Abstract
We report the successful fabrication and testing of 3D printed microfluidic devices with integrated membrane-based valves. Fabrication is performed with a low-cost commercially available stereolithographic 3D printer. Horizontal microfluidic channels with designed rectangular cross sectional dimensions as small as 350 μm wide and 250 μm tall are printed with 100% yield, as are cylindrical vertical microfluidic channels with 350 μm designed (210 μm actual) diameters. Based on our previous work [Rogers et al., Anal. Chem. 83, 6418 (2011)], we use a custom resin formulation tailored for low non-specific protein adsorption. Valves are fabricated with a membrane consisting of a single build layer. The fluid pressure required to open a closed valve is the same as the control pressure holding the valve closed. 3D printed valves are successfully demonstrated for up to 800 actuations.
I. INTRODUCTION
Microfluidics1 is a critical technology for an extremely broad range of biomedical applications including tissue engineering,2,3 drug discovery,4 point-of-care diagnostics and pathogen detection in both developed and developing countries,5–8 and cancer screening using approaches such as cell identification,9 and protein,10–13 DNA,14 and micro-RNA15,16 biomarkers. Microfluidic device prototyping for proof-of-principle demonstration typically utilizes hot embossed or injection molded plastics1,17 or polydimethylsiloxane (PDMS).18,19 In either case, two or more individually fabricated layers are bonded together to form a completed device. The fabrication process typically involves cleanroom microfabrication of molds using photolithography for one or more of the individual layers, followed by molding and release of each layer and then careful layer alignment and bonding. In our experience, this sequence of steps can lead to a delay of a week or more between completing the design of a device and actually having one in hand to test, especially taking into account the inevitable problems that crop up for various fabrication steps in a university environment and the turn-around time to design and procure photolithographic masks. To reduce mask acquisition time, we have a Heidelberg DWL-66FS laser lithography system20 in our cleanroom, with which mask fabrication times range from 2 h for low resolution masks (>5 μm) to ∼5 days for high resolution masks (>0.6 μm). An alternative low cost option for rapid mask fabrication is inkjet printing on transparencies, which takes only a few minutes, but has much lower resolution (>50 μm).21 Moreover, limited material choices for prototyping microfluidic systems also hinder their broad development, as problems such as non-specific adsorption that plague PDMS and other polymers22,23 prevent many potential applications from being tested.
Of course, once the masks and processes are in place for a given design, it usually takes only hours to possibly a day or two to produce additional devices of that design. However, initial microfluidic testing often reveals design or performance deficiencies that necessitate modifying the design and starting the process over, thereby incurring yet another significant delay. Numerous cycles around this loop can be required to develop a successful device, which stretches the development time with a concomitantly large increase in personnel costs. Moreover, this lengthy cycle time discourages trying new approaches when faced with tight development deadlines. This is in direct contrast to the “fail fast and often” strategy successfully employed for web and smartphone application software development where early and rapid user feedback is used to guide project development throughout the development cycle. By analogy, 3D printing of microfluidics offers the opportunity to shrink the time from design to first device to an hour or less because the device is created directly in a single step with no need for layer-by-layer fabrication and assembly as with PDMS. This completely changes the development landscape by not only dramatically reducing the opportunity cost of trying new ideas but also permitting a “fail fast and often” strategy in which early and rapid empirical feedback is used to guide and accelerate device development. Moreover, 3D printing does not require a cleanroom environment with its attendant start up investment and ongoing operational costs. In other words, 3D printed microfluidics can dramatically lower the barrier for creating sophisticated microfluidic devices and offers a true rapid-prototyping ability with its attendant benefits to positively disrupt microfluidic development cycles.
Unfortunately, this promise in 3D printed microfluidics has not yet been realized, although there have been a number of efforts in this direction.24 For example, Kitson et al.25–27 demonstrated fluidic devices 3D printed by extruding plastic through a heated nozzle. However, this fabrication method is inherently unable to produce feature sizes and flow channel dimensions needed for microfluidic (as opposed to macrofluidic or millifluidic) device fabrication. For the reported devices, the flow channels had very large cross sections (∼4 mm diameter).
A more promising approach for microfluidics is stereolithography in which a vector scanned laser beam or a stationary image pattern from a projector is used to photopolymerize an appropriate photosensitive resin layer-by-layer until a full device is completed. For example, Bhargava et al.28 report a system in which discrete ∼1 cm3 3D printed cubes, each with internal plumbing to perform a specific passive elementary function (such as an L-joint, mixer, T-junction, XX-junction, etc.), are assembled into more complex fluidic devices in a 3D geometry. Each cube has standardized fluidic interfaces on 2 or more sides according to the elementary function performed within the cube. The cubes snap together to create precise cube-to-cube fluidic connections. While innovative, the overall system size can be comparatively large depending on how many cubes are needed. Furthermore, since the fluid channel minimum cross section dimension ranges from 500 to 1000 μm, this is more properly termed a millifluidic system. The cubes themselves are fabricated by a contract manufacturer (FineLine Prototyping, Raleigh, NC) using a proprietary, commercially available resin with a scanned laser stereolithographic 3D printer. This approach is appealing in that it is universally available to any customer, but the large flow channels and system size, and lack of control over resin formulation and hence surface and bulk chemistry can be unnecessarily restrictive for many applications.
