Abstract
Background
To identify genome wide single nucleotide variants (SNVs) and mutations in African Americans (AAs) with colorectal cancer (CRC). There is a need of such studies in AAs since they display a higher incidence of aggressive CRC tumors.
Methods
We performed whole exome sequencing (WES) on DNA from 12 normal–tumor pairs of AA CRC patients’ tissues. Data analysis was carried out using GATK (Genome Analysis Tool Kit). Normative population databases (e.g. 1000 Genomes SNP database, dbSNP, and HapMap) were used for comparison. Variants were annotated using Annova and validated by Sanger sequencing.
Results
We identified somatic mutations in genes that are known targets in CRC such as APC, BRAF, KRAS, and PIK3CA. We detected novel alterations in the WNT pathway gene, APC, within its exon 15 of which mutations are highly associated with CRC.
Conclusions
This first WES from AA CRC patients provides insight into the identification of novel somatic mutations in APC. Our data suggest an association between specific mutations in the WNT signaling pathway and increased risk of CRC. The analysis of the pathogenicity of these novel variants might help understand the aggressive nature of CRC in AAs.
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide 1. African Americans (AAs) are more at risk than Caucasians 2, 3. Genetic alterations in oncogenes and tumor suppressor genes (TSGs) lead to CRC development 4, 5. Advances in sequencing technologies allow high throughput data generation 6,7-14.
A large proportion of mutations occurring in colon cancer involves genes encoding proteins of the WNT, EGFR and p53 pathways10-15,16-19,20.Studies have identified candidate genes as driver/recurrent mutational targets in CRC and metastatic CRCs 6, 7. TSGs and oncogenes which are relevant as predictive and prognostic markers in CRC have been established9. The involvement of the majority of these, including APC, mutations in CRC progression is not well known, particularly in AAs 9, 20, 21.
Based on our WES analysis of CRCs from AAs, we identified novel somatic mutations in WNT pathway genes that have an impact on genomic instability leading to CRC. Our analysis revealed alterations in several pathways including WNT, EGFR, and p53 with novel SNVs in APC gene. To our knowledge this is the first CRC WES study in this high risk population.
Materials and Methods
Patients
Twelve pairs of fresh frozen matched normal and adenocarcinoma specimens collected from AA patients were used for WES (Table 1). This study was approved by the Institutional Review Board (IRB) of Howard University. Written consent forms were obtained from all patients. All tissue samples were reviewed by two pathologists (ELL and BS) for microdissection. Subjects with FAP, HNPCC or family history of CRC were excluded.
TABLE 1.
Clinico-pathological Characteristics of the CRC Patients
| Patient ID | Sex/Age | MSI | TNM | Stage | Location | Kras |
|---|---|---|---|---|---|---|
| CC1014 | M/71 | S | pT2pNopMx | I | L | W |
| CC1016 | F/68 | S | pT4bpN2a | IIIc | R | M |
| CC1017 | F/52 | S | pT2pNopMx | I | L | M |
| CC1018 | F/83 | S | pT3pN1b | IIIb | L | M |
| CC1028 | M/41 | H | pT3pN0pMx | IIa | L | W |
| CC1029 | F/50 | H | pT2pNopMx | I | R | M |
| CC1045 | F/57 | S | pT3pN0pMx | IIa | R | M |
| CC1053 | F/49 | S | pT3pN0pMx | IIa | R | W |
| CC1054 | M/52 | S | pT3pN0 | IIa | R | W |
| CC1057 | M/88 | S | pT3pN0pMx | IIa | L | M |
| CC1060 | F/52 | S | pT3pN1pMx | IIIb | L | W |
| CC1062 | M/34 | H | pT3pN1pMx | IIIb | L | W |
H, high; F, female; L, left; M, male, MSI, microsatellite instability; M, mutant; R, right; S, stable; TNM, tumor–node–metastasis, W, wild type.
Whole Exome Sequencing
DNA quantification and quality assessment; Illumina DNA Library Preparation; Single Nucleotide Variations calling; Public Genome Data comparison; Sequencing Validation; Single nucleotide variants’ description; Mutations frequencies; and Copy number alterations are described in the Supplement section.
Results
Clinicopathological Characteristics of patients
The 12 patients’ data is listed in table 1. Twenty five percent (n=3) were stage I, 42% (n=5) stage II, and 33% (n=4) stage III. There were 25% (n=3) MSI tumors, each with stage I, II and III, 2 with wild type Kras and one with mutated kras. Half of the tumors were wild type for Kras. Four patients were positive for lymph nodes involvement, one of which was MSI. There were 9 patients with metastatic tumors including the MSI tumors (Table 1).
