Abstract
Botulinum neurotoxins (BoNTs) are the most lethal toxin known to human. Biodefense requires early and rapid detection of BoNTs. Traditionally, BoNTs can be detected by looking for signs of botulism in mice that receive an injection of human material, serum or stool. While the living animal assay remains the most sensitive approach, it is costly, slow and associated with legal and ethical constrains. Various biochemical, optical and mechanical methods have been developed for BoNTs detection with improved speed, but with lesser sensitivity. Here, we report a novel nanopore-based BoNT type B (BoNT-B) sensor that monitors the toxin’s enzymatic activity on its substrate, a recombinant synaptic protein synaptobrevin 2 derivative. By analyzing the modulation of the pore current caused by the specific BoNT-B-digested peptide as a marker, the presence of BoNT-B at a subnanomolar concentration was identified within minutes. The nanopore detector would fill the niche for a much needed rapid and highly sensitive detection of neurotoxins, and provide an excellent system to explore biophysical mechanisms for biopolymer transportation.
Keywords: botulinum, neurotoxin, nanopore, peptide, single molecules, biosensor
Introduction
Botulinum neurotoxins (BoNTs) are the most poisonous substances known to human, with a median lethal dose (LD50) of 1 ng per kg of body weight,1 and are the cause of the life-threatening neuroparalytic illness botulism.2 BoNTs are regarded as potential biological warfare agents that could be used for bioterrorism attacks on the food chain. Of the seven known serotypes of botulinum neurotoxins (BoNTs), designated A-G, BoNT-A, -B, -E and -F are toxic to humans.3 They are mainly produced by Clostridium botulinum and released from this bacillus as single polypeptide chains. After cleavage by bacterial or host proteases, these polypeptides adopt an active dichain form of toxin, which consists of a 100 kDa heavy (H) chain linked to 50 kDa light (L) chain by a disulfide bond. H chains control cellular internalization of the toxins via receptor-mediated endocytosis, allowing them to preferentially attack the nervous system. Upon endocytosis of the holo-protein, the L chain is translocated from the vesicular lumen into the cytosol, where it acts as a Zn2+-endopeptidase to specifically cleave components of the soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE) complex, and thus hampers exocytosis and neurotransmitter release. Currently, worldwide geographical distributions of BoNTs, which are dictated by the type of foods that serve as the toxins source, have started to blur owing to the ever increasing international trade of food products. Moreover, the majority of new cases of BoNT intoxication have a trend to be associated with drug use, in particular that of black tar heroin.4 Owing to the ease of distribution of BoNTs, along with their potency, the Centers for Disease Control and Prevention (CDC) in Atlanta, Georgia, lists BoNTs as one of the six most dangerous bioterrorist threats.5 On the other hand, due to their specific actions, BoNTs are the first toxins licensed for human use in the United States to treat muscle dysfunctions/spasms1 and associated skin wrinkles.6
Both bioterrorism defense and medical attention require an early and rapid detection of BoNTs. Traditionally, this is accomplished by looking for signs of botulism in mice that receive an injection of human serum or stool samples.5 Although this bioassay remains the most sensitive detection approach, it is costly, slow (takes days), associated with legal and ethical constraints and is restricted to the CDC along with some state health department laboratories. Besides the use of living animals in detection, various biochemical assays, such as electrophoresis and immunoblot analysis have been used to detect the cleavage of specific SNARE proteins.7−9 These in vitro detections are faster (within 1 day), but with reduced sensitivity, compared to the mouse bioassay. The subsequent development of time-resolved fluorescence-based enzyme-linked immunosorbent assays (ELISA)10 allowed utilizing Eu3+-tagged antibodies11 to detect BoNT-A and -B. ELISA sped up the outcome of BoNT assays to hours. Within a similar time-frame, the BoNT-A activity can also be monitored using the substrate SNAPtide, a fluorescently conjugated synthetic peptide that contains the native cleavage site for BoNT-A.12 Recent developments in mechano- and fluorescence resonance energy transfer (FRET)-based sensors offer a reduced detection time and/or increased sensitivity. The micromechanosensor uses force spectroscopy to detect BoNT-B.13 This toxin cleaved synaptobrevin 2 (Sb2) [reviewed in refs (14) and (15)] attached to a bead, which was suspended off a cantilever via Sb2 interaction with syntaxin 1A attached to the cantilever. The detachment of the bead from the cantilever marked the cleavage of Sb2, allowing detection of low nanomolar range of BoNT-B within 15 min.13 Additionally, in vitro detection using FRET probes that contain fragments of SNARE proteins as toxin substrates shows picomolar sensitivity within 4−16 h.16 Although the two latter assays are very promising for practical use, they require expensive and technically complex equipment. Overall, there is a need for further development of assays for detection of BoNTs.
Nanopore technology provides a single-molecule platform for sensitive biodetection.17−20 Target molecules interacting with a nanopore can specifically modulate the pore’s ion current. The resulting single-molecule signatures can be analyzed to report both the target identity and quantity. The nanopore sensor has been extensively explored for nucleic acids structural detections21−25 and is being developed as a next-generation sequencing technology.26,27 We also utilized the nanopore-based sensor to detect biomarker nucleic acids such as microRNAs for disease diagnostics.28,29 Recently, the nanopore approach has demonstrated its power in analyzing peptide/protein processes, from peptide–nanopore interactions,30,31 protein unfolding32,33 and peptide–metal ion interaction,34 to peptide translocation,35 peptide modification36 and protein sensing.37,38 Notably, the nanopore can reveal the function of enzymes through analyzing the enzyme-processed peptides.36,39−41 In this report, we describe a nanopore-based assay for detection of BoNT-B. We utilized the emerging aerolysin pore42−45 to track the BoNT-B digestion of its substrate, a derivative of the synaptic protein Sb2 (also known as vesicle-associated membrane protein 2, VAMP2).7 The dynamic change of the specific digestion product reported the existence of the toxin at subnanomolar concentration within minutes. We also demonstrate the specificity of detection in serum, a blood-derived material that is presently used for detection of the toxin presence in human. This system also demonstrates usefulness in studying biophysical mechanisms for peptide translocation and interaction with the nanopore.
Results
Design of Synaptobrevin 2 Substrate Protein for Nanopore Detection of BoNT-B
The BoNT-B light chain is a Zn2+-endopeptidase that specifically cleaves the synaptic protein Sb2.7 To detect BoNT-B, we designed a recombinant derivative of Sb2, Lp-Sb2(1-93), which contains the truncated Sb2 cytoplasmic domain [amino acids (aa) 1-93] appended to the C-terminus of a 36 aa lead peptide (Lp) (Figures 1a and S1, Supporting Information). Six histidines at the N-terminus of Lp-Sb2(1-93) ease the protein purification. In the presence of Zn2+, the BoNT-B light chain can cleave Lp-Sb2(1-93) into two fragments, the long N-terminal Lp-Sb2(1-76)d and the short C-terminal Sb2(77-93)d. Due to the characteristics of these digests (Table S1, Supporting Information), we posit that they generate different signals in the nanopore current recordings (Figure 1b), which we experimentally tested. For simplicity, we refer to the toxin’s light chain as BoNT-B.
Figure 1.
Schematized principle of BoNT-B detection using aerolysin nanopore. (a) BoNT-B substrate design and its cleavage products. The 116 aa vesicular exocytotic protein synaptobrevin 2 (Sb2, red; linearized form shown to the right) spans the vesicular membrane. Its N-terminus is located in the cytosol with its cytosolic domain being a target for proteolytic (Zn2+-endopeptidase) activity of BoNT-B light chain, which in the presence of Zn2+ cleaves (indicated by scissors, left) the Q76-F77 peptide bond (yellow band, right). The recombinant protein, Lp-Sb2(1-93), contains truncated cytoplasmic domain (left, truncation site indicated by arrow) aa 1–93 appended to the C-terminus of the lead peptide (Lp; right, black); six histidine residues (right, turquoise) in the lead peptide ease the purification of this chimera. The digested products, Lp-Sb2(1-76)d and Sb2(77-93)d, are generated by BoNT-B/Zn2+ digestion of Lp-Sb2(1-93) substrate. Drawings are not to scale (see Figure S1, Supporting Information, for detailed information). (b) Illustration of the principle for nanopore BoNT-B detection. Lp-Sb2(1-93) (based on structure PDB ID code 2KOG in the Protein Data Bank (www.pdb.org)) would not affect the pore current, as shown by the hypothetical trace below the structure. After the cleavage of Lp-Sb2(1-93) by BoNT-B/Zn2+ (arrow), the short digested product Sb2(77-93)d would generate a distinct modulation of pore currents, as shown by downward transient blocks in the hypothetical trace, while the long product Lp-Sb2(1-76)d would not affect the nanopore current. Dotted lines are levels of zero current. Analytes would be applied from the cis (pore vestibule) side of the bilayer that partitions the recording chamber.