Interestingly, another group recently published a paper in which the same contract manufacturer was used for direct fabrication of entire custom microfluidic devices.29 They showed that flow channels with cross sectional features down to 400 μm are possible. However, this approach is still limited to using commercially available resins, and only passive microfluidic components have been demonstrated.
Alternatively, Shallan et al.30 reported the use of an inexpensive commercially available stereolithographic printer (MiiCraft) to fabricate microfluidic devices with flow channel cross sectional dimensions >500 μm. Unfortunately, the two available resin formulations (blue and transparent) are proprietary and supplied by the printer manufacturer. Additionally, the transparent resin exhibits only 60% transmission for a 500 μm thick layer at wavelengths >430 nm and exhibits absorption of small hydrophobic molecules such as rhodamine 6G.
The Fang group and collaborators have built several custom stereolithographic 3D printers that achieve submicron feature sizes for microfluidic devices and use their own resin formulations.3,31,32 The small feature sizes are realized by photoreduction of an image projected by a UV-illuminated dynamic mask (i.e., digital light projector or liquid crystal on silicon microdisplay). However, the required photoreduction reduces the exposed area to only a millimeter or two on a side. To obtain reasonable part sizes (tens of millimeters in each lateral dimension), the image must be stepped many times across each layer using precise translation stages (250 nm positional repeatability). The end result is a complicated and expensive system that does not lend itself to low-cost microfluidic rapid prototyping.
Previously, we developed a custom resin that was UV polymerized into a polyethylene glycol diacrylate (poly-PEGDA) microfluidic material. We initially optimized the resin for conventional microfluidic fabrication techniques in which individual layers are molded and subsequently bonded to each other to create a device. The material was also optimized for low non-specific adsorption of proteins, low bulk background fluorescence (i.e., comparable to PDMS), and high bond strength.33 More recently, we demonstrated that despite having a significantly larger bulk modulus than PDMS (>100 MPa compared to ∼0.5 MPa), our poly-PEGDA material can be configured in a 3-layer design to create a membrane-type valve with compelling characteristics: 19 ms closure time and 115 000 actuations with no degradation in performance.34
Although 3D printed devices have excellent potential for biomedical microfluidic applications, current methods have limitations in terms of resolution, resin versatility, overall device dimensions, and/or prototyping system cost. Moreover, in all cases, the reported 3D printed microfluidic devices are composed of only passive elements. In this paper, we report the first 3D printed active elements in microfluidic systems, showing that both our low-adsorption resin and the basic valve structure can be adapted to successfully create 3D printed valves. We also characterize microfluidic channel fabrication, repeatability, and yield. These results represent the first step toward 3D printed microfluidic devices for integrated analyses of nucleic acids and other molecules in which many active and passive components are incorporated in a single device.35–40
II. EXPERIMENTAL METHODS
A. Materials and methods
PEGDA (M.W. 258), Sudan I, and 3-(trimethoxysilyl) propyl methacrylate were purchased from Sigma Aldrich (Milwaukee, WI). Phenylbis (2,4,6-trimethylbenzoyl) phosphine oxide (Irgacure 819) was acquired from BASF (Vandalia, IL). Prepolymer resin was prepared by mixing 1% (w/w) Irgacure and 0.2% (w/w) Sudan I in PEGDA and sonicated for 35 min. Silanized glass slides were prepared by placing clean slides in a 5% 3-(trimethoxysilyl) propyl methacrylate solution in toluene for 3 h. After deposition, the slides were scribed (to mark the print face), cleaved, rinsed with clean toluene, blown dry with a nitrogen gun, and stored for later use.
B. Experimental setup
We used a B9 Creator 3D printer v1.1 (B9 Creations, Rapid City, SD) to fabricate our devices. To determine feature size fidelity and device yield we used 8 3D printed samples, each with an identical set of horizontal flow channels with different designed cross sections ranging from 300 to 500 μm width and 150–250 μm height in 50 μm increments. Vertical flow channels were 3D printed on one die but with four different vertical holes for each size from 300 to 450 μm and eight holes for 500–800 μm (each in 50 μm increments). The cross-sectional dimensions were measured using digital photographs processed in ImageJ 1.48v.