Exome sequence analysis and coverage
WES covered 201,121 coding exons, splice junctions (31.3 Mb), and adjacent regions (UTRs, promoters: 30.7 Mb). Using massive parallel sequencing on a HiSeq platform (Illumina, San Diego, CA), we generated approximately 850 million bases of effective sequence data with an average read length of 101 bases. After mapping to the human reference genome (NCBI36.1/HG19) using Burrows-Wheeler Alignment tool (BWA) 26, we obtained an average depth of coverage for the target regions of 30× for each sample. The extent of coverage of the target region at >10× depth was more than 73% in all samples. The variants’ calls and analysis are described in the Supplement section.
Copy number variations
CNVs were detected in all samples and on most chromosomes. We found regions of copy number gain or loss that overlapped genes with sequence alterations and plotted the copy number along the genome for each sample. Circos software was used. Somatic variants from all samples were combined to generate a Circos plot (Supplementery Figure 1). For each 10MB section of the genome, we calculated the number of total variants, synonymous (non amino-acid changing) and missense variants. These were each plotted on separate tracks. For “stopgain”, “stoploss” and “frameshift” (insertions or deletions) variants, we marked their locations on separate tracks. For example, alteration at the APC site is primarily displayed on the SNP track at the chromosomal loci level for most of these samples (Supplementary Figure 2).
Comparison of African Americans’ exome data to public databases
We used R software (version 2.15.2, http://www.r-project.org/) to compare the variants in the normal and tumor samples to the 1000 Genomes database (a nominally non-cancerous population; supplementary Figure 3) All samples displayed more or less an equal number of SNVs in their tumors compared to their matched normals. CC1045 Tumor-Normal pair displayed the highest number (with 75% and 88%, respectively) of SNVs followed by CC1014.
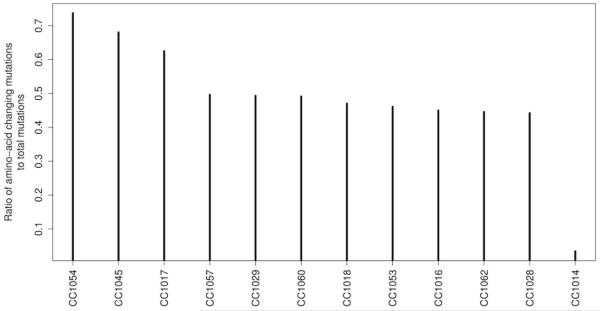
Amino Acid changing ratio
For each sample, we computed the ratio of amino-acid changing variants (mutations) to the total number of variants (Figure 1). We looked at variants that were somatic (not present in normal: heterozygous or homozygous in tumor while homozygous or heterozygous in matched normal. We computed the total number of these variants and the number of variants that are amino-acid changing (variants belonging to the categories “frameshift deletion”, “frameshift insertion”, “non-frameshift deletion”, “non-frameshift insertion”, “nonsynonymous SNV”, “stopgain SNV”, “stoploss SNV”). The ratio of these counts was plotted for each sample. . Tumor CC1014 had the lowest number of amino-acid changing SNVs while tumors CC1017, CC1045, and CC1054 had 50% or more of their variants as amino-acid changing. The rest of the tumors had an average 45%. The MSI tumors were within the 45% average group.
Figure 1.
Amino-acid changes based on gene mutations analysis in African Americans with CRC. We computed the total number of these variants and the number of variants that are amino-acid changing (variants belonging to the categories “frameshift deletion”, “frameshift insertion”, “nonframeshift deletion”, “nonframeshift insertion”, “nonsynonymous SNV”, “stopgain SNV”, “stoploss SNV”). The ratio of these two counts was plotted for each sample.
Single nucleotide substitution
While investigating single nucleotide substitutions (SNS) within the mutations’ spectrum, we found that C: G-.T: A transitions were the most significant changes in individual CRC samples and all CRC samples combined (Supplementary Figure 4). Our data show a significantly higher enrichment of C: G-.T: A in CpG dinucleotides in carcinoma compared to that expected by chance (supplementary, Figure 3). Given the fact that DNA methylation occurs almost exclusively within the context of CpG dinucleotides27, the enrichment of C:G-.T:A in CpG dinucleotides may be associated with the extensive methylation of CpG dinucleotides in CRC11, 28, 29.
