Abstract
Objective:
One of the early hallmarks of osteoarthritis (OA) is a progressive degeneration of the articular cartilage. Early diagnosis of OA-associated cartilage alterations would be beneficial for disease prevention and control, and for the development of disease-modifying treatments. However, early diagnosis is still hampered by a lack of quantifiable readouts in preclinical models.
Design:
In this study, we have shown the potency of contrast-enhanced nanofocus x-ray computed tomography (CE-nanoCT) to be used for virtual 3-dimensional (3D) histopathology in established mouse models for OA, and we compared with standard histopathology.
Results:
We showed the equivalence of CE-nanoCT images to histopathology for the modified Mankin scoring of the cartilage structure and quality. Additionally, a limited set of 3D cartilage characteristics measured by CE-nanoCT image analysis in a user-independent and semiautomatic manner, that is, average and maximum of the noncalcified cartilage thickness distribution and loss in glycosaminoglycans, was shown to be predictive for the cartilage quality and structure as can be evaluated by histopathological scoring through the use of an empirical model.
Conclusions:
We have shown that CE-nanoCT is a tool that allows virtual histopathology and 3D morphological quantification of multitissue systems, such as the chondro-osseous junction. It provides faster and more quantitative data on cartilage structure and quality compared with standard histopathology while eliminating user bias. CE-nanoCT thus should allow capturing subtle differences in cartilage characteristics, carefully mapping OA progression and, ultimately, asses the beneficial changes when testing a candidate disease-modifying treatment.
Keywords: contrast-enhanced nanofocus computed tomography (CE-nanoCT), osteoarthritis, virtual histopathology, modified Mankin scoring, 3D cartilage quantification
Introduction
Musculoskeletal diseases are a major burden on individuals, health systems, and resources. They are the most common cause of severe long-term pain and physical disability, and affect hundreds of millions of people’s quality of life all around the world. Osteoarthritis (OA), a progressive, degenerative disease of the (articular) joints (hips, knees, neck, lower back, or small joints of the hands), represents the most common musculoskeletal disease. The etiology of the disease is not well understood and OA is considered a complex disease to which both genetic and acquired or environmental factors contribute.1 Misalignment of the joint and joint instability resulting from (untreated) cartilage defects, previous surgery (e.g., partial or total meniscectomy) or from other traumas and injuries to the joint, as well as intensive loading due to obesity or intensive sports activities, are all particular risk factors to develop the disease.2,3
OA affects all structures within the joint, but an early hallmark is a progressive degeneration of the articular cartilage accompanied by bony remodeling (subchondral bone sclerosis and osteophyte formation), occasional flares of inflammation, mild synovitis (in some patients), and osteitis. OA affects the joint in a nonuniform and focal manner. Localized loss of cartilage can increase focal stress across the joint leading to further cartilage destruction. This then eventually results in a misalignment and destabilization of the whole joint and progression and aggravation of the disease. Early diagnosis of OA-associated cartilage alterations would be very beneficial for disease prevention and control, not at the least because current treatment of established OA is symptomatic and palliative with an emphasis on control of joint pain and maintenance of function. The lack of a cure and associated huge unmet medical need provide an opportunity for the development of novel approaches and medicines to treat OA, especially for disease-modifying therapies that can halt and/or reverse the onset and progression of the disease. However, the development of therapeutic approaches is largely hampered by the lack of quantifiable readouts in preclinical models. Refinement and validation of the readout technology for OA biology would be tremendously valuable for the animal studies on joint degeneration, and should allow us capturing subtle differences and thus broader hallmarks of the disease, carefully map the disease progression in the different tissues and animal models and, ultimately, asses the beneficial changes of those hallmarks when testing the candidate disease-modifying treatment.4
Currently, preclinical efficacy of novel therapies for cartilage repair in small animal models is commonly assessed by histopathology.5-7 The modified Mankin scoring system6,8,9 is often used for semiquantitative histological assessment of OA because of its already widespread adoption, ease of use, and its similarity to scoring systems used for OA in humans.10 This method focuses, among others, on the structural changes observed, such as lesions or fibrillations and losses in proteoglycan staining. Histopathology has a high discriminative power but is destructive, labor intensive, costly in terms of time and resources, and, in standard settings it only provides 2-dimensional (2D) information because of the loss of data by the thickness of the sections, thus spatial and quantitative 3D assessment of the cartilage structure and quality are not feasible. Additionally, the modified Mankin scoring is currently not annotated automatically, with consequent interreader variability. The said limitations prompt the current need for a 3D imaging technique that is able to quantify the cartilage properties in the murine joint and can replace histopathology with a more efficient, high throughput, longitudinal imaging method.11
Recent advances in 3D imaging techniques, such as high-resolution microfocus x-ray computed tomography (microCT) imaging, have led to successful quantification of the changes in the subchondral bone structure in the mouse knee joint during the progression of OA.5,7 The use of ionic partition equilibrium contrast agents, such as anionic (i.e., Hexabrix; Covidien, Hazelwood, MO)12-16 or cationic contrast agents,17 as well as lead-based contrast agents18 have shown to allow quantification of functional and structural information of the cartilage. However, the limited spatial image resolution of most microCT devices does not allow distinguishing calcified cartilage from the subchondral bone. Additionally, most studies implementing new structural13,15 and functional14,19 readouts refrain to show equivalence to standard histopathological scorings.15,18,19 Moreover, although several 3D cartilage characteristics have been quantified, no validation compared with the golden standard of histopathological scoring was performed yet.