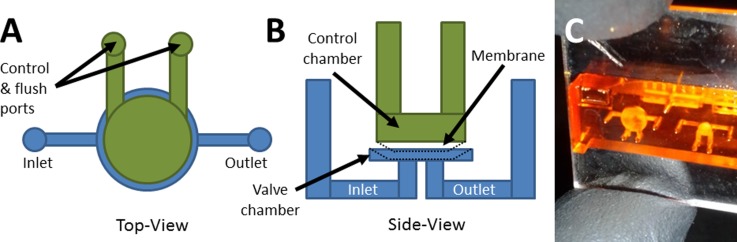
Once feasible channel dimensions were determined, these dimensions were then used to create the channels for the valve design. The valve design, shown in Figs. 1(a) and 1(b), consists of a membrane suspended over a valve chamber, on the bottom of which are inlet and outlet openings. When an external pressure source is applied to the control chamber above the membrane, the membrane is deflected downward until it seals the inlet and outlet openings, thereby closing the valve. When pressure is released, the membrane returns to its original position and the valve opens. A photograph of a fabricated test valve device is shown in Fig. 1(c).
FIG. 1.
Valve schematic and device image. (a) Top view and (b) side view schematics of test valve design. The control chamber (green) and fluidic chamber (blue) regions are voids in the 3D printed device. The control chamber has 2 access ports to enable it to be drained after printing. Pressure can be applied through both ports to actuate the valve, or one channel can be sealed and pressure applied through the other to actuate the valve. Pressurized membrane (black dotted line) shows valve closure. (c) Photograph of a fabricated valve test device looking through the top surface of the device. The left valve has a 3 mm diameter membrane, while the right valve membrane is 1.5 mm diameter.
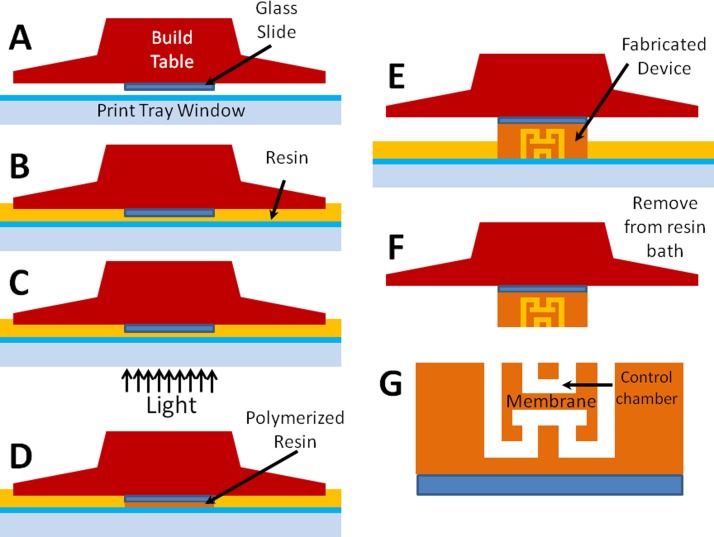
The 3D printing process to fabricate a device with a valve is illustrated schematically in Fig. 2. In brief, we used double-sided tape to affix a methacrylate silane functionalized glass slide to the bottom of the build table before calibrating the build table height for the print. After the resin was introduced into the tray and the projector was focused at the surface of the glass slide, different images were projected for each layer to polymerize each layer and create the desired 3D structure. Once the print was complete, unpolymerized resin was drained from the structure, thereby resulting in a completed device.
FIG. 2.
Fabrication process. (a) An acrylate silane functionalized glass slide is attached to bottom of the build table. (b) Resin is added to the print tray and the slide is positioned above bottom window. (c) Projector image is focused on the bottom surface of the glass slide, which (d) polymerizes resin in the exposed region. (e) The projector image is varied layer-by-layer to create the desired 3D structure. (f) When the device is pulled from the bath after all layers are exposed, the channels contain unpolymerized resin, which must be drained from the structure, resulting in (g) a finished device.
C. Membrane thickness
Membrane thickness as a function of exposure time was evaluated by measuring a 2 mm diameter circular single layer membrane (∼50 μm) suspended between two 250 μm high chambers. Exposure times between 2 s and 10 s were tested. Membrane thicknesses were measured from digital photographs using ImageJ.
D. Valve evaluation and performance
Previously, we demonstrated a successful method for valve evaluation.34 Briefly, two pressure sensors were placed in-line to monitor both the air pressure applied to close a valve and the fluidic pressure applied at the front of the device used to open the valve. A CCD camera was used to track the meniscus at the device outlet which was then converted to volumetric flow rate. The valve was considered open when the flow rate reached 0.2 μl/min. Valves were initially evaluated at air closure pressures of 0, 70, and 140 kPa. Valves were then actuated 400 times at 1 Hz (50% duty cycle) and the pressure tests repeated. This whole process was repeated until a given valve failed.