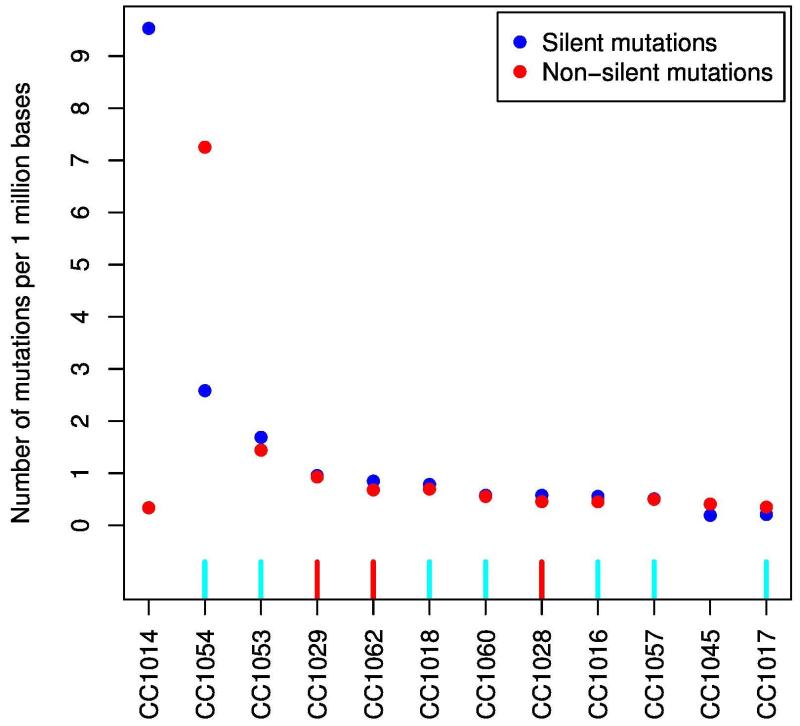
Hypermutated vs. non-hypermutated samples
Based on mutation rates, we stratified the cases in two groups: hypermutated (more than 7 mutations per 106 bases, 2 samples) and non-hypermutated (less than 3 mutations per 106 bases, 10 samples; Fig. 2).The number of silent mutations in one hypermutated sample (CC1014) was noticeably high compared to non-silent-mutations in the other hypermutated sample (CC1054). To assess the basis for the considerably different mutations rates, we evaluated the MSI status in the analyzed samples, the 3 MSI-H tumors were non-hypermutated when compared to MSS tumors (Fig. 2).
Figure 2.
Mutations rate in African American colorectal samples. MSI status was evaluated, 3 non-hypermutated tumors were MSI-H (red), none of the hypermutated tumors were MSI-H. Hypermutated were defined as having >7 mutations per 106 bases (2 samples) and non-hypermutated defined as < 3 mutations per 106 bases (10 samples). Red, MSI high, light blue, MSI low.
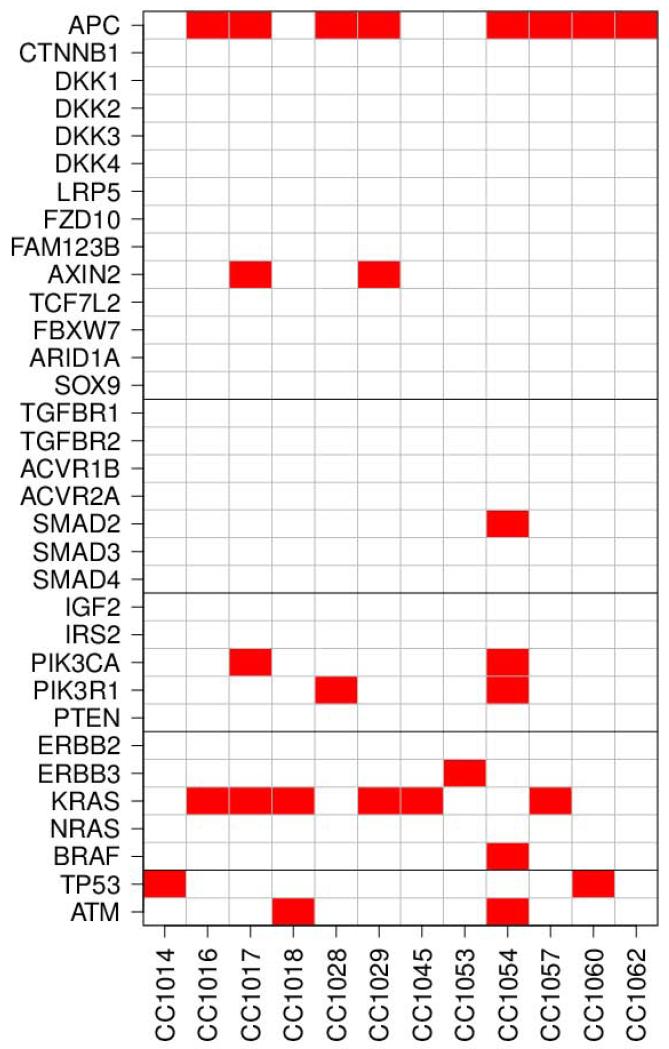
Altered pathways
To explore major affected pathways, we analyzed alterations in the WNT, EGFR and PI3K pathways 9, 16-19, 30-32. A special focus was on APC, BRAF, KRAS, and PIK3CA as major targets in colon tumorigenesis. The WNT pathway was altered in 80% of all CRC patients. Thus, we looked at the major genes of interest in the WNT pathway 30, 33-35. We determined and plotted altered genes and pathways for each sample (Figure 3). For each sample, an altered gene was defined as any gene that has an amino-acid changing somatic SNVs (mutation). An altered pathway was defined as any pathway in which one or more genes are altered. We found 16 genes altered in WNT signaling pathway followed by EGFR signaling (8 mutations), PI3K and p53 pathways (4 mutations each) across all tumors. We also determined TSGs and oncogenes’ alterations based on a recent publication 9 (Supplementary Figure 5). Fifteen oncogenes and 37 TSGs were altered in our samples compared to Vogelstein et al 9. We plotted the altered group of oncogenes or TSGs or individual oncogenes or TSGs in these samples based on the genes that had at-least one amino-acid changing mutation (Supplementary Figure 5). There were 17 TSGs and 8 oncogenes which were common with Voglstein et al.’s recent cancer study 9. The TSG, APC and the oncogenes: Kras, Braf, PIK3CA are topping the list. We then determined and plotted the altered genes that are involved in chromatin assembly, remodeling and silencing that effect genes’ expression and regulation (Figure 5, supplement) based on Gene Ontology categories (GO:0031497: chromatin assembly, GO:0006338–chromatin remodeling, GO:0006342–chromatin silencing) using AmiGO (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi). First we determined the group of genes in chromatin alteration (Supplementary Figure 6A), then individual genes that effect chromatin assembly, reprogramming and silencing (Supplementary Figure 6B). The following chromatin assembly genes: CDAN1, CHAF1B, and TP53, chromatin remodeling genes: BAZ2A, TOP1MT, BRDT, PBRM1, and ACTL6B, and chromatin silencing gene: UBR2 were altered.
Figure 3.
