Abstract
Biosensors are valuable tools used to image the subcellular localization and kinetics of protein activity in living cells. Signaling at the edge of motile cells that regulates cell protrusion and retraction is important in many aspects of cell physiology, and frequently studied using biosensors. However, quantitation and interpretation is limited by the heterogeneity of this signaling behavior; automated analytical approaches are required to systematically extract large data sets from biosensor studies for statistical analysis. Here we describe an automated analysis to relate the velocity at specific points along the cell edge with biosensor activity in adjoining regions. Time series of biosensor images are processed to interpolate a smooth edge of the cell at each time point. Profiles of biosensor activity (‘line scans’) are then calculated along lines perpendicular to the cell edge. An energy minimization method is used to calculate a velocity associated with each line scan. Sorting line scans by the proximal velocity has generated novel biological insights, as exemplified by analysis of the Src merobody biosensor. With the large data sets that can be generated automatically by this program, conclusions can be drawn that are not apparent from qualitative or ‘manual’ quantitative techniques. Our ‘LineScan’ software includes a graphical user interface (GUI) to facilitate application in other studies. It is available at hahnlab.com and is exemplified here in a study using the RhoC FLARE biosensor.
Introduction and Overview
To understand cell signaling, detailed information regarding the spatio-temporal dynamics of protein activity is required. This is quantified in living cells using fluorescent biosensors, which report the localization and kinetics of protein structural changes, including conformational change, phosphorylation, cleavage etc. [1]. Biosensors can be based on different designs and use different mechanisms to generate fluorescence (e.g. FRET, solvatochromic dyes, fluorophore-quencher interactions etc.) but, in general, biosensor studies map the distribution of protein activity across the cell; each pixel in an image is associated with a specific value reporting activation at that pixel. Such maps are obtained at multiple time points to reveal the kinetics of activation. These values are related to the actual level of protein activity, or the activity per molecule (i.e., the percent of molecules in the active state within a given pixel), as in the single chain ratiometric sensor of GTPase activity used here.
In motility, tight regulation of multiple processes at different regions of the cell edge generate coordinated extension, retraction, and directed movement in response to a variety of directional cues [2–4]. For example, it has been shown that Rho GTPases regulate cell edge dynamics differently at the front and rear of migrating cells [3], [5–7], and that these proteins’ distance from the edge determines their role in different aspects of actin and adhesion regulation [4]. Biosensors are particularly useful for quantifying the regulation of cell migration because they can dynamically assay the relationship between protein activity and nearby, local behavior of the cell edge. However, a significant obstacle to experimental interpretation of biosensor data from motile cells is the heterogeneity of cell behavior, arising from both cell-to-cell heterogeneity and variation in a given protein’s behavior within each cell. The majority of this variability is inherent to the biology and can arise from local conditions, cell-cycle state, and cell morphology. How inter-cell heterogeneity arises, and how regulation is managed on a background of such variability, is an open question.
In a previous publication we applied a new tool that automatically quantifies the relationship between line profiles of protein activity and nearby edge velocity. This approach led to novel biological insights, demonstrating relationships between velocity and Src activity [8]. Here we describe this tool and its development in detail, and provide a user-friendly graphical user interface (GUI) to aid in its application by other researchers.
Due to cellular heterogeneity, qualitative observations based on a small number of cells or line scans are difficult to interpret, and are subject to selection bias. To limit the amount of data to something that can be handled ‘manually’, observers have restricted observations to subcellular regions that exemplify specific behaviors. The automated tool supplied here is designed to analyze all frames in a movie, and therefore does not introduce human bias in selectivity. It can readily provide sufficient data for statistical analysis, revealing relationships between protein activity and velocity that are not obvious to the eye, as exemplified by our previous study of Src activation [8].
Line scans are a popular way of quantifying biosensor activity at the edge of cells, where biosensor activity is recorded as a function of distance from the cell edge. Popular imaging software (for example MetaMorph® or ImageJ) will perform this calculation for user-defined (manual) lines. The tool LineScan described here automatically places the lines normal to the cell edge, and couples the line scan with the proximal velocity of the edge. LineScan is written in MATLAB and contains a user friendly GUI. The software package is available at hahnlab.com and http://www.med.unc.edu/pharm/elstonlab/.
