Abstract
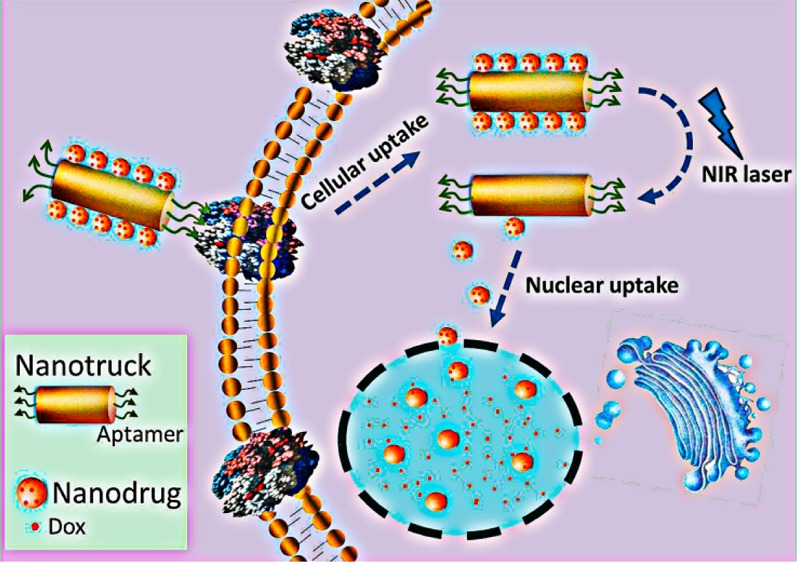
The development of multidrug resistance (MDR) has become an increasingly serious problem in cancer therapy. The cell-membrane overexpression of P-glycoprotein (P-gp), which can actively efflux various anticancer drugs from the cell, is a major mechanism of MDR. Nuclear-uptake nanodrug delivery systems, which enable intranuclear release of anticancer drugs, are expected to address this challenge by bypassing P-gp. However, before entering the nucleus, the nanocarrier must pass through the cell membrane, necessitating coordination between intracellular and intranuclear delivery. To accommodate this requirement, we have used DNA self-assembly to develop a nuclear-uptake nanodrug system carried by a cell-targeted near-infrared (NIR)-responsive nanotruck for drug-resistant cancer therapy. Via DNA hybridization, small drug-loaded gold nanoparticles (termed nanodrugs) can self-assemble onto the side face of a silver–gold nanorod (NR, termed nanotruck) whose end faces were modified with a cell type-specific internalizing aptamer. By using this size-photocontrollable nanodrug delivery system, anticancer drugs can be efficiently accumulated in the nuclei to effectively kill the cancer cells.
Keywords: Nuclear uptake, photocontrollable size, drug-resistant cancer therapy, cell-targeted delivery, aptamer
Chemotherapy, the most effective treatment for metastatic tumors, is essentially powerless against those cancer cells which have developed multidrug resistance (MDR), making the long-term survival of patients with such cancers extremely challenging.1 One major mechanism by which cancer cells can resist a broad range of drugs is associated with the overexpression of cell-membrane transporters, typically P-glycoprotein (P-gp). P-gp acts as an ATP-dependent efflux pump that can actively expel multiple drugs from the cell, thereby reducing the intracellular drug doses to levels below the lethal threshold. Since the efflux channel of P-gp is limited to small substrates (300–2000 Da),2 nanodrug delivery systems could, to some extent, circumvent MDR by bypassing P-gp.3−5 So far, however, most current nanocarriers are designed to target cells, not nuclei, posing an apparent drawback in these systems. Specifically, free drugs released from the nanocarriers into the cytoplasm are re-exposed to the efflux pump, partially reducing therapeutic efficacy.
As the control center of the cell,6−8 the nucleus is the final target location of most anticancer drugs, such as doxorubicin (Dox), cisplatin (CDDP), and camptothecin (CPT)9 Therefore, a nuclear-uptake nanodrug which can deliver anticancer drugs directly to the nucleus would greatly enhance therapeutic efficacy, especially for drug-resistant cells.10 Despite the appeal of these approaches, only a few nuclear-uptake nanodrug delivery systems have been developed.6,11 This primarily results from the difficulty experienced by nanocarriers in passing through the nuclear envelope, which is a double-layered membrane embedded with thousands of nuclear pore complexes (NPCs). NPCs are unique portals for transporting molecules across the nuclear envelope, with a functional diameter of 9–40 nm.12,13 Since the nuclear transport process is size-dependent, the entering particle must be small enough to pass through the NPCs. On the other hand, the small particle size can confound the transport of nanocarriers across the cell membrane, essentially because small nanoparticles (≤8 nm) can be cleared quickly from the bloodstream by the renal system, leading to a reduced circulation half-life and cell-uptake efficiency.14 Meanwhile, for nanoparticles of 60–400 nm, tumor tissue will exhibit the enhanced permeability and retention (EPR) effect, which can enhance tumor uptake by passive targeting.15,16 However, this size range is too large for nuclear internalization. Therefore, to coordinate intracellular and intranuclear delivery, a size-transformable nanodrug delivery system is needed.