Altered pathways and genes plots for the 12 AA samples. The dark horizontal lines separate the genes belonging to different pathways. Gene alteration defined as the presence of at least one protein changing mutation in the gene. The definition of altered pathway is at least one altered gene in the pathway.
An exome sequencing analysis by The Cancer Genome Atlas network (TCGA) 6 sequencing of 276 colon tumors led to the establishment of a catalog of altered genes in these tumors. More than 13% of the tumors were MSI-H in the TCGA compared to 25% in our study. Figure 4 shows the percentage of samples that had amino-acid changing mutations in each gene. We found 6 genes that had mutation rates significantly higher than background using MuSiC software. The most frequently mutated genes were APC, KRAS, FCRL5, OBSCN, PR1L1, DNAH17, ZNF568, CACNA1C, TELO2, and SRMS. KRAS as expected had oncogenic codon 12 and 13 mutations. KRAS, ZNF568, CACNA1C, TEL02, SRMS had a significant (p<0.05) enrichment of amino-acid changing mutations within all recorded somatic variants. Tumor suppressor genes TP53 and APC were mutated in both hypermutated and non-hypermutated samples, but significantly more in non-hypermutated tumors (50% vs. 100%). Other genes that were frequently mutated are KRAS, PIK3CA and TP53 (Supplementary Table 1). Two non-synonymous SNVs were found in tumor suppressor TP53. KRAS gene showed 4 non-synonymous, and 1 non-frameshift deletion. We found 2 non-synonymous mutations in PIK3CA. In addition, our results showed 1 frameshift deletion in AXIN2 and 2 non-synonymous alterations in ATM gene. Three non-synonymous mutations were found in CACNA1C (Table 1, supplement) which is part of MAPK/Ca channel pathway 36. Non-synonymous SNVs in mismatch repair genes MSH6 (two) and MSH3 (one) were also detected.
Figure 4.
Gene Mutations in African Americans with CRC. The genes with top highest frequency in the 12 samples indicated by white and dark bars. White bars indicate 6 genes (APC, KRAS, ZNF568, CACNA1C, TEL02, SRMS) that have a significantly (p<0.05) higher frequency of mutation above background (as determined by MuSiC). Background mutation rate (BMR) is defined as the number of mutations per length of genome with read coverage that is sufficient to call a mutation.
We further focused on APC, a gatekeeper and one of the most important contributors in colorectal carcinogenesis 19. This gene displayed 15 mutations in our samples (Table 2). Validation of the APC mutations: Sanger sequencing validation of APC mutations including 1 missense mutation, 6 frameshift deletions, 5 stopgains, 2 frameshift insertions, and 1 non-frameshift deletion was performed. Of the detected APC mutations, 14 that were confirmed were novel (Table 3) including: frameshift deletions (APC, 112155014 in tumor CC1029; APC,112175723-29, APC,112175746 in tumor CC1017; APC,112116573-82 in tumor CC1060 and APC,112175212 in tumor CC1062), stop codons (APC, 112175207 in tumor CC1028; APC,112175639 in tumor CC1029; APC,112103015 in tumor CC1054; APC,112174763 in tumor CC1060 and APC, 112174658 in tumor CC1062), a frameshift insertion in tumor CC1057 APC,112175993, a non-frameshift deletion in tumor CC1029 APC, 112155017, and a synonymous mutation (APC112177171 rs465899, in tumor CC1028).