Our method is complementary to the approach of Machacek and Danuser [9], which was used to provide detailed insights into spatio-temporal dynamics of RhoGTPases at the edge of migrating cells [4]. Our method, which relies on line scans as opposed to grid generation near the cell edge, can be simpler to implement in some cases, but lacks the sophisticated two-time correlation capabilities of the Machacek and Danuser approach. For thorough literature reviews and further insights into the critical necessity for automated tools in image analysis the reader is referred to Dehmelt and Bastiaens [10] and Danuser [11].
The tool presented here enables automated collection of large amounts of image data coupled to a phenotypic measure of cell behavior (edge velocity). We automate and extend ‘line scan’ analysis, a popular method for analyzing biosensor data. Although manual generation of line scans has been sufficient in many studies [12–15], automating the process allows large amounts of data to be analyzed, enabling new biological insights to be gained even in the presence of heterogeneity and relatively poor signal/noise ratio.
Automated line scan analysis presents several computational challenges: line scans of a fixed length may extend outside the cell interior, a direction perpendicular to the local curvature of the edge needs to be automatically established (on a scale greater than the pixel size), and it is critical to store and manipulate of large amounts of data efficiently. Our approaches to these concerns are outlined below.
For novel biological insights it is essential to couple line scan measurements to a relevant phenotypic read-out. In cell migration, the velocity of the cell edge is one such read-out. Our software package calculates the edge velocity and then groups line scans in terms of this velocity, allowing signaling at different distances from the cell edge to be correlated with edge velocity. This is relevant to motility because proteins are known to have different functions in zones at successive distances from the cell edge.
Method
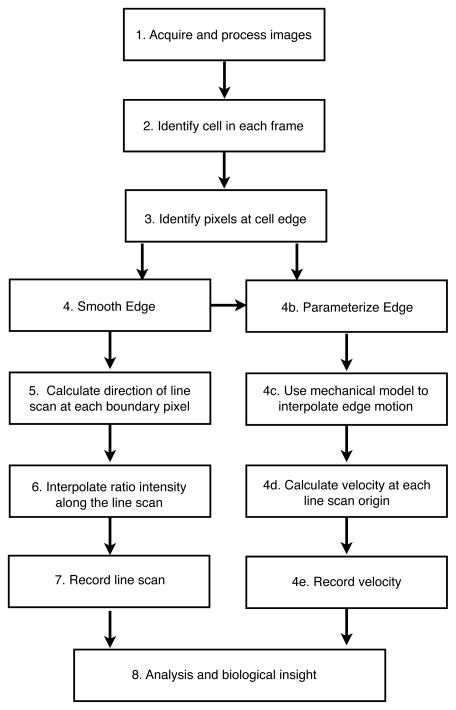
The algorithm for LineScan is outlined in figure 1. The software was developed in Mathworks’ MATLAB and requires the image processing toolbox.
Figure 1. Outline of algorithm for automatically calculating line scans and edge velocity.
Line scans are automatically quantified by an algorithm, which finds the cell, finds the edge of the cell, and calculates the direction and origin of line scans for quantification. A smoothed edge is used to interpolate the velocity in a sequence of image frames (movie). Edge velocity and line scans can be calculated independent of one another (line scans steps 4–7, edge velocity steps 4b–e).
The starting point (figure 1, 1.) is a movie composed of biosensor activity images. Pre-processing of raw images is required to produce biosensor activity maps. This is beyond the scope of this article, but has been well addressed in several recent methods papers [16–19]. Depending on the biosensor and imaging method used, this pre-processing can include background subtraction, shade correction, thresholding, ratio imaging, and correction for photobleaching or signal bleed through.
For the procedure described here, the next step (figure 1, 2.) is to identify the cell in each frame. A well-processed biosensor image will already be thresholded such that regions of intensity below the threshold are determined to be extra-cellular or too noisy for analysis, and so are set to zero. For our purposes, identifying intracellular regions is therefore trivial. The software identifies the cell for analysis as the largest connected region in the image that is above the threshold. This excludes fragments in the image from any further processing and analysis. If two cells are in an image the software will analyze the largest. When analyzing a movie with multiple cells in the image field, care must be taken to exclude cases where different cells become the largest in each frame during the course of the movie. We recommend cropping out unneeded cells before the analysis at the pre-processing step.
