Abstract
Objective:
Lumbar disc degeneration (LDDG), recently reported to have strong genetic determinants, is a major cause of discopathy and lower back pain. However, most studies have only evaluated the effects of a single susceptibility polymorphism. Our purpose was to examine the effect of two susceptibility polymorphism for LDDG in Japanese collegiate athletes.
Design:
We investigated two susceptibility genes for LDDG—cartilage intermediate layer protein (CILP) and asporin (ASPN)—in 516 collegiate athletes and genotyped the risk allele of CILP (1184T/C) and ASPN (D14). LDDG was evaluated using T2-weighted magnetic resonance imaging.
Results:
By using logistic regression analysis, we found that the ASPN D14 allele and CILP genotype were associated with an increased risk of LDDG in male but not female athletes (CILP CT: odds ratios [OR] = 1.77, 95% confidence interval [CI] = 1.07-2.93; CILP CC: OR = 4.38, 95% CI = 1.42-13.54; ASPN D14: OR = 2.17, 95% CI = 1.10-4.28]. We also found that CILP C and ASPN D14 were independent variables. The ORs with more than two risk alleles were largely increased.
Conclusions:
The CILP and ASPN polymorphisms are independent genetic risk factors for LDDG in male but not female Japanese collegiate athletes.
Keywords: disc degeneration, polymorphisms, athlete, CILP, ASPN
Introduction
Lumbar disc degeneration (LDDG) is a degenerative change in the lumbar disc that occurs with aging, affecting from 30% in people younger than 30 years to 50% in those 30 years and older.1 LDDG also occurs in competitive athletes2-5 and is one of the causes of lumbar disc diseases, including lower back pain and discopathy. LDDG is caused by genetic and environmental factors. Cartilage layer intermediate protein (CILP) is abundantly expressed in the intervertebral discs.6 CILP interacts directly with transforming growth factor-β1 (TGF-β1), controlling the TGF-β1-mediated construction of extracellular matrix proteins. Several recent reports have shown that the CILP gene polymorphism (1184T/C) is significantly associated with lumbar disc diseases in Japanese subjects.4-6 Mutated C allele was reported to increase the binding of TGF-β1 and to disturb the regulation of TGF-β1 signaling.
Previous studies have also shown that asporin (ASPN) alleles with 14 aspartic acid repeats (designated D14) in the N-terminal region are associated with osteoarthritis (OA) of the knee.7 Recently, ASPN D14 was also shown to be associated with LDDG in the Asian population.8 ASPN mRNA is abundantly expressed in the articular cartilage, heart, and liver.9 High ASPN mRNA and protein expression were also observed in degenerated lumbar discs, especially in the outer annulus.7,10 Nakajima et al. reported that ASPN inhibits TGF-β1/smad signaling by co-localization with TGF-β and blocking the interaction between TGF-β1 and its receptors.11 Kizawa et al. showed that the ASPN D14 allele results in greater inhibition of TGF-β activity in ATDC5 cells.7 As both CILP and ASPN proteins in some way affect TGF-β signaling, these two susceptible polymorphisms for LDDG may mutually or independently affect the inhibition of TGF-β signaling and LDDG occurrence.
Recently, we showed that there was a significant association between the CILP 1184T/C polymorphism and LDDG occurrence in Japanese collegiate male athletes.4,5 In this study, we first examined whether ASPN D14 is a significant risk factor for LDDG occurrence in collegiate athletes. As a result, we found that ASPN D14 and LDDG occurrence were significantly associated. We then hypothesized that the evaluation of multiple risk alleles of ASPN and CILP shows higher statistical predictability in comparison to only one risk allele, because these two susceptible polymorphisms have similar inhibitory effects on TGF-β signaling.
Materials and Methods
Subjects
We analyzed the relationship between genetic factors and LDDG in 516 (male, 341; female, 175) collegiate athletes from seven types of sports (wrestling, judo, gymnastics, swimming, soccer, American football, and track and field), with an average of more than 5 years of experience. We defined wrestling, judo, soccer, and American football as collision sports and gymnastics, swimming, and track and field as noncollision sports. We obtained written informed consent from all subjects by ethical committees (Permission No. 008-G04).