In a previous study,20 we evaluated the potency of contrast-enhanced nanofocus x-ray computed tomography (CE-nanoCT) to simultaneously image in 3D the subtissue architecture of cartilage at high contrast and spatial image resolution without invasive, labor-intensive sample preparation. In this study, we have shown, in an established mouse model for OA,21 that CE-nanoCT can be used as an imaging tool for virtual 3D histopathology. By direct comparison with standard histopathology we showed equivalence of CE-nanoCT images for the scoring of the structure, quality, and pathophysiology of the chondro-osseous junction. Additionally, the quantitative nature of the 3D CE-nanoCT data sets allowed the development of an empirical model based on a limited set of 3D cartilage characteristics measured by 3D image analysis, which is predictive for the cartilage quality and structure as can be evaluated by histopathological scoring. This model thus could be a potential replacement of or complimentary tool for the modified Mankin scoring when assessing preclinical efficacy of novel therapies for cartilage repair.
Materials and Methods
Mouse Model
Destabilization of the medial meniscus (DMM) by medial meniscotibial ligament transection leads to mild OA with various degrees of cartilage lesions.21 To show the potential of CE-nanoCT for virtual histopathology and 3D quantification of cartilage characteristics, 4 treatment groups were assessed. Right knee joints were harvested from adult (Rj:NMRI-Foxn1nu/Foxn1nu) mice that were sham operated (sham), or that underwent a medial meniscus destabilization procedure (DMM) and were sacrificed 8 weeks later. As a third group (DMM/HA), right knee joints were harvested after 8 weeks from mice that underwent a DMM procedure and were injected after 2 and 5 weeks with hyaluronic acid, which is in clinical use as viscosupplementation treatment. A final group contained age-matched nonsurgical controls (AM). We have worked with the nude mouse model to assess the effect of DMM on the cartilage structure and quality, because it allows testing a cellular therapy in the mouse using human cells (ongoing work).
These 4 research-relevant groups were selected to obtain a broad spectrum of cartilage defects on a per-animal basis (i.e., from bad to unaffected) that one might encounter in preclinical OA research, thus to show the potency of CE-nanoCT to be implemented as a standard technique in the OA field rather than to interpret the biology behind DMM-induced OA. All knee joints were imaged using CE-nanoCT, after which some were randomly sampled to allow a comparison with the histological gold standard.
Animal experiments were performed in accordance with the Belgian legislation under the national authorization number LA1210530 and were approved by the Ethical Committee of the Faculty of Biomedical Sciences of KU Leuven.
Contrast-Enhanced Nanofocus Computed Tomography
To enable CE-nanoCT from the 4 mouse knee joint groups, samples were processed as described in Kerckhofs et al.20 Briefly, the knee joint capsules were dislocated to expose the tibial and femoral cartilage layers. Subsequently, all joints were immersed for 30 minutes in a solution of Hexabrix 320 diluted 4/5 in phosphate-buffered saline, then wrapped in parafilm, and were stably positioned in the nanoCT system. As the negatively charged ioxaglate of which Hexabrix 320 is partially composed, will be locally repelled by the anionic glycosaminoglycans (sGAG) in the cartilage, ioxaglate accumulation in the cartilage is inversely related to the sGAG content.22
The nanoCT system applied in this study was a Phoenix NanoTom S (GE Measurement and Control Solutions, Munich, Germany). It was equipped with a tungsten target and was operated, for all scans, at a voltage of 60 kV and a current of 140 µA. A 1-mm filter of aluminum was used. The exposure time was 500 ms and a frame averaging of 1 and image skip of 0 was applied, resulting in a scanning time of 20 minutes per sample. Reconstruction was performed using Phoenix datos|x CT software. The isotropic voxel size of all scans was 2 µm.
Three-Dimensional Image Visualization and Analysis
As described in Kerckhofs et al.,20 to quantify the 3D characteristics of the noncalcified and underlying calcified cartilage layers, the cartilage region was selected using CTAn software (Bruker micro-CT, Kontich, Belgium) by manually drawing a consistent region of interest (ROI) in every 20th cross-section throughout the whole tibial plateau. Briefly, delineating the total cartilage area (i.e., both noncalcified and calcified) was performed based on (a) the difference in grayscale between the noncalcified cartilage and the surrounding (i.e., liquid) and (b) the difference in structure compared with the subchondral bone and noncalcified cartilage for the calcified cartilage (i.e., having a porous structure). The ROI was automatically interpolated for the 19 intermediate cross-sections. After automatic Otsu segmentation,23 the noncalcified and calcified cartilage were segmented and analyzed in 3D by calculating the volume, the thickness distribution,24 and the average thickness. Dataviewer (Bruker micro-CT, Kontich, Belgium) was used on the CE-nanoCT images to perform the modified Mankin scoring for cartilage structure and quality, as well as to quantify the loss in sGAG content.
Histology
After CE-nanoCT imaging, a set of representative knee joints were processed for histology. They were fixed in 10% formalin, decalcified in 4% formic acid, and embedded in paraffin. Sections of 5 µm were made every 60 µm and were stained with Safranin-O and counterstained with Fast Green.
