Abstract
Copy number alterations (CNAs) are among the most common molecular events in human prostate cancer genomes and are associated with worse prognosis. Identification of the oncogenic drivers within these CNAs is challenging due to the broad nature of these genomic gains or losses which can include large numbers of genes within a given region. Here we profiled the genomes of four genetically engineered mouse prostate cancer models that reflect oncogenic events common in human prostate tumors, with the goal of integrating these data with human prostate cancer datasets to identify shared molecular events. Met was amplified in 67% of prostate tumors from Pten p53 prostate conditional null mice and in approximately 30% of metastatic human prostate cancer specimens, often in association with loss of PTEN and TP53. In murine tumors with Met amplification, Met copy number gain and expression was present in some cells but not others, revealing intratumoral heterogeneity. Forced MET overexpression in non-MET amplified prostate tumor cells activated PI3K and MAPK signaling and promoted cell proliferation and tumor growth, whereas MET kinase inhibition selectively impaired the growth of tumors with Met amplification. However, the impact of MET inhibitor therapy was compromised by the persistent growth of non-Met amplified cells within Met-amplified tumors. These findings establish the importance of MET in prostate cancer progression but reveal potential limitations in the clinical use of MET inhibitors in late state prostate cancer.
Keywords: MET, Prostate Cancer, GEM models, secondary genomic alterations, targeted therapy
Introduction
The development of high-throughput genomic platforms has allowed comprehensive profiling of human malignancies with the goal of identifying oncogenic driver events that may impact our understanding of the biology of cancer and ultimately improve patient management. Studies profiling primary and metastatic prostate cancer have identified a number of established and novel oncogenic events in prostate cancer, including loss of the tumor suppressors PTEN and TP53, genomic rearrangements of ERG, amplification of MYC, focal loss of chromosome 3p, and mutations in SPOP (1-3).
Genomic studies efficiently identify oncogenes or tumor suppressors when specific genes are frequently mutated or have focal gain or loss. However, the utility of this approach is limited when broad regions of genomic gain or loss are present because of the large number of genes impacted. Furthermore, these broad genomic gains and losses could simply be a consequence of genomic instability rather than playing a causal role in the cancer. Interestingly, tumors that develop in genetically engineered mouse (GEM) models often do not have the same complexity of copy number alterations as their human counterparts, unless these mice are crossed into strains with genetically unstable backgrounds (4). The relative simplicity of murine tumor genomes provides a convenient opportunity to conduct integrative mouse-human tumor genomic analysis to identify critical human tumor drivers (5, 6).
Several GEM models of prostate cancer have been developed which faithfully recapitulate the oncogenic driving genomic alterations and histopathology of prostate cancer. While these model systems to date have not faithfully recapitulated the metastatic disease process several of these GEM models indeed model genetic events enriched in both primary and metastatic prostate cancer. We elected to use 4 GEM models most representative of the common genomic alterations present in prostate cancer. PB-MYC mice, model amplification and over-expression of MYC observed in human prostate cancer (7). These mice develop high grade prostatic intra-epithelial neoplasia (HGPIN) by 2 months of age which progresses to established invasive adenocarcinoma by 12 months of age. Pten prostate conditional null mice (Ptenlox/lox PB-Cre) develop HGPIN by 2 months of age which progress to intraductal carcinoma by 6 months of age (8). GEM modeling of prostate conditional loss of both Pten and Tp53 (Ptenlox/lox p53lox/lox PB-Cre), two genomic events enriched in metastatic prostate cancer, results in a rapidly progressive invasive carcinoma at 2 months of age and a lethal phenotype secondary to local invasion of adjacent organs by 6 months of age (9). Finally, mouse models displaying prostate conditional loss of Pten and over-expression of ERG, modeling the ERG genomic rearrangements present in 50% of prostate cancers (Rosa-26lox-stop-lox ERG Ptenlox/lox PB-Cre) demonstrate HGPIN by 2 months of age with progression to invasive adenocarcinoma by 6 months of age (10).
Here we applied this integrative mouse-human tumor genomics strategy to prostate cancer. Using these four genetically engineered mouse models that reflect common driver alterations found in human tumors (7-10), we found that the gene encoding the Met receptor tyrosine kinase was frequently amplified in murine prostate tumors initiated by loss of the tumor suppressors Pten and p53. Analysis of publically available genomic data sets of human prostate cancer and revealed MET amplification in approximately 30% of metastatic cases but rarely in primary tumors. Importantly, Met copy number gain and expression in Met-amplified tumors is heterogeneous. Thus, while MET inhibition impairs the growth of MET-amplified tumors, intratumoral heterogeneity compromises long-term therapeutic efficacy. These findings have implications for ongoing clinical trials of MET inhibitors in advanced prostate cancer.
Materials and Methods
Mouse models of prostate cancer
The GEM models of human prostate cancer (PB-MYC, Ptenlox/lox PB-Cre, Rosa-26lox-stop-lox ERG Ptenlox/lox PB-Cre) used in our experiments were maintained in our animal housing facility in accordance with our IACUC protocol and experiments involving Ptenlox/lox p53lox/lox PB-Cre were conducted in collaboration with Zhenbang Chen, PhD at Meharry Medical College in accordance with the IACUC protocol. All mice were genotype according to established protocols (7-10). Tumor tissues were harvested for molecular profiling at 18 months of age for PB-MYC mice, 12 months of age for Ptenlox/lox mice, 6 months of age for ERG Ptenlox/lox mice, and 6 months of age for Ptenlox/lox p53lox/lox mice. The tissue was macro-dissected to enrich for epithelial cancer cells. Genomic DNA and RNA were isolated from 30 mg of prostate cancer tissue using the DNeasy Blood and Tissue Kit (Qiagen) and TRIzol (Invitrogen) extraction followed by purification with the RNeasy Mini Kit (Qiagen). Protein was harvested from prostate cancer specimens by digestion with RIPA buffer.
Cell Lines
The cell lines used in our experiments included the MYC CaP line derived from a PB-MYC mouse by Sawyers’ lab, the CaP8 line derived from a Ptenlox/lox from the Wu lab, the MPC3 line derived from a Ptenlox/lox p53lox/lox from the Chen lab, and the LAPC4 parental line generated by the Sawyers’ lab. These lines have been maintained by our lab. LAPC4 cells have been authenticated using DNA HapMap genotyping. The MYC CaP, CaP8, and MPC3 lines have been authenticated by DNA genotyping of the transgene and recombination of lox sites.