TABLE 2.
Novel and Known APC Somatic Mutations in African American Patients with CRC
| Sample | N | T | ExonicFunc | Start | End | Reference | Obs | dbSNP135 | Frequencya | Mutation Typeb |
Protein Change |
Exon |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC1016 | 0/0 | 0/1 | Frameshift insertion | 112175993 | 112175993 | – | A | 0 | Novel | p.D1550fs | 15 | |
| CC1017 | 0/1 | 0/0 | Frameshift deletion | 112175723 | 112175729 | AGGGTCC | – | 0 | Novel | p.1460_1462del | 15 | |
| CC1017 | 0/1 | 0/0 | Frameshift deletion | 112175746 | 112175746 | T | – | 0 | Novel | p.A1467fs | 15 | |
| CC1028 | 0/0 | 0/1 | Stopgain SNV | 112175207 | 112175207 | G | T | 0 | Novel | p.E1288X | 15 | |
| CC1028 | 0/0 | 0/1 | Synonymous SNV | 112177171 | 112177171 | G | A | rs465899 | 0.65 | Known | p.P1942P | 15 |
| CC1029 | 0/0 | 0/1 | Frameshift deletion | 112155014 | 112155014 | C | – | 0 | Novel | p.P411fs | 9 | |
| CC1029 | 0/0 | 0/1 | Nonframeshift deletion | 112155017 | 112155028 | GGCATGGACCAG | – | 0 | Novel | p.412_415del | 9 | |
| CC1029 | 0/0 | 0/1 | Stopgain SNV | 112175639 | 112175639 | C | T | 0 | Novel | p.R1432X | 15 | |
| CC1054 | 0/0 | 0/1 | Stopgain SNV | 112103015 | 112103015 | C | A | 0 | Novel | p.S127X | 3 | |
| CC1054 | 0/0 | 0/1 | Missense SNV | 112179011 | 112179011 | C | A | 0 | Novel | p.L2556I | 15 | |
| CC1057 | 0/0 | 0/1 | Frameshift insertion | 112175993 | 112175993 | – | A | 0 | Novel | p.D1550fs | 15 | |
| CC1060 | 0/0 | 0/1 | Frameshift deletion | 112116573 | 112116582 | CTGCCAGGAT | – | 0 | Novel | p.216_219del | 5 | |
| CC1060 | 0/0 | 0/1 | Stopgain SNV | 112174763 | 112174763 | A | T | 0 | Novel | p.R1140X | 15 | |
| CC1062 | 0/0 | 0/1 | Stopgain SNV | 112174658 | 112174658 | C | T | 0 | Novel | p.Q1105X | 15 | |
| CC1062 | 0/0 | 0/1 | Frameshift deletion | 112175212 | 112175216 | AAAAG | – | 0 | Novel | p.1289_1291del | 15 |
Abbreviation: SNV, single nucleotide variant, Obs, Observed.
Frequency in 1000 Genomes database.
Mutation type is novel if no dbSNP135 entry was found and if frequency in 1000 Genomes database was zero.
Mutation spectra of SNS
Nucleotide transition was analyzed across all tumors using R software. The MSI tumors showed more C to T changes compared to MSS tumors. There was no difference in the SNS distribution between right and left colon and tumors of different stage (Supplementary Figure 7).
Discussion
Whole exome sequencing of colorectal carcinogenesis will lead to the discovery of specific driver mutations that are important in this process especially in high risk populations such as African Americans 9. With growing genomic databases across many tumor types, specific and pan-cancer analyses are becoming of great interest 37-39. WNT and EGFR pathways are among the leading CRC pathways 6, 9, 38. In this study, we analyzed alterations in major CRC pathways and associated effectors. This first WES analysis of AA CRCs provides insight into the identification of novel somatic mutations that suggest an association between WNT signaling alterations and risk of CRC. Our results indicate novel somatic nonsynonymous variants in APC alters the function of this protein in the analyzed CRC samples.
WNT and EGFR signaling pathways were the most altered in the analyzed samples indicating their significant contribution in colorectal carcinogenesis. Genes from these two pathways were also primary targets of SNVs in other studies of CRC patients regardless of sex, age or race of the patients 6, 9, 40. A limited number of TSGs and oncogenes have been proposed to be involved in CRC progression 9. Our results define a total of 52 genes that have been cited by Vogelstein et al. recently 9. However, the identified APC mutations are different in AA CRCs compared to public databases and recent studies.