Modified Mankin Scoring
The medial side of the tibial plateau in the histological sections (n = 10 per joint) and coronal CE-nanoCT images (n = 10 with a 60 µm interspacing) was graded using a modified Mankin scoring system8 specifically for cartilage structure and quality. In brief, for both the histological sections and the CE-nanoCT images, articular cartilage structure was graded from 0 to 11. Concerning the cartilage quality, the Safranin-O staining for the histological sections and the Hexabrix staining for the CE-nanoCT images were graded between 0 and 8. For both parameters, grade 0 represented normal/healthy cartilage. A grade of 11 for the cartilage structure was indicative for clefts/loss of articular cartilage extending into the subchondral bone. Grade 8 for cartilage quality implied a severe loss in Safranin-O (or increase in Hexabrix) staining in more than half of the depth of the noncalcified cartilage thickness and involving more than half of the plateau. Grades of each sample (n = 10) were averaged and summed for the 2 scoring parameters, whereby 0 represented completely unaffected hyaline cartilage and the maximum Mankin score of 19 represented a highly degenerated OA knee.
Quantification of the Normalized sGAG content
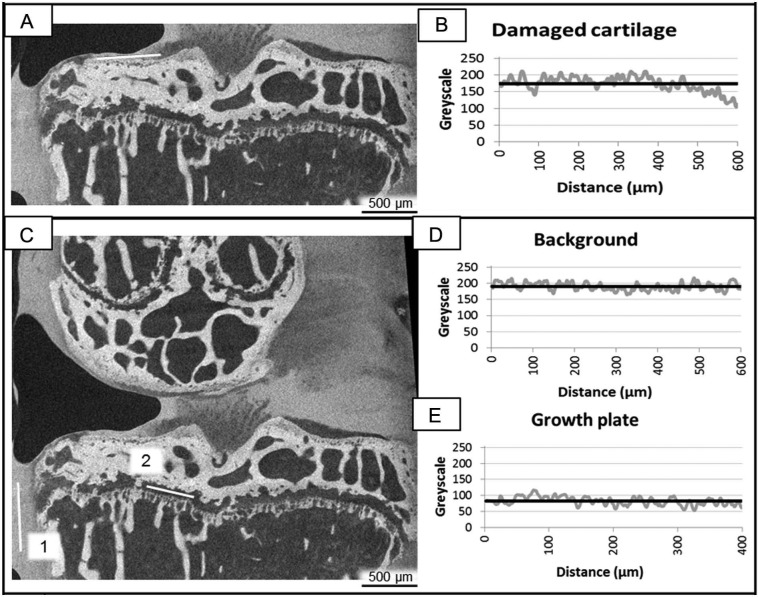
To quantify the sGAG content in the hyaline cartilage, the intensity of the Hexabrix excluded signal in the noncalcified layer of the hyaline cartilage was normalized to the signal of the growth plate cartilage (i.e., in the same coronal CE-nanoCT cross-sections). A grayscale histogram was plotted along an arbitrary 600 µm line through the noncalcified cartilage on the medial side of the tibial plateau (Fig. 1A). The normalized sGAG content in the cartilage compared to the healthy growth plate cartilage was then calculated according to Equation (1). The normalized sGAG content was measured in the same coronal CE-nanoCT cross-sections as used for the modified Making scoring.
Figure 1.
(A) A typical coronal contrast-enhanced nanofocus computed tomography (CE-nanoCT) image and (B) the corresponding grayscale histogram along the white line drawn in (A) through the noncalcified cartilage on the medial (left) side of the tibial plateau (gray curve) with the average grayscale value (black curve). (C) A typical coronal CE-nanoCT image and (D, E) the corresponding grayscale histograms (D) along the white line annotated with “1” drawn in (C) through the background of the image (gray curve) with the average grayscale value (black curve) and (E) along the white line annotated with “2” drawn in (C) through the growth plate cartilage (gray curve) with the average grayscale value (black curve).
with CART the average grayscale value (Fig. 1B) along an arbitrary line through the noncalcified cartilage on the medial side of the tibial plateau (Fig. 1A), BG the average grayscale value (Fig. 1D) along a 600 µm long line through the background in the same image (i.e., liquid, Fig. 1C-1), and GP the average grayscale value (Fig. 1E) along a 400 µm long line through the growth plate cartilage in the same image (Fig. 1C-2).
Partial Least Square Regression Modeling
In this study, a computational model was used to determine whether a combined set of morphological cartilage characteristics could quantitatively and in 3D predict the level of cartilage damage in a comparative manner to histological cartilage pathology scoring (using the modified Mankin scoring as a measure for cartilage structure and quality). A linear modeling formalism was used that was already shown to be capable of relating quantitative contributions of multiple signals to a (single) measured response,25-27 namely partial least square regression (PLSR). PLSR is able to identify the information content within the set of measured characteristics that most closely maps onto the output response (i.e., modified Mankin score). The resulting mapping of lumped signals to corresponding responses allowed to identify the most “important structural cartilage characteristics” predictive for the level of the cartilage damage. PLSR uses linear algebra to reduce the dimensionality of multivariate data sets by defining principal component axes from the original data set that contain the most important information on a specific output variable.28 For the optimization of the number of model dimensions, the root mean squared error (RMSE) between the measured level of cartilage damage and the level predicted by PLSR models with increasing numbers of principal components was investigated. Each cartilage characteristic was individually withheld from the training set to construct a cross-validation model, in which an RMSE of the prediction could be assessed by predicting the withheld sample. The predicted RMSE was minimal in this study when using two principal components. To generate the PLSR model, the software package JMP 8 (SAS Institute Inc., Cary, NC) was used.
Statistical Analysis
Statistical analysis was done using a statistical software add-in for Microsoft Excel for Windows, namely, Analyse-it version 2.26 Excel 12+. To evaluate the significance level between the groups, one-way analyses of variance were used with a confidence level of 95%. All results were expressed as average ± standard deviation.