Molecular profiling
Array CGH
DNA was isolated, quantified using the Thermo Scientific NanoDrop 1000 Spectrophotometer, and submitted to our Genomics Core Laboratory for array CGH analysis for 16 PB-MYC mice, 11 Ptenlox/lox mice, 7 ERG Ptenlox/lox mice, and 18 Ptenlox/lox p53lox/lox mice. Additionally, DNA prepared from the GEM model derived cell lines, MYC CaP (PB-MYC), CaP8 (Ptenlox/lox), and MPC3 (Ptenlox/lox p53lox/lox) were submitted for CGH analysis. Reference DNA was prepared from the prostates of genotype/strain matched wild-type littermate mice. All DNA met the requirements of an A260/280 ratio of 1.6 – 1.8 and a concentration greater than 150 ug/ul. Three micrograms of tumor and reference DNA was digested and labeled by random priming using the Bioprime kit (Invitrogen). Labeled DNA was hybridized to the mouse Agilent 244K CGH array and the slides were scanned and images quantified using Feature Extraction 9.1 (Agilent). Raw data from the Agilent mouse 244K CGH array were normalized as previously described and probe level data was segmented with Circular Binary Segmentation and analyzed with RAE.
Transcriptome profiling
Total RNAs were isolated and quantified using the Thermo Scientific NanoDrop 1000 Spectrophotometer. RNA samples with an A260/280 ratio of >1.8 and concentration greater than >20 ng/ul were submitted to our Genomics Core Laboratory from prostate tumors profiled by CGH of 5 PB-MYC mice, 5 Ptenlox/lox mice, and 14 Ptenlox/lox p53lox/lox mice. Quality of the RNA was assessed by the RNA 6000 picoAssay and a RIN number > 7.0 was considered adequate for labeling. A total of 300 ng of RNA was labeled and hybridized on the Illumina MouseRef-8 v2 bead arrays. Raw data were Log2 transformed and normalized. Gene Set Enrichment Analysis (GSEA) was performed to determine gene sets disproportionally over- or under-expressed in groups stratified by specific copy number alterations. Hierarchial clustering was carried out on genes meeting the criteria of greater than 3 fold change across more than two samples using Pearson correlation with pairwaise complete linkage.
FISH analyses
FISH analysis was performed on formalin fixed paraffin embedded (FFPE) sections using a two-color probe mix. The probe mix consisted of BAC clones containing the full length Met gene (Locus 6qA2; clones RP23-173P9 and RP23-444N4; labeled with Red dUTP) and a proximal centromeric locus which served as the control (locus 6qA1; clones RP23-258F1 and RP23-355D10; labeled with Green dUTP). Probe labeling, hybridization, post-hybridization washing, and fluorescence detection were performed according to standard procedures. Slides were scanned using a Zeiss Axioplan 2i epifluorescence microscope equipped with a megapixel CCD camera (CV-M4+CL, JAI) controlled by Isis 5.2 imaging software (Metasystems Group Inc, Waltham, MA). Entire sections were scanned under 63× objective to assess heterogeneity for Met copy number or amplification and in each representative region, a minimum of 50-100 nuclei scored. Amplification was defined as, Met:6qA1(Control) ratio of >2.2 or >10 copies of Met independent of control locus. Cells with 3~6 copies and 7~10 copies of Met & control locus were considered to be polysomic and high-polysomic respectively.
Human prostate cancer molecular profiling
The human prostate cancer data sets used herein have been previously published (3). The complete genomics dataset and analytic methods is reported separately and is available at: http://cbio.mskcc.org/cancergenomics-dataportal/. The CGH and expression array data was analyzed to evaluate copy-number alteration and expression changes of MET, JUN, YAP1, HGF. Amplification is defined as gain of two or more copies. Data was analyzed independently for primary tumor and metastatic lesions. Student’s t-test was used to identify association between copy number alteration of HGF and MET. GSEA was performed as previously described above.
In vitro experiments
In vitro experiments were conducted using the LAPC4 cell line. The LAPC4 cell lines were infected and selected with RFP control and MET expressing virus and selected with puromycin. Proliferation assays were conducted by plating 1×105 cells per well of a 12 well cell culture plate and counted using Cell Titer Glo on Days 1, 3, and 5. HGF stimulation of LAPC4 cells was conducted over a time course with 50 ng of HGF. Crizotinib (Pfizer), a MET inhibitor, was obtained through a materials and transfer agreement with Pfizer and administered to LAPC4 cells at defined concentrations for 24 hours for protein evaluation and as part of a cell proliferation assay. Cell lysates for western blot analysis were prepared using standard RIPA buffer. All experiments were conducted in triplicate and standard deviations were reported.
Pre-clinical in vivo studies
Xenograft Model
For xenograft experiments, 1×106 LAPC4-RFP and LAPC4-MET cells were injected into the bilateral flanks of SCID mice (10 mice per group) and tumor volumes were measured weekly over 28 days. At the end of study mice were euthanized and tumor tissue was procured for FFPE. Representative slides were evaluated by H&E staining and used for immunohistochemical analyses.
GEM prostate cancer transplant model
Prostate tumors from Pten p53 null mice were harvested at 6 months of age, dissected into 2 × 2 × 2 mm cubes and transplanted into the unilateral flank of athymic mice. Growth of these tumors was demonstrated by measuring tumor volume over 16 days. Animals were euthanized and specimens were procured for FFPE. Transplanted mice were randomized to receive vehicle control or crizotinib (Pfizer, 50 mg/kg/day) and tumor volumes were measured weekly over 21 days. At the end of study mice were euthanized and tumors were procured by FFPE for H&E and immunohistochemical staining. Similar studies were conducted using our Rosa-26-ERG Ptenlox/lox mice, which by aCGH do not demonstrate copy number gain of Met.
Immunohistochemical and western blotting antibodies
The antibodies used for western blot analysis and immunohistochemistry were pAKT Ser473 (Cell Signaling Technology, 1:1000 dilution), pAKT Thr308 (Cell Signaling Technology 1:500 dilution), AKT (Cell Signaling Technology, 1:1000 dilution), pS6 Ser240/244 (Cell Signaling Technology, 1:1000 dilution), pERK Thr202/Tyr204 (Cell Signaling Technology, 1:1000 dilution), ERK (Cell Signaling Technology, 1:1000 dilution), pMET Tyr1234/1235(Cell Signaling Technology, 1:1000 dilution), MET (Cell Signaling Technology, 1:1000 dilution), MET (Santa Cruz Biotechnology, 1:200 dilution) and Actin (Cell Signaling Technology, 1:1000 dilution). All immunohistochemical analyses were conducted by the MSKCC Molecular Cytology core.
