Abstract
c-CBL (CBL) encodes a member of the Cbl family of proteins, which functions as an E3 ubiquitin ligase. We describe a dominant developmental disorder resulting from germline missense CBL mutations, which is characterized by constitutional anomalies that include impaired growth, developmental delay, cryptorchidism, and a predisposition to juvenile myelomonocytic leukemia (JMML). Some individuals experienced spontaneous regression of their JMML but developed vasculitis later in life. Importantly, JMML specimens from affected children show loss of the normal CBL allele through acquired isodisomy. Consistent with these genetic data, the common p.Y371H mutant Cbl protein induces cytokine-independent growth and constitutive phosphorylation of ERK, AKT, and S6 only in hematopoietic cells in which normal Cbl expression is reduced by RNA interference. We conclude that germline CBL mutations have developmental, tumorigenic, and functional consequences that are reminiscent of disorders that are caused by hyperactive Ras/Raf/MEK/ERK signaling and include neurofibromatosis type 1, and Noonan, Costello, cardiofaciocutaneous, and Legius syndromes.
Myeloproliferative neoplasms (MPNs) are clonal malignancies characterized by overproduction of immature and mature myeloid lineage cells. In particular, juvenile myelomonocytic leukemia (JMML) is an aggressive MPN of childhood characterized by malignant transformation in the stem cell compartment with clonal proliferation of progeny that variably retain the capacity to differentiate (reviewed in 1). Hematopoietic stem cell transplantation (HSCT) is the only curative therapy for JMML; however, relapse rates approach 30% 2. While spontaneous remissions occur in some infants 3,4, the underlying mechanism for this is unknown.
Extensive molecular data implicate germline and somatic mutations that deregulate Ras signaling as key initiating events in JMML, with studies showing that 60% of patients harbor an oncogenic mutation in PTPN11, NRAS, or KRAS while another 15% have neurofibromatosis type 1 (NF1) and demonstrate loss of the normal NF1 allele in leukemic cells 5-9. Patients with the myeloproliferative subtype of chronic myelomonocytic leukemia (CMML), a similar MPN of adulthood, frequently acquire NRAS, KRAS, and JAK2 mutations 10,11. Genetically accurate mouse models recapitulate these diseases, supporting the hypothesis that hyperactive Ras is necessary and sufficient to cause MPN 12-15. A hallmark feature of JMML and CMML is the formation of abnormally high numbers of granulocyte-macrophage colony-forming units (CFU-GM) in methylcellulose cultures containing low concentrations of GM-CSF 16,17. Phosphorylation of the βc chain of the GM-CSF receptor creates docking sites for adapters and signal relay molecules, resulting in activation of the Ras pathway.
Recently, we and others used high density single nucleotide polymorphism arrays to analyze blood and bone marrow specimens from patients with MPN 18-21. These studies revealed copy-neutral loss of heterozygosity (acquired isodisomy) of a region on chromosome 11q in some cases, and subsequent studies demonstrated homozygous mutations in CBL. Approximately 10-15% of children with de novo JMML are estimated to harbor homozygous CBL mutations20,22. CBL mutations are acquired somatically in adults with MPN 18,19,21.
Children with NF1 and Noonan syndrome (NS) are predisposed to JMML 8,9,23-25, and we therefore considered the possibility that germline CBL mutations occur in some affected children. A review of the medical records of the 21 children with JMML found to have CBL mutations enrolled in the EWOG-MDS studies or treated at USCF (16 of 21 were previously included in a screen of a larger international cohort20) uncovered an unexpectedly high percentage with developmental delay and other congenital anomalies, which included cryptorchidism, and impaired growth (Tables 1 and 2). All children met diagnostic criteria for JMML26,27 but six patients with a follow-up of more than seven years did not undergo transplantation for various reasons. Of these, one died of progressive JMML (D088), but the MPN improved spontaneously in five others. All of these patients continued to display variable degrees of splenomegaly in the presence of normal blood counts and exhibited the persistence of a homozygous CBL mutation in CD4, CD8, CD14, and CD19 sorted cells from their peripheral blood at last follow-up. In addition, four of these patients have developed clinical signs consistent with vascular pathology, including optic atrophy, hypertension, and an acquired cardiomyopathy; one was diagnosed with Takayasu arteritis, type III by angiography (Figure 1a). Another patient (D256) developed an intracranial germinoma harboring the same homozygous CBL mutation as in his bone marrow. Of note, among the patients treated with HSCT, there was a high rate of conversion to stable mixed chimerism (8/11 patients with available data) (Table 1).