Nine genes are associated with chromatin regulation including CDAN1, CHAF1B, TP53, BAZ2A, TOP1MT, BRDT, PBRM1, ACTL6B, and UBR2. These genes are part of the mTOR signaling pathway that mediates transcriptional silencing via histone ubiquitination 41-44. It is worth noting that this combined set of chromatin regulatory genes were mutated in the analyzed CRC tumors. This finding suggests that the catalog of cancer genes is far from being exhaustive and that many cancer genes are still awaiting their placement on such a catalog as studies, like ours, characterize unique patient cohorts. These genes’ SNVs have far reaching consequences as they govern the expression of many genes through their effect on chromatin structure.
Copy Number Variations (CNVs) for the analyzed samples were generated using the exome data using R software. While exome data based CNVs are not as precise as aCGH CNVs 13, they allow an approximate assessment of CNVs in regions of interest. We detected no copy number shift at the APC locus for any sample consistent with the APC gene as a primary target of SNVs rather than CNV alterations9. The Circos plot clearly illustrates the intricacies between different genomic alterations mechanisms.
The number of mutations was different among samples. Two samples were hypermutated (>7 mutations per 106 bases), while ten samples were not (<3 mutations per 106 bases; Fig. 2). The number of silent mutations in one hypermutated sample (CC1014) was pronounced compared to the other hypermutated sample (CC1054). To assess the basis for the considerably different mutation rates, we evaluated the tumors’ MSI status 12, 45, 46. Three non-hypermutated tumors were MSI-H. No sample was MSI-H in the hypermutated tumors (Fig. 2). The general expectation would be to have higher mutation rate in MSI-H than in MSS tumors. Such is not the case in our analyzed samples. Indeed, MSI-H is known to be expressed in repetitive sequences along the genome, which is why the MSI status is generally investigated through PCR amplification of mono-, di- or tri-nucleotide repeats/microsatellites, which are less prevalent in exomic regions.
Among the CRC genes established by TCGA 6, the most frequently mutated were APC, KRAS, FCRL5, OBSCN, PR1L1, DNAH17, ZNF568, CACNA1C, TELO2, and SRMS. Most samples in the TCGA dataset are from Caucasians. Beyond identifying new non-synonymous mutations and candidate CRC genes, our study demonstrates that we are far from having a complete catalogue of CRC genes, with many genes at clinically important frequencies within individual CRC tumors still awaiting identification. The number of such genes is still increasing steeply with the number of analyzed samples and tumor types. Follow-up studies will be required to confirm and understand the functional impact of the mutations in these genes.
Two non-synonymous SNVs were found in tumor suppressor TP53. KRAS gene showed 4 non-synonymous, and 1 non-frameshift deletion. MSH3 and MSH6 showed 2 and 1 non-synonymous mutations, respectively. The mutations in MSH3 and MSH6 are in line with a high rate of MSI-H in African American CRC patients. Indeed, we have previously reported a 20% rate of MSI-H in AA CRC patients that we assigned primarily to the methylation of MLH1 gene, another DNA MMR gene. Our WES data demonstrate that along with methylation of MLH1, SNVs targeting other DNA MMR genes do participate in the higher MSI rate in this population 13, 14, 45.
Our results showed 1 frameshift deletion in AXIN2 and 2 non-synonymous alterations in ATM gene. We found 2 non-synonymous mutations in PIK3CA gene. Three non-synonymous mutations were detected in CACNA1C that belongs to the MAPK/Ca channel pathway 36.
As for the WNT pathway APC gene, we detected 1 missense mutation, 5 frameshift deletions, 5 stopgains, 2 frameshift insertions, and 1 non-frameshift deletion (Table 2). APC is a large protein with multiple domains that bind to various other proteins in addition to β-catenin. In the nucleus, β-catenin interacts with LEF-1/TCF DNA-binding proteins and activates Wnt target genes through a unique C-terminal activation domain. The N-terminal ARM repeat of β-catenin associates with BCL9/Legless and Pygopus, a PHD finger protein that binds H3K4me2 and promotes H3K4me3 at target gene promoters 30, 48. APC is also recruited to Wnt target genes upon activation of the pathway and regulates the periodic exchange of β-catenin and TLE1/Gro corepressor complexes. Therefore, we think the mutations that we observe in the CRC samples effect both cell proliferation and tumor progression through both gene regulation via chromatin modification and protein functional changes 41-43.
