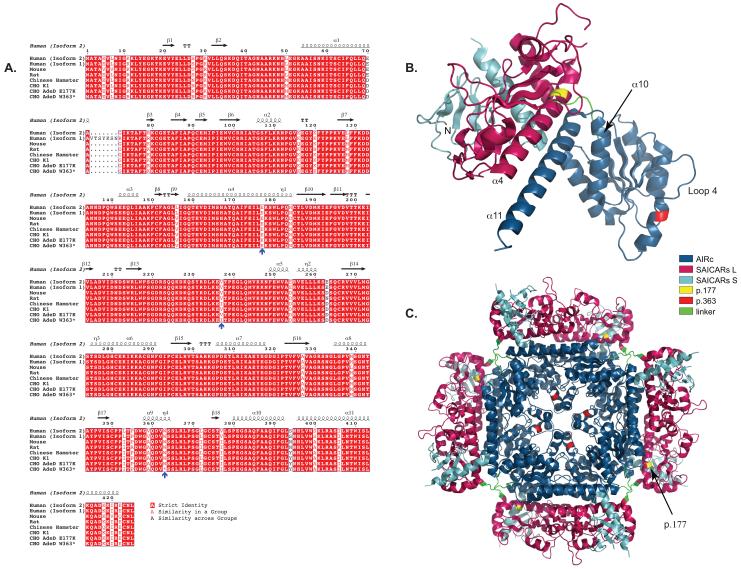
Figure 3.
COBALT alignment of PAICS protein sequences using Genbank IDs. The Genbank IDs for the amino acid sequences are Human (Isoform 2): 119220557, Human (Isoform 1): 119220559, Mouse: 13385434, Rat: 18266726, Chinese Hamster: 354503014. CHO K1, CHO AdeD E177K, and CHO AdeD W363* are described in this report. The E177K, I237V, and W363* mutations are indicated by blue arrows. Secondary sequence is indicated above the aligned sequences. This representation of the aligned sequences was created using the Espript program (http://espript.ibcp.fr/ESPript/ESPript/).
3.2 The mutations in AdeD lead to lack of detectable PAICS protein
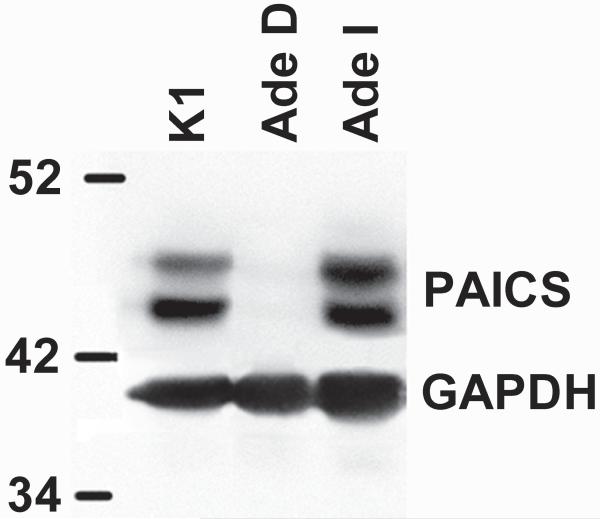
The p.W363* PAICS variant would be expected to be incapable of producing a full-length protein. However, to address the possibility that the p.E177K variant produces a full-length inactive protein, we carried out western blot analysis of protein extracts from CHO-K1 and AdeD. The result demonstrates that CHO-K1 contains PAICS protein as expected. However, AdeD does not contain detectable levels of PAICS protein (Figure 4). We also assessed whether PAICS is present in AdeI, which has a mutation in ADSL, the next enzyme in the de novo purine biosynthetic pathway. PAICS is abundant in AdeI cells. Two bands of similar molecular weight were detected, possibly representing different PAICS isoforms.
Figure 4.
