1. Abstract
Proteomic technologies using mass spectrometry (MS) offer new perspectives in circadian biology, in particular the possibility to study posttranslational modifications (PTMs). To date, only very few studies have been carried out to decipher the rhythmicity of protein expression in mammals with large-scale proteomics. Although signaling has been shown to be of high relevance, comprehensive characterization studies of PTMs are even more rare. This review aims at describing the actual landscape of circadian proteomics and the opportunities and challenges appearing on the horizon. Emphasis was given to signaling processes for their role in metabolic heath as regulated by circadian clocks and environmental factors. Those signaling processes are expected to be better and more deeply characterized in the coming years with proteomics.
Keywords: Circadian rhythms, metabolism, Phosphorylation, Post-translational modifications, SILAC
2. Circadian Biology: Central and Peripheral Clocks
2.1 The Molecular Clock
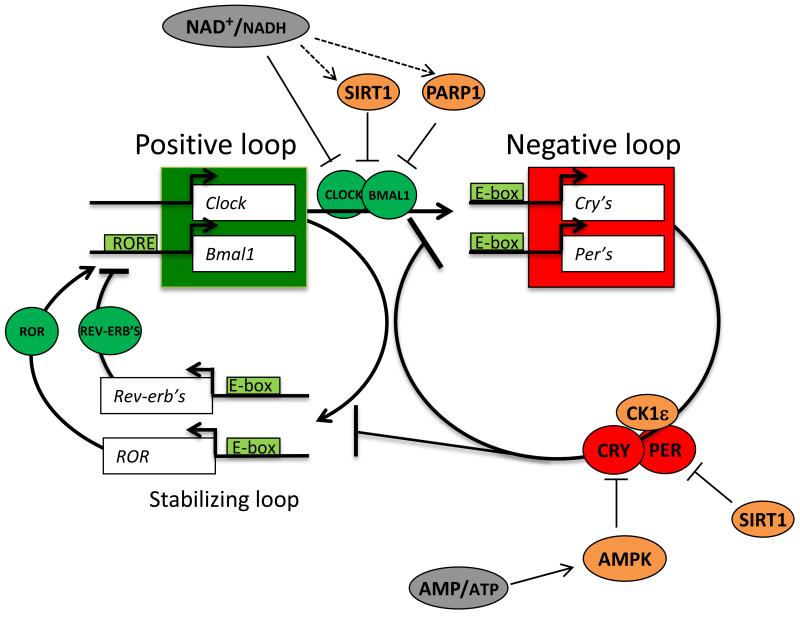
Circadian (from Latin “circa-diem” around a day) clocks operate in all light-sensitive organisms including mammals, but also cyanobacteria, fungi, plants, protozoans and metazoans. Circadian clock allows the organism to adapt to environmental changes on a daily basis. At the molecular level, the cell-autonomous circadian rhythms rely on a signaling network of transcriptional and translational feedback loops. Basically, the activation of a repressor gene results in its later repression by its own protein product that is unstable, thereby starting the cycle again. In mammals the activating loop is composed of CLOCK (circadian locomotor output cycles kaput) and BMAL1 (brain and muscle Arnt-like protein-1) proteins and their homologs. As dimers they activate, via E-box elements, the transcription of the negative feedback loop components, which are PERIOD (PER1-3) and CRYPTOCHROME (CRY1-2). PER and CRY also dimerize which results in the suppression of the CLOCK/BMAL1 activating complex. The CLOCK/BMAL1 complex also activates the expression of an additional loop involving REV-ERB’s and ROR proteins. REV-ERB’s and ROR compete for the access of ROR elements located within the Bmal1 gene promoter. They respectively repress and activate Bmal1 expression thereby reinforcing the oscillator’s stability, the precision and robustness of the circadian clock (for a review see [1]). (Fig.1)
Figure 1. The molecular clock.
Shematic representation of the coupled feedback loop with core clock components of the positive loop (green), the negative loop (red) and their interactions with metabolic regulators. The NAD+/NADH ratio is circadian. It regulates the activity of CLOCK/BMAL1 directly or indirectly (via SIRT1 and PARP1 for example). SIRT1 also promotes PER2 degradation via deacetylation. When activated, AMPK promotes the phosphorylation of CRY directly or indirectly via CK1ε. It leads to the degradation of CRY. See text for details.