To identify the pixels at the cell edge the Matlab function ‘bwboundaries’ is used. This will find the exterior pixels of connected regions in binary images (in this case, the binary image is simply the image where intra-cellular pixels have intensity 1 and extra-cellular pixels have intensity 0), and is based on the Moore-Neighborhood tracing algorithm [20].
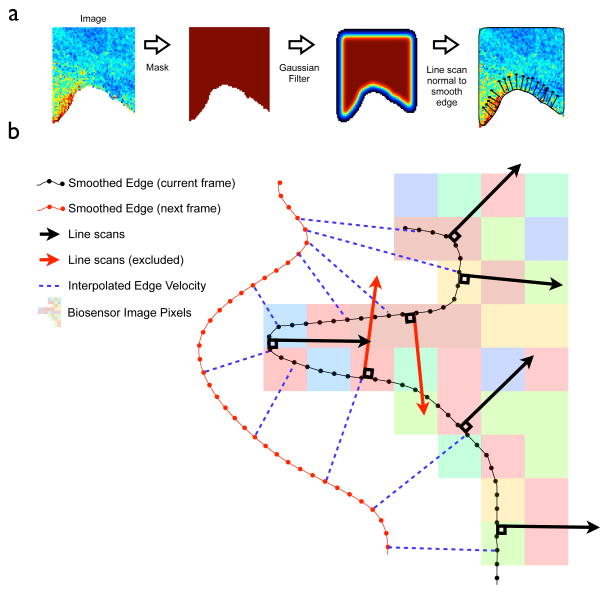
For calculating the velocity and the directions of line-scans it is necessary to interpolate a smooth edge (figure 1, 4.) to overlay onto the edge pixels. Without smoothing, the edge (defined by the edge pixels) can only be in one of 8 directions (N, NE, E, etc.). To generate a smooth-edge a Gaussian filter of the binary image is applied, generating a new image (figure 2). In the filtered image each pixel is a weighted local average of pixels surrounding the corresponding pixels in the original image, figure 2. For pixels in the interior of the image they retain the value 1, but for pixels on the edge they assume a new value between 0 and 1. The smooth-edge is then defined by use of the MATLAB function ‘contour’ – which identifies isolines in the filtered image. By construction these will be on, or near, the cell boundary (figure 2a). Note this can result in a small offset from the edge pixels – but the smooth edge is only used to generate the direction of line scans, not their origin.
Figure 2. Finding the edge and associating line scans with edge velocity.
a: Intermediary Images are used for calculating a smooth edge and line scan direction. Starting with the biosensor image (left), line scans are automatically calculated towards the cell interior (right). Line scans are directed normal to the cell edge, which is interpolated by Gaussian filtering of the cell mask. b: The edge calculated in 2a is used for interpolating velocity and calculating line scan direction. Line scans are calculated along, and normal to, the cell edge (black arrows). Line scans in regions of high curvature that reach out of the cell (red arrows) are excluded from any further analysis. A mapping (blue dotted lines) between the current edge (black line with dots) and the edge in the next frame (red line with dots) defines the current velocity. Line scans are associated with the nearest calculated velocity for future analysis.
The starting position and direction of line scans (figure 1, 5.) is determined by the location of the edge pixels (which defines the starting position) and the normal to the smooth edge (directed to the interior of the cell), figure (2b). The length of the scan is user-defined (interactively in the GUI). Line scans are not recorded for perfectly straight edges (so that cells which are only partially in the frame can be recorded – figure 2a), or if the line is not entirely in the cell (red arrows, figure 2b). The latter case can occur in regions of higher curvature. Line scans are excluded if they cross any pixel where the intensity is at or below the threshold (as mentioned above, for well processed images this will be pixels of zero intensity). Due to this, the tool is not suitable for correlating filopodial dynamics of the membrane with biosensor activity (for the analysis of filopodia dynamics consider a new software CellGeo, available at hahnlab.com). Another rationale for focusing this tool on broader low-curvature regions is the potential for line scans to overlap – potentially over-sampling pixels. In practice, for cells such as fibroblasts, results do not appear to be sensitive to this (tested by under-sampling images with sparser line scanning). Caution is urged for cells with complex boundaries, such as nerve cells.