Western blot of CHO-K1, AdeD, and AdeI protein (20μg/lane). The antibodies for detecting PAICS are.polyclonal anti-PAICS antibody (Sigma-Aldrich HPA035895), and goat anti-rabbit IgG-HRP conjugate (Pierce-Thermo Scientific 31460). The antibodies for detecting GAPDH are monoclonal anti-GAPDH antibody (Sigma-Aldrich G8795), and goat anti-mouse IgM-HRP conjugate antibody (Pierce-Thermo Scientific 31440).
3.3 AdeD produces abundant amounts of PAICS mRNA
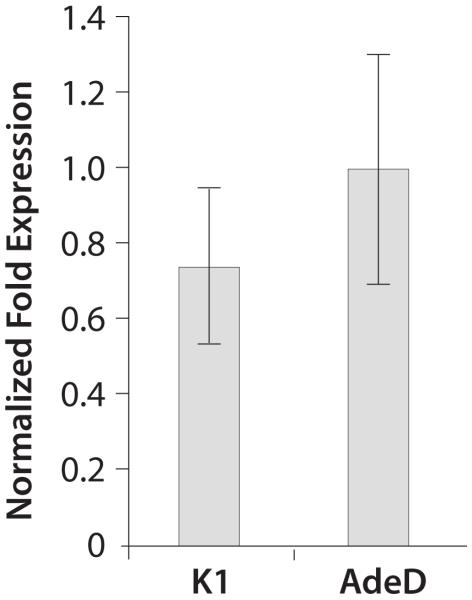
We performed qPCR to determine the level of PAICS mRNA in AdeD and CHO-K1 cells. Figure 5 shows the relative fold expression of PAICS mRNA in CHO-K1 and AdeD cells. There was no significant difference in PAICS transcript levels in AdeD cells compared to CHO-K1.
Figure 5.
qPCR analysis of PAICS mRNA levels in CHO-K1 and AdeD cells. Total RNA was isolated and cDNA was prepared from each cell line. Normalized fold expression levels of PAICS transcript in K1 and AdeD was quantified by qPCR, in triplicate, using CHO PAICS specific primers and beta-actin reference gene primers. Relative expression levels were determined using the ΔΔCt method.
3.4 Alleviation of the purine requirement of AdeD cells by transfection with CHO-PAICS cDNA
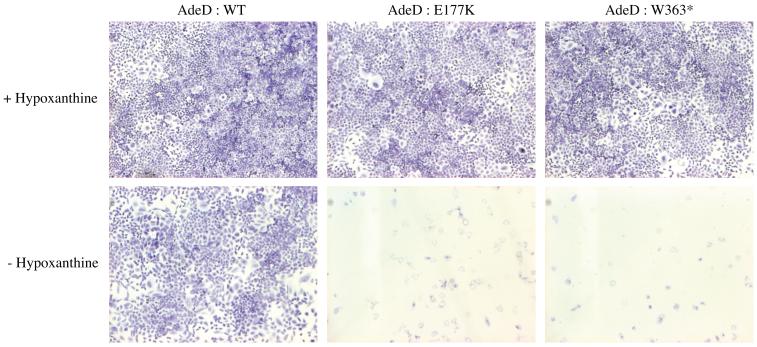
To confirm that the AdeD mutations inactivate PAICS, we constructed vectors with wild type (WT) and both mutant PAICS. We then transfected AdeD cells with each of the PAICS vectors. As expected, transfection with WT PAICS cDNA rescued the AdeD purine requirement, but transfection with either mutant clone did not. This demonstrates that the mutations in AdeD inactivate PAICS (Figure 6).
Figure 6.
Cell growth of AdeD cells stably transfected with WT and mutant PAICS (E177K and W363X) cDNA plasmids. Purine requirement was assessed by plating cells into supplemented (+Hypoxanthine) or purine deficient (−Hypoxanthine) a MEMFCM10 medium.