III. RESULTS AND DISCUSSION
A. Device characterization results
We modified our previous resin formulation34 for use in a B9 Creator 3D printer by replacing the original photoinitiator, 2,2-dimethoxy-2-phenylacetophenone (DMPA), with Irgacure 819 and adding an absorber dye, Sudan I. The B9 Creator's light source is a commercial XGA (1024 × 768 pixels) projector which does not emit UV light. The DMPA UV photoinitiator therefore had to be replaced with a photoinitiator sensitive to the blue end of the visible spectrum emitted by the projector. Likewise, the absorber dye must absorb in the wavelength range covered by the photoinitiator to limit the depth to which the photoinitiator is exposed; otherwise, no voids or overhanging features can be fabricated (nearly all microfluidic components involve voids, i.e., locations in which there is no material in the final device; for example, a flow channel). Our choice of Sudan I fulfills the absorption requirement, although it has absorbance throughout the visible spectrum, resulting in 3D printed parts with an orange color. Although this is not a problem for our initial proof-of-concept microfluidic valve development here, many microfluidics applications will require visible optical transparency. Nonetheless, the material reported here is compatible with non-optical sensing methods such as nanowires, microcantilevers, and electrochemical approaches (for example, amperometry, potentiometry, and impedance measurement).41–46
At its highest resolution setting, the B9 Creator specifies 50 μm × 50 μm resolution in the X-Y plane (i.e., the plane of each polymerized layer). A typical Z step size (layer-to-layer spacing) is also ∼50 μm. Note that the X-Y resolution of the B9 is twice as good as that of the scanning laser 3D printer (100 μm × 100 μm) used by the commercial fabrication service, FineLine Prototyping, mentioned previously, while the Z step size is the same. However, depending on the resin viscosity, actual fabricated flow channel dimensions and yield can be affected more by incomplete draining of uncured resin in the flow channel after 3D printing and prior to final curing of the part than by 3D printer resolution.29 Hence, taking advantage of improved 3D printer resolution requires development of effective techniques for draining voids.
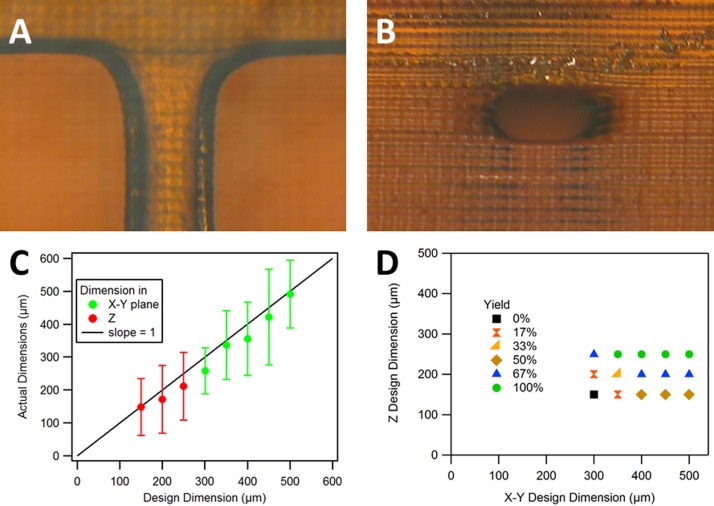
We found that draining flow channels with either deionized water (DI) or 2-propanol was effective for our resin formulation. Microscope images of an example channel are shown in Figs. 3(a) and 3(b), while measurement results for horizontal channels are included in Figs. 3(c) and 3(d). Fig. 3(c) shows the average actual measured size for each designed size for both in-plane (X-Y) and out of plane (Z) dimensions. In most cases, the average fabricated size is nearly equal to or somewhat smaller than the designed size. Fig. 3(d) shows the measured yield as a function of the designed X-Y and Z dimensions, with the smallest design size for 100% yield being 350 μm × 250 μm. Smaller flow channel sizes with high yield are likely feasible with further optimization, such as ensuring that the flow channel Z position and dimensions align with actual fabrication layers as determined by the software that slices a 3D computer-aided design (CAD) file to prepare it for 3D printing.
FIG. 3.
Horizontal channel fabrication, repeatability, and yield. (a) Top view and (b) side view of a flow channel with designed cross section size of 350 μm × 250 μm. The measured cross section of the fabricated flow channel is 316 μm × 217 μm. (c) Average actual (measured) flow channel size as a function of the designed size (error bars show standard deviation based on measurement of 8 samples). (d) Yield as a function of the designed X-Y and Z dimension sizes for 8 devices where yield represents the frequency of a successfully printed open channel.
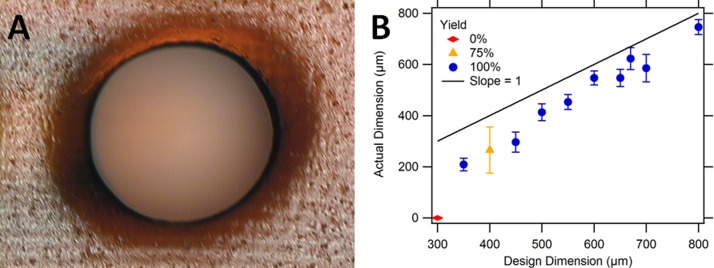
The microscope image shown in Fig. 4(a) shows a typical example of a vertical cylindrical channel. Measurement results for channels designed with diameters ranging from 300 μm to 800 μm are shown in Fig. 4(b). The smallest vertical channel successfully printed with 100% yield had a 350 μm designed diameter and 210 μm average measured diameter. As seen in Fig. 4(b), the as-printed diameters of the holes are smaller than the designed size.