In this work, we have, for the first time, used DNA self-assembly to fabricate a smart size-photocontrollable nanocomplex to coordinate efficient cell-targeted delivery and nuclear uptake of nanodrugs for cancer therapy. As shown in Scheme 1, this nanoassembly consists of one gold–silver nanorod (NR) and multiple small gold nanoparticles (NPs). The NR, which has intense absorption in the near-infrared (NIR) range (700–900 nm) and can efficiently convert the absorbed NIR light into heat,17 functions as a NIR-responsive nanotruck for the cell-targeted transport of the small-sized NPs. The nanotruck is directed to the target cancer cell by cell type-specific internalizing aptamers on the rod ends. Aptamers are artificial oligonucleotides screened from a large random sequence pool via an in vitro SELEX (Systematic Evolution of Ligands by Exponential Enrichment) method on the basis of target-specific binding.18,19 They have been used in biological research because of their intrinsic advantages, including simple synthesis, convenient modification, low immunogenicity, and highly specific affinity. The sides of the nanorod are modified with the capture strand (CS) which is complementary to the 5′-end portion of the drug-loading/anchoring strand (DAS) modified on the NP. The 3′-end portion of the DAS serves as the drug-loading sites, and an 11-mer spacer is located in the middle to reduce the photothermal impact of NR on the drug loading stability of NP. Via CS/DAS hybridization, the drug-loaded NPs (termed nanodrugs) can self-assemble onto the side face of the nanotruck and cross the cell membrane into the cytoplasm using the cell-targeting nanotruck as the guiding carrier. Then, an NIR laser (808 nm) is used to trigger the on-demand release of nanodrugs via heat denaturation of the CS/DAS duplex based on the photothermal effect of the NR. Thus, the nanodrugs can enter the nucleus where they release chemotherapeutic drugs in a sustained way to induce cell apoptosis.
Scheme 1. Schematic Illustration of the Cell-Targeted Photocontrolled Nuclear-Uptake Nanodrug Delivery System for Cancer Therapy.
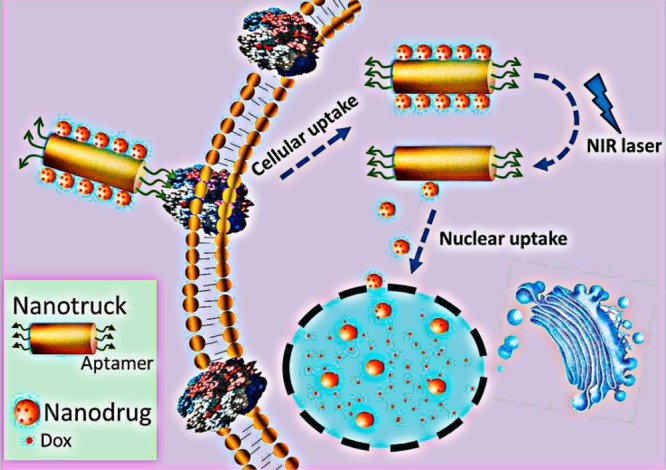
The NP-Dox/NR-aptamer nanocomplexes pass through the cell membrane via the guidance of the cell type-specific aptamer. Subsequently, NIR irradiation triggers the release of nanodrugs (Dox-loaded NPs) from the nanotruck (functionalized NR) via the photothermal effect of the NR, which leads to the dehybridization of DNA-duplex linker between NP and NR. Because of their small size, the released nanodrugs can diffuse into the nuclei where they sustainably release Dox to induce cancer cell apoptosis.
Results and Discussion
Construction of the DNA-Based Nanoassemblies
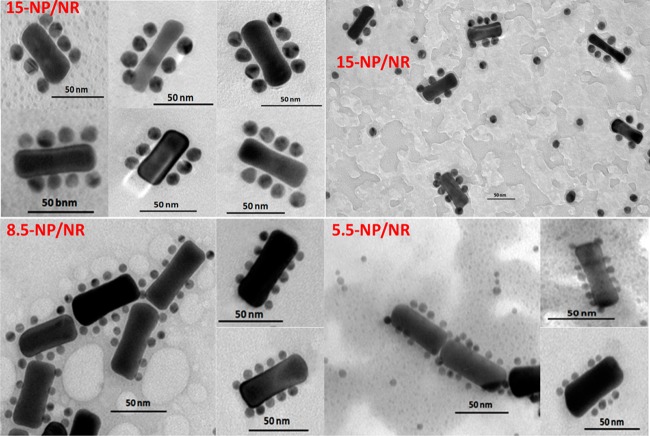
Since the particle size is a key consideration in nuclear transport, three NPs of different sizes (15.0 ± 1.2, 8.5 ± 0.8, and 5.5 ± 0.8 nm, respectively) were synthesized to study the size effect of the nanoparticle on nuclear internalization (see TEM images in Figure S1). After modification with a dense shell of the DAS, the hydrodynamic diameters of these NPs (termed 15-NP, 8.5-NP, and 5.5-NP), as measured by dynamic light scattering (DLS), were 37.8 ± 1.3, 18.2 ± 1.2, and 11.7 ± 1.5 nm, respectively. To prevent mutual interference between tumor targeting and nanodrug capture, a directional modification was performed on the NRs (65 nm × 20 nm), which means the aptamer and the CS were separately modified on the end faces and the side face of the NR, rather than being randomly spread. The principle of this selective modification process was based on the preference of CTAB to accumulate on the side face of NR, which makes its end faces more reactive to thiolated DNAs.20 Specifically, the aptamer at a relatively low concentration was first used to occupy the NR ends. Subsequently, the CS at a high concentration was added to replace CTAB on the NR side face. To make the resulting nanomaterial biocompatible, thiolated PEG was used, followed by several washing steps. Via DNA hybridization, as characterized by TEM (Figure 1), NPs were exactly immobilized onto the NR sides, leaving the NR ends bald, indicating successful fabrication of the side-assembly nanostructures (termed 15-NP/NR, 8.5-NP/NR, and 5.5-NP/NR). The self-assembly process was further investigated with DLS and UV–vis absorption spectroscopy. The results from different angles all demonstrated the successful assembly of NPs and NRs resulting from DNA hybridization (see details in Figures S2A and S2B).