Nanopore Detection of BoNT-B-Digested Sb2 Fragments
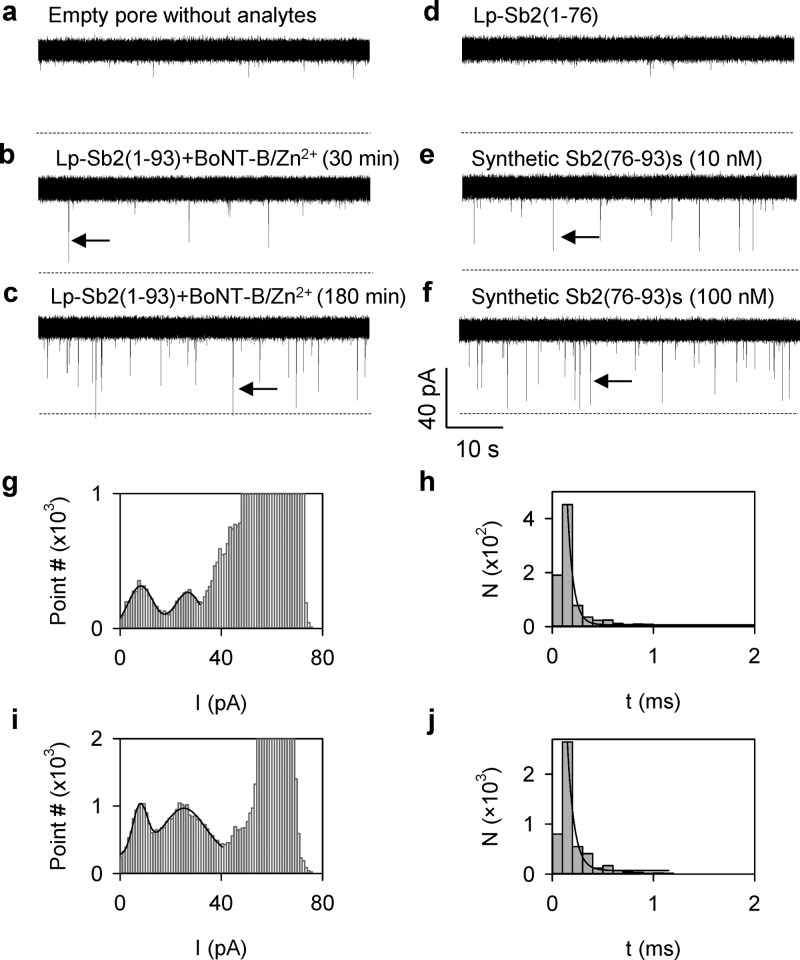
The background ion current through an empty aerolysin pore was 62.7 ± 0.9 pA (IRMS = 1.6 pA) at +120 mV in 1 M KCl (Figure 2a). This background current was unaffected when BoNT-B (500 pM) or Lp-Sb2(1-93) (20 nM) alone or combined with Zn2+ (100 nM) was presented to the cis recording solution (Figure S2, Supporting Information), indicating that each reactant, substrate or toxin protein alone, does not interact with the aerolysin pore. However, when Lp-Sb2(1-93) (20 nM) was mixed with BoNT-B (500 pM) in the presence of Zn2+ (100 nM) in the cis solution for 30 min, we recorded a series of transient blocks of nanopore current (Figure 2b). After incubation for 180 min, these events became more frequent (Figure 2c). Given that each reactant alone cannot block the pore (Figure S2, Supporting Information), we hypothesize that these blocking events are generated by the reaction digests, Lp-Sb2(1-76)d and/or Sb2(77-93)d fragments, generated by the BoNT-B/Zn2+ cleavage of Lp-Sb2(1-93).
Figure 2.
Experimental detection of BoNT-B activity using the aerolysin pore. (a) Current traces for the empty pore without analytes. (b,c) BoNT-B/Zn2+ digestion of Lp-Sb2(1-93) generated transient pore blocks (arrows) after 30 min, and more prominently, after 180 min of incubation, respectively. (d) Current trace showing that the recombinant Lp-Sb2(1-76), identical to the N-terminal digest Lp-Sb2(1-76)d, did not produce blocks of the aerolysin pore. (e,f) Synthetic Sb2(76-93)s peptide (10 and 100 nM, respectively), emulating the C-terminal digest Sb2(77-93)d, generates pore current blocks (arrows) in concentration-dependent manner (also see Figure S3, Supporting Information, for more traces). All analytes were applied into the cis side of the recording chamber, with currents recorded at +120 mV. Dotted lines represent the zero current. (g–j) Current amplitude (g and i) and duration of blocks (h and j) generated by the substrate–toxin [Lp-Sb2(1-93)-BoNT-B/Zn2+] mixture (g and h) or by the synthetic peptide Sb2(76-93)s (i and j). The all-point histograms (g and i) includes instantaneous current values of every sampling time point. Two populations of blocking events were observed based on the current amplitude and fitted with a Gaussian distribution. Block duration (h and j) was fitted with an exponential function.
To determine which digestion fragment plays a role in the observed nanopore current modulation, we studied the effect of each fragment on the current when applied to the cis solution. First, we constructed the recombinant Lp-Sb2(1-76) that is identical to the Lp-Sb2(1-93) N-terminal digestion fragment Lp-Sb2(1-76)d. Lp-Sb2(1-76) cannot be cleaved by BoNT-B, as it lacks the N-terminal portion of Sb2 needed for the action of BoNT-B.14 We found that the recombinant Lp-Sb2(1-76) alone (20 nM) did not produce transient blocks (Figure 2d). Next, we investigated the synthetic peptide Sb2(76-93)s that emulates the Lp-Sb2(1-93) C-terminal digestion fragment Sb2(77-93)d. Unlike Lp-Sb2(1-76), Sb2(76-93)s (10 nM) generated transient pore blocks (Figure 2e) similar to events observed when Lp-Sb2(1-93) was treated with BoNT-B/Zn2+ (compare to Figure 2b,c). The frequency of these blocking events also increased with increasing concentration of the synthetic peptide (Figures 2f and S3, Supporting Information). These results suggest that the nanopore blocks observed in the substrate/toxin mixture are generated by the short C-terminal digestion peptide Lp-Sb2(77-93)d. This conclusion was further confirmed by the similar characteristics of the blocks for the substrate/toxin mixture (Figure 2b,c) and the synthetic peptide Sb2(76-93)s (Figure 2e,f). The current of blocks for the substrate/toxin mixture was distributed broadly with two populations located around 8.2 ± 0.9 pA (lower) and 26.8 ± 1.1 pA (higher), i.e., relative conductance of 13.1% and 42.9%, respectively (Figure 2g). Similarly, synthetic Sb2(76-93)s also reduced the pore currents to 7.9 ± 1.5 pA and 25.3 ± 1.4 pA, corresponding to relative conductance of 12.6% and 40.5%, respectively (Figure 2i). In addition, the duration of both populations for the Lp-Sb2(1-93)/BoNT-B/Zn2+ mixture was 170 ± 40 μs (Figure 2h), similar to 170 ± 40 μs (Figure 2j) for synthetic Sb2(76-93)s events. Both block durations were also similar at various voltages (Figure S4, Supporting Information). We interpret that the population with lower blocking level (13%) could be generated by the peptide translocation to the contralateral (from cis to trans) side of the chamber, while the events with a higher blocking level (40%) could be caused by the peptide partially entering the pore (“bumping-the-pore”) with a subsequent retrieval back to the ipsilateral (cis) side of the chamber. This interpretation is consistent with previous studies on protein interactions with the aerolysin pore. For example, the translocation of unfolded maltose binding protein (370 aa) reduced the current to 20%, while “bumping-the-pore” blocks partially reduced the current to 55%;44 the translocation and “bumping” of an unfolded histidine-containing phosphotransfer protein (86 aa) blocked the current to 14% and 54%, respectively.45 Overall, we conclude that in the BoNT-B/Zn2+–substrate reaction mixture, only the short C-terminal digest peptide (17 amino acids) can specifically block the aerolysin pore current from the cis side. The peptide-generated nanopore signatures are markers that can be used to determine the presence of BoNT-B in the solution.