MRI Examination
The analysis of the distribution of LDDG was based on our previous study.4 We modified the definitions of LDDG presented by Pfirrmann’s classification.12 MRI evidence of LDDG was defined as decreased signal intensity of the intervertebral discs at L1/L2 and L5/S1 in the T-2 weighted midsagittal fast spin-echo images (repetition time, 5,000 ms; echo time, 125 ms). All MRI scans were analyzed and graded independently by two experienced orthopedic surgeons who were blinded to the subject’s status. The grading system for the assessment of LDDG was based on the study of Pfirrmann et al.12 We defined grades 3, 4, and 5 as the LDDG group and grades 1 and 2 as the CONTROL group, as reported by Kaneoka et al.13 If the athlete had multiple degenerations, the highest grade and its location were determined. In the case of any discrepancy in the grading of LDDG between the two orthopedic surgeons, the disagreement was resolved by consensus. The frequency of LDDG in these athletes was 37% (191/516).
Genotyping
DNA was extracted from the buccal cells of the athletes using cotton swabs. The CILP polymorphism was performed as described in our previous study.4 The ASPN D-repeat polymorphism was genotyped by fragment size. The D-repeat region was amplified using the forward primer 5′-TCCTAGACTGGTCTTCTACACT-3′ and the reverse primer 5′-TCTGAGCAATGTACAACTCGTG-3′. The forward primer was 5′ labeled with FAM. The polymerase chain reaction (PCR) cycling reactions, using a thermal cycler (MJ Mini, BioRad, Hercules, CA), were performed at 94°C for 10 minutes, 30 cycles at 94°C for 30 seconds, at 64°C for 30 seconds, and at 72°C for 30 seconds, followed by one cycle at 72°C for 10 minutes. After amplification, the alleles were discriminated using the ABI PRISM 310 genetic analyzer (Applied Biosystems, Foster City, CA) and the fragment analysis GeneMapper V4.0 software (Applied Biosystems).
Statistical Analysis
We used the software R (version 2.8.0) for statistical analyses. The genotype-specific risks were estimated as odds ratios (ORs) and 95% confidence intervals (CIs). Logistic regression analysis was performed to investigate the relationships between LDDG and the two susceptibility genes, as well as with body weight, gender, and collision sport. P values less than 0.05 were considered statistically significant.
Results
Characteristics of the Subjects
The characteristics of the subjects are shown in Table 1. In male athletes, the height, weight, and body mass index of the LDDG group were significantly higher than that of the CONTROL group. There was no significant difference between the LDDG and CONTROL groups in female athletes.
Table 1.
Characteristics of the Subjects.
| LDDG |
CONTROL |
|||
|---|---|---|---|---|
| Variable | Male | Female | Male | Female |
| Age (years) | 19.9 ± 1.2 | 19.9 ± 1.3 | 19.8 ± 1.2 | 19.8 ± 1.1 |
| Height (cm) | 172.7 ± 7.0** | 159.8 ± 6.0 | 170.5 ± 6.6** | 160.3 ± 5.5 |
| Weight (kg) | 74.0 ± 15.6*** | 54.5 ± 6.6 | 68.01 ± 12.2*** | 53.9 ± 5.9 |
| BMI (kg/m2) | 24.7 ± 4.3** | 21.3 ± 2.1 | 23.3 ± 3.6** | 21.0 ± 1.8 |
| Sport experience (years) | 8.4 ± 3.6 | 9.5 ± 3.9 | 7.7 ± 3.1 | 9.6 ± 3.6 |
LDDG = subjects with lumbar disc degeneration; CONTROL = subjects without lumbar disc degeneration; BMI = body mass index.
Unpaired t-test, P < 0.01. ***Unpaired t-test, P < 0.001.
Effect of ASPN D14 Polymorphism on LDDG Occurrence
The association between the ASPN D14 allele and LDDG is shown in Table 2. The frequencies of the genotypes were similar to those of the previous study.8 The frequency of the ASPN D14 allele in the LDDG group was higher than that in the CONTROL group. We found a significant association between the ASPN D14 allele and LDDG (P < 0.01) with an OR of 2.02 (95% CI = 1.21-3.36) in all athletes (Table 2A). By using logistic regression analysis concomitant with the interaction term and the Wald test, we found that weight (OR = 1.03; 95% CI = 1.01-1.05) and ASPN D14 allele (OR = 2.14; 95% CI = 1.1-4.18) were significant risk factors for LDDG (Table 2B). We also confirmed that the effect of D14 was significant only for male athletes (Table 2C).