Intensities are quantified at unit-pixel intervals along the line (figure 1. 6.). If these points do not intercept the center of a pixel exactly, the recorded intensity is bi-linearly interpolated from surrounding pixels. The data is then stored. Analyzing a typical experiment of 10 movies of 100 frames each might generate 1000 line scans per frame - giving a total of 106 line scans. If the line scans are each of length 30 pixels this gives 3 × 107 data points and over 200 mb of data. Given the requirements to store other data (for example information about edge velocity), and the already onerous data overhead associated with imaging experiments, we employ a more compact storage mechanism where the data are finely binned into a histograms by intensity and velocity. Data are stored in a 2-D array; even a 1000 × 1000 (1000 velocity bins, 1000 intensity pins) produces a reduction in data requirements of more than an order of magnitude. This can be highly efficient at storing data with very little, if any, loss in data resolution. In the GUI, the bin number and size can be easily adjusted for the best fit to the specific data.
As discussed above, the program can be used to correlate the line scans with edge velocity. The smoothed edges calculated in figure 1, step 4 are used to interpolate the velocity of the edge between frames. Calculating the velocity is not trivial. Typically 10–30 seconds elapse between frames and the cell edge may have moved a significant distance (for example, over 10 pixels). To find the velocity at a given time and a given point of the edge it is necessary to interpolate a corresponding point on the edge in the subsequent frame. This can be done using sophisticated mathematical tools, such as level-set methods [9], that preserve topology (any set of ordered points on the cell edge in one frame correspond to points in the subsequent frame in the same order). However, computationally efficient level set methods are challenging to implement in general settings. To overcome this technical hurdle, Machacek et. al. [9] introduced a spring-system in which cell edge markers are located at the equilibrium points of the elastic potential. The current equilibrium position of the markers, relative to their equilibrium in the prior frame, gives the velocity between frames (blue dotted line, figure 2b).
For this work we modified this approach and replaced the spring-force between neighboring markers by a Coulomb’s inverse square law: q/r2. The constant q is by default 0.4, with units M.L3/T2 (pixel length - L, marker mass - M, and time between frames – T.) This modification strongly enforces the topological condition that the velocity markers retain their relative order on the cell edge. This system is then integrated until it reaches equilibrium. Specifically, the integration proceeds until the maximum net force on all markers is below a particular threshold of F = 0.5 M.L/T2. The values for q and F were established by simulation using data collected with typical experimental conditions. In cases where the frames have not been captured at high enough resolution (as a rule of thumb where the edge has moved >10 pixels/frame) this integration can become computationally expensive, or even fail in extreme cases, such as when there are large numbers of filopodial protrusions.
To apply either of these techniques it is convenient, and computationally efficient, to parameterize the edge by a parameter s, the distance around the edge, and perform calculations based on s (1D) rather than the absolute 2D distance (figure 1. 4b). Numerically this is achieved by linearly interpolating by a fixed distance (defined by boundary length and the number of required points - typically this fixed distance is on the sub-pixel length) between points on the smoothed-edge. This gives a relationship between s and (x, y) positions on the cell boundary. The evolution of the edge markers is along the cell edge, and is therefore a 1D problem and relatively quick to integrate.
With these steps in place, each line scan can be associated with a local velocity (simply by finding the nearest velocity marker to the origin of the line scan). Thus, every line scan provides values of the biosensor intensity for a determined velocity (of the line) as a function of depth. To obtain the final biosensor activity map we group all the intensity values from all the line scans into two-dimensional (velocity, depth) bins with a specified velocity step and the depth step of one pixel size. Then, the means of intensity values in each bin (as displayed in the GUI) and the corresponding standard deviations are calculated and saved in the data file.
Given the heterogeneity of intracellular processes it is usually desirable to take an average across all the data. There are two ways to do this: 1) assume all the line scans are independent and average across all of these. In this case the number of line scans lends statistical power to the observations - i.e. it is better to have 1000 line scans from one cell than 200 from two cells. In the second case, it is assumed that only observations from individual cells are independent and therefore 200 line scans from two cells is a more powerful observation that 1000 line scans from one cell. Which approach to use is a subtle question – the assumption that all line scans are independent measurements is almost assuredly not valid. However, weighting by cell can skew results in favor of small, dynamic cells. In practice we have observed that these are qualitatively similar. It is recommended that the user examine outputs from individual cells to check for outliers (this can occur if there is debris in the field of view for example).