FIG. 4.
Vertical cylindrical channel fabrication, repeatability, and yield. (a) Top view of a designed 650 μm cylindrical channel. The measured diameter of the channel is 606 μm. (b) Actual (measured) cylindrical channel size as a function of the designed size. Successfully printed open channels (yield) as a function of the designed cylinder diameters. Error bars denote standard deviation based on measurement of four channels (300–450 μm holes) or eight channels (500–800 μm holes).
B. Membrane thickness
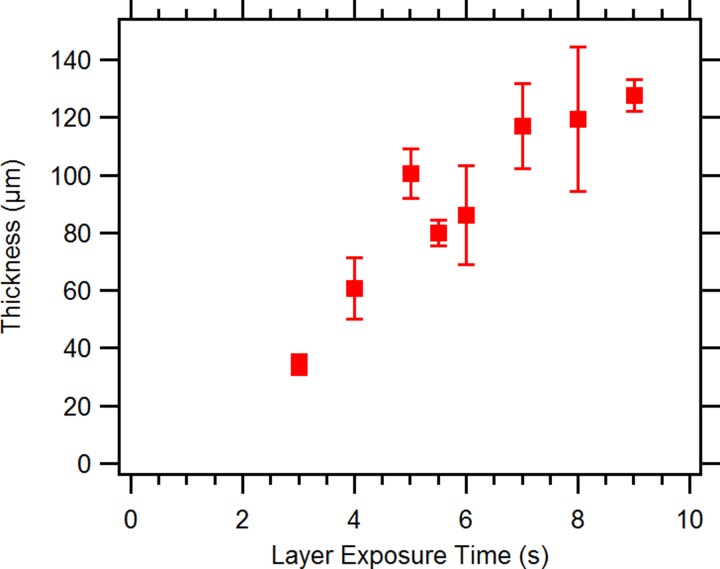
The as-fabricated membrane thickness has a critical effect on valve performance and lifetime. Fig. 5 shows the measured membrane thickness as a function of layer exposure time. As expected, longer layer exposure time results in greater membrane thickness because the valve chamber behind the membrane is filled with unexposed resin. The longer the exposure, the deeper into this region the polymerization front advances. Note that this not only makes the membrane thicker (and therefore stiffer) but also decreases the distance the membrane must deflect to seal the inlet and outlet openings. Our experiments indicate an exposure time in the range of 5.0 to 5.5 s/layer works well. Exposures less than 3 s failed to successfully print due to weak bonding between print layers, and at 3 s the print layers were easily damaged. On the other hand, exposures greater than 9 s resulted in overpolymerized devices which were prone to cracking under internal stress. With a 5 s exposure time, the total 3D printer build time for a typical 5 mm tall × 8.5 mm × 30 mm device is only 35–40 min.
FIG. 5.
Measured membrane thickness as a function of layer exposure time. In the design, the membrane thickness is specified as a single 3D printed layer. Error bars for data points at 3 s or greater exposure time represent standard deviation based on measurement of 4 to 9. There are no error bars for the 3 s data point, which is the average of two samples.
C. Valve evaluation and performance
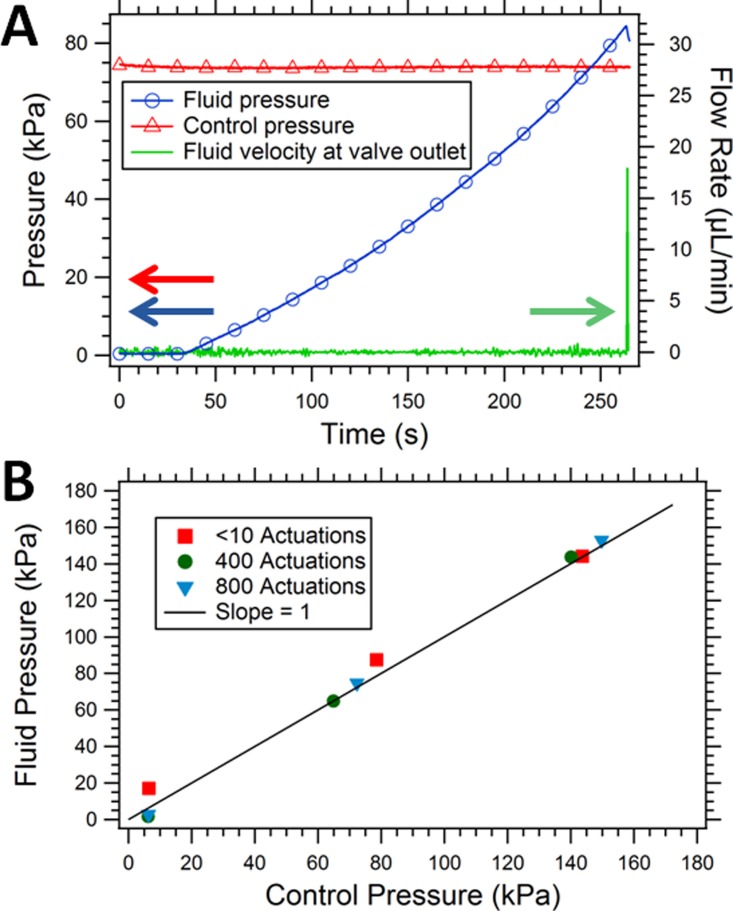
Fig. 6(a) shows the typical performance characteristics of a fabricated valve. The valve is closed by applying ∼74 kPa (∼20 psi, red triangle marked curve, left axis) to deflect the membrane down over the valve inlet and outlet channels. Meanwhile, a syringe pump introduces fluid into the valve inlet while the fluid pressure (blue circle marked curve, left axis) and flow rate at the valve outlet (solid green curve, right axis) are monitored (see Ref. 34 for further details on the measurement method). As expected, the fluid pressure increases within the device as a function of time until it reaches approximately the control pressure that is used to close the valve membrane. At this point, the membrane can no longer remain closed and fluid flows through the valve and out of the device. This performance characteristic is typical of a valve that operates as intended.