Figure 1.
TEM images of 15-NP/NRs, 8.5-NP/NRs, and 5.5-NP/NRs. 15-NPs, 8.5-NPs, and 5.5-NPs mostly attach onto the NR sides, but not the NR ends, indicating the successful fabrication of the side-assembly nanostructures. The diameters of these nanoassemblies are in the range of 60–100 nm. The scale bar represents 50 nm.
Photocontrolled Dissociation and Stable Anticancer Drug Loading of Nanoassemblies
After synthesizing and characterizing these nanoassemblies, we next investigated their NIR-response and drug-carrying capability. The efficient photothermal effect of NRs was verified by the rapid temperature rise of the NR medium when irradiated by the NIR laser (see details in Figure S3A). Then the NIR-activated release of NPs from NRs was demonstrated by the reduced average size (DLS) and the disassembled structure (TEM) of the 15-NP/NRs after exposure to the laser (Figure S3B).
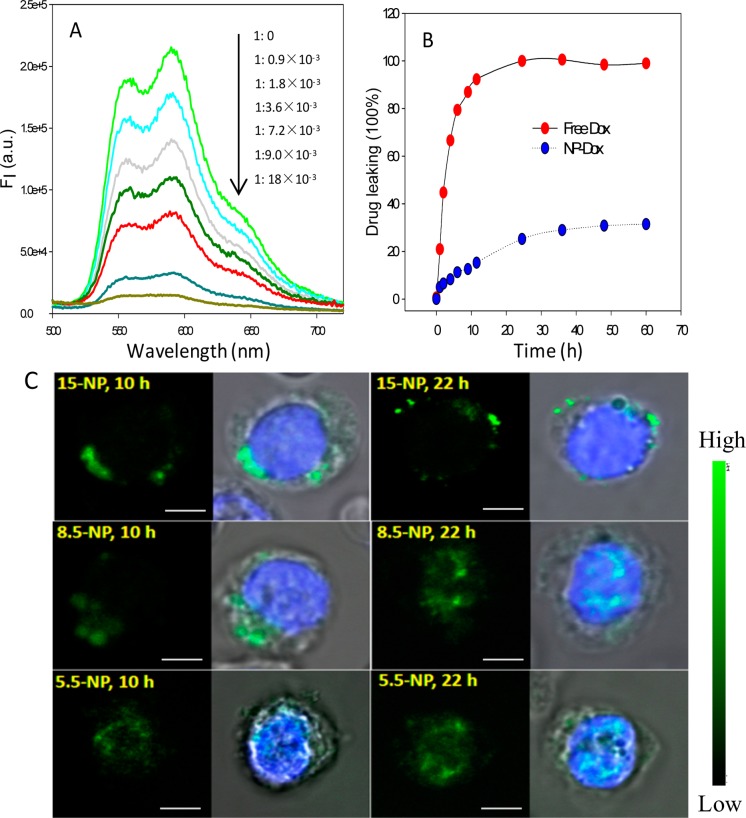
To fabricate the nuclear-uptake nanodrug delivery system, doxorubicin (Dox), a widely used anticancer drug, was chosen as the model and loaded on the NPs by intercalating into the (CGT)6/(ACG)6 duplexes of the DAS (Table S1). The drug payload of NPs was measured by monitoring the Dox fluorescence from the supernatant after NPs were centrifuged down. As shown in Figure 2A, a gradual decrease of Dox fluorescence was observed when increasing the molar ratio of NPs. By interpolating from a standard calibration curve, the Dox payload of each 15-, 8.5-, and 5.5-NP was 450 ± 19, 277 ± 28, and 40 ± 13 molecules, respectively. The stability of NP/Dox complexes was evaluated via a drug leakage experiment using MINI dialysis units. As shown in Figure 2B, the release of Dox from NPs was rather slow, with less than 30% of the entire payload detected in the solution after 60 h, in comparison to the rapid diffusion of free Dox, indicating high stability of the NP/Dox complexes. Furthermore, the nanoasemblies stored at 4 °C for 8 weeks remained mostly intact.
Figure 2.
Potential of the DAS-modified NPs as nuclear-uptake nanodrugs. (A) Dox fluorescence spectra of the supernatant after centrifuging to precipitate the DAS-modified NPs, the Dox concentration was fixed at 2 μM when increasing the 15-NP-DAS/Dox mole ratio. (B) Dox leakage dynamics from NP-Dox complexes (Dox: 10 μM). Equivalent of free Dox was used as a control. The Dox signal was normalized to the percentage of drug payload. (C) CLSM images of CEM cells after incubation with 15-NPs, 8.5-NPs, and 5.5-NPs (which represent the DAS-modified NPs of 15, 8.5, and 5.5 nm, respectively) at 37 °C for 10 or 22 h. The green and blue fluorescence arises from the TAMRA fluorophore labeled on the 3′ end of DAS and Hoechst 33342, respectively. The scale bar represents 5 μm.