Mechanism for Capturing Peptides in the Aerolysin Pore
In the above study, the Sb2 substrate digest can be selectively captured into the aerolysin pore. Understanding the biopolymer capture mechanism is important because optimized capture efficiency can enhance the detection sensitivity.46−49 The capture efficiency is measured by the capture rate constant. Therefore, we investigated the voltage-dependent and the nanopore sensor directionality-dependent capture rate constant for both the synthetic Sb(76-93)s and digest Sb(77-93)d in the toxin/substrate mixture.
The current traces in Figure 3a show that the capture of Sb(76-93)s from the cis side is highly dependent on the voltage polarity: the block was much more prominent at positive than negative voltages (see Figure S5, Supporting Information, for additional traces). The capture rate constant (kon, μM–1·s–1) at +120 mV was 81 ± 11 μM–1·s–1, 9 times higher than that at −120 mV (8.7 ± 3.3 μM–1·s–1, Figure 3b), suggesting that this peptide can be driven by positive voltage. However, such observed voltage-dependence is opposite to the expectation based on the electrostatic analysis. The in silico analysis of protein/peptides (Table S1, Supporting Information) indicates that the substrate Lp-Sb2(1-93) and its N-terminal digest Lp-Sb2(1-76) appeared negatively charged (−2.7e and −6.7e, receptively), while the pore blocking Sb2(77-93)d and synthetic Sb(76-93)s carried positive charge (+3.76e). Thus, due to the static charges alone, the cationic Sb(76-93)s applied to the cis (grounded) side should be attracted to the pore by a negative voltage applied from the trans side. This discrepancy between the predicted and measured voltage-dependence of kon suggests that the peptide capture could be dominated by factors other than the electrostatic attraction. We propose that the peptide capture is enhanced by the electroosmotic effect originating from the ion selectivity of the aerolysin pore. Namely, it has been reported that the aerolysin pore is a moderately anion-selective channel, with its cation/anion permeability ratio (P+/P–) of 0.21.50 We experimentally verified this anion selectivity of the aerolysin pore (Figure S6, Supporting Information). Consequently, the pore current (in 1 M KCl) at a positive voltage (applied from the trans side) is dominated by a cis-to-trans anion flow (Cl–) (Figure 3d). The water molecules accompanying this net anion flow produce an electroosmotic force to drive the peptide presented in the cis solution toward the pore. When the electroosmotic force dominates over the opposite electrostatic force that repulses the cationic peptide from the pore at positive voltage, the overall driving direction is toward the pore, which is what we observed.
Figure 3.
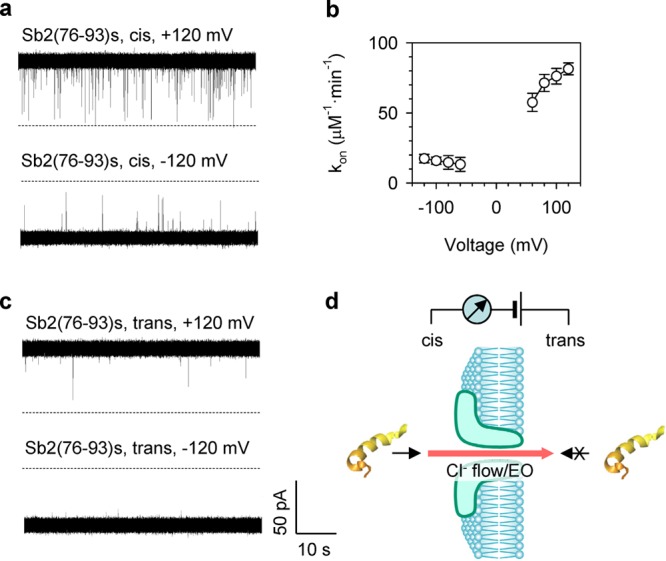
Mechanism for capture of peptide into the aerolysin pore. (a) Current traces showing block events for synthetic Sb2(76-93)s peptide (1 μM) from cis (grounded) side of the pore at ±120 mV (applied from trans side). See Figure S5 (Supporting Information) for the additional currents recorded at ±80 mV and ±100 mV. (b) Voltage-dependence of the capture rate (kon) for Sb2(76-93)s. The blocks were more prominent at positive over negative voltages. Block events at voltages above +120 mV were not analyzed because they were affected by the spontaneous pore current flicking,44 while blocks at voltages above −60 mV and lower than +60 mV were not analyzed because the block recognition at low voltages was affected by the noise of background current (empty pore). (c) Current traces showing fewer blocks for Sb2(76-93)s (1 μM) from trans side at ±120 mV. (d) Cartoon showing the detection layout and directionality of the pore block by the peptide. As the pore is anion selective, under our detection conditions, there is a Cl– flow from cis to trans side, resulting in electroosmosis (EO)-driven peptide translocation. Peptide can interact with the pore from cis (arrow), but not trans side (crossed arrow).
In addition to voltage dependence, the aerolysin pore also displays rectification for the peptide binding. We found that the addition of Sb(76-93)s to the trans side of the chamber rarely blocked the pore at either negative or positive holding potentials (Figures 3c and S5, Supporting Information). Similar sidedness selectivity for peptide translocation has been reported previously.44 The reason is not clear, but this complex process should be related to the peptide structure and its interaction with the pore entry motif such as the flexible loop domain at the trans entrance.51 Overall, the toxin-digested peptide can translocate through the pore driven by an electroosmotic flow from the cis side at a positive voltage with an optimized capture rate at +120 mV.
Detection of BoNT-B Activity
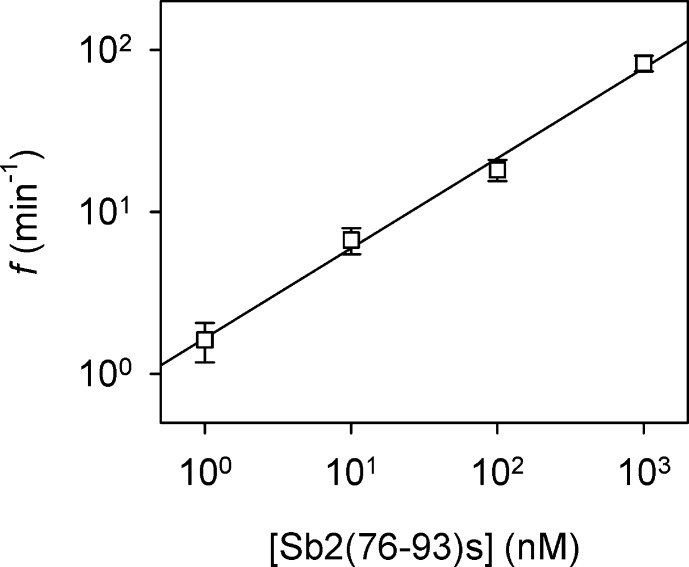
In practical toxin detection, the observation of the blocks for the specific digest peptide can mark the presence of BoNT-B. Furthermore, the dynamic change of the block frequency would not only confirm the presence of the toxin but also reveal the toxin activity. We identified a positive correlation between the frequency of blocking event (f) and the Sb2(76-93)s peptide concentration (Figure 4), which could be linearly fitted on a log–log plot. Given the 1000-fold dynamic range of the linear relationship, this nanopore sensor can be calibrated using the synthetic peptide in order to selectively detect and quantify the BoNT-B digest Sb2(77-93)d. The dynamic change of the digest concentration would indicate the activity of BoNT-B.