LineScan Software and GUI
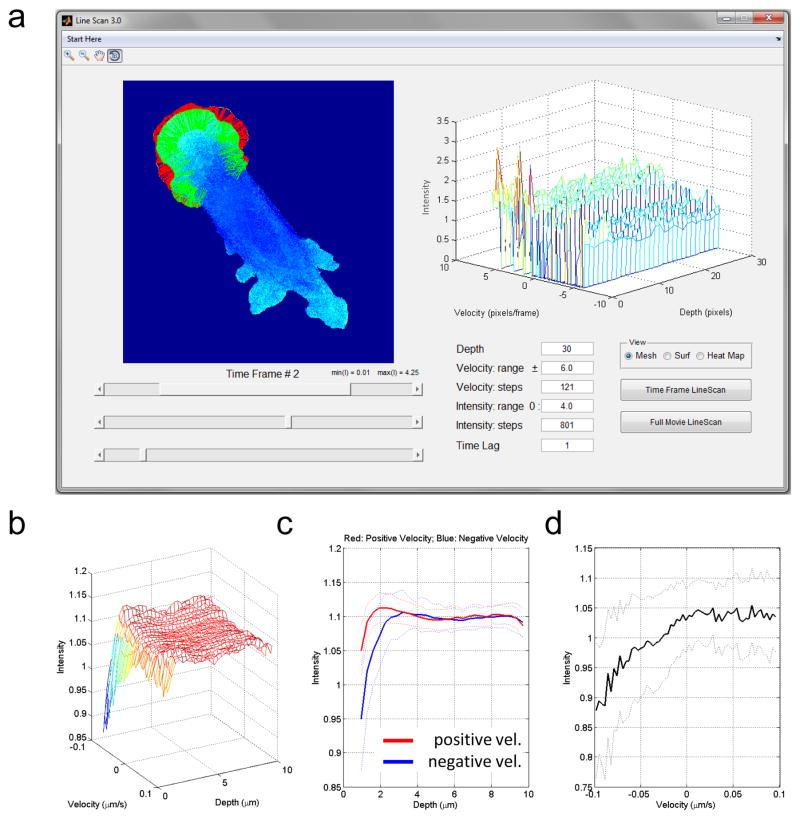
To make our quantitative method accessible to researches without any background in programming we created user-friendly software in the form of a Graphical User Interface (GUI). LineScan can be run on any computer with MATLAB, and no direct interaction with or modification of the MATLAB code is required. The GUI is designed to be self-explanatory and consists of just a few controls (sliders, buttons and editable text fields) for easy adjustment of the parameters and intuitive visual display (Figure 3a).
Figure 3. GUI Screenshot and an example of automated quantification of the biosensor activity at the edge that shows RhoC is positively related to velocity at the edge.
a: The GUI layout with interactive controls for easy adjustments of the parameters. Only a segment of the cell boundary is selected for the quantification in this example. By default the whole cell is processed. b–d: Applying the GUI to a representative cell suggests that RhoC is activated during protrusion and not retraction, at 2 microns from the cell edge. The relationship between velocity and activity is seen only at the edge of cells. c: The average intensity as a function of distance from the edge is shown for positive (red) and negative (blue) velocities separately. d: The average intensity (over the first 5 pixels from the edge) as a function of velocity. c,d: Dashed lines indicate the standard deviations from the mean values.
Using the menu, a user can import a single tif-file with time frames of the biosensor data (a popular and flexible way of storing time-lapsed image data), save results at any stage of the processing, and load previously saved results to continue the work. Upon loading the data are displayed in the left panel of the main window, so that the change of the cell edge and the biosensor signal over time can be visually explored with the time-slider below the panel.
By default, the GUI will process the entire cell automatically; however, in special cases, a researcher might want to exclude certain boundary segments. This could occur, for example, when the cell nucleus happened to be located too close to the edge, bringing biosensor data from the golgi or perinuclear region close to the edge.
As described above, the intensity values of the biosensor signal along line scans are binned into intensity-velocity-depth boxes of a pre-specified size and number. For that purpose, the GUI has editable text fields to enter: the length of lines (maximum depth from the edge in pixels), the range of velocities (in pixels per frame), the number of bins within the range, and the range of intensities (in the units of the image data).
For additional flexibility we also included an option to specify a time lag for velocity calculation. The time lag of L means that velocity is calculated as the boundary displacement between time frames i and (i + L) divided by L, where i = 1, 2, …, (T − L) and T is the length of the movie. This feature is useful when the cell movies very little from a time point to the next, so that noise from boundary detection dominates the analysis. In these situations, choosing L>1 improves the accuracy. However, in most cases, we expect time steps of image recording to be large enough for significant change of the cell edge and, thus, L=1 is adequate and used as the default value.