Functional annotation of the mutated genes showed several pathways including Wnt pathway, cell adhesion pathway and ubiquitin mediated proteolysis pathway were altered genetically in the early stage of colorectal tumorigenesis. Therefore, the biological functional effects of these mutations especially in APC may have profound colorectal carcinogenicity. Functional APC protein annotation of the APC novel mutations are likely to lead to changes in the activity of APC protein since some of them located in exons 5 and 15. None of the novel mutations defined in this study have been reported to be associated with any disease. Since frameshift mutations and stop codons have major effects on the final protein product, they are expected to exert drastic influences on protein function. In addition, most of these SNVs such as APC4664, APC3418 and APC3862 (Table S4), are located in exon 15, which is the portion of APC most highly associated with CRC risk. Why do these mutations differ from other reported exon 15 APC mutations?. These mutations may contribute to major biological alterations of this protein with clinical and pathological implications in this population that might explain the disparity issue. This manuscript reveals new mutations in AA CRC patients of which the functional analysis will determine their pathogenicity.
A number of variants in the analyzed 12 samples were not found in dbSNP135 and considered novel variants as a result (Table 2). We were able to validate the APC mutations in the original samples by Sanger sequencing. These mutations are novel and as such their frequencies are not known in AA and other patients. It will be very relevant to establish such frequencies for personalized treatment approaches and also to establish whether such mutations have any racial relevance. We are aware that CRC incidence in African Americans has other confounding factors such as inequities in health care access and utilization and appropriateness/quality of treatment, as well as other social and epidemiological factors, that likely play far more significant roles. Our data in this study reports novel somatic alterations in CRC that may be another factor in the observed disparities.
The sample size in this study is relatively small and it can therefore be considered as a pilot study. Based on recent publications, providing a comprehensive catalogue of genes in which somatic point mutations in cancer at both high (>20%) and intermediate (2-20%) frequency will require analyzing an average of approximately 2,000 tumors for colorectal tumors 40.There have been other relatively large genome-wide or exome sequencing studies on CRC, but none of them has differentiated the patients populations and whether or not African Americans were part of the study 6, 9, 40.
In summary, exome sequencing is becoming increasingly common in clinical practice especially in the field of oncology and represents a cost effective approach to comprehensively characterize somatic mutations. This application will lead to the discovery of novel targets, driver mutations as well as known and novel colon cancer predisposing mutations. The information of SNVs in African Americans with colorectal cancer should be able to pave the way to reduce health disparity using personalized and precision medicine for diagnostic tumor profiling as well as for the development of targeted therapies in this high risk population.
Supplementary Material
Implications.
There are little genome landscape studies of colorectal cancer (CRC) tumors from African Americans. Correlations between genomic alterations and tumor phenotypes are required. This study’s relevance lies in its potential to identify race specific CRC biomarkers. Such biomarkers will be useful for targeted therapy and better management of the disease.
Acknowledgments
Grant Support
This project was supported (in part) by the National Institute on Minority Health and Health Disparities of the National Institutes of Health under Award Number G12MD007597.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest to disclose.
Authors’ Contributions
Conception and design: HA, HB
Development of methodology: JD, EL, BS, MD, HR
Acquisition of data (acquired and managed patients): HA, JD,
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): SV, JD, MN, RS
Writing, review, and/or revision of the manuscript: HA, HB
Administrative, technical, or material support (i.e., reporting, organizing data): SD, JD, MD, HR
Study supervision: HA
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Desantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 3.Ashktorab H, Paydar M, Namin HH, et al. Prevalence of Colorectal Neoplasia Among Young African Americans and Hispanic Americans. Dig Dis Sci. 2013 doi: 10.1007/s10620-013-2898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 5.Jones S, Chen WD, Parmigiani G, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Network CGA Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wood LD, Parsons DW, Jones S, et al. The Genomic Landscapes of Human Breast and Colorectal Cancers. Science. 2007 doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 8.Bass AJ, Lawrence MS, Brace LE, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43:964–968. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashktorab H, Rahi H, Soltani S, et al. Novel Mutation and Hypomethylation Define Distinct Biological Subgroups of altered KRAS colon tumors. AACR. 2013 [Google Scholar]