2.2 Hierarchical Organization of Molecular Clocks
Based on findings derived from surgical ablation and transplantation of the suprachiasmatic nuclei (SCN), it was thought that the mammalian circadian clock resides only in these bilateral groups of neurons of the brain, which are located in the hypothalamus [2-4]. However, the molecular clock described above exists in most of the cells in the body. Indeed, these molecular oscillators are operative even in established cells lines [5] and, in fact, the clocks are organized in a hierarchical manner with the SCN acting as the “master” clock. The SCN receive light input via the retina and communicate timing with direct (glucocorticoids and innervation by the autonomous nervous system) or indirect (like timing of food intake, temperature changes resulting from activity) signals with peripheral organs acting as “slaves” [6]. As the exact phase of the peripheral clocks varies from organ to organ, under “normal” circumstances the SCN entrain peripheral clocks so that peripheral organs act in a coordinated fashion. On the other hand, during dephasing such as time travel zone, the SCN shift its phase more quickly than the peripheral organs do, resulting in desynchronized internal clock times.
3. Circadian Rhythm and Metabolic Health
When dephasing becomes chronic, as it is the case for rotational shift workers, it deeply disturbs molecular clocks and in the long term, is associated with a wide range of disorders. In humans, perturbations of this clock in shift-workers leads to pathologies including metabolic syndrome [7], vascular diseases [8], and higher risk of cancer [9]. Moreover, sustained and high amplitude rhythms are predictors of weight loss after dietary intervention [10], and survival rate after chemotherapeutic treatment of colorectal [11, 12] or lung [13] cancer patients. One actual objective of the circadian field is to decipher how the (central and peripheral) circadian clocks coordinately regulate metabolism both at the cell-autonomous and organism level without neglecting the effect of environmental factors such as timing and composition of the consumed food.
3.1 Circadian Core Clock and Metabolic Signaling
To be finely tuned, the molecular clock constantly receives feedback from the metabolic signaling in the cell. For example, the CLOCK/BMAL1 complex is known to be regulated by the redox status of the cell reflected by the NAD(P)+/NAD(P)H ratio, which is rhythmic [14]. Indeed, NAD+ and NADH respectively inhibit and stimulate CLOCK/BMAL1 binding capacity to E-Boxes [15]. NAD is a cofactor of SIRT1 and PARP-1. SIRT1 interacts with CLOCK/BMAL1 and mediates BMAL1 and histone H3 deacetylation in order to modulate the transcriptional activity of the complex [16]. SIRT1 also promotes PER2 degradation via its deacetylation [17]. On the other hand, PARP-1 mediated poly ADP-ribosylation of CLOCK leads to the inhibition of CLOCK/BMAL1 DNA binding capacity [18]. Furthermore, BMAL1 can also be activated by ubiquination and SUMOylation [19, 20].
In fact, most of the known clock proteins are phosphoproteins and phosphorylation events play a key role in generating circadian rhythms [21]. AMP-activated protein kinase (AMPK) constitutes an interesting example of interconnection between phosphorylation events of clock components and metabolism status. Indeed, AMPK, which is activated by an elevated AMP/ATP ratio, directly phosphorylates CRY1 [22] and leads to the phosphorylation of PER2 via casein kinase 1 epsilon (CK1ε [23]. In both cases, AMPK activation promotes the degradation of the negative feedback components. (Fig.1) (for further details see [24]).
Clock systems based on post-translational modifications (PTMs), such as phosphorylation, exist and are well characterized in bacteria [25]. Little is known about such systems in mammals, although, clock-based systems have been identified in mammalian cells where transcription and translation is absent [26] (see paragraph 3).