Figure 4.
Correlation between the frequency of blocking events and the concentration of synthetic peptide Sb2(76-93)s. Peptide was applied to the cis chamber at +120 mV. The linear fitting of the log–log plot by the equation log(f) = a·log([Sb2(76-93)s]) + b (a = 0.55, b = 0.22, R2 = 0.98) represents a calibration curve for the BoNT-B nanopore detector.
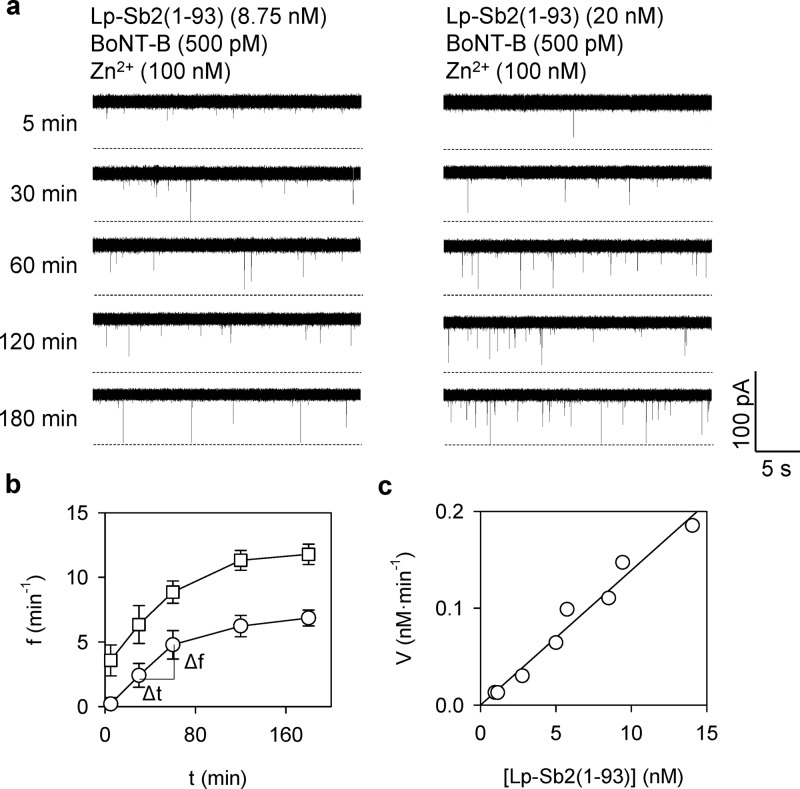
To analyze the activity of BoNT-B digestion, we recorded the nanopore currents for the time-course of the Lp-Sb2(77-93)d formation over 3 h (Figure 5a). The reaction product Lp-Sb2(77-93)d was generated by BoNT-B (500 pM)/Zn2+ (100 nM) mixed with its substrate Lp-Sb2(1-93) at two concentrations (8.75 and 20 nM). From the current traces, we measured the correlation of the product Lp-Sb2(77-93)d to the frequency of the current block, i.e., the block frequency-digesting time (f–t) curves (Figure 5b). Of note, recordings from the empty pore showed the absence of any blocks, i.e., a simple background (Figure S7, Supporting Information). For both substrate concentrations, the block frequency exhibited a rapid initial increase, which tapered off to reach a plateau in about 2 h. The f–t curves allowed for analysis of the BoNT enzymatic activity. Traditionally, the enzyme activity can be analyzed using the Lineweaver–Burk (L-B) plot, which can give the maximal digestion rate (Vmax) and the substrate concentration at the half maximal rate (Km). Of note, the L-B plot is valid for relatively high substrate concentrations (μM to mM levels).39,52 In the nanopore detection, however, the substrate concentration (10 nM level) is at least 2 orders of magnitude below that and also far lower than Km for BoNT-B (100 μM level52). In this case, the L-B plot would be inaccurate. Instead, the simplified Michaelis–Menten rate equation V = Vmax·[Lp-Sb2(1-93)]/Km is valid, where the digestion rate V is the increase in Sb2(77-93)d concentration per unit time (nM·min–1), [Lp-Sb2(1-93)] is the estimated substrate concentration, and the slope of the linear fitting of the V vs [Lp-Sb2(1-93)] gives the ratio Vmax/Km. Specifically, V was obtained from the f–t curves (Figure 5b) in two steps: (1) the frequency of the product Sb2(77-93)d event at each time point was transformed into the Sb2(77-93)d concentration based on the frequency–concentration correlation using Sb2(76-93)s (Figure 4) and (2) the digestion rate was obtained by the increase of Sb2(77-93)d concentration between adjacent time points divided by the time interval. The substrate concentration [Lp-Sb2(1-93)] at each time point during digestion was evaluated by the initial substrate concentration [Lp-Sb2(1-93)]0 (8.75 and 20 nM) minus the estimated product Sb2(77-93)d concentration [Sb2(77-93)d]. Through this process, we obtained the plot of V vs [Lp-Sb2(1-93)] in Figure 5c, which involved data from both digesting curves in Figure 5b. The digestion rates for both substrate concentrations can be fitted together with the single line, showing the slope (Vmax/Km) of 0.012 min–1. The Km for BoNT-B light chain targeting Sb2 has been reported to be 350 μM.53 Thus, Vmax was 0.06 μM·s–1. From Vmax, the maximal turnover rate Vmax/[BoNT-B] can be evaluated, where [BoNT-B] is the toxin concentration, which in our setting was 500 pM. The maximal turnover rate is estimated to be ∼120 s–1, a value comparable to and within the order of magnitude of 40 s–1 as in previous reports.52,54 Overall, the nanopore has been proven useful in detecting the BoNT-B light chain by quantitative analysis of the toxin enzymatic cleavage of the designed Sb2-based probe/substrate.
Figure 5.
Kinetics of BoNT-B enzymatic activity on Lp-Sb2(1-93) substrate. (a) Time-lapse (5 to 180 min) of the nanopore current traces recording the digestion of Lp-Sb2(1-93) substrate at two set initial concentrations (left, 8.75 nM; right, 20 nM), by light chain BoNT-B (500 pM)/Zn2+(100 nM). (b) Frequency of blocking events reporting on the presence of the digestion product Sb2(77-93)d at the two substrate concentrations (squares, 8.75 nM; circles, 20 nM) over digestion time. (c) Simplified Michaelis–Menten plot showing the digestion rate (V) as the function of the evaluated Lp-Sb2(1-93) substrate concentration ([Lp-Sb2(1-93)]) during digestion. See text for definitions of V and [Lp-Sb2(1-93)] and their evaluation methods. The data that was obtained from both digesting curves in panel b were fitted together with a line V = a·[Lp-Sb2(1-93)] + b, where a = 0.012 min–1 is the ratio of V/Km, and b = 9.2 × 10–5 nM·min–1 (R2 = 0.97).
As serum would be an penultimate material received for detection of the toxin in humans, we examined the possibility of detecting the activity of a BoNT-B spike-in in serum. It has been reported (which we confirm) that the lipid bilayer membrane with embedded protein pore can be easily raptured using the original serum sample.38 Therefore, as in the previous study,38 we used diluted serum (see Table S2, Supporting Information, for serum components) as a medium for detection of BoNT-B. Specifically, we mixed 8.75 nM Lp-Sb2(1-93), 500 pM BoNT-B and 100 nM Zn2+ in the prediluted serum (1% v/v in 1 M KCl) for 3 h. The empty pore current was not significantly affected when diluted serum alone was added to the cis side (Figure 6a). In contrast, when the BoNT-B/Zn2+–substrate mixture in diluted serum was added to the cis side, the signature pore blocks, at a frequency of 5.7 min–1, caused by Sb2(77-93)d were evident (Figure 6b). In a positive control experiment, the addition of the 10 nM synthetic peptide Sb2(76-93)s alone to the diluted serum generated similar pore blocks with the frequency of 8.7 min–1 (Figure 6c). These results suggest that the BoNT-B can be detected in diluted serum by detecting the presence of the pore blocks caused by the specific toxin-digested peptide.
