Abstract
APEX is an engineered peroxidase that functions both as an electron microscopy tag, and as a promiscuous labeling enzyme for live-cell proteomics. Because the limited sensitivity of APEX precludes applications requiring low APEX expression, we used yeast display evolution to improve its catalytic efficiency. Our evolved APEX2 is far more active in cells, enabling the superior enrichment of endogenous mitochondrial and endoplasmic reticulum membrane proteins and the use of electron microscopy to resolve the sub-mitochondrial localization of calcium uptake regulatory protein MICU1.
Heme peroxidases are powerful tools for biotechnology due to the great diversity of reactions that they catalyze. For example, horseradish peroxidase (HRP) is used widely to generate chemiluminescent signals for western blots and chromogenic signals for ELISAs.1,2,3 Recently, our lab engineered a new monomeric peroxidase reporter, called APEX (28 kD), derived from dimeric pea4 or soybean5 ascorbate peroxidases (Fig. 1A-C). Unlike HRP, APEX lacks disulfides and calcium binding sites and hence can be expressed in the reducing cytosolic environment of cells without loss of activity.4 Consequently, APEX can be used for two novel cell-based applications: intracellular specific protein imaging by EM,4 and spatially-resolved proteomic mapping.5,6
Figure 1.
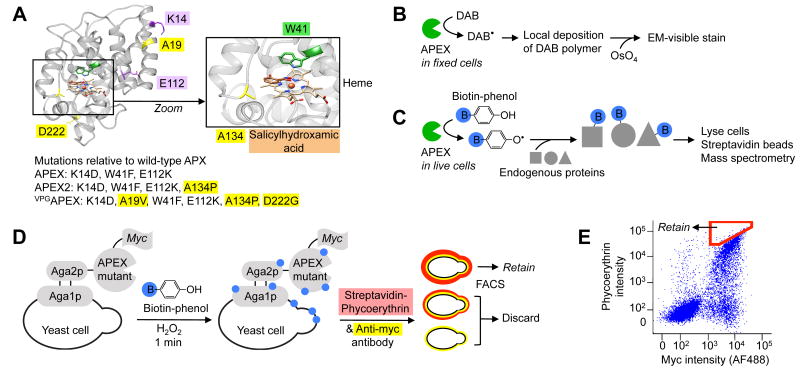
Yeast display evolution of APEX2 for electron microscopy (EM) and proteomics applications. (A) Structure of wild-type soybean ascorbate peroxidase (APX) with mutations present in APEX, APEX2, and VPGAPEX indicated. New mutations discovered in this study are highlighted yellow. The active site is magnified to show the heme cofactor, aromatic substrate (salicylhydroxamic acid, brown) binding site, and A134 position (yellow) that is mutated to proline in APEX2. From PDB ID 1 V0H9. (B) Scheme showing how APEX or APEX2 can be used as a reporter to generate contrast for EM.4 (C) Scheme showing how APEX or APEX2 can be used for proteomic tagging in living cells.5, 6 Blue B, biotin. (D) Labeling and selection scheme used to evolve APEX2 by yeast display. Biotin-phenol was added to a dilute yeast suspension for 1 minute to allow cells displaying highly active APEX variants to promiscuously biotinylate themselves. Minimal inter-cellular labeling was observed under these conditions. Biotinylation sites (blue) were stained with fluorescent streptavidin-phycoerythrin (red), and APEX expression level was quantified via anti-myc antibody staining (yellow). Two-dimensional fluorescence activated cell sorting (FACS) was used to enrich for cells displaying the highest activity/expression ratio (i.e., streptavidin/myc ratio). (E) FACS plot showing the initial APEX mutant library, and the sorting gate used in the first round of selection (red polygon). Single trial, 10,000 cells shown. FACS analyses of enriched populations and individual clones shown in Supplementary Figure 2.
For EM (Fig. 1B), APEX is genetically fused to a protein of interest, and the fusion construct is expressed inside cells. The cells are then fixed and overlaid with a solution of diaminobenzidine (DAB) and H2O2. APEX catalyzes the polymerization and local deposition of DAB, which subsequently recruits electron-dense osmium, giving EM contrast.
For proteomic mapping (Fig. 1C), APEX is genetically targeted to a cellular organelle or protein complex of interest. Then live cells are treated for 1 minute with H2O2 in the presence of biotin-phenol. APEX catalyzes the one-electron oxidation of biotin-phenol to generate a very short-lived biotinphenoxyl radical. This radical covalently tags endogenous proteins proximal to APEX, allowing their subsequent enrichment using streptavidin beads and identification by mass spectrometry.
As a general intracellular peroxidase with similar substrate breadth as HRP, APEX has already enabled some biological discoveries, including the proteomic mapping of the human mitochondrial matrix5 and intermembrane space (IMS),6 and determination of the membrane topology of the mitochondrial calcium uniporter.4 However, in our work with APEX, we have observed that a major limitation is its low sensitivity. Frequently, when APEX is expressed at low levels, its activity with DAB (for EM) and biotin-phenol (for proteomics) becomes undetectable (Supplementary Fig. 1A). This problem can be solved in some cases by increasing expression level, but for many fusion constructs, overexpression is detrimental. For example, mitochondrial outer membrane- and endoplasmic reticulum (ER) membrane-targeted APEX constructs cause organelle aggregation when overexpressed (Supplementary Fig. 1A-B).