Selective Cellular Internalization and Photocontrolled Intracellular Distribution of the Nanoassemblies
Having confirmed the potential of using the NP/NR nanoassemblies as NIR-responsive drug nanocarriers in buffer solution, we proceeded to test their performance in cells. A nondrug-resistant leukemia cell line, CEM, was first used as the cell model. To monitor the cellular uptake and intracellular distribution of NPs of different sizes, the DAS was labeled with a 5′-end TAMRA fluorophore (DAS-TMR). After modification with DAS-TMR, the fluorescent particles were incubated with the CEM cells at 37 °C for different lengths of time. Then confocal laser scanning microscopy (CLSM) measurements were performed, and the results are shown in Figure 2C. NPs of 15 nm were mainly localized in the cytoplasm, even after 22-h incubation, as indicated by the TAMRA fluorescence signal outside the nucleus. In contrast, 5.5-NPs accumulated in the nucleus after incubation for 10 h, demonstrating rapid nuclear uptake. For 8.5-NPs, no obvious signal was observed in the nucleus for 10-h incubation, but the nucleus emitted TAMRA fluorescence after incubation for 22 h, indicating that 8.5-NP-DASs can enter the nucleus starting from 10 h. To strike a balance between drug loading capability and nuclear translocation efficiency, the 8.5-NP was used as a model nuclear-uptake nanoscaffold of Dox in the following study.
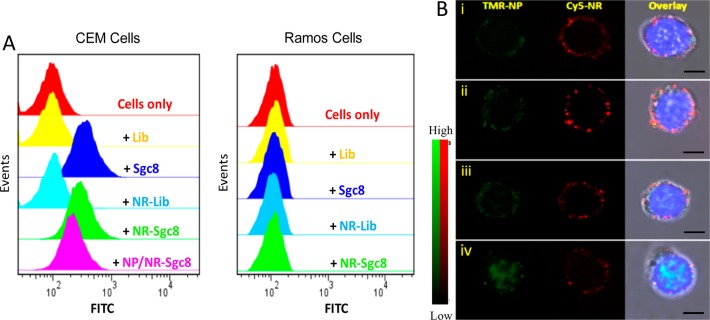
To achieve active tumor targeting, Sgc8, an aptamer that can specifically bind to protein tyrosine kinase 7 (PTK7, Kd = ∼0.8 nM) which is overexpressed on the membrane of CEM cells but not Ramos cells,18 was used as the targeting ligand and conjugated on the ends of NRs. The specific binding of Sgc8, NR-Sgc8, and NP/NR-Sgc8 to the target CEM cells, rather than nontarget Ramos cells, was demonstrated by flow cytometry (Figure 3A). Also, the specific cellular uptake and cytoplasmic location of NR-Sgc8s were visualized with CLSM (Figure S4). Moreover, the amount of the internalized NPs delivered by the NR-Sgc8s was higher than that of equivalent free NPs (Figure S5), which may have resulted from the high payload and the favorable cell-uptake size of the nanoassembly, as well as the promotion of the cell-internalizing aptamer.
Figure 3.
Specific cell binding and photocontrolled intracellular distribution of NP/NR-Sgc8s.(A) Flow cytometry assay proving the specific binding of Sgc8, NR-Sgc8, and NP/NR-Sgc8 to target CEM cells not to nontarget Ramos cells (Lib represents a random library sequence). (B) CLSM images of CEM cells after treatment with 15-NP/NR-Sgc8s without (i) and with (ii) NIR irradiation or after treatment with 8.5-NP/NR-Sgc8s without (iii) and with (iv) NIR irradiation. From left to right: fluorescence image for NP-TMR, NR-Cy5, and overlay of the NP-TMR, NR-Cy5, and Hoechst 33342 fluorescence channels plus the bright field channel. The scale bar represents 5 μm.
The NIR-responsive behavior of 8.5-NP/NR-Sgc8s and 15-NP/NR-Sgc8s in CEM cells was investigated with CLSM. Without laser treatment, the 8.5-NP signal was observed in the cytoplasm and overlapped with the NR-Sgc8 signal (Figure 3B). However, upon NIR irradiation, 8.5-NPs were found in the nuclei, while NR-Sgc8s remained in the cytoplasm, indicating that 8.5-NPs were released from NRs after exposure to the laser and then diffused into the nuclei. The 15-NPs remained in the cytoplasm irrespective of NIR irradiation, corresponding well with their inability to undergo nuclear internalization. The photocontrolled nuclear internalization of 8.5-NPs was further confirmed by using inductively coupled plasma atomic emission spectrometry (ICP-AES, Figure S6).
On the basis of the fluorescence quenching of Dox by intercalating into the GC duplex, the intracellular distribution of Dox was investigated by treating CEM cells with nonfluorophore-labeled 8.5-NP-Dox/NR-Sgc8s. After NIR irradiation and then incubation for another 22 h, the recovered fluorescence of Dox was highly accumulated in the nuclei, with a relatively small amount in the cytoplasm, indicating that most Dox were released in the nuclei (Figure S7). However, without NIR treatment, the intranuclear Dox fluorescence was rather weak, showing a slow and sustained release of Dox in the cytoplasm. As a control, cells were treated with free Dox, and the Dox signal was found throughout the cells, resulting from concentration-gradient diffusion. Furthermore, the negligible influence of the laser irradiation on the stability of the NP-Dox complex was confirmed by the small Dox signal change from the 8.5-NP-Dox/NR-Sgc8s-treated CEM cells before and right after laser exposure (Figure S8). These results show great potential of the 8.5-NP-Dox/NR-Sgc8 system for NIR-controlled intranuclear drug delivery.