3.5 HPLC-EC analysis of AdeI and AdeD intermediate accumulation
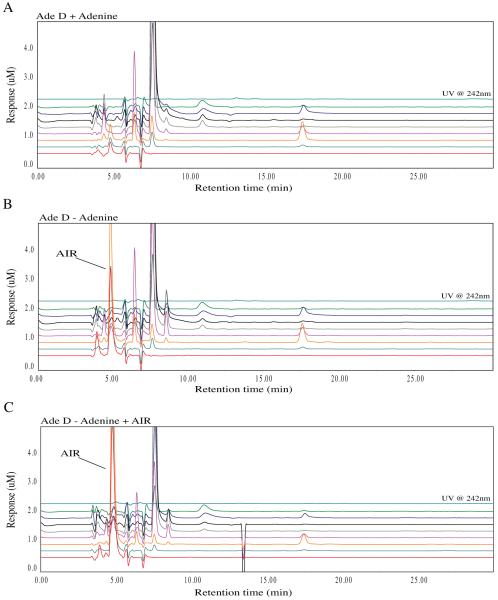
We previously demonstrated that AdeD cells accumulate an intermediate presumed to be AIR [9]. To confirm that this intermediate is indeed AIR, we devised a simple method for detecting AIR that does not depend on radioisotopic labeling. Using HPLC-EC, we found that AdeD accumulates two compounds upon incubation in purine-free medium. To determine whether one of these is AIR, we performed HPLC-EC analyses employing standard AIR (validated by mass spectroscopic analysis, using a method devised in our laboratory to be described elsewhere) and demonstrated that AIR co-elutes, and co-oxidizes, with one of the two compounds. The purine deprived cells show accumulation of a major peak with a retention time of 5 minutes when compared with adenine supplemented cells (Figure 7 A, B). The addition of AIR to the AdeD sample from cells incubated in purine-free media does not yield a new peak, but instead increases the intensity of an existing peak. AIR standard alone yields a significant peak with the same 5 minute retention time and the same oxidation pattern.
Figure 7.
Metabolomic analysis of AdeD (PAICS) mutant CHO cell starvation. The traces show the baseline profile of AdeD unstarved (A) and the accumulation of a peak at 5 minutes with a maximum peak height in the 400 mV EC channel in the starved sample (B). This peak corresponds to the appearance of a peak at 5 minutes with the same peak profile in the Ade D sample spiked with AIR standard (isolated by Zikanova) (C).
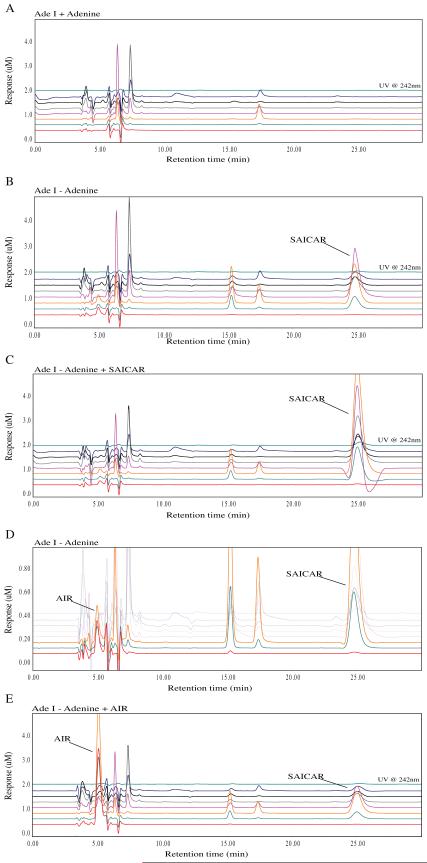
Similarly, we examined the accumulation of purine intermediates in AdeI cells. When cultured in purine-free media, AdeI cells accumulate a compound identified as SAICAR (Figure 8 B, C). They also accumulate an additional compound that appears from retention time and oxidation characteristics to be the same compound accumulated by purine-depleted AdeD. Co-chromatography of AdeI extracts with standard AIR confirms that AdeI, like AdeD, accumulates AIR (Figure 8 D). There are no differences detected in the intermediate accumulation of starved versus unstarved CHO-K1 cells.
Figure 8.