Interestingly, the dimeric form of APEX, W41FAPX, is a more sensitive peroxidase inside cells (Supplementary Fig. 1A), despite equivalent catalytic constants measured in vitro.4 Poulos and co-workers have shown that monomerizing ascorbate peroxidase (APX) decreases its thermal stability.7 Hence, we hypothesized that the low sensitivity of APEX may result from sub-optimal folding/stability, poor heme binding, or some combination of these factors.
Since improved stability is a difficult parameter to engineer by rational design, we employed directed evolution to improve the sensitivity of APEX. We utilized a yeast display platform coupled with fluorescence activated cell sorting (FACS). A library of 106 APEX variants was displayed on the surface of yeast cells (Fig. 1D-E). Using FACS, we selected for yeast displaying the most active APEX mutants based on their ability to promiscuously biotinylate the surface of yeast cells to which they were bound (Supplementary Fig. 2). We further increased selective pressure for efficient heme incorporation by preincubating cells with succinyl acetone, a heme biosynthesis inhibitor.8 Over three rounds of selection, we observed a striking increase in the activity of the yeast pool (Supplementary Fig. 2A). Sequencing of clones after round three revealed two predominant mutants (Supplementary Fig. 3): the A134P mutant of APEX, and a triple mutant: A19V, A134P, and D222G (“VPGAPEX”).
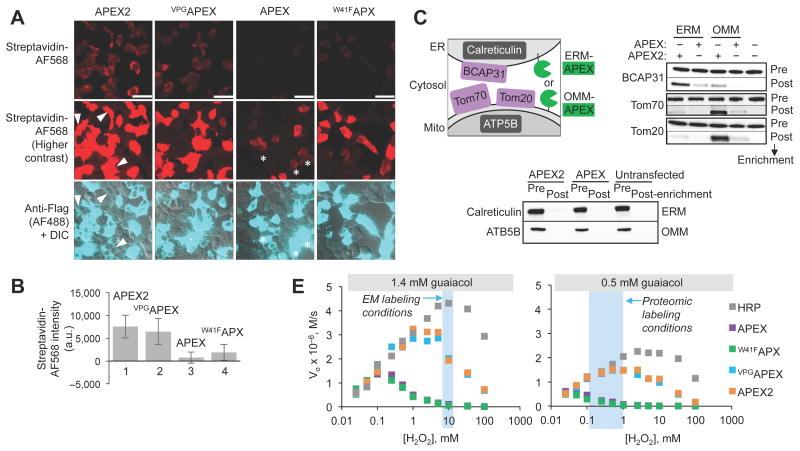
In subsequent assays, we could not discern a difference between A134PAPEX and VPGAPEX. Therefore we focused on the simpler single mutant, which we rename “APEX2”. We first determined if APEX2 was a more sensitive peroxidase than APEX for proteomic mapping applications. Live HEK cells expressing either APEX or APEX2 in the cytosol were treated with H2O2 in the presence of biotin-phenol for 1 minute, then fixed and stained with streptavidin-AlexaFluor 568 for imaging (Fig. 2A-B and Supplementary Fig. 4A), or lysed and run on a gel for streptavidin blot analysis (Supplementary Fig. 4B). By both readouts, APEX2 was much more active than APEX. Surprisingly, APEX2 was also much more active than dimericW41F APEX. By microscopy, we observed many cells expressing APEX2 at very low levels that nevertheless had high biotinylation activity.
Figure 2.
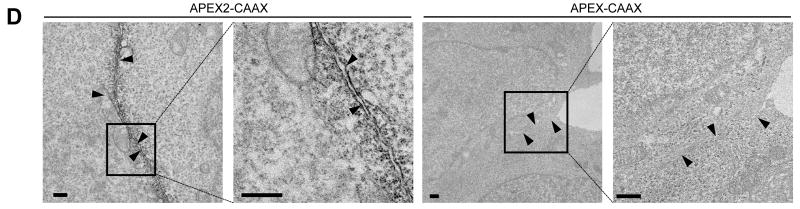
APEX2 has improved cellular activity and sensitivity for proteomic tagging and electron microscopy. (A) HEK cells expressing the indicated APEX variant were labeled with biotin-phenol for 1 minute before fixation and staining with streptavidin-AlexaFluor568 (red) to visualize biotinylation sites, and anti-Flag antibody (cyan) to visualize APEX expression. Arrowheads, cells with low APEX2 expression and strong biotinylation. Asterisks, cells with high APEX expression and low biotinylation. DIC, differential interference contrast. Images representative of 25 fields of view. Scale bars, 50 μm. (B) Quantitation of experiment shown in (A). >50 single cells were analyzed across >16 fields of view for each APEX variant. Mean streptavidin intensities are plotted ± 1 s. d. (C) Comparison of proximity dependent biotinylation by APEX2 and APEX. Live HEK cells expressing APEX or APEX2 targeted to the endoplasmic reticulum (ER) membrane (ERM) or outer mitochondrial membrane (OMM) were labeled with biotin-phenol as in (A). After cell lysis, biotinylated proteins were enriched using streptavidin beads and blotted for the endogenous ER and mitochondrial proteins shown. (D) Comparison of APEX2 and APEX for EM imaging of HEK cells expressing plasma membrane-targeted constructs. Arrowheads point to plasma membrane. Images representative of >3 fields of view. Scale bars, 500 nm. For (C) and (D), controls showed that APEX and APEX2 construct pairs were expressed at similar levels (data not shown). (E) Purified peroxidases were incubated with 1.4 mM or 0.5 mM guaiacol (a model aromatic substrate24, 25) to mimic EM and proteomic tagging conditions, respectively. Initial rates (Vo) were measured for a range of H2O2 concentrations. Each plot is representative of 2-5 trials.