Results
CE-nanoCT Images Are Equivalent to Histological Sections for the Modified Mankin Scoring
In Kerckhofs et al.,20 it was already shown that CE-nanoCT has a similar discriminative power as histology with Safranin-O staining to differentiate bone, noncalcified and calcified cartilage in mouse knee joints, and to identify and assess local alterations in cartilage sGAG content (i.e., a decrease in sGAG content vs. chondral lesions).
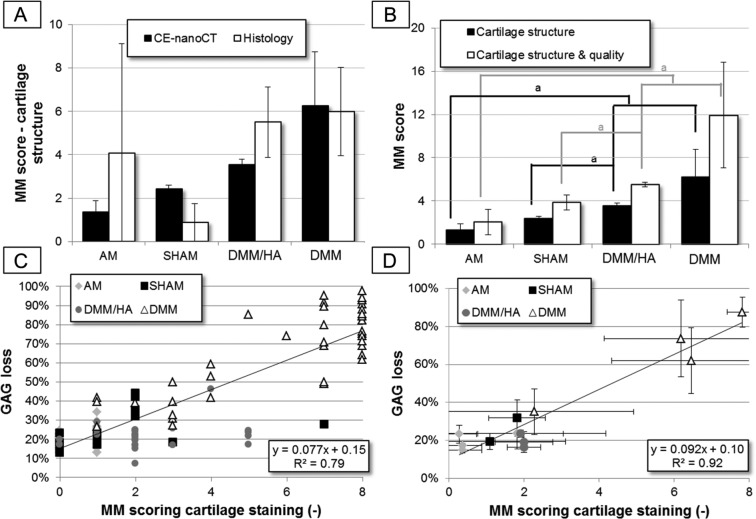
To validate the potency of the CE-nanoCT images to be used for the modified Mankin scores, that is, the gold standard for histological assessment with Safranin-O staining, these scores were determined on the histological sections and on the CE-nanoCT coronal sections (Fig. 2A). Since full histological scoring was only performed on animals that were not imaged by CE-nanoCT, no comparison for the same animal was possible. However, Figure 2A shows that, within one treatment group, there were no significant differences between the histology-based and the CE-nanoCT-based scoring for cartilage structure. This supports the equivalence of CE-nanoCT images to Safranin-O stained histological sections for modified Mankin score determination. The apparent difference between both scores was rather a result of the large variability in manifestations of the symptoms between different animals within one treatment group.
Figure 2.
(A) The modified Mankin (MM) scoring for the cartilage structure, determined on histological and contrast-enhanced nanofocus computed tomography (CE-nanoCT) sections for the different treatments groups. The animals used for the histological scoring (n = 3 or 4 per group) were not the same individuals that than the ones assessed by the CE-nanoCT scoring (n = 3 or 4 per group). (B) The MM scoring of 2-dimensional CE-nanoCT images. The score for the cartilage structure only and the sum for cartilage structure and quality are shown for the different treatments groups. (C) Correlation plot between the percentage of sGAG (anionic glycosaminoglycans) loss normalized to the growth plate cartilage and the MM scoring for the cartilage quality (as measured by Hexabrix staining intensity), both determined on the individual 2-dimensional sectional CE-nanoCT images throughout the different treatments groups and (D) the same correlation plot as (C) but the % sGAG loss and MM score were averaged over all scored sections of each animal (n = 10).
The modified Mankin subscore for the cartilage structure on the 2D CE-nanoCT images, as well as the sum of the cartilage structure and quality (Fig. 2B), showed significant differences between the AM group and the animals that had undergone a DMM procedure (i.e., DMM and DMM/HA), as well as between the sham-operated group and the group that had received a 2-fold injection of hyaluronic acid after DMM (i.e., DMM/HA). This indicates that for the 4 different groups, having a broad spectrum of cartilage defects (i.e., from bad in case of the DMM group to almost unaffected for the AM and sham-operated groups), CE-nanoCT provides valid readouts over this whole range, thus showing the potency of CE-nanoCT to be implemented as a standard technique for evaluation and quantification of cartilage damage in the OA field.
Next, instead of visually scoring in a semiquantitative way the degree of noncalcified cartilage staining in the CE-nanoCT images, as was done for the modified Mankin subscore for cartilage quality, the normalized sGAG loss was quantified by image analysis on the same images (Fig. 2C). Every data point in Figure 2C represents one CE-nanoCT image. For the DMM group, CE-nanoCT images with both a low and a high modified Mankin scoring (and sGAG loss) were present and could be distinguished. Although a strong correlation (R2 = 0.79) was found between this analysis and the cartilage quality score, the latter sometimes varied for the same percentage of normalized sGAG loss. Moreover, for the same cartilage quality score, a range in percentage of normalized sGAG loss was found, thus showing the higher sensitivity of the quantification of the normalized sGAG loss by CE-nanoCT image analysis compared to the modified Mankin scoring for cartilage quality. When plotting the averaged modified Mankin score per animal (n = 10), an even higher correlation was found (R2 = 0.92). Within the DMM group, some animals had a high modified Mankin scoring (and sGAG loss), while there was one animal that had a modified Mankin scoring (and sGAG loss) comparable to animals in the sham and DMM/HA groups. CE-nanoCT scoring thus allows not only to discriminate intact versus highly damaged cartilage (Fig. 2C) but also to detect differences within a particular treatment group (Fig. 2D).