Table 2.
Association between the Presence of the ASPN D14 Allele and LDDG in 516 Athletes.
A. Frequency of LDDG and ASPN D14 Allele in 516 Athletes.
| LDDG (%) | CONTROL (%) | |
|---|---|---|
| D14+ | 34 (8.9) | 30 (4.6) |
| D14− | 348 (91.1) | 620 (95.4) |
| P value | 0.006** | |
| OR (95% CI) | 2.02 (1.21-3.36) |
OR = odds ratio; CI = confidence interval.
P < 0.01.
B. Logistic Regression Analysis.
| Crude |
Adjusted |
||||
|---|---|---|---|---|---|
| Group | OR | 95% CI | OR | 95% CI | P Value |
| Weight | 1.02 | 1.01-1.04 | 1.03 | 1.01-1.05 | <0.001*** |
| Gender | 0.87 | 0.59-1.27 | 1.48 | 0.89-2.44 | .129 |
| Collision | 1.33 | 0.93-1.9 | 0.94 | 0.62-1.42 | .755 |
| D14 | 2.05 | 1.2-3.52 | 2.14 | 1.1-4.18 | .025* |
| Sex:D14 | 0.9 | 0.28-2.87 | .864 | ||
OR = odds ratio; CI = confidence interval; D14 = allele of ASPN gene.
The formulated logistic model is as follows:
LDDG = exp(β0 + β1W + β2G + β4COL + β5D14 + β6G × D14),
where W, G, COL, and D14 denote weight, gender, collision, and ASPN D14 allele, respectively. βj (j = 1-6) are parameters to be estimated.
P < 0.05. ***P < 0.001.
C. Wald Test.
| Test Statistics | P Value | |
|---|---|---|
| D14 + D14:sex | 1.884 | 0.17 |
The null hypothesis is composed of the following restrictions: D14:β5 + D14:gender:β6 = 0.
β5 and β6 are coefficients in the multinomial logistic model as shown in Table 2B.
Logistic Regression Analysis on LDDG Occurrence in CILP and ASPN Genotypes
We further evaluated the clinical and genetic risk factors for LDDG. The logistic regression model was constructed with six variables, as shown in Table 3. LDDG was significantly correlated with body weight, gender, ASPN D14, CILP CT, and CC. However, collision sports were not correlated with LDDG (Table 3A). Furthermore, the two genotypes had no effect on LDDG occurrence in female athletes (Table 3B).
Table 3.
Logistic Regression Analysis for LDDG in Two Susceptible Genes
A. Logistic Regression Analysis.
| Crude |
Adjusted |
||||
|---|---|---|---|---|---|
| Group | OR | 95% CI | OR | 95% CI | P Value |
| Weight | 1.02 | 1.01-1.04 | 1.03 | 1.02-1.05 | <0.001*** |
| Gender | 0.87 | 0.59-1.27 | 2.1 | 1.14-3.87 | 0.018* |
| Collision | 1.33 | 0.93-1.9 | 0.93 | 0.61-1.42 | 0.723 |
| D14 | 2.05 | 1.2-3.52 | 2.17 | 1.10-4.28 | 0.026* |
| TC | 1.07 | 0.73-1.57 | 1.77 | 1.07-2.93 | 0.027* |
| CC | 2.79 | 1.28-6.06 | 4.38 | 1.42-13.54 | 0.010* |
| D14:gender | 0.89 | 0.28-2.86 | 0.842 | ||
| TC:gender | 0.39 | 0.17-0.92 | 0.032* | ||
| CC:gender | 0.4 | 0.08-2.11 | 0.281 | ||
OR = odds ratio; CI = confidence interval; D14 = allele of ASPN gene; TC = genotype of CILP gene; CC = genotype of CILP gene.
The formulated logistic model is as follows:
LDDG = exp(β0 + β1W + β2G + β3COL + β4D14 + β5TC + β6CC + β7D14 × G + β8TC × G + β9CC × G),
where W, G, COL, D14, TC, and CC denote weight, gender, collision, ASPN D14 allele, CILP TC genotype, and CILP CC genotype, respectively. βj (j = 1-9) are parameters to be estimated.