Figure 6.

Detection of BoNT-B in diluted serum. Sterile filtered fetal bovine serum (1% v/v) was diluted in 1 M KCl. (a) Current trace through the aerolysin pore for diluted serum alone. No signature blocks were observed. (b) Current trace in the presence of toxin BoNT-B (500 pM), Zn2+ (100 nM) and substrate protein Lp-Sb2(1-93) (8.75 nM) in the serum-containing solution after incubation for 3 h. Signature blocks for the digested short peptide Sb2(77-93)d were identified at a frequency of 5.7 min–1. (c) Current trace for synthetic Sb2(76-93)s (10 nM) as the positive control. Signature blocks for the synthetic Sb2(77-93)s were identified at 8.7 min–1. All analytes were applied to the cis chamber at +120 mV.
Discussion
We have used the aerolysin protein pore to develop a novel BoNT-B sensor with subnanomolar sensitivity achieved within minutes. The neurotoxin is detected through measuring the frequency of the nanopore current blocks generated by the specific BoNT-B cleavage product of the recombinant substrate. Hence, the aerolysin pore selectively detects dynamic levels of the C-terminal 17 aa digest peptide Sb2(77-93)d, while it is insensitive to other analytes, i.e., BoNT-B/Zn2+, the toxin’s substrate Lp-Sb2(1-93) and its longer N-terminal digest Lp-Sb2(1-76)d. Of note, α-hemolysin, another protein pore, has been used for distinction of enzymatically cleaved/modified peptides.36,39,40 However, the α-hemolysin pore has proved inadequate for detection of BoNT-B itself or its activity based on digest products (Figure S8, Supporting Information). Also, the selective pore blocking action of short Sb2(77-93)d/Sb2(76-93)s on the aerolysin pore differs from previous studies, which have showed that long peptides/proteins can translocate through the aerolysin pore.44,45,55 A possible explanation for these seemingly disparate findings perhaps rests with the protein state. Namely, the peptides/proteins in the previous studies were denatured (unfolded) prior to their translocation through the pore, while here all the proteins/peptides we studied were in their native (folded) state. Thus, it appears that the aerolysin pore might be inaccessible to such large proteins/peptides in their native state, which could also explain the ability to detect BoNT-B activity in serum (Figure 6), as the aerolysin pore is insensitive to serum components (Figure S8, Supporting Information). Taken together, we attained accuracy for detection of BoNT-B activity in serum, a bodily fluid derivative, which is the most common material that would be analyzed in medical practice when diagnosing botulism.
We have identified the electroosmotic effect that can enhance the capture efficiency of the digest peptide in the aerolysin pore. In our detection, the cationic Sb2(77-93)d and Sb2(76-93)s are driven both by the electroosmotic and electrophoretic forces, albeit in the opposite direction. The electrophoretic effect is relatively weak because the charge of the peptide is shielded in high salt solution (1 M KCl). As a result, the electroosmotic effect that originates from the anion selectivity of the pore overpowers that of the opposing electrophoretic drive. Therefore, the overall effect is that the peptide is attracted to the pore against the electric field direction. The nanopore electroosmosis originates from the ion selectivity of the pore.56 This effect has been found to be able to enhance the binding of small compounds57 and influence the capture of nucleic acids46,48 in the nanopore. Because the peptides/proteins used in those studies were negatively charged, the electrophoretic and electroosmotic forces were in the same detection and hence indistinguishable in the recordings.
The nanopore BoNT-B sensor has an inherently built-in amplification of the product as each BoNT-B molecule can cleave many molecules of substrate. Accordingly, the frequency of blocking events can be increased many-fold, as seen in f–t curves (Figure 5b). Thus, the nanopore approach greatly shortens the analytical time, while retaining high detection accuracy. We have demonstrated the detection of 500 pM or 25 ng/mL BoNT-B (light chain) by measuring the dynamic levels of the digest peptide in the nanopore. This sensitivity is similar to the detection levels of flow cytometry-based assays (73 ng/mL for BoNT-B)58 and FRET endopeptidase assays (a Sb2-based sensor, 30 ng/mL of BoNT-B),16,59 but lower than the sensitivity of the “gold” standard mouse lethality assay, improved ELISA assays, and immuo-PCR (see refs (60)–63); of note, the U.S. Environmental Protection Agency (EPA) lists the lethal dose concentration for botulinum type A and B at 300 ng/mL. The nanopore performance in BoNT-B detection could be greatly enhanced by increasing the concentration of the substrate peptide. The substrate concentration in this study (∼10 nM) is much lower than Km of BoNT-B (100 μM level).53 At low substrate concentration, the actual digestion rate by 500 pM BoNT-B is only 0.2 nM min–1, which is equivalent to a turnover rate of 0.4 s–1, almost 3 orders of magnitude slower than that at the maximal cleavage speed (120 s–1). The actual low turnover rate is associated with two issues: (1) the detection time is prolonged when the BoNT-B level is extremely low and (2) due to the low concentration of the digestion product, the frequency of blocking events is lowered, thus lowering the detection sensitivity. We could partly resolve this issue by simply increasing the concentration of the substrate. Incidentally, miniaturized nanopore systems allow for the presence of high substrate concentration. For example, the droplet-interfaced bilayer64−66 system has been reported to be able to detect hundreds of molecules in its small volume sample (less than 1 μL).67 With small volume (as compared to presently used 2 mL), it is possible to achieve high substrate concentration at low cost to elevate the digest rate. This allows generating sufficient digest peptides for nanopore detection at a low toxin concentration, therefore increasing the sensitivity. In addition, more digest peptides can be produced in a shorter time, enhancing the time-efficiency of the detection.
Conclusion
The aerolysin protein pore can be used to elucidate the interaction of the nanopore with a synaptic protein Sb2 derivative and its BoNT-B cleavage products. We determined that only the small C-terminal BoNT-B digest peptide in its native state can interact with the pore to generate current blocks, while other native peptides/proteins in the reaction mixture and serum cannot produce observable blocking events. By specifically tracking the dynamic change in frequency of the small peptide-generated signatures, we determined the existence of the BoNT-B toxin in the subnanomolar range within minutes. This research is motivated by danger posed by BoNT-B, which is one of the most potent biotoxins. Rapid and sensitive detection of this toxin, be that due to drug-use, food-poising or terrorism, is a priority in toxicology and biodefense. This is an area of future interest and a fertile ground for improvements of the present BoNT-B nanopore detection scheme. The above described approach has potential to be adapted to the selective peptide detection for biosensing and investigating biological processes, including the detection of activity of Clostridial toxins, and biophysical mechanisms for biopolymers interaction and translocation through the nanopore.
Methods
Plasmids
We used a plasmid (generously provided by Edwin R. Chapman, University of Wisconsin, Madison, WI) encoding the most of the cytoplasmic domain, i.e., amino acids (aa) 1–93, of rat synaptobrevin 2 (Sb2) fused to the C-terminus of the vector (pTrcHisA) sequence encoding the 36 aa lead peptide (MGGSHHHHHHGMASMTGGQQMGRDLYDDDDKDRWGS), which contains a six histidine (H6) tag near the N-terminus. We termed the recombinant fusion protein, produced using this plasmid, as Lp-Sb2(1-93) (see Figure S1, Supporting Information). This plasmid served as the PCR template for subcloning of another plasmid, which encodes the fusion protein containing the lead peptide followed by aa 1–76 of Sb2 (see Figure S1, Supporting Information); we termed the resulting recombinant protein as Lp-Sb2(1-76). The subcloning was done using the In-Fusion HD Cloning Kit (Clontech Laboratories, Inc., Mountain View, CA) and the following four primers with the 15 bp overlapping sequences: Insert Fwd, 5′- AGCACCGCCTACATACCTCGCTCTGCTAATCCTG -3′; Insert Rev, 5′- TCATTACTATTATCACTGGGAGGCCCCTGCCTGGAG -3′; Vector Fwd, 5′- TGATAATAGTAATGAATTCGAAGCTTGGCTGTTTTGGCGGATG -3′, Vector Rev: 5′- TATGTAGGCGGTGCTACAGAGTTCTTGAAGTGGTG -3′. DNA Sequencing of constructs was used to confirm the above-described encoding of Lp-Sb2(1-93) and Lp-Sb2(1-76).