Table 1.
Hematological Features at Diagnosis, Hematopoietic Stem Cell Transplantation and Current Status in 21 Children with homozygous CBL Mutations in JMML Cells
| ID | Sex | Age and Hematological Features at Diagnosis | HSCT | Last fo low-up | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (years) | WBC (×109/L) | % blasts PB/BM | Platelets (×109/L) | Spleen Sizea | Karyotype | Age (years) | MC / CC | Alive/Dead | Age (years) | ||
| A053 | M | 0.1 | 112 | 1 / 2 | 20 | 9 | normal | - | - | A | 18.2 |
| A054 | F | 1.1 | 36 | 1 / 4 | 95 | 7 | normal | - | - | A | 17.7 |
| D256 | M | 3.6 | 13 | 1 / 6 | 18 | 10 | normal | - | - | A | 13.0 |
| D048 | F | 1.6 | 23 | 0 / 2 | 46 | 9 | normal | - | - | D | 9.9 |
| D389 | M | 0.6 | 14 | 0 / 6 | 67 | 8 | normal | - | - | A | 7.5 |
| D088 | M | 0.8 | 46 | 3 / nd | 32 | 12 | normal | - | - | D | 7.3 |
| NL075 | F | 1.5 | 55 | 5 / 2 | 55 | 12 | normal | 1.8 | MC | A | 7.3 |
| CZ039 | M | 1.1 | 29 | 3 / 4 | 284 | 6 | normal | 1.2 | MC/CCc | A | 7.0 |
| D451 | M | 0.7 | 27 | 2 / 3 | 86 | 6 | normal | 1.2 | MC/CCd | A | 6.2 |
| D647 | F | 2.5 | 67 | 1 / 0 | 117 | 6 | normal | 3.9 | CC | A | 6.1 |
| I066 | F | 5.0 | 21 | 2 / 10 | 33 | 5 | normal | 5.7 | MC | D | 6.0 |
| SC084 | M | 1.4 | 33 | 0 / 0 | 33 | 13 | normal | 2.0 | NA | A | 4.8 |
| D104 | M | 2.2 | 27 | 0 / 2 | 232 | 5 | normal | - | - | D | 3.9 |
| D347 | M | 0.9 | 72 | 4 / 6 | 48 | 10 | 45,XY,−16 | 1.2 | MC | D | 3.4 |
| D703 | F | 1.4 | 27 | 2 / 1 | 15 | 4 | 46,XX+der (8) | 1.7 | MC | A | 3.0 |
| UPN1333 | M | 0.6 | 36 | 3 / 4 | 46 | 4 | normal | 1.1 | MC | D | 2.6 |
| D251 | F | 0.6 | 31 | 0 / 20 | 37 | 4 | normal | 1.1 | CC | D | 2.4 |
| UPN1778 | M | 1.3 | 22 | 0 / 1.5 | 61 | 5 | normal | - | - | A | 2.0 |
| UPN1125 | F | 1.3 | 196 | 4 / 5 | 47 | 4 | normal | 1.7 | CC | D | 2.1 |
| D774 | F | 1.5 | 22 | 0 / 4 | 266 | 3 | normal | - | - | A | 1.5 |
| UPN1241 | F | 0.6 | 43 | 5 / 3 | 62 | 4 | normal | 1.1 | MC | D | 1.5 |
ID, patient identification. M, male. F, female. WBC, white blood count. PB, peripheral blood. BM, bone marrow. HSCT, -hematopoietic stem cell transplantation. A, alive. D, dead. MC, mixed chimerism. CC, complete chimerism. NA, not analyzed.
in cm below costal margin in left midclavicular line
b All patients had received a myeloablative preparative regimen prior to HSCT. One patient with MC died of progressive disease (UPN 1333), the other deaths were due to transplant related toxicities.
patient converted back to CC after donor lymphocyte infusion
patient converted back to CC without therapy
Table 2.