CE-nanoCT Allows 3D Quantification of the Cartilage Structure at the Subtissue Level
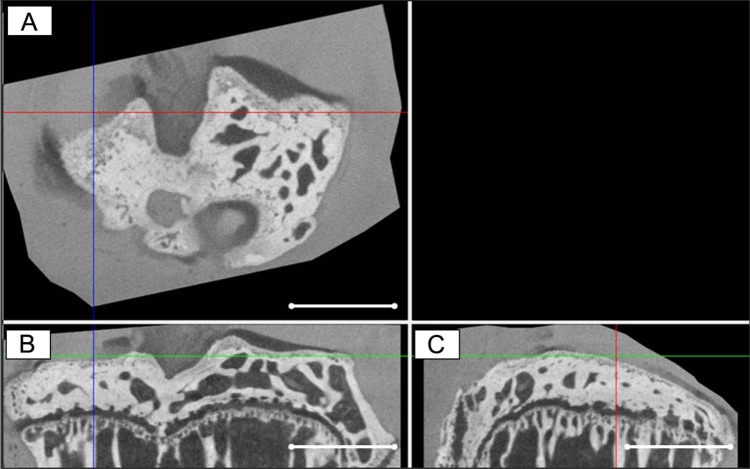
The in vivo medial meniscus destabilization, which serves as a model for OA,21 is characterized by cartilage lesions of various degrees of severities on the medial side (Fig. 3A-C). As shown in Kerckhofs et al.,20 creating 3D CE-nanoCT models of the different cartilaginous layers allowed a straightforward quantification of the defects.
Figure 3.
A representative (A) cross-sectional, (B) coronal, and (C) sagittal contrast-enhanced nanofocus computed tomography (CE-nanoCT) image of a tibial plateau of a mouse of which 8 weeks in vivo medial meniscus destabilization (DMM) resulted in a local chondral defect (indicated by the crossed lines). Scale bars = 1 mm.
While no significant differences were found between the different groups for both calcified cartilage volume (Fig. 4A) and average thickness of the calcified cartilage (Fig. 4B), the overlying noncalcified hyaline cartilage showed significant differences between the DMM group and all other groups.
Figure 4.
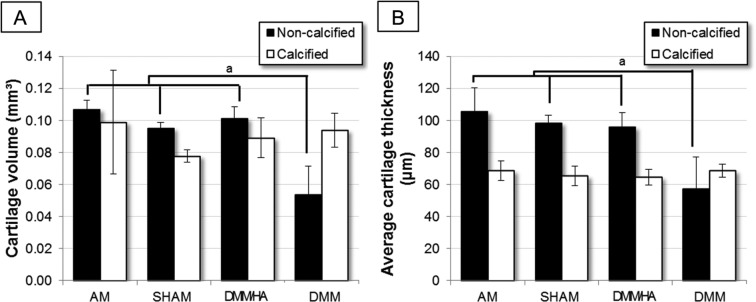
(A) The volume and (B) average thickness of noncalcified and calcified cartilage on the medial tibial plateau of the 4 different treatments groups (n = 3 or 4) (aP < 0.05 = significantly different), based on quantitative analysis of 3-dimensional CE-nanoCT images. The data for the sham and DMM (destabilization of the medial meniscus) group have already been presented in Kerckhofs et al.20
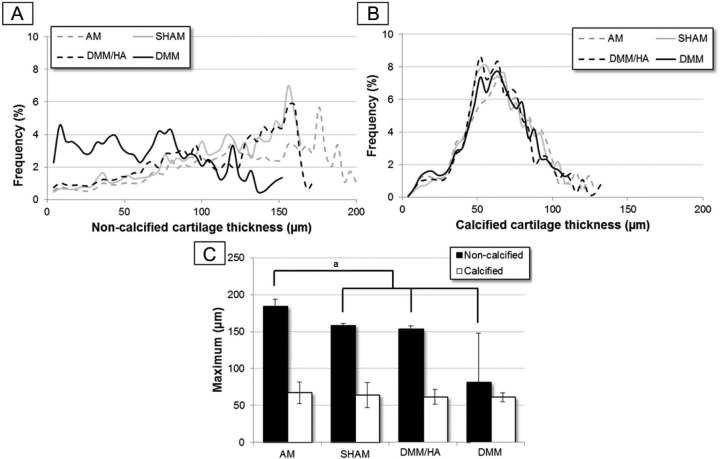
The averaged thickness distribution plots (n = 3 or 4 per group) of noncalcified and calcified cartilage (Fig. 5A and B, respectively) showed similar findings. For the calcified cartilage thickness, the distribution plots for all different treatment groups coincided, and the distribution maxima were not significantly different (Fig. 5C). The noncalcified cartilage thickness of the DMM group displayed a wide spread distribution with a higher frequency of small thicknesses compared with the other groups (Fig. 5A), reflecting a higher presence of thinner cartilage (in particular lesions) and loss of sGAGs throughout the noncalcified cartilage. Consequently, the “maximum of the distribution” plot for the AM group was significantly different compared with the other groups.
Figure 5.
The (A) noncalcified and (B) underlying calcified cartilage thickness distribution on the medial tibial plateau of the different treatments groups (n = 3 or 4). The data for the sham and DMM (destabilization of the medical meniscus) group have already been presented in Kerckhofs et al.20 (C) The “maximum of the thickness distribution” plot averaged per animal group (n = 3 or 4), both for the noncalcified and calcified cartilage layers (aP < 0.05 = significantly different).