Selective Cytotoxicity of Anticancer Drug-Loaded Nanoassemblies
The therapeutic effect of this 8.5-NP-Dox/NR-Sgc8 system on CEM and Ramos cells was tested by MTS assay. As shown in Figure 4 and Figure S9, the nanomaterials themselves and the pure NIR irradiation had little negative impact on either CEM or Ramos cells (the cell viability of both cell lines remained above 95%), indicating excellent biocompatibility of these nanomaterials and the laser. For free Dox, a dose-dependent cytotoxicity was observed on both CEM and Ramos cells. However, when treated with 8.5-NP-Dox/NR-Sgc8s, only CEM cells showed dose-dependent cell inactivity, indicating the selective cytotoxicity of Dox delivered by this nanoassembly platform. After NIR irradiation, a dramatic decrease of cell viability on CEM cells was caused by 8.5-NP-Dox/NR-Sgc8s with a nearly 3-fold lower IC50 of 0.36 μM compared to that without NIR irradiation (1.22 μM). To verify whether the enhanced therapeutic efficacy originated from the synergy of Dox, photothermal effect, and NP/NR nanocomplexes, CEM cells were incubated with free Dox and non-Dox-loaded 8.5-NP/NR-Scg8c (the Dox loading site of DAS was replaced with a common DNA duplex) together and then irradiated with the NIR laser. The therapeutic effect of this case was lower than that of the NIR-activated 8.5-NP-Dox/NR-Scg8c, indicating that the synergistic effect was not an important consideration in this system (Figure S10). Thus, it is reasonable to attribute the enhanced killing efficiency to the released 8.5-NP-Dox from the nanotruck. To ensure that the nuclear accumulation of nanodrug induced higher cell apoptosis, the NIR-activated 15-NP-Dox/NR-Sgc8 system was used as a non-nuclear-uptake control. As shown in Figure S11, with NIR irradiation, the cytotoxicity of 8.5-NP-Dox/NR-Sgc8s was 24% higher than that of 15-NP-Dox/NR-Scg8c, while no obvious difference was observed in either case without NIR irradiation. These results demonstrate that this nuclear-uptake nanodrug delivery system can greatly enhance the therapeutic efficacy on the target cancer cells.
Figure 4.
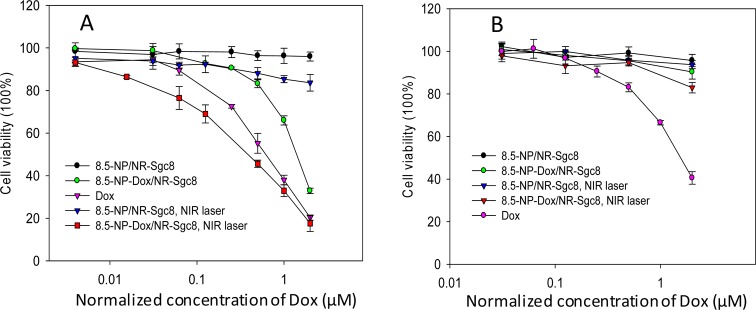
Cytotoxicity assay. Viability of CEM cells (A) and Ramos cells (B) with different treatments. The error bars represent the standard deviation of three independent experiments.
Recovery of Chemotherapeutic Sensitivity in Drug-Resistant Cancer Cells
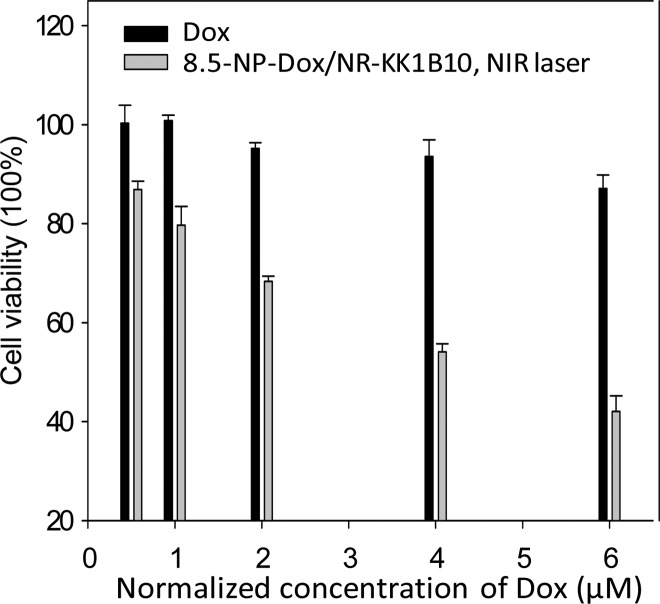
To investigate the ability of this nuclear-uptake nanodrug delivery system to address the MDR problem, K562/D, a drug-resistant cancer cell line with overexpression of P-gp (Figure S12), was used, while its specific internalizing aptamer, KK1B10,21 was used as the targeting ligand to functionalize the nanotruck. The specific binding of KK1B10, NR-KK1B10, and NP/NR-KK1B10 with K562/D cells was proven by flow cytometry (Figure S13). As shown in Figure 5 and Figure S14, enhanced killing efficiency was achieved by incubating K562/D cells with 8.5-NP-Dox/NR-KK1B10s with NIR irradiation, while a much lower therapeutic effect of free Dox was detected for this cancer cell line. To confirm that the enhanced therapeutic effect originates from the intracellular accumulation of Dox by this NIR-responsive nanodrug delivery system, flow assay was performed to measure the Dox signal in K562/D cells under different treatments. Upon NIR irradiation, the Dox fluorescence from the sample incubated with 8.5-NP-Dox/NR-KK1B10s was ∼2.5-fold higher than that from the sample incubated with free Dox (Figure S15). In contrast, without laser treatment, the cells incubated with 8.5-NP-Dox/NR-KK1B10s produced a modest Dox signal. These results demonstrate that this nuclear-uptake nanodrug delivery system can recover the chemotherapeutic sensitivity of K562/D to Dox by bypassing cell membrane-expressed P-gp.