3.2 The Mammalian Clock in Peripheral Organs and Optimized Metabolic Process
Evidence suggests that the mammalian clock in peripheral organs such as the liver plays a substantial role in optimizing the timing of metabolic process. Indeed, the expression of hepatic enzymes involved in metabolism robustly fluctuates during the day [27]. Consequently, inactivating the liver clock decreases the number of cycling transcripts [28].Clock disruption in the liver or in the pancreas impairs glucose metabolism while keeping behavioral rhythms intact [29-31]. Moreover, timing of the circadian clock in tissue is controlled by feeding rather than by the SCN [32, 33]. Remarkably, the rhythmicity of a large part of the rhythmic liver proteome we recently identified persists in clock-deficient mice under night restricted feeding [34].
On top of their role in core clock machinery, clock proteins have additional functions in regulating metabolism. In liver, CRY proteins inhibit fasting-induced gluconeogenesis by influencing glucocorticoid signaling: CRY1 and CRY2 interact directly with the glucocorticoid receptor [35], and also by mediating circadian regulation of cAMP signaling. As a result, the overexpression of CRY1 improves glucose tolerance and insulin sensitivity in diabetic mice [36]. In contrast, Bmal1 KO mice and clock-mutant mice present impaired gluconeogenesis [37] and in liver-specific Bmal1 KO mice, hepatic glucose export is deregulated [29]. The role of BMAL1 is also important for β-cell function [30] and this protein is, like PER2, a regulator of adipocyte differentiation [38, 39]. REV-ERB-α is another regulator of gluconeogenic gene expression [40] and has been shown to have an important role in skeletal muscle formation and adipogenesis [41, 42]. One of its well characterized targets is the sterol regulatory element binding protein (SREBP). Indeed, rhythmic activation of SREBP is the result of both transcriptional regulation orchestrated by the clock [43] and post-translational regulation orchestrated by feeding [44] or clock regulated metabolism [45]. The intersection of the two processes finally results in the rhythmic transcriptional landscape of SREBP [46].
Proteomic technologies offer many perspectives in circadian biology and in particular the possibility to study PTMs. Large-scale proteomic studies can reveal many regulation events at a time, enabling a broader characterization of signal processes and definition of interactions.
4. Circadian Proteomics
To date, most of the large-scale circadian expression studies rely on genomic technologies. Considering the relevance of the rhythmicity of protein expression and regulation by PTMs, the extension of these genomic analyses to large-scale in-depth quantitative proteomic analysis is as essential as it is still missing. Cutting-edge shotgun proteomic approaches using high accuracy and high resolution mass spectrometry (MS) render the study of several thousands of proteins in cell extracts within a few hours of analysis time feasible. Multiplexed isotopic labeling enables relative quantification of several experimental conditions at the same time in one single LC-MS/MS run, with enhanced trueness and precision, and thereby facilitates the revelation of quantitative changes occurring, for instance, during the day and night. Proteomics approaches in circadian biology have recently been reviewed by Robles and Mann [47].
For historical reasons, proteomics roots in and has for a long time relied on 2-dimensional gel electrophoresis (2D-GE) to separate complex protein mixtures and visualize expression pattern changes in different conditions by protein spot staining. As a consequence, many of the circadian proteomic studies reported so far have used 2D-GE associated with MS for identification of the day/night-regulated proteins, although 2D-GE suffers from several limitations such as the restricted coverage of the soluble proteome and the cumbersome, sequential gel processing, image analysis, spot excision, digestion and MS analyses.
Møller and co-workers compared the superficial pineal proteome in rats sacrificed during day time at zeitgeber time (ZT) 6 and during night time at ZT 18 [48]. 60 protein spots showed significant differences between day and night. From these spots, 37 proteins were identified with matrix-assisted laser desorption/ionization (MALDI) time-of-flight (TOF) MS. While up-regulation of proteins during the night accorded with the melatonin secretion of the pineal gland, protein up-regulation during the day deserved and required further investigation towards better understanding of the changes associated to metabolism, protein translation and protein synthesis. Proteomic analysis of the mouse liver was performed using 2D differential gel electrophoresis (DIGE) to compare every 4 hours the pooled soluble proteomes from 6 animals [49]. Across the 2D-DIGE analyses, 642 proteins spots were consistently detected, of which 60 showed significant rhythmicity. Considering isoforms, MS identified 49 rhythmic protein products from 39 unique genes. Aldolase, catalase, and arginase rhythmicity was further validated using Western blotting. The data revealed a complex regulation of circadian processes at transcriptional, post-transcriptional and post-translational level. The same group of researchers conducted a systematic proteomic exploration of the SCN from mice using again 2D-DIGE [50]. Among the 871 protein spots consistently detected across the 4 time points, 115 protein spots displayed significant changes. 53 proteins exhibited significant rhythmic patterns. Protein accumulation happened preferentially during the circadian day than night. Circadian proteins were associated to metabolism, protein trafficking, and synaptic vesicle recycling.