Accession number
NIH NCBI Gene Expression Omnibus (GEO)
Agilent aCGH GEO accession number is GSE61382
Illumina microarray expression GEO accession number is GSE61379
Results
Recurrent amplification of Met in Pten/ p53 null murine prostate tumors
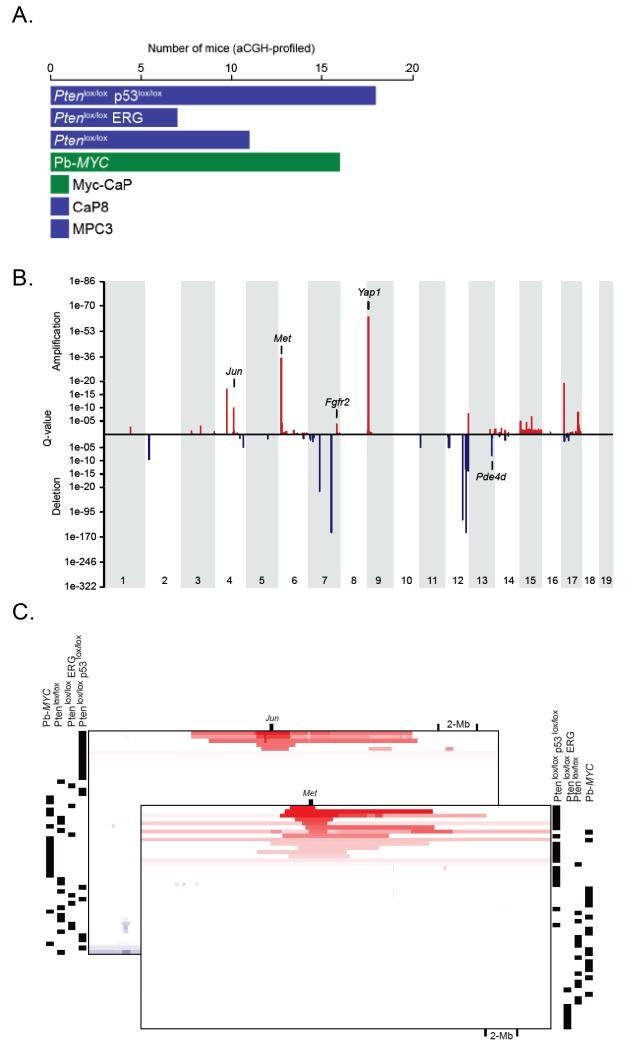
Prostate cancer specimens harvested from PB-MYC, Ptenlox/lox, Rosa26-ERG Ptenlox/lox, and Ptenlox/lox p53lox/lox mice were profiled by array CGH (Agilent) to identify secondary acquired genomic alterations (Figure 1A, Table S1). Copy number alterations (CNAs) were analyzed using the RAE method and revealed recurrent amplifications and deletions across a number of chromosomal regions (Figure 1B, Table S2). Unsupervised clustering of CNAs demonstrated that the majority of genomic changes were in specimens derived from Ptenlox/lox p53lox/lox mice (Figure S1). Regions of focal amplifications spanning known or putative oncogenes included 4qC5 (Jun), 6qA2 (Met), 7qF3 (Fgfr2), 9qA1 (Yap1, Mmp3, Mmp7) (Figure 1B, 1C). Copy number gains spanning Met and Jun were focal in some tumors (Figure 1C). Overall, 12/18 (67%) prostate cancer specimens from Pten p53 null mice demonstrated gain of Met. Additionally, broad regions of amplification were observed for mouse chromosome 5 and 15 which are syntenic for human chromosome 7 and 8q and are broadly amplified in human prostate cancer (Figure S1, S2). Recent work from Ding et al reported that re-expression of mTert in the prostates of Pten p53 null mice resulted in the accumulation of secondary genomic alterations (4). In a direct comparison of our copy number data with their published data, the CNAs were significantly concordant with the exception of 3 chromosomal regions (Figure S2). Therefore, mTert over-expression is not required for the development of secondary genomic alterations in this model, likely due to the impact of p53 loss.
Figure 1. Genomic profiling of GEM models of prostate cancer.
(A) Prostate tumor specimens were harvested from 6 PB-MYC mice, 11 Ptenlox/lox mice, 7 ERG Ptenlox/lox mice, and 18 Ptenlox/lox p53lox/lox mice and analyzed by aCGH. Additionally, cell lines derived from the GEM models, MYC CaP (PB-MYC), CaP8 (Ptenlox/lox), and MPC3 (Ptenlox/lox p53lox/lox) were analyzed by aCGH. (B) RAE analysis of copy number changes from GEM models of prostate cancer showing focal regions of copy number gains and losses. (C) Integrative genomics view of focal amplification of Met and Jun across the various GEM models of prostate cancer analyzed by aCGH.