Recombinant Proteins
The above plasmids were utilized in recombinant protein production and purified with nickel–Sepharose beads (Qiagen, Valencia, CA) as per manufacturer procedure. H6-tagged proteins were quantified using the Bradford reagent (Pierce Biotechnology, Rockford, IL, USA) and bovine serum albumin as a standard. Determined protein concentrations were as 0.1 μg/μL for Lp-Sb2(1-93) and 0.175 μg/μL for Lp-Sb2(1-76). Purified recombinant proteins represented 84–97% of the total protein content.68
Western Blotting
Generation of the recombinant proteins was confirmed by Western blotting, as previously described.68,69 Samples with purified proteins were loaded at 1 μg per lane and subjected to 15% SDS-PAGE, followed by transfer to nitrocellulose membranes. Membranes were probed with specific antibodies against the H6 tag (Chemicon, catalogue no. MAB3114, 1:500 dilution). Immunoreactivity of the bands was detected using enhanced chemiluminescence (Amersham Biosciences). All proteins showed single immunoreactive bands with appropriate molecular weights.
Peptide Synthesis
We obtained custom synthesized 18 aa peptide with Sb2(76-93) sequence (APeptide Co., Ltd., ShangHai, China) having purity of 95% (HPLC). Note that this Sb2(76-93)s peptide contains Q76 otherwise not present in Sb2(77-93)d, the Lp-Sb2(1-93) C-terminal fragment after its cleavage by BoNT-B light chain/Zn2+.
In Silico Protein/Peptide Analysis
We carried out an in silico peptide analysis using the Vector NTI Suite v.6 BioPlot module (Informax, Inc., Bethesda, MD) to obtain various predicted characteristics (Table S1, Supporting Information), which assisted us with the nanopore experimental design and results interpretation.
BoNT-B
The light chain of botulinum neurotoxin type B (50 kDa, List Biological Laboratories, Campbell, CA) was used to cleave recombinant Sb2.13 Hence, a mixture of Lp-Sb2(1-93) with the Zn2+-endopeptidase BoNT-B (500 pM) and zinc chloride (100 nM) in solution (1 M potassium chloride, 10 mM HEPES, pH = 7.35) results in two fragments, the N-terminal digest, annotated as Lp-Sb2(1-76)d to discriminate it from the otherwise identical recombinant protein Lp-Sb2(1-76) produced above, and the 17 aa C-terminal digest, annotated as Sb2(77-93)d.
Serum
Sterile filtered fetal bovine serum was purchased from HyClone, Thermo Fisher Scientific Inc., Waltham, MA (catalogue No. SH30910, lot No. AWB98615; see Table S2 (Supporting Information) of biochemical analysis provided by the manufacturer). The serum sample (1% v/v) for the nanopore detection was prepared by diluting it 100 times in 1 M KCl. Specifically, 200 μL of fetal bovine serum was mixed with 20 mL of 1 M KCl buffered with 10 mM HEPES (pH = 7.35), to obtain 1% (v/v) serum solution.
Digestion
2.5 μL of 7 μM (0.1 μg/μL) recombinant Lp-Sb2(1-93), 1 μL of 1 μM (0.05 μg/μL) BoNT-B light chain and 2 μL of 100 μM ZnCl2, were mixed with 2 mL of 1 M KCl solution or prediluted serum solution. The concentrations of various species in the mixture were: Lp-Sb2(1-93), 8.75 nM (initial concentration); BoNT-B light chain, 0.5 nM; and Zn2+, 100 nM. After incubation for 3 h at room temperature, the mixture was released to the cis chamber for nanopore recording.
Aerolysin and Recordings Using Single Aerolysin Pores
Proaerolysin was purchased from Aerohead Scientific, SK, Canada. According to the product information, to activate proaerolysin, 2 μM proaerolysin was incubated with 1 μg/mL trypsin (final concentration in the mixture) for 1.5 h to cleave a short fragment from the carboxyl terminal. The activated aerolysin was stored at −80 °C. The electrical setup for recording single protein pore or channel have been described in previous reports such as reference.24 Briefly, the recording apparatus was composed of two chambers (cis and trans) that were partitioned with a Teflon film. The planar lipid bilayer of 1,2-diphytanoyl-sn-glycerophosphatidylcholine (Avanti Polar Lipids) was formed spanning a 100–150 μm hole in the center of the partition. 1–5 μL of the aerolysin stock solution was released in the cis solution. After a couple of minutes, single aerolysin proteins can be inserted in the lipid bilayer to form molecular pores. Both cis and trans chambers were filled with 1 M salt solutions (KCl) buffered with 10 mM HEPES and titrated to pH = 7.35. For detection of digestion peptides, the incubation solution (described above) was presented in the cis chamber. For the α-hemolysin experiment, the α-hemolysin heptameric protein was synthesized using in vitro transcription and translation and obtained as described previously.23,24 1–5 μL of the α-hemolysin stock solution was released in the cis solution to form the pores in the lipid bilayer. To increase the capture rate constant, the cis and trans chambers were filled with 1 and 0.2 M asymmetric KCl solutions, respectively.46 All saline solutions (without protein addition) were filtered with MillexGP 0.22 uM membrane (EMD Millipore Corporation, Billerica, MA) before use. Single-channel currents were recorded with an Axopatch 200A patch-clamp amplifier (Molecular Device Inc., former Axon Inc.), filtered with a built-in four-pole low-pass Bessel Filter at 5 kHz, and acquired with Clampex 9.0 software (Molecular Device Inc.) through a Digidata 1332 A/D converter (Molecular Device Inc.) at a sampling rate of 20 kHz. All reactants were applied to the cis chamber, with the sole exception of experiments in Figures 3b and S3b (Supporting Information) where Sb2(76-93)s was alternatively applied to the trans chamber. Data were based on at least four separate experiments and obtained by Single Channel Search function pClamp provided for events that blocked the current more than 50%. The current amplitude and duration histograms of blocking events were fitted by the Gaussian and exponential functions, respectively. The electrophysiology experiments were conducted at 22 ± 1 °C. Data were represented as mean ± SD for at least three independent experiments.
Acknowledgments
We thank Edwin R. Chapman for providing the Lp-Sb2(1-93) plasmid. The Heflin Center Genomics Core at UAB provided sequencing information for the DNA plasmids used in this work. This investigation was supported by grants from NSF CAREER 0546165 (L.-Q.G.), NIH GM079613 (L.-Q.G.), and NIH HD078678 (V.P.).
Supporting Information Available
Principle for design of a recombinant protein substrate for BoNT-B, control experiments for detection of BoNT-B activity using the aerolysin pore, current traces showing transient aerolysin pore blocks generated by the synthetic peptide Sb2(76-93)s at different concentration, time constant histograms for Sb2(77-93)d or Sb2(76-93)s aerolysin pore current blocks, current traces showing detection directionality and voltage dependence of Sb2(76-93)s in the aerolysin nanopore assay, current–voltage (I–V) curve for the aerolysin pore recorded in asymmetric 1 M/0.2 M (cis/trans) KCl solutions, long current trace obtained with the empty aerolysin pore at +120 mV applied from the trans side, current traces showing inability to detect BoNT-B activity by using the α-hemolysin pore, table for in silico protein/peptide analysis and table for biochemical analysis of the sterile filtered fetal bovine serum. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
⊥ These authors contributed equally to this work.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Schantz E. J.; Johnson E. A. Properties and Use of Botulinum Toxin and Other Microbial Neurotoxins in Medicine. Microbiol. Rev. 1992, 56, 80–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembek Z. F.; Smith L. A.; Rusnak J. M. Botulism: Cause, Effects, Diagnosis, Clinical and Laboratory Identification, and Treatment Modalities. Disaster Med. Public Health Prep. 2007, 1, 122–134. [DOI] [PubMed] [Google Scholar]
- Hatheway C. L. Toxigenic Clostridia. Clin. Microbiol. Rev. 1990, 3, 66–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passaro D. J.; Werner S. B.; McGee J.; Mac Kenzie W. R.; Vugia D. J. Wound Botulism Associated with Black Tar Heroin among Injecting Drug Users. JAMA 1998, 279, 859–863. [DOI] [PubMed] [Google Scholar]
- Bioterrorism Agents/Diseases; Center for Disease Control and Prevention, Atlanta, GA, 2003; http://www.bt.cdc.gov/agent/agentlist-category.asp.