Three-Dimensional Cartilage Characteristics Determined by CE-nanoCT Are Predictive for the Cartilage Function and Structure
To determine whether a specific combination of quantitative 3D structural and quality cartilage parameters could quantitatively predict the severity of cartilage damage as determined by the modified Mankin scoring for cartilage structure and quality, a PLSR model was developed. The resulting 2-component PLRS model was used to examine the correlation between the measured modified Mankin score, as was assessed by multiple blinded readers on histological sections, and the predictions for each 3D cartilage parameter (Fig. 6A and B). The PLSR model combined the (a) noncalcified hyaline cartilage volume, (b) maximum of the noncalcified cartilage thickness distribution, and (c) averaged normalized sGAG loss (Fig. 6C). A high predictability of 90.26% was obtained for the model. Including more measured characteristics (e.g., total cartilage volume [noncalcified and calcified cartilage], average thickness of the total cartilage layer, and calcified cartilage volume and average thickness) led to a lower predictability of the model due to inclusion of irrelevant information (noise). Inclusion of less measurement characteristics than the 3 mentioned above also led to a reduction in predictability due to the omission of relevant information.
Figure 6.
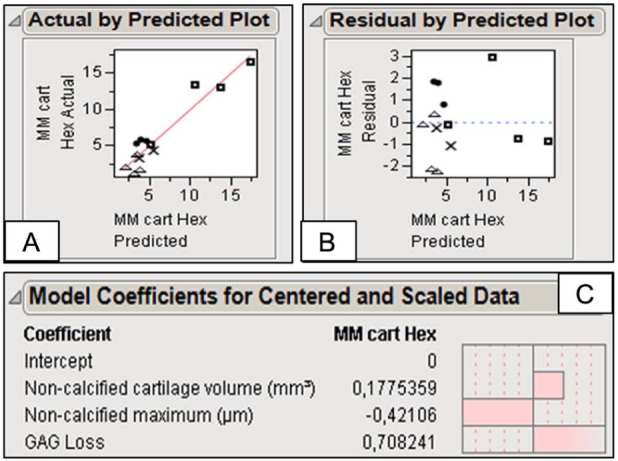
(A) The measured modified Mankin (MM) score versus the cross-validated predictions for each 3-dimensional cartilage parameter, (B) the residuals for the cross-validated predictions, and (C) the three 3-dimensional cartilage parameters that comprise the partial least square regression model, resulting in the following predictive equation: MM scoring = 3.55 + 33.70 * (noncalcified cartilage volume in mm3) − 0.0375 * (maximum of the noncalcified cartilage thickness distribution plot in µm) + 14.41 * (average sGAG loss, nondimensional value).
Discussion
Current diagnostics for OA usually do not discover the disease until it is in advanced stages when joint damage may already have occurred and the patient presents with pain. A method for early diagnosis could open a window of opportunity for prevention, thereby reducing further damage. Measuring subtle alterations in cartilage structure and quality is equally needed to evaluate the (pre)clinical efficacy of new treatment options for cartilage damage. Small animal models of OA are essential and thus quantitative readouts with functional relevance are key for advancing skeletal research. Nowadays, preclinical efficacy of novel therapies for cartilage repair in small animal models is still commonly assessed by histopathology, and quantified by using scoring systems such as the modified Mankin scoring. Such scoring systems are already in clinical use to assess the severity of the cartilage damage. Although validated for being an accurate and user-independent technique,10 histopathology remains a visual, subjective, and nonquantitative scoring approach and might not allow one to distinguish small differences in therapeutic response because of the restricted 2D character.
As demonstrated in Kerckhofs et al.,20 CE-nanoCT provides equivalent images compared to Safranin-O staining on histological sections, with a high contrast between noncalcified and calcified cartilage. In this study, we have shown that this allows performing the modified Mankin scoring also on CE-nanoCT images of a murine knee joint, which can be assessed immediately as it avoids time-consuming and destructive histoprocessing steps, such as decalcification, embedding, or sectioning, which may take several weeks or, for larger species, even months. Moreover, CE-nanoCT generates much more sections throughout the tibial plateau than histology and allows objective and quantitative scoring. Grayscale quantification of the noncalcified cartilage has shown a good correlation with the modified Mankin scoring for cartilage quality, with the advantage of being user independent and quantitative. Although in this study the calculation of the normalized sGAG content was still limited to manual image analysis, by implementation of more sophisticated image analysis algorithms this could evolve toward a semiautomated tool. Therefore, CE-nanoCT is a valuable tool for virtual histopathology. It has to be mentioned that, when the noncalcified cartilage is fully depleted of sGAGs, the CE-nanoCT images might not allow detection of the difference with a cartilage lesion, and histological sectioning might be required. However, as shown by the quantitative measurements of the sGAG loss based on the Hexabrix staining, very low concentrations of sGAGs can still be discriminated from the conditions where cartilage is absent or fully depleted of sGAGs. Additionally, CE-nanoCT does not yet allow visualization of the cartilage at the cellular level. When this information is required, histological sectioning needs to be performed.