Enrichment of MET/HGF copy number gain in castration resistant human prostate cancer
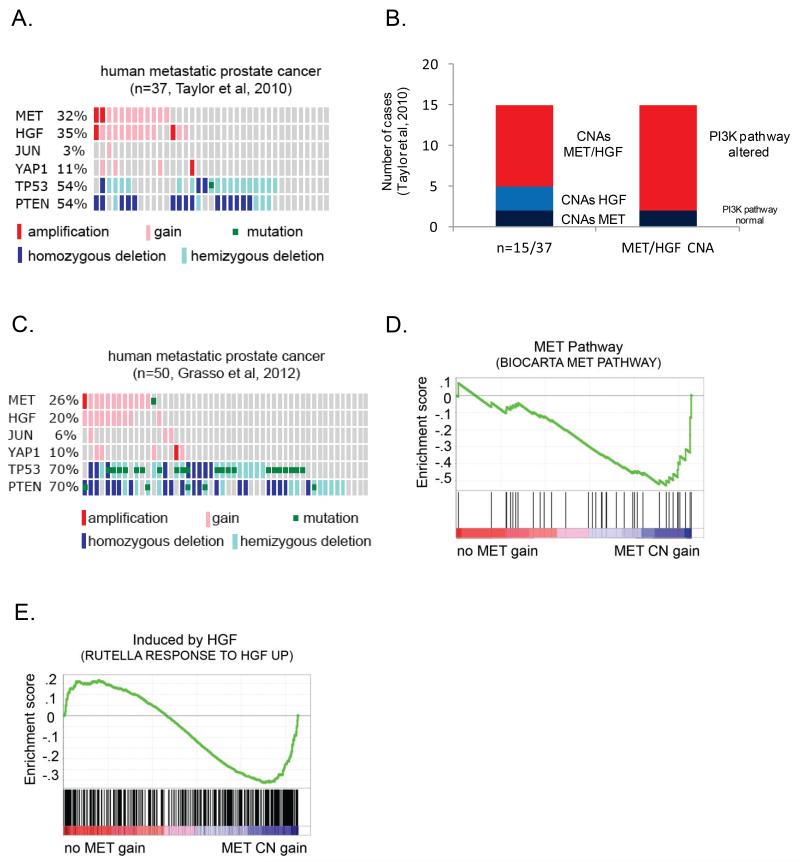
Several genomic profiling studies of human prostate cancer failed to recognize MET amplification as a frequent event (2, 3). Based on the association of Met amplification with Pten/ p53 loss in murine prostate cancer, we reanalyzed the copy number data from these studies, taking a focused look in metastatic prostate cancer cases when PTEN and TP53 are frequently deleted. Remarkably, copy number gain involving MET was present in 32% of metastatic prostate cancer (3). MET amplification was significantly associated with concomitant copy number gain of the MET ligand HGF (present in 35% of all cases and in 83% of MET amplified cases; p-value <0.001) (Figure 2A). 13/15 (87%) of cases with MET/HGF gain also had alterations of the PI3K pathway (PTEN loss, INPP4B loss, PHLPP1/2 loss, AKT1/2/3 copy number gain, PIK3CA/B copy number gain/mutation) (Figure 2B). These findings were validated in an independent dataset of castration resistant metastatic prostate cancers (Figure 2C) (2). MET gain or mutation was present in 25% of cases and was significantly associated with gain of HGF (p<0.001). Importantly, cases with MET amplification were enriched for gene expression signatures of MET pathway activation (FDR<0.1) and HGF stimulation (FDR<0.1), signifying that tumors with MET amplification are associated with a biologic signature of MET activation (Figure 2D, 2E). In contrast to metastatic disease, copy number gain of MET or HGF were rarely observed (4%) in primary prostate cancers.
Figure 2. MET and HGF copy number gains are frequent events in metastatic prostate cancer.
(A) Frequency of MET, HGF, JUN, and YAP1 copy number gain, and PTEN, TP53 copy number loss in metastatic prostate cancer specimens (Taylor et al, 2010). (B) Bar graph demonstrating the frequency of concomitant MET and/or HGF copy number gains and associated alterations in the PI3K signaling pathway (Taylor et al, 2010). (C) Frequency of MET, HGF, JUN, and YAP1copy number gain, and PTEN, TP53 copy number loss in castrate resistant metastatic prostate cancer specimens (Grasso et al, 2012). (D) and (E) Metatstatic prostate cancer specimens (Taylor et al, 2010) stratified by MET CNAs demonstrated enrichment for gene signatures of MET pathway activation.
Since copy number gain of HGF was associated with MET amplification in human metastatic prostate cancer, we asked if this same association was present in mouse tumors. 3 of 11 mouse tumors with Met amplification had regional gain of Hgf and others had increased expression of Hgf in the absence of Hgf gene amplification (Figure S3A). Interestingly, 2 tumors had copy number gain of Hgf without Met amplification.
We also found that two other focally amplified genes in mouse tumors, Jun and Yap1, were also amplified in 3-5% and 10-11% of human metastatic prostate cancers, respectively (Figure 2A, 2C). Thus analysis of murine prostate cancer genomes resulted in the identification of molecular events in human prostate cancer that are easily overlooked by “human only” genome studies.
Met amplification is heterogeneous within individual tumors
To evaluate the impact of Met amplification on Met expression, protein and RNA were harvested from our murine prostate tumor specimens previously analyzed by array CGH. Expression analysis (Illumina) was performed in a subset of prostate cancer specimens analyzed for CNAs. As expected, unsupervised clustering of genes differentially regulated across the GEM tumor specimens was primarily driven by genotype (Figure S3B). In accordance with the loss of PTEN and TP53 being enriched in human metastatic prostate cancers, modeling loss of Pten and p53 in GEM models of prostate cancer revealed enrichment of gene signatures associated with metastasis, although these murine tumors do not display a metastatic phenotype (Table S3). While this is likely secondary to differences in the biology of mouse and humans, and potentially the acquirement of secondary drivers of metastasis, our study reveals that there is molecular similarity when modeling metastatic alterations in primary murine prostate cancers, and thus may provide a pre-clinical model system to study therapies targeting metastatic prostate cancer.