Metabolomic analysis of the AdeI (ADSL) CHO cell starvation. The traces show the baseline profile of AdeI unstarved (A) and the accumulation of a peak at 25 minutes in the starved sample (B). This corresponds to a peak in the unstarved sample spiked with a SAICAR standard (C). Additionally, a small peak at 5 minutes reminiscent of AIR accumulated in small concentrations in the AdeI starved samples. This suggests that accumulation of SAICAR may inhibit the function of PAICS resulting in AIR accumulation (D). This peak corresponds to AIR in starved AdeI cells spiked with AIR (E).
4. Discussion
Analysis of AdeD PAICS cDNA sequences revealed two sequence variants, p.E177K and p.W363* which map to separate alleles. AdeD was produced by treatment of CHO cells with EMS, which induces point mutations, consistent with our characterized (point) mutants. The CHO-K1 variants c.G-5A and c.A153G probably have no consequence for PAICS protein structure and function. One is upstream of the start codon and the other is silent. The c.G-5A and c.A153G variants differ from the predicted CHO PAICS sequence (NCBI XM_003513528.1, 2168 bp).
Except for p.W363* (AdeD), p.E177K (AdeD), and p.I237V [AdeD (the p.I237V allele) and CHO-K1], no other differences were found comparing our AdeD and CHO-K1 PAICS sequences to JP049561.1. These three variants were identified both by RT-PCR and by sequencing the inserts of pTarget clones containing the PAICS cDNA from either CHO-K1 or AdeD cells. Transfection of AdeD cells with WT and mutant PAICS vectors demonstrates that WT PAICS cDNA rescues the AdeD purine requirement, but transfection with either mutant cDNA does not. Western blot analysis demonstrated expression of PAICS protein in CHO-K1 and in AdeI, but not in AdeD. These results and the observed abundance of PAICS mRNA in AdeD cells strongly suggests that both the mutations observed in PAICS lead to production of truncated and/or unstable protein.
The p.E177K substitution is located in the SAICARs domain of PAICS in an alpha helical (α4) region of the protein important for interaction of the SAICARs and AIRc domains as well as maintenance of SAICARs structure (Figure 3). It may also play a role in substrate channeling within PAICS [26]. The fly, frog, chicken, cattle, mouse, and human PAICS all have p.E177 in the α4 helix [26]. At physiological pH, glutamate (E) is negatively charged. In the structure for human PAICS (2H31.pdb, Li et al. 2007), p.E177 is extremely close to p.R102 (arginine), which is located next to β6, between β6 and α2. Arginine is positively charged, and it is possible that attraction between p.E177 and p.R102 in the wild type protein may play a role in stabilizing PAICS monomer structure. If this is the case, p.K177 (lysine) would be disruptive; it is positively charged, so rather than attraction between it and p.R102, it would most likely repel p.R102. In addition, K has a longer side chain than E, so steric hindrance may also be a destabilizing factor. This is consistent with the possibility that the mutation affects the folding of AdeD PAICS, thus leading to an unstable protein. Indeed, analysis of this mutation using PoPMuSiC 2.1 calculates a ΔΔG of 0.41 and predicts that this is a destabilizing mutation for PAICS [27], [28].
It is unlikely that the p.W363* mutant monomer could participate in the octameric form of the PAICS protein since structures beyond the carboxyl end of p.362, such as the α10 and α11 helices believed to form the dyad symmetry interface crucial for assembly of the octamer, and loop 4, which forms part of the AIRc active site [26], would be absent. The seven-turn α11 helix interacts with the six-turn α4 helix in a coiled coil, so this interaction would be absent as well (Figure 3). AIRc activity and assembly of an octamer would be highly unlikely [26].
Previously, we characterized mutations in AdeI (A291V in ADSL) and AdeC and AdeG (both deficient in trifunctional GART activity) [10], [29]. Here we report characterization of two sequence variants, p.E177K and p.W363*, in two PAICS alleles that are responsible for the purine auxotrophy observed in AdeD. Thus, defined mutants exist in 3 of the 6 proteins required for de novo purine biosynthesis in mammalian cells. We are currently characterizing the mutations in the other mutant CHO cell lines (Figure 1).