Figure 5.
Cytotoxicity assay. Viability of K562/D cells treated with free Dox or NIR-activated 8.5-NP-Dox/NR-KK1B10s. The error bars represent the standard deviation of three independent experiments.
Conclusions
In summary, we have developed a DNA-based nanoassembly platform for cancer therapy. Unlike traditional intranuclear transport strategies of nanoparticles,11,22 no nuclear localization signal (NLS) peptides are required in our design, and the nuclear uptake of nanodrugs is mainly attributed to small particle size, thus avoiding complicated NLS modification processes and maintaining the valid occupancy of drug-loading probes on the NP surface. By using this photocontrolled, size-transformable nanosystem, nanodrugs can be efficiently transported across the cell membrane and enter the nucleus in a coordinated and harmonious manner. Furthermore, this DNA-based nanoassembly platform can accumulate chemotherapeutic drugs in the nuclei, thus greatly enhancing their therapeutic efficacy against drug-resistant cancer cells by effectively bypassing P-gp. This proof-of-concept structure also opens a new door in the use of nanoassemblies for the design of drug delivery systems for biological and clinical research. To comprehensively evaluate the superiority of this nuclear-uptake nanodrug delivery system, further efforts are being made on the testing in tumor-bearing mice models. On the other hand, since the tissue penetration of the NIR laser is limited to around ten millimeters, an alternative strategy for activatable dissociation of the nanoassemblies are needed to apply this system to treatment of drug-resistant metastatic tumors.
Materials and Methods
DNA Synthesis and HPLC Purification
All DNA strands were synthesized on an ABI 3400 DNA synthesizer (Applied Biosystems, Foster City, CA, USA), and the specific sequences are listed in Table S1. Both the synthesis and the deprotection processes were conducted as described by the reagent manufacturers. Then, the DNAs were precipitated by high-salt ethanol in a freezer at −20 °C for 30 min and collected by centrifugation at 4000 rpm for 30 min. Subsequently, the DNA precipitates were dissolved with 400 μL of 0.2 M triethylamine/acetate (Glen Research Corp). The purification step was performed by HPLC (ProStar, Varian, Walnut Creek, CA, USA) with a C18 column (5 μm, 250 mm × 4.6 mm, Alltech) using acetonitrile and 0.1 M triethylammonium acetate (TEAA) aqueous solution as the mobile phase. After being dried by a rotary evaporator, the purified DNAs were detritylated with 80% acetic acid, precipitated with cold salted ethanol, collected by centrifugation, and dried by vacuum. Finally, the DNA products were obtained, and their concentrations were measured with a UV–vis spectrometer (Cary Bio-300, Varian).
Synthesis and Modification of Nanomaterials and Fabrication of the NP/NR Nanoassemblies
NPs of 15 nm were synthesized by citrate reduction of HAuCl4.23 NPs of 5.5 and 8.5 nm were synthesized by a seed-mediated growth method24 and then washed by centrifugation to remove hexadecyltrimethylammonium bromide (CTAB). The modification of NPs with the drug-loading/attaching DNA strand (DAS) was conducted following a reported protocol.23 The thiolated DAS (0.2 mM, 20 μL) was deprotected by 10 mM tris(2-carboxyethyl) phosphine (TCEP, neutral pH, Thermo Scientific) at room temperature for 60 min and then mixed with 1 mL of NPs (20 nM). After incubation for 16 h, the mixture was salt-aged by slowly adding 200 μL of NaCl (1 M) and allowed to incubate for 16 h. Then, the excess DNAs were twice removed by centrifugation, and the precipitate was resuspended in 250 μL of 1 × PBS.
Gold–silver nanorods were synthesized and washed according to the procedure in our previous report,25 and the concentration was determined through the longitudinal absorption band of the UV–vis spectrum.26 The selective modification of NR was performed according to a reported protocol with some adjustment.20 Briefly, NRs were incubated with targeting aptamers (NR/aptamer ratio was 1:100) in 2 mM CTAB solution for 12 h. Then, the capture strands (CS) were added (NR/CS ratio was 1:500) and allowed to incubate for another 12 h. The adsorbed CTAB was further displaced with thiolated PEG. After that, the modified NR was salt aged by slowly adding 1 M NaCl to give a final 0.3 M concentration of Na+, and the mixture was held at room temperature for at least 12 h. After washing 5 times by centrifugation, the resultant NRs were resuspended in 1× PBS for further use.