- Guerrissi J.; Sarkissian P. Local Injection into Mimetic Muscles of Botulinum Toxin a for the Treatment of Facial Lines. Ann. Plast. Surg. 1997, 39, 447–453. [DOI] [PubMed] [Google Scholar]
- Schiavo G.; Benfenati F.; Poulain B.; Rossetto O.; Polverino de Laureto P.; DasGupta B. R.; Montecucco C. Tetanus and Botulinum-B Neurotoxins Block Neurotransmitter Release by Proteolytic Cleavage of Synaptobrevin. Nature 1992, 359, 832–835. [DOI] [PubMed] [Google Scholar]
- Schiavo G.; Rossetto O.; Benfenati F.; Poulain B.; Montecucco C. Tetanus and Botulinum Neurotoxins Are Zinc Proteases Specific for Components of the Neuroexocytosis Apparatus. Ann. N. Y. Acad. Sci. 1994, 710, 65–75. [DOI] [PubMed] [Google Scholar]
- Sudhof T. C.; De Camilli P.; Niemann H.; Jahn R. Membrane Fusion Machinery: Insights from Synaptic Proteins. Cell 1993, 75, 1–4. [PubMed] [Google Scholar]
- Keller J. E.; Nowakowski J. L.; Filbert M. G.; Adler M. Rapid Microplate Assay for Monitoring Botulinum Neurotoxin B Catalytic Activity. J. Appl. Toxicol. 1999, 19Suppl 1S13–S17. [DOI] [PubMed] [Google Scholar]
- Peruski A. H.; Johnson L. H. 3rd; Peruski L. F. Jr. Rapid and Sensitive Detection of Biological Warfare Agents Using Time-Resolved Fluorescence Assays. J. Immunol. Methods 2002, 263, 35–41. [DOI] [PubMed] [Google Scholar]
- Baldwin M. R.; Bradshaw M.; Johnson E. A.; Barbieri J. T. The C-Terminus of Botulinum Neurotoxin Type A Light Chain Contributes to Solubility, Catalysis, and Stability. Protein Expression Purif. 2004, 37, 187–195. [DOI] [PubMed] [Google Scholar]
- Liu W.; Montana V.; Chapman E. R.; Mohideen U.; Parpura V. Botulinum Toxin Type B Micromechanosensor. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 13621–13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeau Y.; Doussau F.; Grant N. J.; Poulain B. How Botulinum and Tetanus Neurotoxins Block Neurotransmitter Release. Biochimie 2000, 82, 427–446. [DOI] [PubMed] [Google Scholar]
- Schiavo G.; Matteoli M.; Montecucco C. Neurotoxins Affecting Neuroexocytosis. Physiol. Rev. 2000, 80, 717–766. [DOI] [PubMed] [Google Scholar]
- Dongo M.; Tepp W. H.; Johnson E. A.; Chapman E. R. Using Fluorescent Sensors to Detect Botulinum Neurotoxin Activity in Vitro and in Living Cells. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 14701–14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasianowicz J. J.; Robertson J. W.; Chan E. R.; Reiner J. E.; Stanford V. M. Nanoscopic Porous Sensors. Annu. Rev. Anal. Chem. 2008, 1, 737–766. [DOI] [PubMed] [Google Scholar]
- Bayley H.; Cremer P. S. Stochastic Sensors Inspired by Biology. Nature 2001, 413, 226–230. [DOI] [PubMed] [Google Scholar]
- Gu L. Q.; Shim J. W. Single Molecule Sensing by Nanopores and Nanopore Devices. Analyst 2010, 135, 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howorka S.; Siwy Z. Nanopore Analytics: Sensing of Single Molecules. Chem. Soc. Rev. 2009, 38, 2360–2384. [DOI] [PubMed] [Google Scholar]
- Clarke J.; Wu H. C.; Jayasinghe L.; Patel A.; Reid S.; Bayley H. Continuous Base Identification for Single-Molecule Nanopore DNA Sequencing. Nat. Nanotechnol. 2009, 4, 265–270. [DOI] [PubMed] [Google Scholar]
- An N.; Fleming A. M.; Middleton E. G.; Burrows C. J. Single-Molecule Investigation of G-Quadruplex Folds of the Human Telomere Sequence in a Protein Nanocavity. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 14325–14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J. W.; Gu L. Q. Encapsulating a Single G-Quadruplex Aptamer in a Protein Nanocavity. J. Phys. Chem. B 2008, 112, 8354–8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J. W.; Tan Q.; Gu L.-Q. Single-Molecule Detection of Folding and Unfolding of the G-Quadruplex Aptamer in a Nanopore Nanocavity. Nucleic Acids Res. 2009, 37, 972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Luan B. Q.; Yang Z.; Zhang X.; Ritzo B.; Gates K.; Gu L. Q. Single Molecule Investigation of Ag+ Interactions with Single Cytosine-, Methylcytosine- and Hydroxymethylcytosine-Cytosine Mismatches in a Nanopore. Sci. Rep. 2014, 4, 5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherf G. M.; Lieberman K. R.; Rashid H.; Lam C. E.; Karplus K.; Akeson M. Automated Forward and Reverse Ratcheting of DNA in a Nanopore at 5-Å Precision. Nat. Biotechnol. 2012, 30, 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo A. H.; Derrington I. M.; Ross B. C.; Brinkerhoff H.; Adey A.; Nova I. C.; Craig J. M.; Langford K. W.; Samson J. M.; Daza R.; Doering K.; Shendure J.; Gundlach J. H. Decoding Long Nanopore Sequencing Reads of Natural DNA. Nat. Biotechnol. 2014, 32, 829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Zheng D.; Tan Q.; Wang M. X.; Gu L.-Q. Nanopore-based Detection of Circulating Micrornas in Lung Cancer Patients. Nat. Biotechnol. 2011, 6, 668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Tian K.; Hunter L. L.; Ritzo B.; Gu L. Q. Probing Molecular Pathways for DNA Orientational Trapping, Unzipping and Translocation in Nanopores by Using a Tunable Overhang Sensor. Nanoscale 2014, 6, 11372–11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movileanu L.; Schmittschmitt J. P.; Scholtz J. M.; Bayley H. Interactions of Peptides with a Protein Pore. Biophys. J. 2005, 89, 1030–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereuta L.; Roy M.; Asandei A.; Lee J. K.; Park Y.; Andricioaei I.; Luchian T. Slowing Down Single-Molecule Trafficking through a Protein Nanopore Reveals Intermediates for Peptide Translocation. Sci. Rep. 2014, 4, 3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Larrea D.; Bayley H. Multistep Protein Unfolding During Nanopore Translocation. Nat. Nanotechnol. 2013, 8, 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oukhaled G.; Mathe J.; Biance A. L.; Bacri L.; Betton J. M.; Lairez D.; Pelta J.; Auvray L. Unfolding of Proteins and Long Transient Conformations Detected by Single Nanopore Recording. Phys. Rev. Lett. 2007, 98, 158101. [DOI] [PubMed] [Google Scholar]
- Asandei A.; Schiopu I.; Iftemi S.; Mereuta L.; Luchian T. Investigation of Cu2+ Binding to Human and Rat Amyloid Fragments Αβ (1–16) with a Protein Nanopore. Langmuir 2013, 29, 15634–15642. [DOI] [PubMed] [Google Scholar]
- Wolfe A. J.; Mohammad M. M.; Cheley S.; Bayley H.; Movileanu L. Catalyzing the Translocation of Polypeptides through Attractive Interactions. J. Am. Chem. Soc. 2007, 129, 14034–14041. [DOI] [PubMed] [Google Scholar]
- Rosen C. B.; Rodriguez-Larrea D.; Bayley H. Single-Molecule Site-Specific Detection of Protein Phosphorylation with a Nanopore. Nat. Biotechnol. 2014, 32, 179–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem D.; Jayasinghe L.; Salichou M.; Bayley H. Protein Detection by Nanopores Equipped with Aptamers. J. Am. Chem. Soc. 2012, 134, 2781–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Haque F.; Rychahou P. G.; Evers B. M.; Guo P. Engineered Nanopore of Phi29 DNA-Packaging Motor for Real-Time Detection of Single Colon Cancer Specific Antibody in Serum. ACS Nano 2013, 7, 9814–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q.; de Zoysa R. S.; Wang D.; Jayawardhana D. A.; Guan X. Real-Time Monitoring of Peptide Cleavage Using a Nanopore Probe. J. Am. Chem. Soc. 2009, 131, 6324–6325. [DOI] [PubMed] [Google Scholar]