We have reported previously that isolates of AdeC and AdeG produce markedly reduced or undetectable levels of trifunctional GART (triGART) protein and also of monofunctional GARS [30]. In addition we have published evidence that CHO-K1 AdeB mutants produce undetectable levels of FGAMS (phosphoribosylformylglycinamidine) [31], [32]. We have also reported a mutant CHO-K1 cell that overproduces FGAMS [31]. Deng et al. [2] recently presented evidence supporting the hypothesis that triGART and FGAMS are core components of the purinosome and that PPAT and FGAMS interact intracellularly, a hypothesis we proposed previously on the basis of somatic cell genetic evidence [33]. These mutations, resulting in altered levels of de novo purine biosynthesis proteins, serve as important model systems for analysis of purinosome formation and function. We show here that AdeD cells produce undetectable levels of PAICS protein. This collection of mutants should be helpful in analyzing the formation and functioning of the purinosome. For example, Deng et al. [2] hypothesize that PAICS, ADSL, and ATIC interact individually with the core purinosome but may also interact with each other. Clearly, interactions between ADSL and PAICS cannot take place in AdeD cells since there is no detectable PAICS protein. It would be important to know what the consequences are for other protein-protein interactions relevant to the purinosome.
One difficulty in analysis of de novo purine biosynthesis is that robust methods that are not reliant on the use of radioisotopes to detect and quantify pathway intermediates from small samples of cells are not readily available. Our analysis of AdeD and AdeI demonstrates the utility of HPLC-EC for these studies. The method has provided convincing evidence that AdeD cells incubated in purine-free conditions accumulate AIR, the initial substrate of the bifunctional enzyme PAICS (Figure 7 B, C). Similarly, HPLC-EC readily detects accumulation of SAICAR in AdeI cells (Figure 8 B, C).
HPLC-EC has provided new insights into intermediate accumulation under conditions of purine depletion. For example, AdeI cells incubated in purine-free media accumulate AIR in addition to SAICAR. AdeI cells have virtually undetectable levels of ADSL activity. It is not clear whether cells from ADSL deficiency patients accumulate AIR or not, since these cells all have significant residual ADSL activity. If AIR does accumulate in these cells, it may potentially play a role in pathogenetic mechanisms of ADSL deficiency.
HPLC-EC analysis demonstrates that AdeD and AdeI cells accumulate a second compound that was not identified. The de novo pathway suggests that this compound may be carboxy-AIR (CAIR), the substrate of the second sequential step catalyzed by PAICS. However, the lack of detectable PAICS protein argues against this interpretation. Other possibilities are that the compound may be aminoimidazole riboside, or aminoimidazole, or an earlier intermediate in the pathway.
HPLC-EC offers many advantages for these analyses. Samples undergo redox reactions with 100% efficiency, and the series of incremental voltages allows for high specificity and resolution of co-eluting compounds. HPLC-EC is also extremely sensitive, capable of detecting 1-10 pg of a given compound. Detection of SAICAR is about 200 times more sensitive by EC than by UV. Detection of AIR by UV is possible, but absorption is relatively low. The molar extinction coefficient for AIR (250 nm, pH 6) is 4170 M−1 cm−1 [34]. Our results demonstrate the utility of HPLC-EC as a detection method for investigating the de novo purine biosynthesis pathway and detecting its intermediates. We are currently investigating detection of other de novo purine biosynthesis intermediates using HPLC-EC.
Current clinical methods used to test for purine intermediate accumulation generally detect dephosphorylated compounds in bodily fluids. HPLC-EC is sufficiently sensitive that it can likely be used to detect the true pathway intermediates in clinically relevant samples such as skin biopsies, fibroblast cultures, or small blood samples. Additionally, HPLC-EC methods could be devised to detect dephosphorylated compounds. These possibilities are currently under study.
Acknowledgments
This work was supported by grants from the Bonfils-Stanton Foundation and the Ludlow-Griffith Foundation to DP, and Partners in Scholarship and Summer Research Grants to KL. VB, VS, SK and MZ were supported by the by Charles University institutional programs PRVOUK-P24/LF1/3, UNCE 204011 and SVV2012/ 2645, and by grants LH11031 from The Ministry of Education of Czech Republic and P305/12/P419 from the Czech Science Foundation. The University of Colorado Cancer Center DNA Sequencing and Analysis Core is supported by NIH-NCI grant P30 CA 046934.