Multiplexed and higher throughput MS-based proteomic workflows, in particular quantitative shotgun proteomics, has offered new perspectives in circadian proteomics where typically many time points need to be compared in order to draw solid conclusions on protein rhythmicity. Martino et al. used surface-enhanced laser desorption ionization (SELDI) and reversed-phase liquid chromatography (RP-LC) tandem MS (MS/MS) after 1-dimensional (1D) GE to follow diurnal protein expression in the blood of mice [51]. The authors did not report a comprehensive characterization of cycling proteins in the blood but their proof-of-concept study stressed the importance to work with accessible and minimally invasive sample types. These can be used for the determination of actual “body time” and may find applications in molecular diagnostics, and clinical and personalized medicine and nutrition (“chrono-nutrition”). More recent studies have fully leveraged modern proteomic technology using large-scale shotgun approaches with extensive sample fractionation before RP-LC MS/MS. Figeys and co-workers used an automated and integrated proteomics platform, so-called AutoProteome, to study the effect of light stimulation on the murine SCN proteome [52]. The AutoProteome workflow can handle minute amounts of protein samples and includes protein pre-concentration, buffer exchange, reduction, alkylation, and digestion based on the principles of the previously reported proteomic reactor [53]. The protein digests were then analyzed with 2D LC-MS/MS, with the first LC dimension consisting of strong-cation exchange separation in ten fractions. The SCN expression proteome of mice receiving a 15-minute light pulse after 2 days exposure to dark was compared to that of mice kept in the dark using spectral counting quantification. From 2131 identified proteins, 387 were shown to be light-dependent. Vasopressin-neurophysin 2-copeptin, casein kinase 1 delta, Ras-specific guanine nucleotide-releasing factor 1, ubiquitin protein ligase E3A, and X-linked ubiquitin specific protease 9 were confirmed to be implicated in clock processes. The distribution of the light-inducible proteins into 155 different pathways and the tight connection between these canonical pathways relevant to circadian rhythm, as bioinformatically determined, underlined the relevance of these findings with regard to mammalian circadian rhythm. Moreover, protein interaction network analysis revealed the high biological correlation between the light-inducible proteins. Of great interest, these identified light inducible signaling pathways such as PI3K/Akt and ERK, were also identified as clock dependent rhythmic signaling pathway in mouse liver [54].