P < 0.05. ***P < 0.001.
B. Wald Test.
| Test Statistics | P Value | |
|---|---|---|
| D14 + D14:gender = 0 | 4.21 | 0.24 |
| CILP CT + CILP CT:gender = 0 | ||
| CILP CC + CILP CC:gender = 0 |
The null hypothesis is composed of the following restrictions:
D14:β4 + D14:gender: β7 = 0
TC:β5 + TC:gender:β8 = 0
CC:β6 + TC:gender:β9 = 0
β4 to β9 are coefficients in the multinomial logistic model as shown in Table 3A.
We also calculated the ORs of the susceptibility genes by using logistic regression analysis from the result of each genotype. The OR of LDDG occurrence was increased by the risk alleles of the two susceptibility genes (Table 4).
Table 4.
Odds Ratios by Logistic Regression Analysis.
| Risk Alleles | OR | 95% CI | P Value |
|---|---|---|---|
| CILP CT + D14 | 3.84 | 1.61-9.15 | <0.01** |
| CILP CC + D14 | 9.51 | 2.54-35.64 | <0.001*** |
OR = odds ratio; CI = confidence interval.
P < 0.01. ***P < 0.001.
Interaction Curve of Susceptibility Genes
The interaction curve of the two susceptibility genes is presented in Figure 1. Although the proportion of LDDG occurrence increased with the number of risk alleles, there was no significant interaction between these two susceptibility genes (P > 0.05; Fig. 1).
Figure 1.
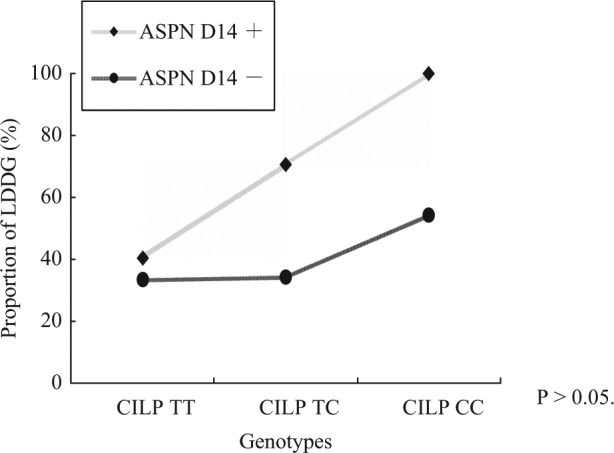
Interaction curve of CILP and ASPN polymorphisms.
There was no significant interaction between these two susceptibility genes.
Discussion
We previously reported that CILP (1184T/C) was significantly associated with LDDG in collegiate male athletes. In this study, we also found a significant association between the ASPN D14 allele and LDDG in collegiate male athletes. Except for gender difference, this finding is consistent with the results in the Asian population as reported by Song et al.8 Although significant interactions of these two susceptibility genes have not been observed, we also found that the addition of the risk alleles CILP 1184T/C and ASPN D14 increased the odds ratio.
The D14 allele of ASPN has been reported as a susceptibility gene for OA of the knee.7 Recently, Song et al. found that ASPN D14 was also associated with LDDG in a Chinese and Japanese cohort.8 We also found that ASPN D14 was significantly associated with LDDG occurrence in collegiate athletes. Song et al. also showed that there was a trend for an association between ASPN D14 and severe LDDG.8 In their study, the Chinese cohort was defined only by radiological abnormality, but the Japanese cohort was dependent on painful disc herniations. In this study, the LDDG group was defined by the MRI findings and the definition was independent of pain. Thus, we think that the ASPN D14 allele is associated with LDDG, independent of symptoms such as pain or discomfort.
It is very important to evaluate whether different susceptibility genes produce a combination effect. We previously found that CILP 1184T/C was significantly associated with LDDG occurrence in collegiate athletes.4,5 In this study, we also found that there was a significant association between ASPN D14 and LDDG. As both these genes are related to TGF-β1 signaling and their high expression was observed in degenerated lumbar discs, we hypothesized that there might be a significant association between them. Surprisingly, we found that CILP C and ASPN D14 were independent variables. Thus, we believe that these two genes affect different portions of the TGF-β signaling pathway. In addition, there is no evidence suggesting that these two molecules interact with each other. As we further confirmed that each additional risk allele increased the odds of LDDG, we think that these two risk alleles have a cumulative effect on the occurrence of LDDG.