When we targeted APEX2 to the mitochondrial outer membrane (OMM) or ER membrane (ERM) using lentiviral transduction to maintain low expression levels, we observed robust biotinylation by cell imaging. In contrast, APEX completely fails in these contexts, since it either gives undetectable biotinylation at low expression levels, or gross organelle perturbation at high expression levels (Supplementary Fig. 5).
Using these same constructs, we tested if APEX2 has a greater ability to enrich proximal endogenous proteins via live cell biotinylation and streptavidin pull-down than APEX, a critical measure of its utility for proteomic mapping. Figure 2C shows that APEX2 targeted to the OMM strongly enriches the endogenous OMM proteins Tom70 and Tom20. Spatial specificity is indicated by the lack of enrichment of endogenous mitochondrial matrix protein ATP5B, and by the only very slight enrichment of endogenous ER membrane protein BCAP31. By contrast, the APEX version of this construct barely enriches Tom70 and Tom20. Similar results were obtained when comparing the ability of the APEX and APEX2 ER membrane fusion constructs to selectively enrich the endogenous ER membrane protein BCAP31 over OMM and ER lumen proteins. From these results, we conclude that APEX2 is superior to APEX for proteomic enrichment.
To test whether APEX2 is also a more sensitive reporter than APEX for EM applications, we prepared matched samples with APEX or APEX2 targeted to the cytosol, plasma membrane, and microtubules of HEK cells, and PSD95 in the post-synaptic density of cultured rat hippocampal neurons (Supplementary Fig. 6). DAB staining was performed on fixed cells, which were then imaged by brightfield microscopy. In this mode, cellular regions that appear dark due to absorbance of visible light reflect the presence of DAB polymer deposits, and hence elevated APEX or APEX2 activity. Supplementary Figure 6 shows that APEX2 fusions consistently give stronger DAB staining than their APEX counterparts. One sample set, a fusion to the CAAX prenylation sequence, was further stained with osmium and imaged by EM. Figure 2D shows that APEX2-CAAX gives strong contrast at the plasma membrane, whereas APEX-CAAX is not detectable above background. To illustrate the generality of APEX2 as an EM tag, several more fusion constructs were imaged by EM in fibroblasts and cultured neurons (Supplementary Fig. 7). Together, our results show that APEX2 is superior to APEX for protein imaging in cells by EM.
We wished to understand the mechanistic basis for APEX2's superior performance in cells. The A134P mutation is in a loop adjacent to both heme and the aromatic substrate binding site (Fig. 1A).9 We first confirmed that this mutation does not revert APEX2 back to a dimer (Supplementary Fig. 8A-B). Next, we performed measurements of thermal stability (Supplementary Fig. 8C) and heme binding (Supplementary Fig. 9). We found that APEX2 was slightly better than APEX in both respects, but inferior to dimericW41F APX, despite being much more active than both inside cells. Thus, the improvements in stability and heme binding only partially explain the superior intracellular performance of APEX2.
When we performed kinetic measurements of phenol substrate oxidation by purified APEX and APEX2, we observed a striking effect at different H2O2 concentrations. Figure 2E shows that the initial rates for APEX2-catalyzed oxidation of the model aromatic substrate guaiacol increase steadily as a function of H2O2 concentration until ∼2.5mM H2O2 (with 1.4 mM guaiacol) or 0.5 mM H2O2 (with 0.5 mM guaiacol), at which point the rate begins to decrease. H2O2-induced peroxidase inhibition has been previously documented.10,11 Interestingly, APEX becomes inhibited at much lower H2O2 concentrations - 0.1 mM or 0.05 mM H2O2 - which are below the concentrations used for both proteomic and EM experiments. Therefore, the different susceptibilities of APEX and APEX2 to H2O2-induced inhibition may be the major reason that APEX2 is superior in cells (see Supplementary Discussion).
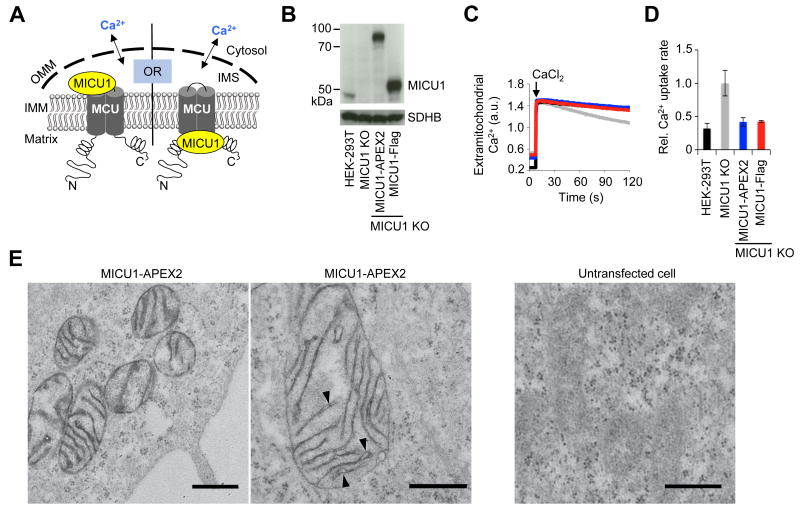
The improved properties of APEX2 allowed us to investigate a biological problem that was previously intractable to APEX. Mitochondria uptake calcium from the cytosol via the mitochondrial calcium uniporter (MCU) channel in the inner membrane.12,13,14 The regulation of MCU, important for cellular signaling and Ca2+ homeostasis, occurs through a soluble Ca2+ binding protein called MICU1.15 Conflicting reports have localized MICU1 either to the intermembrane space (IMS)16 or the mitochondrial matrix,17 resulting in very different mechanistic models of regulation (Fig. 3A). IMS localization implies that MICU1 senses and responds to rises in cytosolic Ca2+, because the porous OMM enables free exchange of Ca2+ between the IMS and cytosol.18 In contrast, matrix localization suggests that MICU1 senses and responds to matrix Ca2+ alone, since the inner membrane is impermeant to Ca2+.