- 11.Ashktorab H, Rahi H, Wansley D, et al. Toward a comprehensive and systematic methylome signature in colorectal cancers. Epigenetics. 2013;8 doi: 10.4161/epi.25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashktorab H, Smoot DT, Farzanmehr H, et al. Clinicopathological features and microsatellite instability (MSI) in colorectal cancers from African Americans. Int J Cancer. 2005;116:914–919. doi: 10.1002/ijc.21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brim H, Hasszadeh H, Schaffer AA, et al. Genomic Aberrations in an African American Colorectal Cancer Cohort Reveals a MSI-Specific Profile and Chromosome X Amplification in Male Patients. PLoS One. 2012 doi: 10.1371/journal.pone.0040392. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar K, Brim H, Mokarram P, et al. Distinct BRAF (V600E) and KRAS mutations in high microsatellite instability sporadic colorectal cancer in African Americans. Clin Cancer Res. 2009:68. doi: 10.1158/1078-0432.CCR-08-1029. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chun SY, Johnson C, Washburn JG, Cruz-Correa MR, Dang DT, Dang LH. Oncogenic KRAS modulates mitochondrial metabolism in human colon cancer cells by inducing HIF-1alpha and HIF-2alpha target genes. Mol Cancer. 2010;9:293. doi: 10.1186/1476-4598-9-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Femia AP, Dolara P, Giannini A, Salvadori M, Biggeri A, Caderni G. Frequent mutation of Apc gene in rat colon tumors and mucin-depleted foci, preneoplastic lesions in experimental colon carcinogenesis. Cancer Res. 2007;67:445–449. doi: 10.1158/0008-5472.CAN-06-3861. [DOI] [PubMed] [Google Scholar]
- 17.Moreno-Bueno G, Hardisson D, Sanchez C, et al. Abnormalities of the APC/beta-catenin pathway in endometrial cancer. Oncogene. 2002;21:7981–7990. doi: 10.1038/sj.onc.1205924. [DOI] [PubMed] [Google Scholar]
- 18.Nagel R, le Sage C, Diosdado B, et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 19.Segditsas S, Rowan AJ, Howarth K, et al. APC and the three-hit hypothesis. Oncogene. 2009;28:146–155. doi: 10.1038/onc.2008.361. [DOI] [PubMed] [Google Scholar]
- 20.Bacolod MD, Barany F. Molecular profiling of colon tumors: the search for clinically relevant biomarkers of progression, prognosis, therapeutics, and predisposition. Ann Surg Oncol. 2011;18:3694–3700. doi: 10.1245/s10434-011-1615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seshagiri S, Stawiski EW, Durinck S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dees ND, Zhang Q, Kandoth C, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC. TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci Publ. 2004:247–270. [PubMed] [Google Scholar]
- 29.Costello JF, Fruhwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 30.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 31.Vogelstein B, Kinzler KW. Winning the war: science parkour. Sci Transl Med. 2012;4:127ed122. doi: 10.1126/scitranslmed.3004019. [DOI] [PubMed] [Google Scholar]
- 32.Diaz LA, Jr., Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perreault N, Katz JP, Sackett SD, Kaestner KH. Foxl1 controls the Wnt/beta-catenin pathway by modulating the expression of proteoglycans in the gut. J Biol Chem. 2001;276:43328–43333. doi: 10.1074/jbc.M104366200. [DOI] [PubMed] [Google Scholar]
- 34.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 35.van Es JH, Jay P, Gregorieff A, et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 36.Choi K, Chen J, Mitra S, Sarna SK. Impaired integrity of DNA after recovery from inflammation causes persistent dysfunction of colonic smooth muscle. Gastroenterology. 2011;141:1293–1301. 1301 e1291–1293. doi: 10.1053/j.gastro.2011.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 38.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamborero D, Gonzalez-Perez A, Perez-Llamas C, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chevillard-Briet M, Quaranta M, Grezy A, et al. Interplay between chromatin-modifying enzymes controls colon cancer progression through Wnt signaling. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt604. [DOI] [PubMed] [Google Scholar]
- 42.Fatemi M, Brim H, Ashktorab H. Epigenetic silencing of CHD5, a novel tumor suppressor gene, occurs in early colorectal cancer stages. Cancer. 2013 doi: 10.1002/cncr.28316. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashktorab H, Belgrave K, Hosseinkhah F, et al. Global histone H4 acetylation and HDAC2 expression in colon adenoma and carcinoma. Dig Dis Sci. 2009;54:2109–2117. doi: 10.1007/s10620-008-0601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kume K, Iizumi Y, Shimada M, et al. Role of N-end rule ubiquitin ligases UBR1 and UBR2 in regulating the leucine-mTOR signaling pathway. Genes Cells. 2010;15:339–349. doi: 10.1111/j.1365-2443.2010.01385.x. [DOI] [PubMed] [Google Scholar]
- 45.Brim H, Mokarram P, Naghibalhossaini F, et al. Impact of BRAF, MLH1 on the incidence of microsatellite instability high colorectal cancer in populations based study. Mol Cancer. 2008;7:68. doi: 10.1186/1476-4598-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashktorab H, Schaffer AA, Daremipouran M, Smoot DT, Lee E, Brim H. Distinct genetic alterations in colorectal cancer. PLoS One. 2010;5:e8879. doi: 10.1371/journal.pone.0008879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smits R, Kielman MF, Breukel C, et al. Apc1638T: a mouse model delineating critical domains of the adenomatous polyposis coli protein involved in tumorigenesis and development. Genes Dev. 1999;13:1309–1321. doi: 10.1101/gad.13.10.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. Embo J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.