Finally, the user has an option to quickly explore the results for individual frames using the “Time Frame LineScan” button or processing the entire movie using “Full Movie LineScan”, which might take some time, depending on the movie length and the length of the cell boundary. We recommend starting the analysis of new data sets by exploring individual frames to get a sense of the range for velocities and intensities and to properly adjust binning parameters before processing the whole movie and all the movies. In any case, the GUI displays the scanning lines in green and the interpolated velocity in red (see the Method section), as a means to visual quality control. The right panel is used to display the resulting intensity profile as a function of velocities and depth. The results can be shown as a mesh or a surface for the 3D view and as a heat map for 2D view. The final intensity profile averaged over all time frames is displayed in a separate window. This window allows the user to zoom in/out and rotate the view, add custom legends and texts, and save it in any of the standard image formats (jpg, png, eps, tif, etc.). For further statistical analysis and amalgamating data from multiple cells, the processed data are saved as .mat data file. An example of how to automatically collate this data is supplied with the GUI.
Analysis of RhoC
The tool can quickly ascertain if there is a relationship between biosensor activity, proximity to the edge and proximal velocity of the edge. As an example of this we analyzed data generated using a novel biosensor for the small GTPase protein RhoC (RhoC FLARE biosensor) [21]. RhoC plays a role in normal motility and it has been implicated in generating cancer metastasis [22]. As shown in Figure 3b–d, the overall behavior of many line scans within a cell can be analyzed to assess the dynamics of RhoC activation and its role in protrusion versus retraction.
A stable, inducible mouse embryonic fibroblast (MEF) cell line expressing RhoC FLARE was created by retroviral infection as described previously [7]. RhoC MEFs were maintained in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% FBS. For imaging, cells were plated on fibronectin-coated (5 μg/ml) glass coverslips in Ham’s F12K media without phenol red (Biosource), supplemented with 10 mM HEPES and 2% FBS. Cells were imaged in a heated closed chamber using an Olympus IX81 inverted microscope with a 40X 1.3 NA objective fitted with an objective heater (Bioptech). CFP and FRET images were acquired simultaneously at 10 sec time intervals through two CoolSNAP HQ2 CCD cameras (Photometrics) using a TuCam camera adapter (Andor Technology). Image processing, ratio calculations and photobleaching correction was performed as described [7]. CFP and FRET images acquired using the two-camera system were aligned pixel-by-pixel using a priori calibration and morphing [23].
The data can be viewed as a function of distance from the edge and velocity, (figure 3b), or as a function of one of these variables (figure 3c,d). This analysis suggests that RhoC is positively correlated with velocity (figure 3b–d), with activity peaking at approximately 2 μm from the edge. This is consistent with recent work that suggests RhoC promotes Rac1-initiated protrusions via the formin-like FMNL3 [24]. This analysis is presented as an example; further work will be required to fully characterize RhoC dynamics, as statistical analysis of behavior across many cells will be required.
Conclusion
We have presented a novel tool for automatically and quickly interrogating biosensor images. This tool can identify relationships between activity, distance from the edge, and velocity. Identifying these relationships is valuable for elucidating the spatio-temporal dynamics of signaling that regulates morphology at the edge of migrating cells. The strength of this method is the quantification of protein activity coupled to a phenotypic measure of cell behavior. In principle, this analysis could be modified such that activity measurements were coupled to cell parameters other than edge velocity, including the localization/trafficking of another protein, local curvature of the edge, or focal adhesion size/location. The tool parses large volumes of complex imaging data into high-resolution quantitative summaries which identify biologically meaningful relationships.