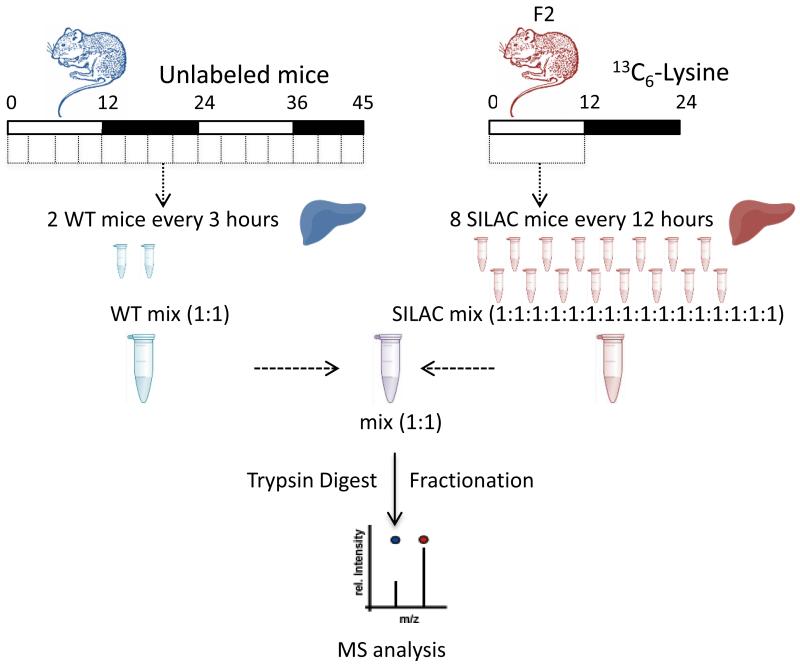
Stable-isotope labelling by amino acids in cell culture (SILAC) presents many advantages in term of quantitative trueness and precision obtained with MS [55]. We have used in vivo SILAC in mice [56] to decipher the diurnal oscillations of the liver proteome (Fig.2) [34]. Mice were fed with a diet containing the 13C6-substituted version of lysine. Labelling was shown to be completed over 2 generations. In the experimental design, pooled SILAC mice served as quantitative reference while normal mice, under restricted feeding during the dark phase, were sampled every three hours during two days. By mixing normal samples with the heavy reference pool, MS-based proteomic analysis of the 16 different time points provided temporal variation of proteins in the collected organ. All protein samples were fractionated using off-gel electrophoresis (OGE) before MS analysis using a hybrid linear ion trap-orbitrap (LTQ-OT) mass spectrometer. In total, 5827 proteins were identified in those experiments, of which 70% were relatively quantified in at least 8 of 16 samples. About 6% of the covered proteome was shown to be rhythmic. These rhythmic proteins preferentially accumulated in the morning and during the night. Half of the rhythmic proteome identified did not display corresponding rhythmic mRNA, appeared clock-independent and was highly enriched in secreted proteins, indicating that feeding behavior might determine the rhythmic functions of circulating proteins in the blood [34].
Figure 2. The circadian SILAC workflow.
Design of the SILAC proteomics experiment. Workflow followed for SILAC MS analysis of mouse liver protein extracts. As described in the text, so-called heavy mice were obtained by feeding the animals with a diet containing the 13C6-substituted version of lysine over 2 generations. Heavy mice were used as quantitative reference to be compared to normal mice, under restricted feeding during the dark phase, which were sampled every three hours during two days. This approach allows following quantitative proteome expression measurement with MS at protein and PTM level over the period under study.
A study by Robles et al. [57] used a comparable in vivo SILAC approach to assess the temporal protein changes in mouse liver in constant darkness condition. Similar conclusions were drawn with respect to the previous study [34] and these are: (i) among 3000 quantified proteins across two consecutive cycles, 6% showed circadian oscillation; (ii) half of the cycling proteome was “delayed” by more than 6 hours with respect to its corresponding rhythmic mRNA; and (iii) the absence of rhythmicity at mRNA level for nevertheless cyclic proteins also suggested post-transcriptional and post-translational mechanisms to play a key role (see next paragraph).
Circadian proteomics is still at its early stage but the field has rapidly grown in the last few years with, for instance, the application of the SILAC technology and shotgun proteomics. In particular, the study of additional tissues and organs other than SCN and liver, as well as body fluids, is expected to complete the circadian proteome landscape, as governed by central and peripheral clocks. Other MS-based quantitative techniques such as isobaric tagging have not yet been but are expected to be applied to the field because of their multiplexing capabilities [58, 59].
5. Circadian Signaling: The Acetylome and Phosphoproteome
In their 2006 study, Reddy et al. pinpointed the relevance of post-transcriptional mechanisms for circadian processes [49]. The authors also predicted PTMs, such as phosphorylation, to play an important role. Indeed, with phospho-protein staining applied to 2D-DIGE, a large proportion of the rhythmic proteins was shown to be phosphorylated. Peroxiredoxin 6 displayed 2 isoforms that were anti-phasic, one being in phase with the transcript. Based on this observation, authors elegantly discover an entirely new post-transcriptional clock with a period of 24 hours that occurs without transcription [26, 60] and is conserved in all domains of life, probably as a readout of rhythmic metabolism [61]. As recently reported, half of the rhythmic proteins identified in the mouse liver could not be explained by the rhythmicity of mRNAs, suggesting that translation and/or protein stability might play a pivotal role in controlling rhythmic protein accumulation [34]. Phosphorylation, acetylation, ubiquitination, and SUMOylation of key circadian proteins have been previously shown to have an important regulatory role [62] (see paragraph 2).