To fabricate the self-assembly nanocomplexes, the DAS-modified NPs of 5.5, 8.5, and 15 nm were mixed with the modified NRs at a NP/NR ratio of 20:1, 15:1, and 10:1, respectively, and incubated at room temperature for at least 24 h. The mixture was then gently centrifuged to remove unbound NPs.
Drug Loading and Leaking of NPs
For Dox loading, NPs were modified with DAS whose 3′-end portion can form a (CGT)6/(ACG)6 duplex with a corresponding cDNA. The Dox loading was conducted by mixing Dox with NP-DASs, incubating at room temperature for 30 min, and then centrifuging to collect the NP/Dox complexes. The number of Dox per NP was determined by measuring the fluorescence intensity of unbound Dox in the supernatant and then interpolating from a standard linear calibration curve.
For the Dox leakage assay, 150 μL of NP/Dox complexes was added to a MINI dialysis unit [3.5 molecular weight cutoff (MWKO), Thermo Scientific], and the equivalent of free Dox was used as a control. Each unit was immersed in 3 mL of 1× PBS in a 5 mL beaker with gentle stirring at 450 rpm. At each given time point, a 100-μL aliquot of the dialysis solution was collected for Dox fluorescence measurement. After that, the collected solution was returned to the corresponding beaker.
Cell Lines and Cell Culture
CEM (human acute lymphoblastic leukemia) and Ramos (human Burkitt’s lymphoma) were purchased from American Type Culture Collection. K562/D (doxorubicin-resistant chronic myelogenous leukemia) was generously provided by Dr. Ruoping Tang and Prof. Troy A. A. Harkness of the Department of Anatomy and Cell Biology, College of Medicine, University of Saskatchewan. CEM, Ramos, and K562/D cells were cultured in RPMI 1640, RPMI 1640, and IMDM medium, respectively, supplemented with 10% FBS (heat-inactivated; Gibco) and 100 IU/mL penicillin–streptomycin (Cellgro).
Confocal Laser Scanning Microscopy Imaging
Cells (5 × 104 in 100 μL of medium) were incubated with free Dox, fluorescent nanoparticles, or nanocomplexes at 37 °C with 5% CO2 for different time lengths. After several centrifugation/washing steps, the cells were suspended in 1× PBS. For photoactivation, cells were irradiated with NIR laser (600 mW/cm2) for 10 min. Bisbenzimide Hoechst 33342 (Sigma-Aldrich) was used for nuclear staining by incubating with cells at 37 °C for 20 min. Fluorescence imaging was performed on a Leica TCS SP5 confocal microscope (Leica Microsystems) with a 63× oil-immersion objective. In most cases, the optical slice thickness was adjusted to 0.5 μm. In CLSM images, the red color represents the fluorescence of Cy5 (Em = 670 nm), the pink color represents the fluorescence of Dox (Em = 600 nm), and the green color represents the fluorescence of TAMRA (Em = 570 nm).
ICP-AES Analysis
CEM cells (1 × 108) were incubated with 8.5-NP/NR-Sgc8s or 15-NP/NR-Sgc8s at 37 °C with 5% CO2 for 6 h. Then the cells were washed with PBS three times, irradiated with NIR laser for 0 or 10 min, and incubated at 37 °C with 5% CO2 for another 22 h. To extract nuclei, the cells were collected by centrifugation, resuspended in 10 mM Tris-HCl buffer (pH 7.4) containing 100 mM NaCl, 1 mM EDTA, and 1% Triton X-100 at 4 °C for 10 min and finally centrifuged at 1000 g for 3 min. After several centrifugation/washing rounds to remove the adsorbed nanoparticles on the nuclear membrane, the collected nuclei were lysed by a lysis solution containing 0.5% Triton X-100 and 1 M NaOH with sonication. Subsequently, the nanoparticles from the nuclei were digested by incubating with aqua regia at 65 °C overnight and diluted in 2% HNO3 solution. NPs accumulated in nuclei were measured by quantifying Au element by inductively coupled plasma atomic emission spectrometry (ICP-AES).
Flow Cytometry Analysis
To evaluate the cell binding affinity of different nanocarriers, the aptamer was labeled with a fluorescein (FITC). Cells were incubated with free aptamers, NR-aptamers, or 15-NP/NR-aptamers at 4 °C for 30 min. After removal of unbound materials by several centrifugation/washing steps, the cells were analyzed on a FACScan cytometer (Accuri C6) by counting 20 000 events.
Cytotoxicity Assay
The cell viability under different treatments was determined by CellTiter 96 cell proliferation assay (Promega). Cells were incubated with free Dox, NP/NRs, or NP-Dox/NRs at 37 °C with 5% CO2 for 2 h and then centrifuged to precipitate. The supernatant (80%) was removed, followed by adding equivalent fresh culture medium (10% FBS). For NIR-responsive regulation, cells were irradiated with a NIR laser (808 nm, 600 mW/cm2) for 10 min, followed by additional incubation to allow further growth for 48 h. Then, 80% of the medium was removed and replaced with 20 μL of MTS reagent diluted in 100 μL of RPMI 1640. The resulting cell samples were incubated at 37 °C for 1–2 h. Finally, the absorbance at 490 nm was collected using a Tecan Safire microplate reader, and cell viability was calculated using the equation provided by the manufacturer.