References
- [1].An S, Kumar R, Sheets ED, Benkovic SJ. Reversible Compartmentalization of de Novo Purine Biosynthetic Complexes in Living Cells. Science. 2008 Apr;320(5872):103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- [2].Deng Y, Gam J, French JB, Zhao H, An S, Benkovic SJ. Mapping protein-protein proximity in the purinosome. Journal of Biological Chemistry. 2012 Sep; doi: 10.1074/jbc.M112.407056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Baresova V, Skopova V, Sikora J, Patterson D, Sovova J, Zikanova M, Kmoch S. Mutations of ATIC and ADSL affect purinosome assembly in cultured skin fibroblasts from patients with AICA-ribosiduria and ADSL deficiency. Human Molecular Genetics. 2012 Mar;21(7):1534–1543. doi: 10.1093/hmg/ddr591. [DOI] [PubMed] [Google Scholar]
- [4].Jurecka A. Inborn errors of purine and pyrimidine metabolism. Journal of Inherited Metabolic Disease. 2009 Mar;32(2):247–263. doi: 10.1007/s10545-009-1094-z. [DOI] [PubMed] [Google Scholar]
- [5].Jurecka A, Zikanova M, Tylki-Szymanska A, Krijt J, Bogdanska A, Gradowska W, Mullerova K, Sykut-Cegielska J, Kmoch S, Pronicka E. Clinical, biochemical and molecular findings in seven Polish patients with adenylosuccinate lyase deficiency. Mol. Genet. Metab. 2008 Aug;94(4):435–442. doi: 10.1016/j.ymgme.2008.04.013. [DOI] [PubMed] [Google Scholar]
- [6].Sempere Á, Arias A, Farré G, García-Villoria J, Rodríguez-Pombo P, Desviat LR, Merinero B, García-Cazorla A, Vilaseca MA, Ribes A, Artuch R, Campistol J. Study of inborn errors of metabolism in urine from patients with unexplained mental retardation. Journal of Inherited Metabolic Disease. 2010 Jan;33(1):1–7. doi: 10.1007/s10545-009-9004-y. [DOI] [PubMed] [Google Scholar]
- [7].Zikanova M, Skopova V, Hnízda A, Krijt J, Kmoch S. Biochemical and structural analysis of 14 mutant adsl enzyme complexes and correlation to phenotypic heterogeneity of adenylosuccinate lyase deficiency. Hum. Mutat. 2010 Feb;31(4):445–455. doi: 10.1002/humu.21212. [DOI] [PubMed] [Google Scholar]
- [8].Tu AS, Patterson D. Biochemical Genetics of Chinese Hamster Cell Mutants with Deviant Purine Metabolism. VI. Enzymatic Studies of Two Mutants Unable to Convert Inosinic Acid to Adenylic Acid. Biochemistry. 1977 Jul;15:195–210. doi: 10.1007/BF00484561. [DOI] [PubMed] [Google Scholar]
- [9].Patterson D. Biochemical Genetics of Chinese Hamster Cell Mutants with Deviant Purine Metabolism: Biochemical Analysis of Eight Mutants. Somatic Cell Genetics. 1975 Aug;1:91–110. doi: 10.1007/BF01538734. [DOI] [PubMed] [Google Scholar]
- [10].Vliet LK, Wilkinson TG, Duval N, Vacano G, Graham C, Zikanova M, Skopova V, Baresova V, Hnízda A, Kmoch S, Patterson D. Molecular characterization of the AdeI mutant of Chinese hamster ovary cells: a cellular model of adenylosuccinate lyase deficiency. Mol. Genet. Metab. 2011 Jan;102(1):61–68. doi: 10.1016/j.ymgme.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pinson Benoit, Vaur Sabine, Sagot Isabelle, Coulpier F, Lemoine S, Daignan-Fornier Bertrand. Metabolic intermediates selectively stimulate transcription factor interaction and modulate phosphate and purine pathways. Genes & Development. 2009 Jun;23(12):1399–1407. doi: 10.1101/gad.521809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tang Y-C, Williams BR, Siegel JJ, Amon A. Identification of Aneuploidy-Selective Antiproliferation Compounds. Cell. 2011 Feb;144(4):499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rattan R, Giri S, Singh AK, Singh I. 5-Aminoimidazole-4-carboxamide-1-- Journal of Biological Chemistry. 2005 Nov;280:39582–39593. doi: 10.1074/jbc.M507443200. [DOI] [PubMed] [Google Scholar]