So far large scale PTM proteomics studies of the circadian proteome are very few. Sassone-Corsi and co-workers compared the acetylome of wild type and clock-deficient mouse livers with MS [63]. In total, 179 proteins were found to be acetylated, revealing 306 unique acetylation sites, of which 10% showed rhythmicity. A significant number of rhythmic acetylation sites in the wild-type animals appeared to be abolished in their clock-deficient littermates. At the opposite, acetylation rhythmicity highlighted only in the clock-deficient mice suggested a dominant effect of food intake. Finally, the acetylome was linked to the circadian metabolome previously obtained by the same authors [64]. Correlation between the oscillations in acetylation sites from enzymes and corresponding metabolites was shown to be positive or inverted in equal proportion. The integration of multi-omics analyses was demonstrated to help to a better understanding of the clock regulation and function.
Our group’s phosphoproteomics study based on the SILAC technology and the related workflow were previously described [34]. In our ongoing PTM study, phosphorylated peptides are being enriched with TiO2 magnetic beads and analyzed with RP-LC MS/MS. In order to decipher circadian regulation at protein phosphorylation level we are combining this new phosphoproteome dataset to the proteome data previously obtained, with both sets derived from the same biological samples. Indeed, we do anticipate different regulation scenarios to occur as described in Fig.3. In particular, we aim at identifying rhythmic regulation that occurs at post-translational level and maybe due to specific phosphorylation sites. Specific bioinformatic tools to mine the MS data are key for such analysis. It is worth to consider that, in our experimental design, the reference sample derived from SILAC mice may lack or present very low levels of some phosphorylation events. As a consequence, those events/sites cannot be quantified efficiently over the circadian cycle, despite our state-of-the-art multiplexed proteomic quantification workflow. On top of phosphorylation, the proteomics analysis of additional oscillating PTMs is expected to bring additional insights into the understanding of circadian clock-dependent or -independent rhythmicity.
Figure 3. Regulation at the protein and/or at the PTM site level.
Schematic representation of regulation at the protein and/or at the PTM site level. Our successive proteomic studies enable to link the expression changes at the protein level to those occurring at the PTM site level. This approach is essential to obtain accurate information on signaling processes.
6. Conclusion and Perspectives
Adequate circadian regulation of physiology and metabolism are key for metabolic health. A review and analysis of proteomic analyses in mammals over the day/night cycle reveals mainly GE-based studies at parent protein level. These are increasingly being complemented by more high-throughput, multiplexed LC-MS/MS-based methods that incorporate stable-isotope labeling. Protein signaling events during circadian cycle, as they typically manifest at PTM level, for example phosphorylation and acetylation, are beginning to emerge and expected to provide key molecular insights into these rhythmic regulatory processes. Proteomic and PTM data sets generated from the same biological samples taken across the 24h cycle plus sophisticated correlative bioinformatics are anticipated to be the enablers of such information.