CE-nanoCT also allows, because of its 3D character, analysis of different morphological parameters of the bone–cartilage unit.20 Although many studies have used EPIC-CT (i.e., equilibrium partitioning of an ionic contrast agent via microCT) images to quantify 3D cartilage parameters,13-15,19 their correlation to validated histopathological scoring techniques has not been shown. Nor has it been shown which of the measured 3D cartilage parameters should be included to correlate with validated histological scores. In this study, we have shown, using PLSR modeling, that a limited set of easily quantifiable 3D morphological parameters (volume and maximum thickness of the noncalcified cartilage and the averaged normalized sGAG loss) is predictive for the MM scoring of the cartilage structure and quality. These 3D parameters are thus minimally required to draw conclusions on the severity of the damage of the articular cartilage in the knee joint. The model could potentially replace the MM scoring based on visual inspection as the latter does not involve any quantification of cartilage quality or structure. The 3D measurements give more information on size and shape (and thus severity) of the cartilage damage compared with 2D histopathology.
In this study, the analysis of the underlying subchondral bone was not included. However, as microCT images (and thus also CE-nanoCT) do allow for this quantification,5,7 including these analyses in the PLSR model could even enhance the correlation of the CE nanoCT analysis with the gold standard MM scoring above 90%.
Finally, this study assessed 4 treatment groups using CE-nanoCT that were selected because they serve in many studies as control groups to assess the efficacy of a cell therapy for cartilage repair, and they represent a broad range of cartilage defects (i.e., from bad to unaffected). Mice with a destabilized meniscus displayed lesions with different degrees of severity and loss in sGAG content. Twofold injection of hyaluronic acid after DMM (DMM/HA) was proven to be a valid positive control group for this model and follow-up period, since the cartilage characteristics, both determined in 2D and 3D, were similar to the ones of nonoperated or sham-operated animals. CE-nanoCT-based virtual histopathology and 3D morphological quantification of a limited set of 3D cartilage parameters allowed us to distinguish the 4 groups, and thus it is a valid tool to be implemented as a standard technique for the evaluation and quantification of cartilage damage in the OA field.
Conclusions
This study has shown the potential of a noninvasive 3D imaging methodology, namely CE-nanoCT, for virtual 3D histopathology as it provides images equivalent to Safranin-O-stained histological sections of murine knee joints. Because of its noninvasive and 3D character, and because it reveals differences in structure and composition between calcified, noncalcified cartilage, and the underlying bone in small animal models with high spatial and contrast resolution, CE-nanoCT provides faster and more quantitative data on cartilage structure and quality compared with standard histopathology. Moreover, it was shown that a limited set of these quantitative 3D measures is able to reproduce the standard histopathological scoring, with the advantage of being user independent and semiautomated, thus being a potential replacement of the histopathological scoring. To conclude, CE-nanoCT has demonstrated its equivalence to standard histopathology in preclinical small animal studies addressing cartilage-related pathologies and therapeutic interventions.
Footnotes
Acknowledgments and Funding: This study was financed by the Agency for Innovation by Science and Technology in Flanders (IWT/OZM/090655), and acknowledges the European Research Council under the European Union’s Seventh Framework Program (FP7/2007-2013)/ERC grant agreement number 279100. We are very grateful to Gert Vanderlinden, Joyce Op De Beeck, Johan Neys, and Muriel Hemelaers for the excellent technical assistance in the histological sectioning and staining, and in the drawing of the regions of interest in the CE-nanoCT images. This work is part of Prometheus, the Leuven Research & Development Division of Skeletal Tissue Engineering of the KU Leuven: http://www.kuleuven.be/prometheus. The x-ray computed tomography images have been generated on the x-ray computed tomography facilities of the Department of Metallurgy and Materials Engineering at the KU Leuven, financed by the Hercules Foundation (project AKUL 09/001: Micro- and nano-CT for the hierarchical analysis of materials).
Authors’ Note: The biological work of this study was performed and financed by TiGenix NV. The CE-nanoCT imaging and image analysis was done at the Department of Metallurgy and Materials Engineering, KU Leuven, Belgium.
Declaration of Conflicting Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of thisarticle: J.S, M.M and T.V.d.P. are former employees and shareholders of Tigenix NV.
Ethical Approval: Animal experiments were performed in accordance with the Belgian legislation under the national authorization number LA1210530 and were approved by the Ethical Committee of the Faculty of Biomedical Sciences of the University of Leuven.