Figure 3.
EM analysis of MICU1 using APEX2 supports intermembrane space (IMS) localization. (A) Scheme showing conflicting models that localize MICU1 to the IMS16 or matrix space17, 26 of mitochondria. Due to porins in the outer mitochondrial membrane (OMM), the IMS is permeable to cytosolic Ca2+, in contrast to the matrix. (B) Western blot characterization of MICU1-APEX2 stable HEK cells. Endogenous MICU1 was knocked out (MICU1 KO) and replaced with either MICU1-APEX2 or MICU1-Flag. Blotting was performed for MICU1 or succinyl dehydrogenase subunit B (SDHB), another IMM-localized protein loading control. (C) Representative traces (n=3) showing mitochondrial calcium uptake in response to a CaCl2 pulse resulting in ∼1 μM [Ca2+]free by monitoring Fluo4 fluorescence. Color coding explained in (D). (D) Graph reporting the linear fits of calcium uptake data from (C) between 50-60 sec after CaCl2 addition. Values are normalized to that of MICU1 KO (n=3). Error bars, ± 1 s.d. (E) EM images of HEK cells stably expressing MICU1-APEX2 (characterized in (B)-(C)). Two fields of view are shown. Dark stain generated by APEX2 is observed exclusively in the IMS, and not matrix space of mitochondria. Arrowheads point to wider cristae regions where dark stain is associated with the IMM rather than filling the IMS. Mitochondria of untransfected HEK cells processed under identical conditions are shown at right. Images representative of > 9 fields of view. Scale bars, 500 nm.
These previous studies determined the sub-mitochondrial localization of MICU1 using classical approaches: protease accessibility tests on purified mitochondria,16 and time-lapse protein imaging following an apoptotic trigger.17 Such assays can be prone to artifacts, such as mitochondrial lysis or nonrelease from the IMS due to protein-protein interactions. To obtain a direct readout of MICU1 localization in intact mitochondria in intact cells under physiological (non-apoptotic) conditions, we fused MICU1 to APEX2 and performed EM imaging. Previously, we found that transiently overexpressed MICU1-APEX gave aberrant localization on the outside of mitochondria (data not shown), while in the present study we found that lower expression via lentivirus gave undetectable staining (Supplementary Fig. 10). MICU1-APEX2, on the other hand, gave clear contrast that was specific to transduced cells.
To further reduce expression, we generated HEK cells that had endogenous MICU1 knocked out using TALEN technology19 and MICU1-APEX2 stably expressed. Recombinant MICU1-APEX2 in these cells was able to fully restore gatekeeping of calcium entry (Fig. 3B-D). EM imaging showed APEX2 staining exclusively in the IMS, and not in the matrix (Figure 3E). Due to the high spatial resolution, we also observed that MICU1-APEX2 was associated with the IMM rather than filling the entire IMS space (Fig. 3E), as expected for a protein that forms a complex with MCU. This result, coupled with recent proteomic evidence,6 strongly suggests that MICU1 is exclusively localized to the IMS.
In summary, three rounds of yeast display evolution produced a single point mutation (A134P) that greatly enhances the cellular activity and hence sensitivity of APEX. Consequently, APEX2 could be used for EM studies and proteomic tagging experiments not previously possible with APEX. As an EM reporter, APEX2 is easier to use than other tags (miniSOG,20 ReAsH,21 fluorescent proteins22,23) and can provide staining across large fields of view without special equipment, because contrast generation does not require light. The directness of the approach makes it an attractive alternative to indirect assays such as subcellular fractionation followed by protease accessibility tests or western blotting to examine the sub-organellar localization and membrane topology4 of important proteins.
The A134P mutation in APEX2 confers improvements in kinetics, thermal stability, heme binding, and most strikingly, resistance to high H2O2 concentrations. Interestingly, though 134P is largely absent from the cytosolic Class I peroxidases from which APEX is derived, it is the predominant amino acid at position 134 in secreted Class II and III peroxidases, including HRP (Supplementary Fig. 11). Perhaps nature independently converged upon this solution to bolster the H2O2 tolerance of peroxidases in non-reducing environments.
Online Methods
Mammalian cell culture
HEK 293T, HeLa, or COS-7 cells from ATCC (passage number < 20) were cultured in growth media (either MEM (Cellgro) or 1:1 DMEM:MEM mixture (Cellgro)) supplemented with 10% fetal bovine serum, 50 units/mL penicillin, and 50 μg/mL streptomycin) at 37°C under 5% CO2. Mycoplasma testing was not performed prior to experiments. For imaging experiments, cells were grown on 7 × 7 mm glass cover slips in 48-well plates. To improve the adherence of HEK293T cells, glass slides were pretreated with 50 μg/mL fibronectin (Millipore) for 15 min at 37 °C before cell plating and washed three times with DPBS, pH 7.4. Cells were transfected at 60–80% confluence using Lipofectamine 2000 (Life Technologies), typically with 0.7 μL Lipofectamine 2000 and 100 ng plasmid per 300,000 cells. Cells were labeled and/or fixed 18–24 h after transfection.