FIG. 6.
Valve operation and evaluation. (a) Operation of a 2 mm diameter valve membrane where the control pressure (red triangles) is the external pressure supplied to deflect the membrane and close the valve, fluid pressure (blue circles) is the pressure that builds up in the inlet channel as the syringe pump pushes fluid into the device, and fluid velocity (green solid line) is the volumetric flow rate at the valve outlet. (b) Fluid pressure at which the valve opens as a function of applied control pressure for a 3 mm valve, before and after 400 actuations and after 800 total valve actuations.
Fig. 6(b) shows the fluid pressure at which flow through a single device occurs as a function of the control pressure applied to close the membrane for an as-fabricated valve and the same valve after it has undergone 400 and 800 open/close actuation cycles. Each data point represents one measurement similar to what is shown in Fig. 6(a). The data shown in Fig. 6(b) is representative of what we measured for several devices. The data indicate that when the fluid pressure rises above the control pressure, the valve opens, as expected. Note that there is essentially no difference in valve performance before and after 400 or 800 actuations. We find that the valve membrane typically breaks sometime after 800 actuations. Given our earlier results for poly-PEGDA microfluidic valves where over 100 000 actuations resulted in little change in performance,34 we are confident that lifetimes of 3D printed microfluidic valves can be dramatically increased.
IV. CONCLUSIONS
We have successfully demonstrated readily fabricated 3D printed microfluidic channels with valves, within devices that take less than 1 h to print. Moreover, the yield for horizontal flow channels with cross sections as small as 350 μm × 250 μm is 100%. Vertical channels were 3D printed successfully as small as 350 μm diameter with 100% yield as well. Undoubtedly, flow channel size can be decreased through further optimization. Valve diameters as small as 2 mm have been shown to be viable and behave as expected with opening fluid pressure approximately equal to the control air pressure applied to close the valve. Switching to a higher resolution printer will likely decrease channel dimensions and valve sizes, while further processing steps such as post-print thermal or UV curing may improve the lifetime of the valves.
Owing to the absorber in our initial resin formulation, the fabricated devices are not fully optically transparent in the visible wavelength region and may also have bulk fluorescence. Although these current devices may be incompatible with biosensing based on optical absorbance or fluorescence measurements, ongoing future work to evaluate resin formulations with alternate photoinitiators and absorbers will address these issues. Development of a non-proprietary resin will allow for greater flexibility in modifying polymer properties such as surface chemistry to enable subsequent modification for application in immunoassays or nucleic acid assays, for example. Furthermore, the ability to print these devices directly onto glass surfaces opens up the potential for direct integration with a range of substrates (e.g., glass, silicon, or materials with patterned electrodes) which could dramatically lower the barrier-to-entry to explore lab-on-a-chip biosensors, thereby expanding the lab-on-a-chip research and development community and enabling accelerated biomedical sensor innovation.
ACKNOWLEDGMENTS
We are grateful to the National Institutes of Health (R01 EB006124) for partial support of this work.