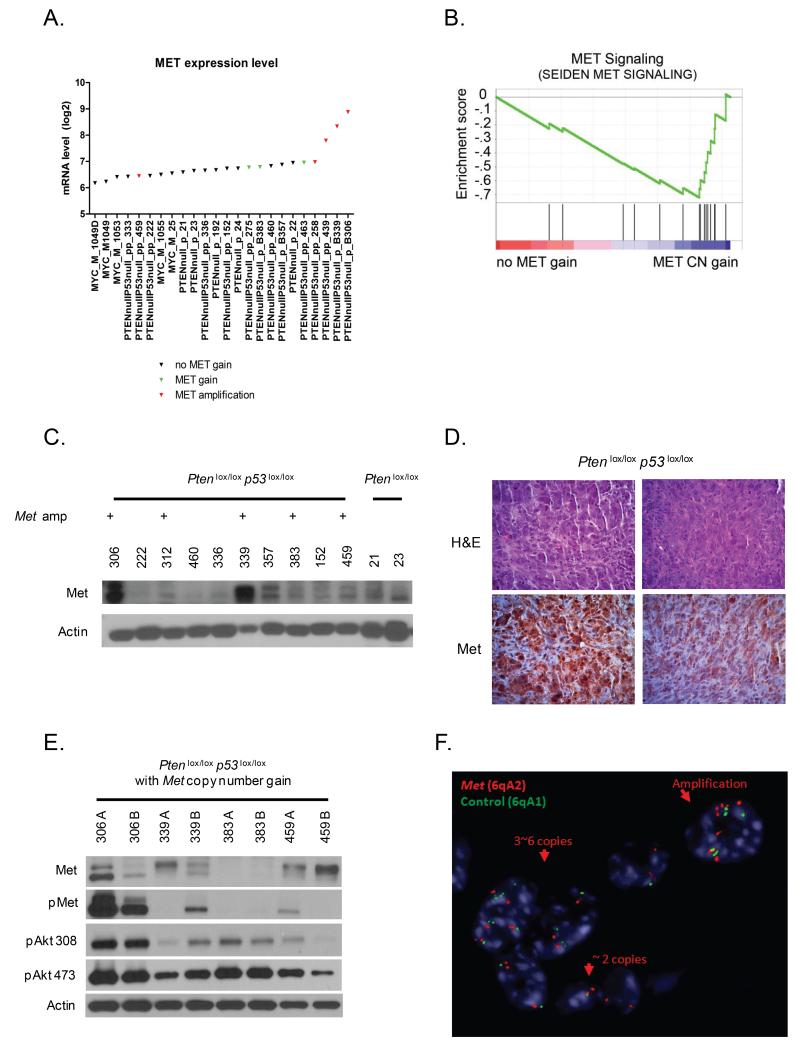
Tumors with high level Met copy number gain had increased Met gene expression when compared to mice without high level copy number gain (Figure 3A, p=0.0002) and were enriched by GSEA for a Met signaling gene signature (Figure 3B, FDR<0.15). Western blot analysis of the prostate cancer specimens from Pten p53 null mice revealed up-regulation of Met in some but not all cases with copy number gains (Figure 3C). We hypothesized that the lack of increased MET protein expression in some tumors with Met copy number gain might be explained by tumor heterogeneity. To evaluate this, we initially performed immunohistochemistry for Met in Met amplified cases and observed heterogeneous Met expression both within and between different tissue sections of the same tumor (Figure 3D). We then analyzed different regions of tumor specimens from Pten p53prostate condition null mice harboring Met amplification by western blot analysis and observed inter- and intra-tumor variability in Met protein expression (Figure 3E). Downstream PI3K signaling, as measured by phosphoAKT levels, correlated with levels of Met phosphorylation. To directly evaluate intra-tumor heterogeneity with regards to Met copy number alterations, we conducted FISH analysis. In concordance with Met expression findings, we also observed intra-tumor heterogeneity in Met copy number gain (Figure 3F). Of 8 tumors analyzed, 2 were primarily diploid for Met, 3 showed foci of low-level copy number gain, and 3 showed foci of high-level copy number gain (amplification).
Figure 3. Genomic gain of Met is a heterogenous selected secondary event.
(A) Met mRNA expression was annotated by Met copy number status for individual mice. Met mRNA up-regulation was observed in individual cases with Met amplification. (B) Pten p53 null prostate tumor specimens stratified by Met copy number gain demonstrated enrichment for gene signatures of MET pathway activation. (C) Western blot analysis of prostate cancer specimens from Pten p53 null mice demonstrate up-regulation of Met in individual cases associated with high-level copy number gains. (D) Heterogeneity of Met expression by immunohistochemistry. (E) Western blot analysis of Met amplified tumor specimens from Pten p53 null mice demonstrating heterogeneity of Met expression between and within individual tumors. (F) Representative FISH analysis for Met copy number in tumors from Pten p53 null mice demonstrating copy number heterogeneity. Red color = Met, Green color = control, Blue color = centromeres, heterochromatin of mouse chromosomes.
Overexpression of MET activates PI3K and enhances proliferation
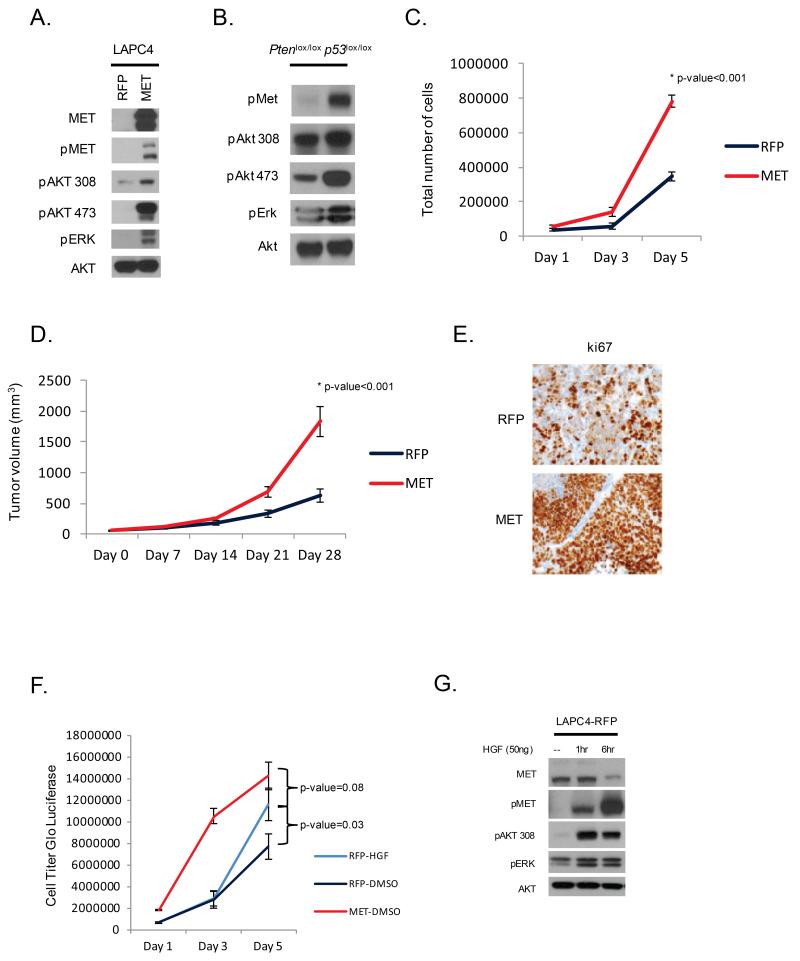
MET plays a critical role in regulating cellular pathways influencing cell proliferation, cell migration, invasion and morphogenesis (11-17). To determine the effect of MET overexpression in prostate tumors, we engineered LAPC4 cells, which lack MET amplification, to express elevated levels of MET. MET over-expression resulted in activation of the PI3K and MAPK downstream signaling pathways as measured by pAKT and pERK (Figure 4A). Concordant with these findings, Met activation was associated with increased pAKT in prostates from Pten p53 null mice (Figure 4B). Over-expression of MET in LAPC4 cells also promoted cell proliferation in vitro (p-value<0.001) (Figure 4C). To explore the impact of MET overexpression in vivo, LAPC4 xenografts were established in SCID mice. Tumors derived from MET over-expressing cells grew faster than controls (p-value<0.001) (Figure 4D), consistent with increased staining for Ki67 by IHC (Figure 4E).