Biotin-phenol labeling in live cells
Genes were introduced into HEK 293T or COS-7 cells either through transient transfection with Lipofectamine 2000 or lentiviral infection. For lipofection, 100 ng of the APEX-fusion plasmid and 0.7 μL Lipofectamine 2000 in MEM (without serum) was used per well of a 48-well plate (0.95 cm2) of cells at 60-80% confluence. 3-6 hours later, the cell culture medium was changed back to fresh growth media. After 24 hours, biotin-phenol labeling was initiated by changing the medium to 200 μL of fresh growth media containing 500 μM biotin-phenol. This was incubated at 37°C under 5% CO2 for 30 minutes according to previously published protocols 1. Afterwards, 2 μL of 100 mM H2O2 was added to each well, for a final concentration of 1 mM H2O2, and the plate gently agitated for 1 minute. The reaction was then quenched by addition of 200 μL of 10 mM Trolox and 20 mM sodium ascorbate in DPBS (for a final concentration of 5 mM Trolox and 10 mM sodium ascorbate). Cells were washed with DPBS containing 5 mM Trolox and 10 mM sodium ascorbate three times and fixed with 3.7% paraformaldehyde in DPBS at room temperature for 10 min. Cells were then washed with DPBS three times and permeabilized with cold methanol at -20°C for 5 min. Cells were washed again three times with DPBS and blocked for 1 hour with 3% BSA in DPBS (“blocking buffer”) at 4°C. To detect APEX-fusion expression, cells were incubated with either mouse-α-FLAG antibody (Agilent, 1:500 dilution) or mouse-α-V5 antibody (Invitrogen, 1:500 dilution) for 1 hour to overnight at 4°C. After washing three times with 0.2% Tween in DPBS, cells were simultaneously incubated with secondary Alexa Fluor 488 goat anti-mouse IgG (Life Technologies, 1:750 dilution) and homemade streptavidin-Alexa Fluor 568 conjugates for 1 hour at 4°C. Cells were then washed three times with 0.2% Tween in DPBS and maintained in DPBS on ice for imaging.
Confocal imaging was performed using a Zeiss Axio Observer. Z1 microscope equipped with a Yokogawa spinning disk confocal head and a Cascade II: 512 camera. The confocal head contained a Quad-band notch dichroic mirror (405/488/568/647 nm). Samples were excited by solid state 491 nm (∼20 mW) or 561 nm (∼20 mW) lasers. Images were acquired using Slidebook 5.0 software (Intelligent Imaging Innovations), through a 48 × oil-immersion objective for YFP/AF488 (528/38 emission filter), AF568 (617/73 emission filter), and differential interference contrast (DIC) channels. Acquisition times ranged from 10 to 1000 milliseconds. Imaging conditions and intensity scales were matched for each dataset presented together unless otherwise noted.
Western blot analysis of biotin-phenol labeling in mammalian cells
For streptavidin blotting, HEK293T cells in wells of a 6-well plate were transfected with APXNES variants and labeled under the same conditions described for biotin-phenol imaging above. After 1 minute of labeling, the cells were washed 3 times with quencher solution (10 mM sodium azide, 10 mM sodium ascorbate, and 5 mM Trolox), then scraped and pelleted by centrifugation at 200 rpm for 5 minutes. The pellet was stored at -80 °C, then lysed with RIPA lysis buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 1 × protease cocktail (Sigma Aldrich, catalog no. P8849), 1 mM PMSF (phenylmethylsulfonyl fluoride), 10 mM sodium azide, 10 mM sodium ascorbate, and 5 mM Trolox) for 1 minute at 4 °C. The cell pellet was resuspended by gentle pipetting. Lysates were clarified by centrifugation at 13,000 rpm for 10 minutes at 4 °C before separation on a 9% SDS-PAGE gel. For blotting analysis, gels were transferred to nitrocellulose membrane, stained by Ponceau S (10 minutes in 0.1% w/v Ponceau S in 5% acetic acid/water), and blocked with “blot blocking buffer” (3% w/v BSA and 0.1% Tween-20 in Tris-buffered saline) at 4 °C overnight. The blots were immersed in streptavidin-HRP in blot blocking buffer (1:3000 dilution, Thermo Scientific) at room temperature for 60 minutes, then rinsed with blot blocking buffer 5 × 5 minutes before development with Clarity™ reagent (Bio-Rad) and imaging on an Alpha Innotech gel imaging system. For assessing comparative enzyme expression level, an identical gel and blot was prepared in parallel, and immunoblotted with α-FLAG-M2-HRP (1:3000, Sigma).