Supplementary Material
Acknowledgments
The authors thank the NIH for funding (GM057464 (KMH) and NIH GM079271 and NCI 200079604 (TE)).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Newman RH, Fosbrink MD, Zhang J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chemical reviews. 2011 May;111(5):3614–66. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sastry SK, Burridge K. Focal adhesions: a nexus for intracellular signaling and cytoskeletal dynamics. Experimental Cell Research. 2000 Nov;261(1):25–36. doi: 10.1006/excr.2000.5043. [DOI] [PubMed] [Google Scholar]
- 3.Jilkine A, Marée AFM, Edelstein-Keshet L. Mathematical model for spatial segregation of the Rho-family GTPases based on inhibitory crosstalk. Bulletin of Mathematical Biology. 2007 Jul;69(6):1943–1978. doi: 10.1007/s11538-007-9200-6. [DOI] [PubMed] [Google Scholar]
- 4.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009 Sep;461(7260):99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science (New York, NY) 2003 Dec;302(5651):1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 6.Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005 Oct;33(Pt 5):891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- 7.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006 Apr;440(7087):1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 8.Gulyani A, Vitriol E, Allen R, Wu J, Gremyachinskiy D, Lewis S, Dewar B, Graves LM, Kay BK, Kuhlman B, Elston T, Hahn KM. A biosensor generated via high-throughput screening quantifies cell edge Src dynamics. Nature chemical biology. 2011 Jan;7(7):437–44. doi: 10.1038/nchembio.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machacek M, Danuser G. Morphodynamic profiling of protrusion phenotypes. Biophysical journal. 2006 Feb;90(4):1439–52. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dehmelt L, Bastiaens PIH. Spatial organization of intracellular communication: insights from imaging. Nature reviews Molecular cell biology. 2010 Jun;11(6):440–52. doi: 10.1038/nrm2903. [DOI] [PubMed] [Google Scholar]
- 11.Danuser G. Computer vision in cell biology. Cell. 2011 Nov;147(5):973–8. doi: 10.1016/j.cell.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. Cortactin promotes cell motility by enhancing lamellipodial persistence. Current biology : CB. 2005 Jul;15(14):1276–85. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 13.Verschure PJ, van der Kraan I, Manders EMM, Hoogstraten D, Houtsmuller AB, van Driel R. Condensed chromatin domains in the mammalian nucleus are accessible to large macromolecules. EMBO reports. 2003 Sep;4(9):861–6. doi: 10.1038/sj.embor.embor922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nature cell biology. 2007 Mar;9(3):299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- 15.Hodgson L, Chan EWL, Hahn KM, Yousaf MN. Combining surface chemistry with a FRET-based biosensor to study the dynamics of RhoA GTPase activation in cells on patterned substrates. Journal of the American Chemical Society. 2007 Aug;129(30):9264–5. doi: 10.1021/ja072900m. [DOI] [PubMed] [Google Scholar]
- 16.Hodgson L, Shen F, Hahn K. Biosensors for characterizing the dynamics of rho family GTPases in living cells. In: Bonifacino Juan S, et al., editors. Current protocols in cell biology. Unit 14.11. Chapter 14. 2010. Mar, pp. 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spiering D, Hodgson L. Multiplex Imaging of Rho Family GTPase Activities in Living Cells. Methods Mol Biol. 2012;827(9):215–234. doi: 10.1007/978-1-61779-442-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welch CM, Elliott H, Danuser G, Hahn KM. Imaging the coordination of multiple signalling activities in living cells. Nature Reviews Molecular Cell Biology. 2011;12(11):749–756. doi: 10.1038/nrm3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vilela M, Halidi N, Besson S, Elliott H, Hahn K, Tytell J, Danuser G. Fluctuation analysis of activity biosensor images for the study of information flow in signaling pathways. 1. Vol. 519. Elsevier Inc; 2013. pp. 253–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez R, Woods R, Eddins S. Digital image processing using MATLAB. New Jersey: Pearson Prentice Hall; 2004. [Google Scholar]
- 21.Zawistowski JS, Sabouri-Ghomi M, Danuser G, Hahn KM, Hodgson L. A RhoC biosensor reveals differences in the activation kinetics of RhoA and RhoC in migrating cells. PLoS One. 2013 Nov 5;8(11):e79877. doi: 10.1371/journal.pone.0079877. doi: 10.1371/journal.pone.0079877. eCollection 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000 Aug;406(6795):532–5. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 23.Danuser G. Photogrammetric calibration of a stereo light microscope. Journal of Microscopy. 1999 Jan;193(1):62–83. doi: 10.1046/j.1365-2818.1999.00425.x. [DOI] [PubMed] [Google Scholar]
- 24.Vega FM, Fruhwirth G, Ng T, Ridley AJ. RhoA and RhoC have distinct roles in migration and invasion by acting through different targets. The Journal of cell biology. 2011 May;193(4):655–65. doi: 10.1083/jcb.201011038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.