Figure 4. MET up-regulation is associated with PI3K signaling and cell proliferation.
(A) Western blot analysis of LAPC4 cells engineer to over-express MET demonstrates active downstream kinase signaling through PI3K. (B) Western blot analysis of prostate tumors from Pten p53 null mice reveals MET activation is associated with increased PI3K signaling. (C) MET over-expression increases LAPC4 cell proliferation compared to vector control. (D) LAPC4-MET cells grafted into SCID mice (n=10) demonstrate increased tumor growth in vivo compared to vector control (n=10) (E) with associated increased ki67 staining on immunohistochemistry. (F) Stimulatoin with HGF promoted cell proliferation and (G) downstream kinase signaling in LAPC4-RPF cells.
HGF, the only known ligand of MET, has become increasingly recognized as a mediator of disease progression even in the absence of MET or HGF amplification. For example, autocrine or paracrine stimulation of MET by HGF confers resistance to tyrosine kinase inhibitors (18-20). Given that in human prostate cancer we observed a small subset of cases with copy number gain of HGF without gain of MET, we asked if excess HGF was sufficient to promote cell proliferation. Stimulation with HGF promoted downstream signaling and proliferation in the setting of endogenous low levels of Met expression (p-value=0.03) (Figure 4F, 4G). Furthermore, stimulation of MET low cells with HGF conferred a growth advantage nearly comparable to that seen with MET over-expression (p-value=0.08).
MET kinase inhibitors impair the growth of Met-amplified tumors
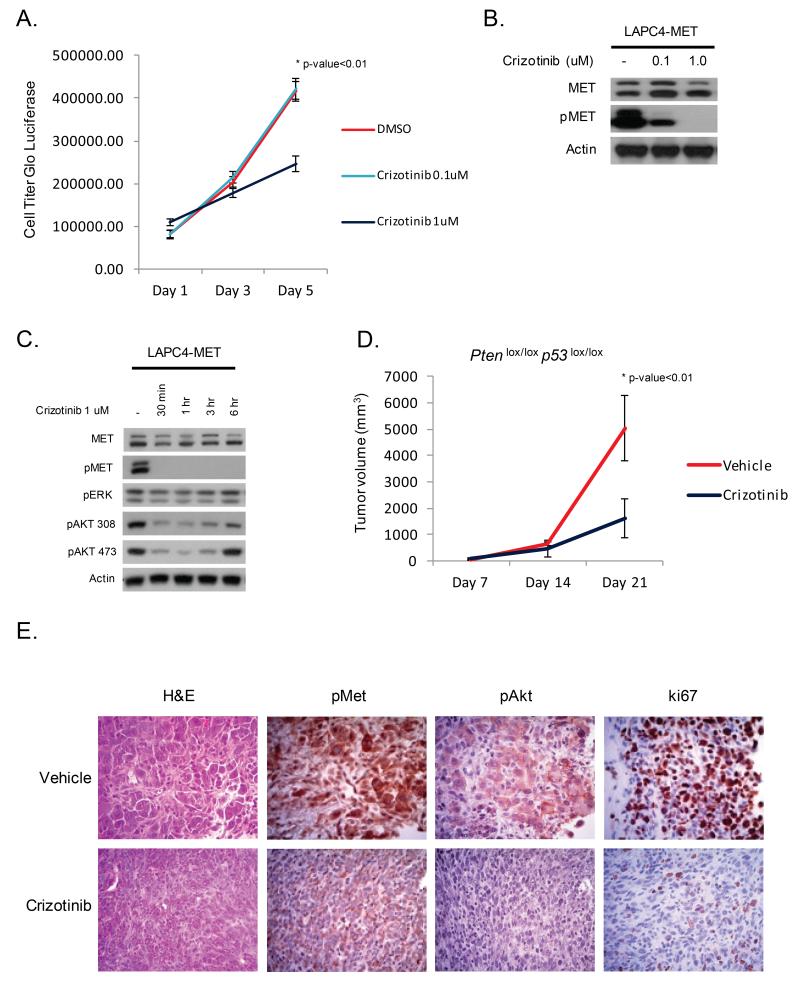
Several MET inhibitors have entered clinical trials across a variety of malignancies (21, 22). We elected to use the MET/ALK inhibitor crizotinib since this drug is relatively selective compared to other clinical MET inhibitors such as cabozantinib (21, 23). To evaluate the therapeutic efficacy of MET inhibition, we treated LAPC4-MET cells with increasing concentrations of crizotinib and observed that cell proliferation was significantly reduced at a dose of 1 uM (p-value<0.01) (Figure 5A). Furthermore, a dose of 1 uM of crizotinib resulted in potent inhibition of MET and downstream kinase signaling (Figure 5B, 5C).
Figure 5. Met is a therapeutic target in prostate cancer.
(A) Inhibition of MET in LAPC4-MET cells reduces cell proliferation. (B,C) Western blot analysis of LAPC4-MET cells treated with increasing concentration of the MET inhibitor crizotinib (0.1uM, 1uM, Pfizer) demonstrates reduction in MET and PI3K signaling. (D) Prostate cancer specimens were harvested from Pten p53 null mice, dissected, and transplanted into athymic mice to establish tumors. Mice with tumors were treated with vehicle (n=8) or crizotinib (n=8, 50 mg/kg/day, Pfizer) for a total of 21 days and tumor volumes were measured on a weekly basis. Mice treated with crizotinib had a significant reduction in tumor growth. (E) At the end of study, prostate tumors from mice treated with crizotinib had lower levels of pMet, pAkt, and ki67 staining by immunohistochemistry.
We then turned to our Pten p53 null prostate cancer model where endogenous Met is frequently amplified. Given the heterogeneous expression of MET in these Met amplified tumors, we predicted that MET inhibition would impact tumor growth but that low MET expressing cells may display relative resistance. To generate sufficient animals to conduct this preclinical trial, we established a protocol for tissue grafting prostate cancer specimens directly from the GEM model (Figure 4SA). These tumors were harvested and dissected into 2 × 2 × 2 mm3 sections and grafted into the flank of athymic mice (Figure 5D). Over a 14 day period the tumors engrafted and grew rapidly. These tumors were further passaged into athymic mice, then treated with the MET inhibitor crizotinib (50 mg/kg/day) or vehicle control. Tumors from Pten ERG mice, which lack Met amplification, were also studied to explore whether the effect of crizotinib was context specific. Pten/ p53 tumors treated with crizotinib had a significant reduction in growth rate whereas Pten/ERG tumors were unaffected (p-value<0.01) (Figure 5D, 5E, S4B). Inhibition of MET in Pten/p53 tumors was associated with reduced levels of pAkt and ki67 (Figure 5E). Thus, inhibition of MET specifically impairs the growth of murine prostate cancers known to harbor amplification or overexpression of Met.