Footnotes
Conflict of interest: The authors have declared no conflict of interest
References
- [1].Abraham U, Granada AE, Westermark PO, Heine M, et al. Coupling governs entrainment range of circadian clocks. Molecular systems biology. 2010;6:438. doi: 10.1038/msb.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain research. 1972;42:201–206. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- [3].Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proceedings of the National Academy of Sciences of the United States of America. 1972;69:1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- [5].Nagoshi E, Brown SA, Dibner C, Kornmann B, Schibler U. Circadian gene expression in cultured cells. Methods in enzymology. 2005;393:543–557. doi: 10.1016/S0076-6879(05)93028-0. [DOI] [PubMed] [Google Scholar]
- [6].Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annual review of physiology. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- [7].Garaulet M, Gomez-Abellan P. Chronobiology and obesity. Nutricion hospitalaria. 2013;28(Suppl 5):114–120. doi: 10.3305/nh.2013.28.sup5.6926. [DOI] [PubMed] [Google Scholar]
- [8].Vyas MV, Garg AX, Iansavichus AV, Costella J, et al. Shift work and vascular events: systematic review and meta-analysis. BMJ. 2012;345:e4800. doi: 10.1136/bmj.e4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Haus EL, Smolensky MH. Shift work and cancer risk: potential mechanistic roles of circadian disruption, light at night, and sleep deprivation. Sleep medicine reviews. 2013;17:273–284. doi: 10.1016/j.smrv.2012.08.003. [DOI] [PubMed] [Google Scholar]
- [10].Bandin C, Martinez-Nicolas A, Ordovas JM, Madrid JA, Garaulet M. Circadian rhythmicity as a predictor of weight-loss effectiveness. Int J Obes. 2013 doi: 10.1038/ijo.2013.211. [DOI] [PubMed] [Google Scholar]
- [11].Innominato PF, Giacchetti S, Bjarnason GA, Focan C, et al. Prediction of overall survival through circadian rest-activity monitoring during chemotherapy for metastatic colorectal cancer. International journal of cancer. Journal international du cancer. 2012;131:2684–2692. doi: 10.1002/ijc.27574. [DOI] [PubMed] [Google Scholar]
- [12].Innominato PF, Giacchetti S, Moreau T, Bjarnason GA, et al. Fatigue and weight loss predict survival on circadian chemotherapy for metastatic colorectal cancer. Cancer. 2013;119:2564–2573. doi: 10.1002/cncr.28072. [DOI] [PubMed] [Google Scholar]
- [13].Sephton SE, Lush E, Dedert EA, Floyd AR, et al. Diurnal cortisol rhythm as a predictor of lung cancer survival. Brain, behavior, and immunity. 2013;30(Suppl):S163–170. doi: 10.1016/j.bbi.2012.07.019. [DOI] [PubMed] [Google Scholar]
- [14].Ramsey KM, Yoshino J, Brace CS, Abrassart D, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- [16].Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Asher G, Gatfield D, Stratmann M, Reinke H, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- [18].Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, et al. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- [19].Cardone L, Hirayama J, Giordano F, Tamaru T, et al. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309:1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- [20].Lee J, Lee Y, Lee MJ, Park E, et al. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Molecular and cellular biology. 2008;28:6056–6065. doi: 10.1128/MCB.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Reischl S, Kramer A. Kinases and phosphatases in the mammalian circadian clock. FEBS letters. 2011;585:1393–1399. doi: 10.1016/j.febslet.2011.02.038. [DOI] [PubMed] [Google Scholar]
- [22].Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Um JH, Yang S, Yamazaki S, Kang H, et al. Activation of 5′-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. The Journal of biological chemistry. 2007;282:20794–20798. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- [24].Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- [25].Johnson CH, Mori T, Xu Y. A cyanobacterial circadian clockwork. Curr Biol. 2008;18:R816–R825. doi: 10.1016/j.cub.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Panda S, Antoch MP, Miller BH, Su AI, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- [28].Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sadacca LA, Lamia KA, deLemos AS, Blum B, Weitz CJ. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia. 2011;54:120–124. doi: 10.1007/s00125-010-1920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Damiola F, Le Minh N, Preitner N, Kornmann B, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- [34].Mauvoisin D, Wang J, Jouffe C, Martin E, et al. Circadian clock-dependent and - independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:167–172. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lamia KA, Papp SJ, Yu RT, Barish GD, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhang EE, Liu Y, Dentin R, Pongsawakul PY, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rudic RD, McNamara P, Curtis AM, Boston RC, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shimba S, Ishii N, Ohta Y, Ohno T, et al. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Grimaldi B, Bellet MM, Katada S, Astarita G, et al. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab. 2010;12:509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yin L, Wu N, Curtin JC, Qatanani M, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- [41].Chawla A, Lazar MA. Induction of Rev-ErbA alpha, an orphan receptor encoded on the opposite strand of the alpha-thyroid hormone receptor gene, during adipocyte differentiation. The Journal of biological chemistry. 1993;268:16265–16269. [PubMed] [Google Scholar]