- [14].Narkar VA, Downes M, Yu RT, Embler E, Wang Y-X, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, Kang H, Shaw RJ, Evans RM. AMPK and PPARδ Agonists Are Exercise Mimetics. Cell. 2008 Aug;134(3):405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Swain JL, Hines JJ, Sabina RL, Harbury OL, Holmes EW. Disruption of the Purine Nucleotide Cycle by Inhibition of Adenylosuccinate Lyase Produces Skeetal Muscle Dysfunction. Journal of Clinical Investigation. 1984 Aug;74:1422–1427. doi: 10.1172/JCI111553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ng A, Uribe RA, Yieh L, Nuckels R, Gross JM. Zebrafish mutations in gart and paics identify crucial roles for de novo purine synthesis in vertebrate pigmentation and ocular development. Development. 2009 Jul;136(15):2601–2611. doi: 10.1242/dev.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zaza G, Zaza G, Yang W, Kager L, Cheok M, Downing J, Pui C-H, Cheng C, Relling MV, Evans WE. Acute lymphoblastic leukemia with TEL-AML1 fusion has lower expression of genes involved in purine metabolism and lower de novo purine synthesis. Blood. 2004 Sep;104(5):1435–1441. doi: 10.1182/blood-2003-12-4306. [DOI] [PubMed] [Google Scholar]
- [18].Sun W, Zhang K, Zhang X, Lei W, Xiao T, Ma J, Guo S, Shao S, Zhang H, Liu Y, Yuan J, Hu Z, Ma Y, Feng X, Hu S, Zhou J, Cheng S, Gao Y. Identification of differentially expressed genes in human lung squamous cell carcinoma using suppression subtractive hybridization. Cancer Letters. 2004 Aug;212(1):83–93. doi: 10.1016/j.canlet.2004.03.023. [DOI] [PubMed] [Google Scholar]
- [19].Firestine SM, Paritala H, Mcdonnell JE, Thoden JB, Holden HM. Identification of inhibitors of N5-carboxyaminoimidazole ribonucleotide synthetase by high-throughput screening. Bioorganic & Medicinal Chemistry. 2009 May;17(9):3317–3323. doi: 10.1016/j.bmc.2009.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jackson M, Phalen SW, Lagranderie M, Ensergueix D, Chavarot P, Marchal G, McMurray DN, Gicquel B, Guilhot C. Persistence and Protective Efficacy of a Mycobacterium tuberculosis Auxotroph Vaccine. Infection and Immunity. 1999 May;67:2867–2873. doi: 10.1128/iai.67.6.2867-2873.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Taha TS, Deits TL. Detection of Glycinamide Ribonucleotide by HPLC with Pulsed Amperometry: Application to the Assay for Glutamine:5-Phosphoribosyl-1-pyrophosphate Amidotransferase. Analytical Biochemistry. 1993 Feb;213:323–328. doi: 10.1006/abio.1993.1428. [DOI] [PubMed] [Google Scholar]
- [22].Matson WR, Langlais P, Volicer L, Gamache PH, Edward Bird AKAM. n-ElectrodeThree-DimensionalLiquidChromatographywith Electrochemical Detectionfor Determinationof Neurotransmitters. Clinical Chemistry. 1984 Oct;30(9):1477–1488. [PubMed] [Google Scholar]