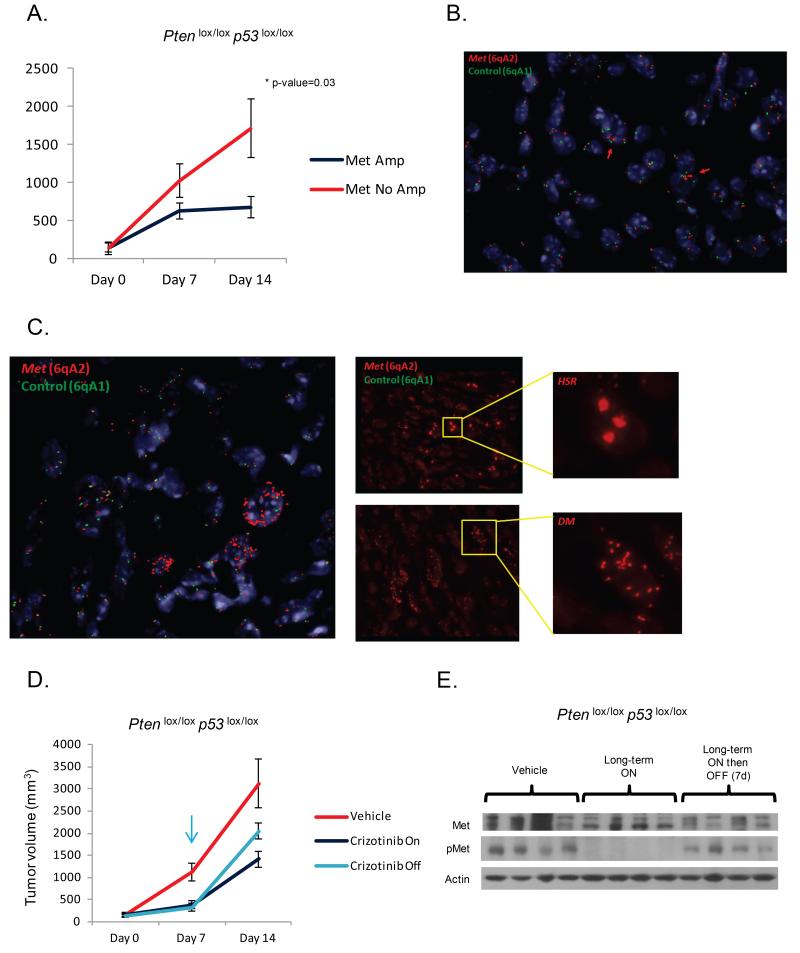
To directly evaluate the impact of Met amplification on tumor response, we conducted an additional experiment in which FISH analysis for Met amplification was performed. Pten p53 tumors were grafted into mice and treated with crizotinib (Figure 6A). Tumors harboring predominantly diploid copies of Met had the least response, while tumors harboring Met amplification had the greatest response (Figure 6A, 6B, 6C). Of note, withdrawal of crizotinib resulted in accelerated tumor progression and reactivation of Met signaling, indicating that the Met-amplified cells may grow faster than the non-Met-amplified tumor cells and importantly that crizotinib does not deplete the Met-amplified cells of Met pathway dependency (Figure 6D, 6E).
Figure 6. Amplification of Met is associated with a cytostatic therapeutic response.
(A) Mice with tumors grafted from Pten p53 null mice were treated with crizotinib (n=8, 50 mg/kg/day, Pfizer) for a total of 14 days, and profiled for Met copy-number by FISH. Tumors harboring Met amplification demonstrated a superior response to crizotinib. (B) FISH analysis from a non-Met amplified tumor demonstrating a predominantly diploid pattern with a few cells with low level copy number gain. Red color = Met, Green color = control, Blue color = centromeres, heterochromatin of mouse chromosomes. (C) Met FISH analysis demonstrating a dominant pattern of Met amplification with regions of HSR and DM. (D) Mice bearing Pten p53 null tumor grafts harboring Met amplification were treated with vehicle (n=8), crizotinib for 2 weeks (n=8), or crizotinib for 7 days followed by drug withdrawal (n=8). Tumors progressed following drug withdrawal. (E) Western blot analysis demonstrating reactivation of Met signaling following withdrawal of crizotinib.
Discussion
Through the genomic profiling of prostate cancer GEM models we identified several recurrent molecular events that develop during the evolution of murine prostate cancer. Integrating this murine data with genomic data from human prostate cancer led us to identify MET amplification as a molecular alteration with a high probability of playing a functional role in prostate cancer progression. Importantly, MET amplification did not emerge as a prostate cancer driver lesion using traditional human tumor genome analysis pipelines. However, reevaluation of the human tumor data based on our discovery of Met amplification in murine tumors revealed that approximately 30% of metastatic tumors have MET copy number gain (2, 3). Further evidence for a biologic role of MET in prostate cancer is the fact that tumors with MET gain are statistically enriched for copy number gains of the MET ligand HGF. Our biological studies of forced MET overexpression and of pharmacological MET inhibition further implicate MET as a driver of prostate cancer progression and a therapeutic target. However, the significant heterogeneity of MET expression within individual Met- amplified tumors may limit the effectiveness of MET inhibitors in advanced prostate cancer.
In this context, it is worth noting recent clinical results with the dual MET/VEGFR2 inhibitor cabozantinib (XL184) showing dramatic resolution of bone scan abnormalities in 86% of patients but soft tissue responses and serum PSA declines in only 25- 30% of patients (22, 24, 25). This discrepancy suggests the bone scan effect of cabozantib may occur through inhibition of a target in the bone microenvironment whereas the antitumor effect may be a consequence of target inhibition in tumor cells. Our data offer a potential explanation for these discrepant clinical response rates, specifically that MET amplification could be associated with tumor response but not bone scan response. One prediction from our data is that MET inhibition would inhibit tumor growth only in the 20-30% of patients with MET amplification, a number consistent with the ~25% of patients with clinical soft tissue responses and serum PSA declines. Our data also suggest that these responses may be short lived due to the intratumoral heterogeneity of Met-amplification we observed in the mouse tumors.