Acknowledgments
C.W. acknowledges support from the American Chemical Society, Division of Analytical Chemistry Fellowship, sponsored by the Society for Analytical Chemists of Pittsburgh. This work is supported by the National Key Scientific Program 2011YQ03012412, and by the National Institutes of Health (GM079359 and CA133086).
Supporting Information Available
DNA sequences and supplementary data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Gottesman M. M.; Fojo T.; Bates S. E. Nat. Rev. Cancer 2002, 2, 48–58. [DOI] [PubMed] [Google Scholar]
- Ambudkar S. V.; Kimchi-Sarfaty C.; Sauna Z. E.; Gottesman M. M. Oncogene 2003, 22, 7468–7485. [DOI] [PubMed] [Google Scholar]
- Wang F.; Wang Y.-C.; Dou S.; Xiong M.-H.; Sun T.-M.; Wang J. ACS Nano 2011, 5, 3679–3692. [DOI] [PubMed] [Google Scholar]
- Dong X.; Mattingly C. A.; Tseng M. T.; Cho M. J.; Liu Y.; Adams V. R.; Mumper R. J. Cancer Res. 2009, 69, 3918–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R.; Wu R. A.; Zhao L.; Wu M.; Yang L.; Zou H. ACS Nano 2010, 4, 1399–1408. [DOI] [PubMed] [Google Scholar]
- Pan L.; He Q.; Liu J.; Chen Y.; Ma M.; Zhang L.; Shi J. J. Am. Chem. Soc. 2012, 134, 5722–5725. [DOI] [PubMed] [Google Scholar]
- Kang B.; Mackey M. A.; El-Sayed M. A. J. Am. Chem. Soc. 2010, 132, 1517–1519. [DOI] [PubMed] [Google Scholar]
- Cooper G. M.; Hausman R. E.. The cell; ASM press: Washington, DC, 2000. [Google Scholar]
- Liu J.; Bu W.; Pan L.; Zhang S.; Chen F.; Zhou L.; Zhao K.; Peng W.; Shi J. Biomaterials 2012, 33, 7282–7290. [DOI] [PubMed] [Google Scholar]
- Goren D.; Horowitz A. T.; Tzemach D.; Tarshish M.; Zalipsky S.; Gabizon A. Clin. Cancer Res. 2000, 6, 1949–1957. [PubMed] [Google Scholar]
- Tkachenko A. G.; Xie H.; Coleman D.; Glomm W.; Ryan J.; Anderson M. F.; Franzen S.; Feldheim D. L. J. Am. Chem. Soc. 2003, 125, 4700–4701. [DOI] [PubMed] [Google Scholar]
- Hetzer M. W. The nuclear envelope. Cold Spring Harbor Perspect. Biol. 2010, 2, 10.1101/cshperspect.a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D.; Kutay U. Annu. Rev. Cell Dev. Biol. 1999, 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Hoshino Y.; Koide H.; Furuya K.; Haberaecker W. W.; Lee S.-H.; Kodama T.; Kanazawa H.; Oku N.; Shea K. J. Proc. Natl. Acad. Sci. 2012, 109, 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel A. E.; Mädler L.; Velegol D.; Xia T.; Hoek E. M.; Somasundaran P.; Klaessig F.; Castranova V.; Thompson M. Nat. Mater. 2009, 8, 543–557. [DOI] [PubMed] [Google Scholar]
- O’Neal D. P.; Hirsch L. R.; Halas N. J.; Payne J. D.; West J. L. Cancer Lett. 2004, 209, 171–176. [DOI] [PubMed] [Google Scholar]
- Huang X.; El-Sayed I. H.; Qian W.; El-Sayed M. A. J. Am. Chem. Soc. 2006, 128, 2115–2120. [DOI] [PubMed] [Google Scholar]
- Shangguan D.; Li Y.; Tang Z.; Cao Z. C.; Chen H. W.; Mallikaratchy P.; Sefah K.; Yang C. J.; Tan W. Proc. Natl. Acad. Sci. 2006, 103, 11838–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefah K.; Shangguan D.; Xiong X.; O’Donoghue M. B.; Tan W. Nat. Protocols 2010, 5, 1169–1185. [DOI] [PubMed] [Google Scholar]
- Xu L.; Kuang H.; Xu C.; Ma W.; Wang L.; Kotov N. A. J. Am. Chem. Soc. 2012, 134, 1699–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefah K.; Tang Z.; Shangguan D.; Chen H.; Lopez-Colon D.; Li Y.; Parekh P.; Martin J.; Meng L.; Phillips J. Leukemia 2009, 23, 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrobattista E.; van der Aa M. A.; Hennink W. E.; Crommelin D. J. Nat. Rev. Drug Discovery 2006, 5, 115–121. [DOI] [PubMed] [Google Scholar]
- Liu J.; Lu Y. Nat. Protocols 2006, 1, 246–252. [DOI] [PubMed] [Google Scholar]
- Jana N. R.; Gearheart L.; Murphy C. J. Langmuir 2001, 17, 6782–6786. [Google Scholar]
- Yasun E.; Gulbakan B.; Ocsoy I.; Yuan Q.; Shukoor M. I.; Li C.; Tan W. Anal. Chem. 2012, 84, 6008–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orendorff C. J.; Murphy C. J. J. Phys. Org. Chem. 2006, 110, 3990–3994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.