- Kukwikila M.; Howorka S. Electrically Sensing Protease Activity with Nanopores. J. Phys.: Condens. Matter 2010, 22, 454103. [DOI] [PubMed] [Google Scholar]
- Harrington L.; Cheley S.; Alexander L. T.; Knapp S.; Bayley H. Stochastic Detection of Pim Protein Kinases Reveals Electrostatically Enhanced Association of a Peptide Substrate. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, E4417–E4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefureac R.; Long Y. T.; Kraatz H. B.; Howard P.; Lee J. S. Transport of Alpha-Helical Peptides through Alpha-Hemolysin and Aerolysin Pores. Biochemistry 2006, 45, 9172–9179. [DOI] [PubMed] [Google Scholar]
- Schatz G.; Dobberstein B. Common Principles of Protein Translocation across Membranes. Science 1996, 271, 1519–1526. [DOI] [PubMed] [Google Scholar]
- Pastoriza-Gallego M.; Rabah L.; Gibrat G.; Thiebot B.; van der Goot F. G.; Auvray L.; Betton J. M.; Pelta J. Dynamics of Unfolded Protein Transport through an Aerolysin Pore. J. Am. Chem. Soc. 2011, 133, 2923–2931. [DOI] [PubMed] [Google Scholar]
- Stefureac R.; Waldner L.; Howard P.; Lee J. S. Nanopore Analysis of a Small 86-Residue Protein. Small 2008, 4, 59–63. [DOI] [PubMed] [Google Scholar]
- Wanunu M.; Morrison W.; Rabin Y.; Grosberg A. Y.; Meller A. Electrostatic Focusing of Unlabelled DNA into Nanoscale Pores Using a Salt Gradient. Nat. Nanotechnol. 2010, 5, 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian K.; He Z.; Wang Y.; Chen S. J.; Gu L. Q. Designing a Polycationic Probe for Simultaneous Enrichment and Detection of Micrornas in a Nanopore. ACS Nano 2013, 7, 3962–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglia G.; Restrepo M. R.; Mikhailova E.; Bayley H. Enhanced Translocation of Single DNA Molecules through α-Hemolysin Nanopores by Manipulation of Internal Charge. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 19720–19725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Wang Y.; Fricke B. L.; Gu L. Q. Programming Nanopore Ion Flow for Encoded Multiplex Microrna Detection. ACS Nano 2014, 8, 3444–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T.; Schmid A.; Notermans S.; Benz R. Aerolysin of Aeromonas Sobria: Evidence for Formation of Ion-Permeable Channels and Comparison with Alpha-Toxin of Staphylococcus aureus. Infect. Immun. 1990, 58, 2127–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degiacomi M. T.; Iacovache I.; Pernot L.; Chami M.; Kudryashev M.; Stahlberg H.; van der Goot F. G.; Dal Peraro M. Molecular Assembly of the Aerolysin Pore Reveals a Swirling Membrane-Insertion Mechanism. Nat. Chem. Biol. 2013, 9, 623–629. [DOI] [PubMed] [Google Scholar]
- Gilsdorf J.; Gul N.; Smith L. A. Expression, Purification, and Characterization of Clostridium Botulinum Type B Light Chain. Protein Expression Purif. 2006, 46, 256–267. [DOI] [PubMed] [Google Scholar]
- Shone C. C.; Roberts A. K. Peptide Substrate Specificity and Properties of the Zinc-Endopeptidase Activity of Botulinum Type B Neurotoxin. Eur. J. Biochem. 1994, 225, 263–270. [DOI] [PubMed] [Google Scholar]
- Schmidt J. J.; Stafford R. G. Fluorigenic Substrates for the Protease Activities of Botulinum Neurotoxins, Serotypes A, B, and F. Appl. Environ. Microbiol. 2003, 69, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madampage C.; Tavassoly O.; Christensen C.; Kumari M.; Lee J. S. Nanopore Analysis: An Emerging Technique for Studying the Folding and Misfolding of Proteins. Prion 2012, 6, 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L.-Q.; Cheley S.; Bayley H. Prolonged Residence Time of a Noncovalent Molecular Adapter, B-Cyclodextrin, within the Lumen of Mutant A-Hemolysin Pores. J. Gen. Physiol. 2001, 118, 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L. Q.; Cheley S.; Bayley H. Electroosmotic Enhancement of the Binding of a Neutral Molecule to a Transmembrane Pore. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 15498–15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly D.; Kirchner S.; Stoermann B.; Schreiber T.; Kaulfuss S.; Schade R.; Zbinden R.; Avondet M. A.; Dorner M. B.; Dorner B. G. Simultaneous Quantification of Five Bacterial and Plant Toxins from Complex Matrices Using a Multiplexed Fluorescent Magnetic Suspension Assay. Analyst 2009, 134, 2028–2039. [DOI] [PubMed] [Google Scholar]
- Parpura V.; Chapman E. R. Detection of Botulinum Toxins: Micromechanical and Fluorescence-Based Sensors. Croat. Med. J. 2005, 46, 491–497. [PubMed] [Google Scholar]
- Cai S.; Singh B. R.; Sharma S. Botulism Diagnostics: From Clinical Symptoms to in Vitro Assays. Crit. Rev. Microbiol. 2007, 33, 109–125. [DOI] [PubMed] [Google Scholar]
- Čapek P.; Dickerson T. J. Sensing the Deadliest Toxin: Technologies for Botulinum Neurotoxin Detection. Toxins 2010, 2, 24–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström M.; Korkeala H. Laboratory Diagnostics of Botulism. Clin. Microbiol. Rev. 2006, 19, 298–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. K.; Whiting R. C. Methods for Detection of Clostridium Botulinum Toxin in Foods. J. Food Prot. 2005, 68, 1256–1263. [DOI] [PubMed] [Google Scholar]
- Funakoshi K.; Suzuki H.; Takeuchi S. Lipid Bilayer Formation by Contacting Monolayers in a Microfluidic Device for Membrane Protein Analysis. Anal. Chem. 2006, 78, 8169–8174. [DOI] [PubMed] [Google Scholar]
- Bayley H.; Cronin B.; Heron A.; Holden M. A.; Hwang W. L.; Syeda R.; Thompson J.; Wallace M. Droplet Interface Bilayers. Mol. Biosyst. 2008, 4, 1191–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portonovo S. A.; Schmidt J. Masking Apertures Enabling Automation and Solution Exchange in Sessile Droplet Lipid Bilayers. Biomed. Microdevices 2012, 14, 187–191. [DOI] [PubMed] [Google Scholar]
- Fischer A.; Holden M. A.; Pentelute B. L.; Collier R. J. Ultrasensitive Detection of Protein Translocated through Toxin Pores in Droplet-Interface Bilayers. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 16577–16581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.; Montana V.; Bai J.; Chapman E. R.; Mohideen U.; Parpura V. Single Molecule Mechanical Probing of the Snare Protein Interactions. Biophys. J. 2006, 91, 744–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montana V.; Liu W.; Mohideen U.; Parpura V. Single Molecule Measurements of Mechanical Interactions within Ternary Snare Complexes and Dynamics of Their Disassembly: Snap25 vs. Snap23. J. Physiol. 2009, 587, 1943–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.