- [23].Kristal BS, Shurubor YI, Kaddurah-Daouk R, Matson WR. High-Performance Liquid Chromatography Separations Coupled With Coulometric Electrode Array Detectors. In: Weckworth W, editor. Metabolomics: Methods and Protocols. 10. 358th ed. 2007. pp. 159–174. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- [24].Becker J, Hackl M, Rupp O, Jakobi T, Schneider J, Szczepanowski R, Bekel T, Borth N, Goesmann A, Grillari J, Kaltschmidt C, Noll T, Pühler A, Tauch A, Brinkrolf K. Unraveling the Chinese hamster ovary cell line transcriptome by next-generation sequencing. Journal of Biotechnology. 2011 Dec;156(3):227–235. doi: 10.1016/j.jbiotec.2011.09.014. [DOI] [PubMed] [Google Scholar]
- [25].Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. Journal of Molecular Biology. 1990 May;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- [26].Li S-X, Tong Y-P, Xie X-C, Wang Q-H, Zhou H-N, Han Y, Zhang Z-Y, Gao W, Li S-G, Zhang XC, Bi R-C. Octameric Structure of the Human Bifunctional Enzyme PAICS in Purine Biosynthesis. Journal of Molecular Biology. 2007 Mar;366(5):1603–1614. doi: 10.1016/j.jmb.2006.12.027. [DOI] [PubMed] [Google Scholar]
- [27].Dehouck Yves, Grosfils A, Folch B, Gilis D, Bogaerts P, Rooman M. Fast and accurate predictions of protein stability changes upon mutations using statistical potentials and neural networks: PoPMuSiC-2.0. Bioinformatics. 2009 Sep;25(19):2537–2543. doi: 10.1093/bioinformatics/btp445. [DOI] [PubMed] [Google Scholar]
- [28].Dehouck Y, Kwasigroch JM, Gilis D, Rooman M. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinformatics. 2011 May;12(1):151. doi: 10.1186/1471-2105-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Knox AJ, Graham C, Bleskan J, Brodsky G, Patterson D. Mutations in the Chinese hamster ovary cell GART gene of de novo purine synthesis. Gene. 2008 Oct;429(1):23–30. doi: 10.1016/j.gene.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brodsky G, Barnes T, Bleskan J, Becker L, Cox M, Patterson D. The human GARS-AIRS-GART gene encodes two proteins which are differentially expressed during human brain development and temporally overexpressed in cerebellum of individuals with Down syndrome. Human Molecular Genetics. 1997 Oct;6:2043–2050. doi: 10.1093/hmg/6.12.2043. [DOI] [PubMed] [Google Scholar]
- [31].Barnes TS, Bleskan JH, Hart IM, Walton KA, Barton JW, Patterson D. Purification of, Generation of Monoclonal Antibodies to, and Mapping of Phosphoribosyl N-Formylglycinamide Amidotransferase. Biochemistry. 1994 Jan;33:1850–1860. doi: 10.1021/bi00173a031. [DOI] [PubMed] [Google Scholar]
- [32].Patterson D, Bleskan J, Gardiner K, Bowersox J. Human phosphoribosylformylglycineamide amidotransferase (FGARAT): regional mapping, complete coding sequence, isolation of a functional genomic clone, and DNA sequence analysis. Gene. 1999 Oct;239:381–391. doi: 10.1016/s0378-1119(99)00378-9. [DOI] [PubMed] [Google Scholar]
- [33].Oates DC, Vannais D, Patterson D. A Mutant of CHO-K1 Cells Deficient in Two Nonsequential Steps of de Novo Purine Biosynthesis. Cell. 1980 Jun;20:797–805. doi: 10.1016/0092-8674(80)90326-8. [DOI] [PubMed] [Google Scholar]
- [34].Meyer E, Leonard NJ, Bhat B, Stubbe J, M SJ. Purification and Characterization of the purE, purK, and purC Gene Products: Identification of a Previously Unrecognized Energy Requirement in the Purine Biosynthetic Pathway. Biochemistry. 1992 Jun;31(21):5022–5032. doi: 10.1021/bi00136a016. [DOI] [PubMed] [Google Scholar]