In summary, our work demonstrates the potential of integrated human and GEM model genomic profiling of prostate cancer to provide insight into new biologic drivers acquired during tumor progression, as illustrated by our discovery of Met amplification. Additional genomic alterations uncovered by this analysis, such as Jun and Yap1, also merit further evaluation.
Supplementary Material
Acknowledgments
The authors would like to thank the Genetically Engineered Mouse Facility for maintaining our mouse colonies. We would like to thank P.P. Pandolfi and Z. Chen for providing the Ptenlox/lox p53lox/lox PB-Cre GEM model. A special thanks to all members of the Sawyers lab providing informative discussion. B.S.C. and C.L.S. designed the study. B.S.C., J.W., J.W., Y.C., Y., S. L., W. L., Q. Y., Z.C., and C.C. performed the mouse experiments. B.S.C., C.C., and J.W. performed the in vitro experiments. H.H. performed the aCGH and expression array analyses in human and mouse prostate cancers. B.T. and N.S. conducted the aCGH analyses in mouse prostate cancers. B.S.C. and C.L.S. prepared the manuscript. All authors approved of the final manuscript.
Financial Information: B.S. Carver is funded in part through RP-2 NIH Prostate SPORE P50-CA92629. B. S. Carver has a Prostate Cancer Foundation Young Investigator Award. C. L. Sawyers is a Howard Hughes Medical Institute Investigator. Z.Chen is supported by Meharry Medical College MD004038.
Abbreviation List
- (CNA)
Copy number alteration
- (GSEA)
Gene set enrichment analysis
- (FFPE)
Formalin fixed paraffin embedded
- (GEM)
Genetically engineered mouse
Footnotes
Conflict of Interest Disclosure Statement: The authors have no conflict of interest to disclose
References
- 1.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding Z, Wu CJ, Jaskelioff M, Ivanova E, Kost-Alimova M, Protopopov A, et al. Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell. 2012;148:896–907. doi: 10.1016/j.cell.2012.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–81. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–71. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–38. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 8.Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med. 2013;19:1023–9. doi: 10.1038/nm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai Y, Siemann DW. Constitutively active c-Met kinase in PC-3 cells is autocrine-independent and can be blocked by the Met kinase inhibitor BMS-777607. BMC Cancer. 2012;12:198. doi: 10.1186/1471-2407-12-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer J, Palmedo G, von Knobloch R, Bugert P, Prayer-Galetti T, Pagano F, et al. Duplication and overexpression of the mutant allele of the MET proto-oncogene in multiple hereditary papillary renal cell tumours. Oncogene. 1998;17:733–9. doi: 10.1038/sj.onc.1201983. [DOI] [PubMed] [Google Scholar]
- 13.Liang TJ, Reid AE, Xavier R, Cardiff RD, Wang TC. Transgenic expression of tpr-met oncogene leads to development of mammary hyperplasia and tumors. J Clin Invest. 1996;97:2872–7. doi: 10.1172/JCI118744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu P, Cheng H, Santiago S, Raeder M, Zhang F, Isabella A, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17:1116–20. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy J, Curry GW, Hillan KJ, McKay IC, Mallon E, Purushotham AD, et al. Hepatocyte growth factor/scatter factor expression and c-met in primary breast cancer. Surg Oncol. 1996;5:15–21. doi: 10.1016/s0960-7404(96)80017-x. [DOI] [PubMed] [Google Scholar]
- 16.Pisters LL, Troncoso P, Zhau HE, Li W, von Eschenbach AC, Chung LW. c-met proto-oncogene expression in benign and malignant human prostate tissues. J Urol. 1995;154:293–8. [PubMed] [Google Scholar]
- 17.Ponzo MG, Lesurf R, Petkiewicz S, O’Malley FP, Pinnaduwage D, Andrulis IL, et al. Met induces mammary tumors with diverse histologies and is associated with poor outcome and human basal breast cancer. Proc Natl Acad Sci U S A. 2009;106:12903–8. doi: 10.1073/pnas.0810402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yano S, Takeuchi S, Nakagawa T, Yamada T. Ligand-triggered resistance to molecular targeted drugs in lung cancer: Roles of hepatocyte growth factor and epidermal growth factor receptor ligands. Cancer Sci. 2012;103:1189–94. doi: 10.1111/j.1349-7006.2012.02279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yano S, Yamada T, Takeuchi S, Tachibana K, Minami Y, Yatabe Y, et al. Hepatocyte growth factor expression in EGFR mutant lung cancer with intrinsic and acquired resistance to tyrosine kinase inhibitors in a Japanese cohort. J Thorac Oncol. 2011;6:2011–7. doi: 10.1097/JTO.0b013e31823ab0dd. [DOI] [PubMed] [Google Scholar]
- 20.Minuti G, Cappuzzo F, Duchnowska R, Jassem J, Fabi A, O’Brien T, et al. Increased MET and HGF gene copy numbers are associated with trastuzumab failure in HER2-positive metastatic breast cancer. Br J Cancer. 2012;107:793–9. doi: 10.1038/bjc.2012.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camidge DR, Bang YJ, Kwak EL, Iafrate AJ, Varella-Garcia M, Fox SB, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol. 2012 doi: 10.1016/S1470-2045(12)70344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurzrock R, Sherman SI, Ball DW, Forastiere AA, Cohen RB, Mehra R, et al. Activity of XL184 (Cabozantinib), an oral tyrosine kinase inhibitor, in patients with medullary thyroid cancer. J Clin Oncol. 2011;29:2660–6. doi: 10.1200/JCO.2010.32.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodig SJ, Shapiro GI. Crizotinib, a small-molecule dual inhibitor of the c-Met and ALK receptor tyrosine kinases. Curr Opin Investig Drugs. 2010;11:1477–90. [PubMed] [Google Scholar]
- 24.Tu WH, Zhu C, Clark C, Christensen JG, Sun Z. Efficacy of c-Met inhibitor for advanced prostate cancer. BMC Cancer. 2010;10:556. doi: 10.1186/1471-2407-10-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yakes FM, Chen J, Tan J, Yamaguchi K, Shi Y, Yu P, et al. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol Cancer Ther. 2011;10:2298–308. doi: 10.1158/1535-7163.MCT-11-0264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.