For blotting of endogenous proteins, HEK293T cells were grown in 6-well plates and infected at ∼50% confluency with 500 μL lentivirus prepared as described above. After 48 hours, cells were split into T25 flasks. From the same suspensions, cells were also plated into 48-well plates for side-by-side biotinphenol and immunofluorescence to check for enzyme activity and comparable enzyme expression levels. Biotin phenol labeling in the T25 flasks was performed with 30 minutes preincubation of 500 μM biotinphenol in 4 mL of a 1:1 mixture of DMEM:MEM (both from Cellgro) at 37°C. Flasks were quickly inverted so biotin-phenol media was on the ceiling of the flask, and 40 μL of 100 mM H2O2 was added (for a final concentration of 1 mM) to the media and agitated to mix. Flasks were again inverted and cells were exposed to the 1 mM H2O2 and 500 μM biotin-phenol solution for 1 minute at room temperature. After the 1 minute duration, the flask was inverted and the solution replaced with 5 mL of ice cold DPBS with quenchers (5 mM Trolox, 10 mM sodium ascorbate, 10 mM sodium azide) and the flask re-inverted for 1 minute. The flask was then washed with 5 mL DPBS + quenchers two more times. Cells were then resuspended by electronic pipette in 5 mL DPBS + quenchers and pelleted at 3000g for 10 minutes at 4°C. The supernatant was removed and the pellet was frozen at -80°C overnight.
Enrichment of endogenous biotinylated proteins was performed according to previous protocols. Cell pellets were lysed in 600 μL RIPA buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, pH 7.5), with 1 × protease cocktail (Sigma Aldrich, catalog no. P8849), 1 mM PMSF (phenylmethylsulfonyl fluoride), 5 mM Trolox, 10 mM sodium ascorbate, and 10 mM sodium azide. The lysates were centrifuged at 15,000g for 10 minutes at 4°C and the supernatant was transferred to a new 1.5 mL microcentrifuge tube as whole cell lysate (WCL). Protein concentrations of each WCL were determined by the Pierce 660 nm Protein Assay (catalog no. 22660).
Streptavidin-coated magnetic beads (Pierce, catalog no. 88817) were washed twice with RIPA buffer and 1750 μg of each WCL sample was incubated with 120 μL of magnetic bead slurry in separate 1.5 mL microcentrifuge tubes with rotation for 1 hour at room temperature. The beads were subsequently washed twice with 1 mL RIPA lysis buffer, once with 1 mL of 1 M KCl in ddH2O, once with 1 mL of 0.1M Na2CO3 in ddH2O, once with 1 mL of 2 M urea in 10 mM Tris-HCl pH 8.0, and twice with 1 mL RIPA lysis buffer. Biotinylated proteins were eluted by boiling the beads in 75 μl 3 × protein loading buffer supplemented with 20 mM dithiothreitol (DTT) and 2 mM free biotin. 17.5 μg of each WCL and 25 μl of each streptavidin eluate (SAE) were separated on 9% SDS-PAGE gels.
Nitrocellulose blot transfer and Ponceau S staining were performed as described above. Membranes were blocked with blot blocking buffer overnight at 4°C, cut into strips based on MW, and incubated facedown into 500 μl of blot blocking buffer containing the specified primary antibodies (see table below) for 1 hour at room temperature. Blots were washed 5 × 5 minutes in blot blocking buffer before incubation in 10 mL of secondary antibody in blot blocking buffer for 1 hour at room temperature. The membranes were then washed again 5 × 5 minutes in blot blocking buffer before imaging with Clarity™ reagent (Bio-Rad) as described above.
| Antibody | Source | Company | Catalog Number | Dilution |
|---|---|---|---|---|
| SA-HRP | -- | Pierce | 21126 | 1:3000 |
| αFLAG-M2-HRP | Mouse | Sigma | A8592 | 1:3000 |
| αTom70 | Rabbit | Proteintech | 14528-1-AP | 1:500 |
| αTom20 | Rabbit | Santa Cruz Biotechnology | sc-11415 | 1:200 |
| αATP5B | Mouse | Santa Cruz Biotechnology | sc-55597 | 1:150 |
| αBCAP31 | Rabbit | Proteintech | 11200-1-AP | 1:500 |
| αCalreticulin | Mouse | Calbiochem | 208912 | 1:1000 |
| αV5 | Mouse | Life Technologies | R960-25 | 1:2000 |
| αMo-HRP | Goat | Pierce | 1858413 | 1:2500 |
| αRb-HRP | Goat | Pierce | 1858415 | 1:2500 |
Generation of a stable HEK 293T MICU1-APEX2 line
HEK 293T cells lacking endogenous MICU1 were made using TALE nuclease as described previously 2 by targeting the 5′ most exon. After single cell cloning, colonies with a premature stop codon were identified by DNA sequencing and knock out was additionally verified by western blot. C-terminally FLAG-tagged MICU1 with or without a C-terminal APEX2 fusion was cloned into the pLJM5 vector3. MICU1 KO cells stably expressing MICU1-FLAG or MICU1-APEX2 (NM_001195518.1) were generated by lentiviral infection, followed by selection with 200 μg/ml hygromycin for three days. Compared with previous studies, we note that there are differences in cell types (HeLa4 vs HEK293T5) and splice variants of MICU1 used (isoform 1 4 vs isoform 25), though the two splice variants differ by only two residues which do not affect any interaction domains or the mitochondrial targeting sequence. Hence we do not anticipate the different localizations to arise from these experimental differences. Immunoblot of whole cell lysates was performed using antibodies from the following sources: Sigma (MICU1, 1:1000), and Mitosciences (SDHB, 1:2000).