References
- 1.Nge P. N., Rogers C. I., and Woolley A. T., Chem. Rev. 113, 2550 (2013). 10.1021/cr300337x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khademhosseini A., Langer R., Borenstein J., and Vacanti J. P., Proc. Natl. Acad. Sci. U. S. A. 103, 2480 (2006). 10.1073/pnas.0507681102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia C. and Fang N. X., Biomed. Microdevices 11, 1309 (2009). 10.1007/s10544-009-9350-4 [DOI] [PubMed] [Google Scholar]
- 4.Neuži P., Giselbrecht S., Länge K., Huang T. J., and Manz A., Nat. Rev. Drug Discovery 11, 620 (2012). 10.1038/nrd3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin C. D., Laksanasopin T., Cheung Y. K., Steinmiller D., Linder V., Parsa H., Wang J., Moore H., Rouse R., Umviligihozo G., Karita E., Mwambarangwe L., Braunstein S. L., van de Wijgert J., Sahabo R., Justman J. E., El-Sadr W., and Sia S. K., Nat. Med. 17, 1015 (2011). 10.1038/nm.2408 [DOI] [PubMed] [Google Scholar]
- 6.Foudeh A. M., Fatanat Didar T., Veres T., and Tabrizian M., Lab Chip 12, 3249 (2012). 10.1039/c2lc40630f [DOI] [PubMed] [Google Scholar]
- 7.Mulvaney S. P., Myers K. M., Sheehan P. E., and Whitman L. J., Biosens. Bioelectron. 24, 1109 (2009). 10.1016/j.bios.2008.06.010 [DOI] [PubMed] [Google Scholar]
- 8.Sin M. L., Mach K. E., Wong P. K., and Liao J. C., Expert Rev. Mol. Diagn. 14, 225 (2014). 10.1586/14737159.2014.888313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saliba A.-E., Saias L., Psychari E., Minc N., Simon D., Bidard F.-C., Mathiot C., Pierga J.-Y., Fraisier V., Salamero J., Saada V., Farace F., Vielh P., Malaquin L., and Viovy J.-L., Proc. Natl. Acad. Sci. U. S. A. 107, 14524 (2010). 10.1073/pnas.1001515107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chikkaveeraiah B. V., Mani V., Patel V., Gutkind J. S., and Rusling J. F., Biosens. Bioelectron. 26, 4477 (2011). 10.1016/j.bios.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu M., Yan J., He Y., Lu H., Weng L., Song S., Fan C., and Wang L., ACS Nano 4, 488 (2010). 10.1021/nn901404h [DOI] [PubMed] [Google Scholar]
- 12.Malhotra R., Patel V., Chikkaveeraiah B. V., Munge B. S., Cheong S. C., Zain R. B., Abraham M. T., Dey D. K., Gutkind J. S., and Rusling J. F., Anal. Chem. 84, 6249 (2012). 10.1021/ac301392g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziober B. L., Mauk M. G., Falls E. M., Chen Z., Ziober A. F., and Bau H. H., Head Neck 30, 111 (2008). 10.1002/hed.20680 [DOI] [PubMed] [Google Scholar]
- 14.Mulvaney S. P., Cole C. L., Kniller M. D., Malito M., Tamanaha C. R., Rife J. C., Stanton M. W., and Whitman L. J., Biosens. Bioelectron. 23, 191 (2007). 10.1016/j.bios.2007.03.029 [DOI] [PubMed] [Google Scholar]
- 15.Moltzahn F., Olshen A. B., Baehner L., Peek A., Fong L., Stoppler H., Simko J., Hilton J. F., Carroll P., and Blelloch R., Cancer Res. 71, 550 (2011). 10.1158/0008-5472.CAN-10-1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vorwerk S., Ganter K., Cheng Y., Hoheisel J., Stähler P. F., and Beier M., New Biotechnol. 25, 142 (2008). 10.1016/j.nbt.2008.08.005 [DOI] [PubMed] [Google Scholar]
- 17.Mei Q., Xia Z., Xu F., Soper S. A., and Fan Z. H., Anal. Chem. 80, 6045 (2008). 10.1021/ac800843v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friend J. and Yeo L., Biomicrofluidics 4, 026502 (2010). 10.1063/1.3259624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sia S. K. and Whitesides G. M., Electrophoresis 24, 3563 (2003). 10.1002/elps.200305584 [DOI] [PubMed] [Google Scholar]
- 20.See http://www.himt.de/index.php/dwl-series-overview.html for Heidelberg laser lithography system description.