References
- 1. Bijlsma JW, Berenbaum F, Lafeber FP. Osteoarthritis: an update with relevance for clinical practice. Lancet. 2011;377:2115-26. [DOI] [PubMed] [Google Scholar]
- 2. Gelber AC, Hochberg MC, Mead LA, Wang NY, Wigley FM, Klag MJ. Joint injury in young adults and risk for subsequent knee and hip osteoarthritis. Ann Intern Med. 2000;133:321-8. [DOI] [PubMed] [Google Scholar]
- 3. Messner K, Maletius W. The long-term prognosis for severe damage to weight-bearing cartilage in the knee: a 14-year clinical and radiographic follow-up in 28 young athletes. Acta Orthop Scand. 1996;67:165-8. [DOI] [PubMed] [Google Scholar]
- 4. Maréchal M, Van Hauwermeiren H, Neys J, Vanderlinden G, Van de, Putte T. In vivo evaluation of different surgical procedures for autologous chondrocyte implantation. Cartilage. 2013;4:83-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Christiansen BA, Anderson MJ, Lee CA, Williams JC, Yik JH, Haudenschild DR. Musculoskeletal changes following non-invasive knee injury using a novel mouse model of post-traumatic osteoarthritis. Osteoarthritis Cartilage. 2012;20:773-82. [DOI] [PubMed] [Google Scholar]
- 6. Kozawa E, Nishida Y, Cheng XW, Urakawa H, Arai E, Futamura N, et al. Osteoarthritic change is delayed in a Ctsk-knockout mouse model of osteoarthritis. Arthritis Rheum. 2012;64:454-64. [DOI] [PubMed] [Google Scholar]
- 7. Botter SM, Glasson SS, Hopkins B, Clockaerts S, Weinans H, van Leeuwen JPet al. ADAMTS5-/- mice have less subchondral bone changes after induction of osteoarthritis through surgical instability: implications for a link between cartilage and subchondral bone changes. Osteoarthritis Cartilage. 2009;17:636-45. [DOI] [PubMed] [Google Scholar]
- 8. Furman BD, Strand J, Hembree WC, Ward BD, Guilak F, Olson SA. Joint degeneration following closed intraarticular fracture in the mouse knee: a model of posttraumatic arthritis. J Orthop Res. 2007;25:578-92. [DOI] [PubMed] [Google Scholar]
- 9. Mankin HJ, Dorfman H, Lippiello L, Zarins A. Biochemical and metabolic abnormalities in articular cartilage from osteo-arthritic human hips. II. Correlation of morphology with biochemical and metabolic data. J Bone Joint Surg Am. 1971;53:523-37. [PubMed] [Google Scholar]
- 10. Glasson SS, Chambers MG, Van Den Berg WB, Little CB. The OARSI histopathology initiative—recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthritis Cartilage. 2010;18(Suppl 3):S17-23. [DOI] [PubMed] [Google Scholar]
- 11. Marenzana M, Hagen CK, Das Neves Borges P, Endrizzi M, Szafraniec MB, Ignatyev K, et al. Visualization of small lesions in rat cartilage by means of laboratory-based x-ray phase contrast imaging. Phys Med Biol. 2012;57:8173-84. [DOI] [PubMed] [Google Scholar]
- 12. Xie L, Lin ASP, Levenston ME, Guldberg RE. Quantitative assessment of articular cartilage morphology via EPIC-microCT. Osteoarthritis Cartilage. 2009;17:313-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Piscaer TM, Waarsing JH, Kops N, Pavljasevic P, Verhaar JA, van Osch GJ, et al. In vivo imaging of cartilage degeneration using microCT-arthrography. Osteoarthritis Cartilage. 2008;16:1011-7. [DOI] [PubMed] [Google Scholar]
- 14. Palmer AW, Guldberg RE, Levenston ME. Analysis of cartilage matrix fixed charge density and three-dimensional morphology via contrast-enhanced microcomputed tomography. Proc Natl Acad Sci U S A. 2006;103:19255-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kotwal N, Li J, Sandy J, Plaas A, Sumner DR. Initial application of EPIC-µCT to assess mouse articular cartilage morphology and composition: effects of aging and treadmill running. Osteoarthritis Cartilage. 2012;20:887-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Silvast TS, Jurvelin JS, Tiitu V, Quinn TM, Töyräs J. Bath concentration of anionic contrast agents does not affect their diffusion and distribution in articular cartilage in vitro. Cartilage. 2013;4:42-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bansal PN, Stewart RC, Entezari V, Snyder BD, Grinstaff MW. Contrast agent electrostatic attraction rather than repulsion to glycosaminoglycans affords a greater contrast uptake ratio and improved quantitative CT imaging in cartilage. Osteoarthritis Cartilage. 2011;19:970-6. [DOI] [PubMed] [Google Scholar]
- 18. Gu XI, Palacio-Mancheno PE, Leong DJ, Borisov YA, Williams E, Maldonado N, et al. High resolution micro arthrography of hard and soft tissues in a murine model. Osteoarthritis Cartilage. 2012;20:1011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xie L, Lin ASP, Guldberg RE, Levenston ME. Nondestructive assessment of sGAG content and distribution in normal and degraded rat articular cartilage via EPIC-mu CT. Osteoarthritis Cartilage. 2010;18:65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kerckhofs G, Sainz J, Wevers M, Van de, Putte T, Schrooten J. Contrast-enhanced nanofocus computed tomography images the cartilage subtissue architecture in three dimensions. Eur Cell Mater. 2013;25:179-89. [DOI] [PubMed] [Google Scholar]
- 21. Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage. 2007;15:1061-9. [DOI] [PubMed] [Google Scholar]
- 22. Stewart RC, Bansal PN, Entezari V, Lusic H, Nazarian RM, Snyder BD, et al. Contrast-enhanced CT with a high-affinity cationic contrast agent for imaging ex vivo bovine, intact ex vivo rabbit, and in vivo rabbit cartilage. Radiology. 2013;266:141-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otsu N. Threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9:62-6. [Google Scholar]
- 24. Hildebrand T, Rüegsegger P. A new method for the model-independent assessment of thickness in three-dimensional images. J Microsc Oxford. 1997;185:67-75. [Google Scholar]
- 25. Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646-53. [DOI] [PubMed] [Google Scholar]
- 26. Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448:604-8. [DOI] [PubMed] [Google Scholar]
- 27. Platt MO, Wilder CL, Wells A, Griffith LG, Lauffenburger DA. Multipathway kinase signatures of multipotent stromal cells are predictive for osteogenic differentiation: tissue-specific stem cells. Stem Cells. 2009;27:2804-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Janes KA, Kelly JR, Gaudet S, Albeck JG, Sorger PK, Lauffenburger DA. Cue-signal-response analysis of TNF-induced apoptosis by partial least squares regression of dynamic multivariate data. J Comput Biol. 2004;11:544-61. [DOI] [PubMed] [Google Scholar]