We previously reported that CILP 1184T/C was a LDDG-susceptible gene for men, but not women.5 In this report, we also found that ASPN D14 is only a significant risk factor for men. Even in the logit model concomitant with two susceptibility genes, genetic variance was only a significant risk factor for men. As these two proteins interact with the TGF-β pathway, a similar sex dependency might be observed. Valdes et al. found that a haplotype of the CILP gene gave the strongest association in men with knee OA.14 Song et al. reported that there is a significant difference in the allele frequency of ASPN D13 between female OA patients and female controls.15 Although the molecular mechanisms of this phenomenon are ambiguous, we think that the effect of the CILP and ASPN polymorphisms is gender dependent.
We evaluated not only genetic factors but also intrinsic and extrinsic factors, such as gender, weight, and collision. As a result, we found that weight and gender were significant risk factors for LDDG. We previously reported that high body weight is a risk factor for LDDG occurrence in collegiate athletes.4,5,16 We also reported that women are at risk for LDDG occurrence.5 As high body weight causes greater stress on the lumbar disc, it is reasonable that high body weight is significantly associated with LDDG occurrence. On the contrary, it is difficult to fathom why gender difference is a risk factor for LDDG. There are several reports showing that more women complained of lower back pain and hip joint stress than men.17,18 Although the molecular mechanisms are still unclear, we think that the occurrence of LDDG might be gender-dependent after adjusting for body weight, collision, and several genetic factors, as discussed before.
The predictive power of tests can be evaluated by the area under the receiver operating characteristic curve (AUC).19 In this study, we calculated AUC for our logit model. A perfect model would have an AUC of 1, whereas a model with no discriminative power has an AUC of 0.5. The AUC of our model was 0.63, indicative of a poor predictive power. Cauchi et al. evaluated 15 polymorphisms associated with type 2 diabetes and found that a good predictive model would have an AUC of 0.98.20 As in type 2 diabetes, LDDG is a polygenic disease caused by genetic and environmental factors. Additional genetic and clinical information is needed to obtain a highly predictive model.
Conclusion
The present study demonstrated that the CILP and ASPN polymorphisms are independent genetic factors for LDDG in Japanese male collegiate athletes. We also raise the possibility that the relationship between these genetic factors and LDDG occurrence is a gender-dependent phenomenon.
Footnotes
Acknowledgments and Funding: We express our gratitude to the participants of the study. This research was partially supported by Grant-in-Aid for Scientific Research (C), 22500615, 2010, from the Ministry of Education, Science, Sports and Culture, Japan.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: This study does not require institutional review board approval.
References
- 1. Powell MC, Wilson M, Szypryt P, Symonds EM, Worthington BS. Prevalence of lumbar disc degeneration observed by magnetic resonance in symptomless women. Lancet. 1986;2:1366-7. [DOI] [PubMed] [Google Scholar]
- 2. Hangai M, Kaneoka K, Hinotsu S, Shimizu K, Okubo Y, Miyakawa S, et al. Lumbar intervertebral disk degeneration in athletes. Am J Sports Med. 2009;37:149-55. [DOI] [PubMed] [Google Scholar]
- 3. Iwai K, Nakazato K, Irie K, Fujimoto H, Nakajima H. Trunk muscle strength and disability level of low back pain in collegiate wrestlers. Med Sci Sports Exerc. 2004;36:1296-300. [DOI] [PubMed] [Google Scholar]
- 4. Min SK, Nakazato K, Okada T, Ochi E, Hiranuma K. The cartilage intermediate layer protein gene is associated with lumbar disc degeneration in collegiate judokas. Int J Sports Med. 2009;30:691-4. [DOI] [PubMed] [Google Scholar]