- 21.Duffy D. C., McDonald J. C., Schueller O. J. A., and Whitesides G. M., Anal. Chem. 70, 4974 (1998). 10.1021/ac980656z [DOI] [PubMed] [Google Scholar]
- 22.Huang B., Wu H. K., Kim S., and Zare R. N., Lab Chip 5, 1005 (2005). 10.1039/b509251e [DOI] [PubMed] [Google Scholar]
- 23.Shah J. J., Geist J., Locascio L. E., Gaitan M., Rao M. V., and Vreeland W. N., Electrophoresis 27, 3788 (2006). 10.1002/elps.200600118 [DOI] [PubMed] [Google Scholar]
- 24.Gross B. C., Erkal J. L., Lockwood S. Y., Chen C., and Spence D. M., Anal. Chem. 86, 3240 (2014). 10.1021/ac403397r [DOI] [PubMed] [Google Scholar]
- 25.Kitson P. J., Rosnes M. H., Sans V., Dragone V., and Cronin L., Lab Chip 12, 3267 (2012). 10.1039/c2lc40761b [DOI] [PubMed] [Google Scholar]
- 26.Kitson P. J., Symes M. D., Dragone V., and Cronin L., Chem. Sci. 4, 3099 (2013). 10.1039/c3sc51253c [DOI] [Google Scholar]
- 27.Symes M. D., Kitson P. J., Yan J., Richmond C. J., Cooper G. J. T., Bowman R. W., Vilbrandt T., and Cronin L., Nat. Chem. 4, 349 (2012). 10.1038/nchem.1313 [DOI] [PubMed] [Google Scholar]
- 28.Bhargava K. C., Thompson B., and Malmstadt N., Proc. Natl. Acad. Sci. U. S. A. 111, 15013 (2014). 10.1073/pnas.1414764111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Au A. K., Lee W., and Folch A., Lab Chip 14, 1294 (2014). 10.1039/c3lc51360b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shallan A. I., Smejkal P., Corban M., Guijt R. M., and Breadmore M. C., Anal. Chem. 86, 3124 (2014). 10.1021/ac4041857 [DOI] [PubMed] [Google Scholar]
- 31.Sun C., Fang N., Wu D. M., and Zhang X., Sens. Actuators, A 121, 113 (2005). 10.1016/j.sna.2004.12.011 [DOI] [Google Scholar]
- 32.Zheng X., Deotte J., Alonso M. P., Farquar G. R., Weisgraber T. H., Gemberling S., Lee H., Fang N., and Spadaccini C. M., Rev. Sci. Instrum. 83, 125001 (2012). 10.1063/1.4769050 [DOI] [PubMed] [Google Scholar]
- 33.Rogers C. I., Pagaduan J. V., Nordin G. P., and Woolley A. T., Anal. Chem. 83, 6418 (2011). 10.1021/ac201539h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers C. I., Oxborrow J. B., Anderson R. R., Tsai L. F., Nordin G. P., and Woolley A. T., Sens. Actuators, B 191, 438 (2014). 10.1016/j.snb.2013.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crabtree H. J., Lauzon J., Morrissey Y., Taylor B., Liang T., Johnstone R., Stickel A., Manage D., Atrazhev A., Backhouse C., and Pilarski L., Microfluid. Nanofluid. 13, 383 (2012). 10.1007/s10404-012-0968-9 [DOI] [Google Scholar]
- 36.Kim J., Jensen E. C., Stockton A. M., and Mathies R. A., Anal. Chem. 85, 7682 (2013). 10.1021/ac303767m [DOI] [PubMed] [Google Scholar]
- 37.Collino R. R., Reilly-Shapiro N., Foresman B., Xu K., Utz M., Landers J. P., and Begley M. R., Lab Chip 13, 3668 (2013). 10.1039/c3lc50481f [DOI] [PubMed] [Google Scholar]
- 38.Godwin L. A., Pilkerton M. E., Deal K. S., Wanders D., Judd R. L., and Easley C. J., Anal. Chem. 83, 7166 (2011). 10.1021/ac201598b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu F., Horowitz M. A., and Quake S. R., Lab Chip 13, 1911 (2013). 10.1039/c3lc41394b [DOI] [PubMed] [Google Scholar]
- 40.Shackman J. G., Reid K. R., Dugan C. E., and Kennedy R. T., Anal. Bioanal. Chem. 402, 2797 (2012). 10.1007/s00216-012-5755-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.García M., García-Carmona L., and Escarpa A., “ Microfluidic system for enzymeless electrochemical determination of inulin using catalytically active metal nanowires,” Microchim. Acta 2014, 1–8 10.1007/s00604-014-1384-5 [DOI] [Google Scholar]
- 42.McClellan M. S., Domier L. L., and Bailey R. C., Biosens. Bioelectron. 31, 388 (2012). 10.1016/j.bios.2011.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson R. R., Hu W., Noh J. W., Dahlquist W. C., Ness S. J., Gustafson T. M., Richards D. C., Kim S., Mazzeo B. A., Woolley A. T., and Nordin G. P., Lab Chip 11, 2088 (2011). 10.1039/c1lc20025a [DOI] [PubMed] [Google Scholar]
- 44.Ness S. J., Anderson R. R., Hu W. S., Richards D. C., Oxborrow J., Gustafson T., Tsai B., Kim S., Mazzeo B., Woolley A., and Nordin G. P., IEEE Sens. J. 13, 959 (2013). 10.1109/JSEN.2012.2225613 [DOI] [Google Scholar]
- 45.Anderson K. B., Lockwood S. Y., Martin R. S., and Spence D. M., Anal. Chem. 85, 5622 (2013). 10.1021/ac4009594 [DOI] [PubMed] [Google Scholar]
- 46.Vázquez M., Frankenfeld C., Coltro W. K. T., Carrilho E., Diamond D., and Lunte S. M., Analyst 135, 96 (2010). 10.1039/b908985c [DOI] [PubMed] [Google Scholar]