Mitochondrial calcium uptake assays
HEK 293T cells were permeabilized in a KCl buffer (125 mM KCl, 2 mM K2HPO4, 10 μM EGTA,1 mM MgCl2, 20 mM HEPES, pH 7.2) along with 0.005% digitonin (Sigma-Aldrich), 5 mM glutamate, 5 mM malate, and 1 μM cell impermeable Fluo4 (Life Technologies). Fluorescence of Fluo4 was monitored with excitation at 485 nm and emission at 535 nm using a Perkin-Elmer Envision plate reader. 10 μM CaCl2 was injected at 4 s, resulting in about 1 μM free Ca2+. Relative rate of calcium uptake is reported using a linear fit of Fluo4 fluorescence from 50-60 s (n=3).
EM imaging of MICU1-APEX2
DAB staining was performed on transiently transfected or stable HEK293T as previously described6. For analysis of DAB activity by brightfield imaging (Supplementary Fig. 6), cells were grown on 7 × 7 mm glass cover slips placed inside 48-well cell culture plates. Cells were fixed on ice using pre-chilled 2% glutaraldehyde (Electron Microscopy Sciences) in DPBS buffer, pH 7.4, for 30 minutes. All subsequent work was performed using pre-chilled buffers and reagents. Cells were rinsed with DPBS three times for two minutes each before addition of 20 mM glycine in phosphate buffered saline (PBS), pH 7.4 to quench unreacted fixative. Cell were again washed with DPBS three times. DAB staining was initiated with the addition of freshly diluted 0.5 mg/mL (1.4 mM) DAB (Sigma; from a stock of the free base dissolved in 0.1 M HCl) and 10 mM H2O2 in DPBS. After five minutes, the reaction was stopped with the removal of the DAB solution and the cells were again washed with DPBS three times for two minutes each. Cells were then imaged by brightfield microscopy. Acquisition times ranged from 50-100 milliseconds.
For MICU1-APEX2 EM shown in Figure 3E, stable cells were grown in plastic six-well plates to 90% confluency and fixed using room temperature 2% glutaraldehyde (Electron Microscopy Sciences) in “buffer” (100 mM sodium cacodylate with 2 mM CaCl2, pH 7.4), then quickly moved to ice. Cells were kept between 0 and 4 °C for all subsequent steps until resin infiltration. After 30–60 min, cells were rinsed five times for 2 minutes each in chilled buffer, then treated for 5 min in buffer containing 20 mM glycine to quench unreacted glutaraldehyde, followed by another five 2-minute rinses in chilled buffer. A freshly diluted solution of 0.5 mg/mL (1.4 mM) DAB free base (Sigma) was combined with 10 mM H2O2 in chilled buffer, and the solution was added to cells for 5 minutes. To halt the reaction, the DAB solution was removed, and cells were rinsed 5 × 2 min with chilled buffer. Post-fixation staining was performed with 2% (w/v) osmium tetroxide (Electron Microscopy Sciences) for 30 min in chilled buffer. Cells were rinsed five times for 2 minutes each in chilled distilled water, then placed in chilled 2% (w/v) uranyl acetate in ddH2O (Electron Microscopy Sciences) overnight. Cells were brought to room temperature, washed in distilled water, then carefully scraped off the plastic, resuspended, and centrifuged at 700 g for 1 minute to generate a cell pellet. The supernatant was removed, and the pellet was dehydrated in a graded ethanol series (50%, 75%, 90%, 95%, 100%, 100%, 100%), for 10 minutes each time, then infiltrated in EMBED-812 (Electron Microscopy Sciences) using 1:1 (v/v) resin and anhydrous ethanol overnight, followed by two changes into 100% resin before letting sit overnight. Finally, the sample was exchanged once more with 100% resin before transfer to fresh resin and polymerization at 60 °C for 48 hours. Embedded cell pellets were cut with a diamond knife into 50 nm sections and imaged on a FEI-Tecnai™ G2 Spirit BioTWIN transmission electron microscope operated at 80 kV.
Supplementary Material
Acknowledgments
We gratefully acknowledge funding from the NIH (DP1 OD003961 to A. Y. T.; P41 GM103412, R01GM086197 to M.H.E.; 5R01GM077465-08 to V. K. M.), and the Howard Hughes Medical Institute Collaborative Initiative Award (A. Y. T.) for funding. S. S. L. and J. D. M. were both supported by National Science Foundation Graduate Research Fellowships and National Defense Science and Engineering Graduate Fellowships. N. Watson (Whitehead Institute W. M. Keck Microscopy Facility) acquired EM images of MICU1-APEX2. V. Hung (MIT) provided APEX2-Stx17 and ATP5JAPEX2 EM images. K. Cox (MIT) provided plasmids. FACS experiments were performed at the Koch Institute Swanson Biotechnology Center Flow Cytometry Facility. Color brightfield imaging was performed at the Koch Institute Microcopy Core Facility. We thank C. Drennan for use of her Cary 300 spectrophotometer. D. Mc Swiggen assisted with enzyme purification. We thank T. Poulos, K. White, and the laboratory of D. Wittrup for advice. V. K. M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Authors Contributions: S.S.L., J.D.M., and A.Y.T. designed the research, analyzed the data, and wrote the paper. All authors edited the paper. K.J.K. and V.K.M. prepared MICU1 stable cells and performed calcium uptake assays. S.S.L. and J.D.M. performed EM sample preparation and T.J.D. and M.H.E. performed EM imaging. J.D.M. performed enzyme kinetic assays and analysis. S.S.L. performed all other experiments.
Competing Financial Interests: The Massachusetts Institute of Technologyhas submitted a patent application on the peroxidase technology.