- 5. Min SK, Nakazato K, Yamamoto Y, Gushiken K, Fujimoto H, Fujishiro H, et al. Cartilage intermediate layer protein gene is associated with lumbar disc degeneration in male, but not female, collegiate athletes. Am J Sports Med. 2010;38:2552-7. [DOI] [PubMed] [Google Scholar]
- 6. Seki S, Kawaguchi Y, Chiba K, Mikami Y, Kizawa H, Oya T, et al. A functional SNP in CILP, encoding cartilage intermediate layer protein, is associated with susceptibility to lumbar disc disease. Nat Genet. 2005;37:607-12. [DOI] [PubMed] [Google Scholar]
- 7. Kizawa H, Kou I, Iida A, Sudo A, Miyamoto Y, Fukuda A, et al. An aspartic acid repeat polymorphism in asporin inhibits chondrogenesis and increases susceptibility to osteoarthritis. Nat Genet. 2005;37:138-44. [DOI] [PubMed] [Google Scholar]
- 8. Song YQ, Cheung KM, Ho DW, Poon SC, Chiba K, Kawaguchi Y, et al. Association of the asporin D14 allele with lumbar-disc degeneration in Asians. Am J Hum Genet. 2008;82:744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lorenzo P, Aspberg A, Onnerfjord P, Bayliss MT, Neame PJ, Heinegard D. Identification and characterization of asporin. A novel member of the leucine-rich repeat protein family closely related to decorin and biglycan. J Biol Chem. 2001;276:12201-11. [DOI] [PubMed] [Google Scholar]
- 10. Gruber HE, Ingram JA, Hoelscher GL, Zinchenko N, Hanley EN Jr, Sun Y. Asporin, a susceptibility gene in osteoarthritis, is expressed at higher levels in the more degenerate human intervertebral disc. Arthritis Res Ther. 2009;11:R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nakajima M, Kizawa H, Saitoh M, Kou I, Miyazono K, Ikegawa S. Mechanisms for asporin function and regulation in articular cartilage. J Biol Chem. 2007;282:32185-92. [DOI] [PubMed] [Google Scholar]
- 12. Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine. 2001;26:1873-8. [DOI] [PubMed] [Google Scholar]
- 13. Kaneoka K, Shimizu K, Hangai M, Okuwaki T, Mamizuka N, Sakane M, et al. Lumbar intervertebral disk degeneration in elite competitive swimmers: a case control study. Am J Sports Med. 2007;35:1341-5. [DOI] [PubMed] [Google Scholar]
- 14. Valdes AM, Van Oene M, Hart DJ, Surdulescu GL, Loughlin J, Doherty M, et al. Reproducible genetic associations between candidate genes and clinical knee osteoarthritis in men and women. Arthritis Rheum. 2006;54:533-9. [DOI] [PubMed] [Google Scholar]
- 15. Song JH, Lee HS, Kim CJ, Cho YG, Park YG, Nam SW, et al. Aspartic acid repeat polymorphism of the asporin gene with susceptibility to osteoarthritis of the knee in a Korean population. Knee. 2008;15:191-5. [DOI] [PubMed] [Google Scholar]
- 16. Okada T, Nakazato K, Iwai K, Tanabe M, Irie K, Nakajima H. Body mass, nonspecific low back pain, and anatomical changes in the lumbar spine in judo athletes. J Orthop Sports Phys Ther. 2007;37: 688-93. [DOI] [PubMed] [Google Scholar]
- 17. Boyer KA, Beaupre GS, Andriacchi TP. Gender differences exist in the hip joint moments of healthy older walkers. J Biomech. 2008;41:3360-5. [DOI] [PubMed] [Google Scholar]
- 18. Ochsmann E, Rüger H, Kraus T, Drexler H, Letzel S, Münster E. Gender-specific risk factors for acute low back pain: starting points for target-group-specific prevention. Schmerz. 2009;23:377-84. [DOI] [PubMed] [Google Scholar]
- 19. Janssens AC, Pardo MC, Steyerberg EW, van Duijn CM. Revisiting the clinical validity of multiplex genetic testing in complex diseases. Am J Hum Genet. 2004;74:585-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cauchi S, Meyre D, Durand E, Proença C, Marre M, Hadjadj S, et al. Post genome-wide association studies of novel genes associated with type 2 diabetes show gene-gene interaction and high predictive value. PLoS One. 2008;3:e2031. [DOI] [PMC free article] [PubMed] [Google Scholar]