- [42].Pircher P, Chomez P, Yu F, Vennstrom B, Larsson L. Aberrant expression of myosin isoforms in skeletal muscles from mice lacking the rev-erbAalpha orphan receptor gene. American journal of physiology. Regulatory, integrative and comparative physiology. 2005;288:R482–490. doi: 10.1152/ajpregu.00690.2003. [DOI] [PubMed] [Google Scholar]
- [43].Le Martelot G, Claudel T, Gatfield D, Schaad O, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Brewer M, Lange D, Baler R, Anzulovich A. SREBP-1 as a transcriptional integrator of circadian and nutritional cues in the liver. Journal of biological rhythms. 2005;20:195–205. doi: 10.1177/0748730405275952. [DOI] [PubMed] [Google Scholar]
- [45].Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 2010;11:47–57. doi: 10.1016/j.cmet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- [46].Gilardi F, Migliavacca E, Naldi A, Baruchet M, et al. Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals. PLoS genetics. 2014;10:e1004155. doi: 10.1371/journal.pgen.1004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Robles MS, Mann M. Proteomic approaches in circadian biology. Handbook of experimental pharmacology. 2013:389–407. doi: 10.1007/978-3-642-25950-0_17. [DOI] [PubMed] [Google Scholar]
- [48].Moller M, Sparre T, Bache N, Roepstorff P, Vorum H. Proteomic analysis of day-night variations in protein levels in the rat pineal gland. Proteomics. 2007;7:2009–2018. doi: 10.1002/pmic.200600963. [DOI] [PubMed] [Google Scholar]
- [49].Reddy AB, Karp NA, Maywood ES, Sage EA, et al. Circadian Orchestration of the Hepatic Proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- [50].Deery MJ, Maywood ES, Chesham JE, Sládek M, et al. Proteomic Analysis Reveals the Role of Synaptic Vesicle Cycling in Sustaining the Suprachiasmatic Circadian Clock. Curr Biol. 2009;19:2031–2036. doi: 10.1016/j.cub.2009.10.024. [DOI] [PubMed] [Google Scholar]
- [51].Martino TA, Tata N, Bjarnason GA, Straume M, Sole MJ. Diurnal protein expression in blood revealed by high throughput mass spectrometry proteomics and implications for translational medicine and body time of day. American journal of physiology. Regulatory, integrative and comparative physiology. 2007;293:R1430–1437. doi: 10.1152/ajpregu.00183.2007. [DOI] [PubMed] [Google Scholar]
- [52].Tian R, Alvarez-Saavedra M, Cheng HY, Figeys D. Uncovering the proteome response of the master circadian clock to light using an AutoProteome system. Molecular and cellular proteomics. 2011;10:M110 007252. doi: 10.1074/mcp.M110.007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ethier M, Hou W, Duewel HS, Figeys D. The proteomic reactor: a microfluidic device for processing minute amounts of protein prior to mass spectrometry analysis. Journal of proteome research. 2006;5:2754–2759. doi: 10.1021/pr060312m. [DOI] [PubMed] [Google Scholar]
- [54].Jouffe C, Cretenet G, Symul L, Martin E, et al. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11:e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular and cellular proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- [56].Krüger M, Moser M, Ussar S, Thievessen I, et al. SILAC Mouse for Quantitative Proteomics Uncovers Kindlin-3 as an Essential Factor for Red Blood Cell Function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- [57].Robles MS, Cox J, Mann M. In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS genetics. 2014;10:e1004047. doi: 10.1371/journal.pgen.1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Dayon L, Hainard A, Licker V, Turck N, et al. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Analytical chemistry. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- [59].Ross PL, Huang YN, Marchese JN, Williamson B, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular and cellular proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- [60].O’Neill JS, van Ooijen G, Dixon LE, Troein C, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Edgar RS, Green EW, Zhao Y, van Ooijen G, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends in biochemical sciences. 2009;34:483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Masri S, Patel VR, Eckel-Mahan KL, Peleg S, et al. Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3339–3344. doi: 10.1073/pnas.1217632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]