References
- 1.Porstmann B, Porstmann T, Nugel E, Evers U. Which of the commonly used marker enzymes gives the best results in colorimetric and fluorimetric enzyme immunoassays: Horseradish peroxidase, alkaline phosphatase or β-galactosidase? J Immunol Methods. 1985;79:27–37. doi: 10.1016/0022-1759(85)90388-6. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Wang Y, Chiu SL, Cline HT. Membrane targeted horseradish peroxidase as a marker for correlative fluorescence and electron microscopy studies. Front Neural Circuits. 2010;4:6. doi: 10.3389/neuro.04.006.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorner J, et al. Chimeric molecules employing horseradish peroxidase as reporter enzyme for protein localization in the electron microscope. Methods Enzymol. 2000;327:35–45. doi: 10.1016/s0076-6879(00)27265-0. [DOI] [PubMed] [Google Scholar]
- 4.Martell JD, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–8. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhee HW, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–31. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hung V, et al. Proteomic Mapping of the Human Mitochondrial Intermembrane Space in Live Cells via Ratiometric APEX Tagging. Mol Cell. 2014;55:332–341. doi: 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandelman D, Schwarz FP, Li H, Poulos TL. The role of quaternary interactions on the stability and activity of ascorbate peroxidase. Protein Sci. 1998;7:2089–98. doi: 10.1002/pro.5560071005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebert PS, Hess RA, Frykholm BC, Tschudy DP. Succinylacetone, a potent inhibitor of heme biosynthesis: Effect on cell growth, heme content and δ-aminolevulinic acid dehydratase activity of malignant murine erythroleukemia cells. Biochem Biophys Res Commun. 1979;88:1382–1390. doi: 10.1016/0006-291x(79)91133-1. [DOI] [PubMed] [Google Scholar]
- 9.Sharp KH, Moody PCE, Brown KA, Raven EL. Crystal structure of the ascorbate peroxidase-salicylhydroxamic acid complex. Biochemistry. 2004;43:8644–51. doi: 10.1021/bi049343q. [DOI] [PubMed] [Google Scholar]
- 10.Nicell JA, Wright H. A model of peroxidase activity with inhibition by hydrogen peroxide. Enzyme Microb Technol. 1997;21:302–310. [Google Scholar]
- 11.Arnao MB, Acosta M, del Río JA, García-Cánovas F. Inactivation of peroxidase by hydrogen peroxide and its protection by a reductant agent. Biochim Biophys Acta. 1990;1038:85–9. doi: 10.1016/0167-4838(90)90014-7. [DOI] [PubMed] [Google Scholar]
- 12.Baughman JM, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–5. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Stefani D, Raffaello A, Teardo E, Szabò I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–40. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamer KJ, Mootha VK. MICU1 and MICU2 play nonredundant roles in the regulation of the mitochondrial calcium uniporter. EMBO Rep. 2014;15:299–307. doi: 10.1002/embr.201337946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perocchi F, et al. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature. 2010;467:291–6. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Csordás G, et al. MICU1 Controls Both the Threshold and Cooperative Activation of the Mitochondrial Ca2+ Uniporter. Cell Metab. 2013;17:976–987. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman NE, et al. MICU1 Motifs Define Mitochondrial Calcium Uniporter Binding and Activity. Cell Rep. 2013;5:1576–1588. doi: 10.1016/j.celrep.2013.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vander Heiden MG, et al. Outer mitochondrial membrane permeability can regulate coupled respiration and cell survival. Proc Natl Acad Sci U S A. 2000;97:4666–71. doi: 10.1073/pnas.090082297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plovanich M, et al. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS One. 2013;8:e55785. doi: 10.1371/journal.pone.0055785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu X, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaietta G, et al. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–7. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- 22.Grabenbauer M, et al. Correlative microscopy and electron tomography of GFP through photooxidation. Nat Methods. 2005;2:857–62. doi: 10.1038/nmeth806. [DOI] [PubMed] [Google Scholar]
- 23.Horstmann H, Vasileva M, Kuner T. Photooxidation-Guided Ultrastructural Identification and Analysis of Cells in Neuronal Tissue Labeled with Green Fluorescent Protein. PLoS One. 2013;8:e64764. doi: 10.1371/journal.pone.0064764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lad L, Mewies M, Raven EL. Substrate Binding and Catalytic Mechanism in Ascorbate Peroxidase: Evidence for Two Ascorbate Binding Sites †. Biochemistry. 2002;41:13774–13781. doi: 10.1021/bi0261591. [DOI] [PubMed] [Google Scholar]
- 25.H A, et al. Kinetic study of the inactivation of ascorbate peroxidase by hydrogen peroxide. 2000 [PMC free article] [PubMed] [Google Scholar]
- 26.Mallilankaraman K, et al. MICU1 Is an Essential Gatekeeper for MCU-Mediated Mitochondrial Ca2+ Uptake that Regulates Cell Survival. Cell. 2012;151:630–644. doi: 10.1016/j.cell.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhee HW, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–31. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanjana NE, et al. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–92. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sancak Y, et al. Ragulator-Rag Complex Targets mTORC1 to the Lysosomal Surface and Is Necessary for Its Activation by Amino Acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman NE, et al. MICU1 Motifs Define Mitochondrial Calcium Uniporter Binding and Activity. Cell Rep. 2013;5:1576–1588. doi: 10.1016/j.celrep.2013.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Csordás G, et al. MICU1 Controls Both the Threshold and Cooperative Activation of the Mitochondrial Ca2+ Uniporter. Cell Metab. 2013;17:976–987. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martell JD, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–8. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.