CBL Mutation and Non-Hematological features in 21 Children with JMML
| ID | CBL Mutation | Café au lait spotsm | JXG | Cryptorchism | Growth < 3rd percentile | Dev. delay | Hearing loss | Optic atrophy | Hypertensionn | Cardiomyopathy | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | Germ | Mo | Fa | ||||||||||
| A053a | p.C396R | + | - | NA | - | - | - | - | + | + | + | + | + |
| A054 | p.Y371H | + | + | NA | - | + | female | - | - | - | - | - | - |
| D256b | p.W408R | + | - | - | + | + | - | + | + | - | + | + | - |
| D048c | p.C384R | NA | NA | NA | - | - | female | + | + | - | + | + | + |
| D389d | p.Y371C | + | - | + | + | - | + | - | + | + | + | + | + |
| D088 | p.Y371D | NA | NA | NA | - | - | + | + | - | - | - | - | - |
| NL075 | p.Y371H | + | - | - | - | + | female | - | - | - | - | - | - |
| CZ039 | p.C404R | + | - | - | - | - | - | - | - | - | - | - | - |
| D451 | p.Y371H | + | - | - | - | - | + | - | + | - | - | - | - |
| D647 | c.1228-2A>G splice site | + | - | - | - | - | female | + | + | - | - | - | - |
| I066 | c.1228-2A>G splice site | NA | NA | NA | - | - | female | - | - | - | - | - | - |
| SC084 | p.Y371H | + | + | - | - | - | - | - | + | - | - | - | - |
| D104e | p.C384R | NA | NA | NA | - | + | - | - | - | - | - | - | - |
| D347f | c.1096-1G>C splice site | + | - | NA | + | + | + | + | + | - | - | - | - |
| D703 | p.C384R | + | - | + | - | - | female | - | - | - | - | - | - |
| UPN1333 | p.Y371H | + | + | - | - | - | - | - | + | + | - | - | - |
| D251g | p.L380P | + | NA | NA | - | + | female | + | + | - | - | - | - |
| UPN1778h | p.Y371N | + | - | NA | - | - | - | - | - | - | - | - | - |
| UPN1125 | p.Y371H | + | + | - | - | - | female | - | - | - | - | + | - |
| D774k | p.H398R | + | + | - | - | - | female | - | - | - | - | - | - |
| UPN1241 | p.Y371H | + | - | - | + | - | female | - | - | - | - | - | -- |
ID, patient identification, Germ, germline. Mo, mother. Fa, Father. NA, not analyzed. JXG, juvenile xanthogranuloma. Dev., developmental.
autoimmune thyreoditis with anti-thyreoglobulin antibodies; echocardiography with mitral insufficiency grade II, myocardial hypertrophy, hypoelastic and hypoplastic ascending aorta and arch of aorta
Diagnosis of intracranial germinoma at age 9 years with homozygous CBL mutation in the brain tumor
Diagnosed with Takayasu arteritis, type III, died 9 months after diagnosis of vasculitis and stenting of major arteries
Father with history of Hodgkin lymphoma
Sudden death at home
Died of cerebral hypoxia, also had a history of supraventricular tachycardia
Prior to HSCT, was diagnosed with erythroderma, post-HSCT: toxic epidermal necolysis, acute liver failure, liver Tx, subtotal brain infarction, apallic syndrome
Pectus excavatum
Older sister with heterozygous mutation
Patients indicated by + have 1-2 café au lait spots, there was no patient with 6 or more café au lait spots
Measurements of blood pressure following HSCT were excluded from analysis
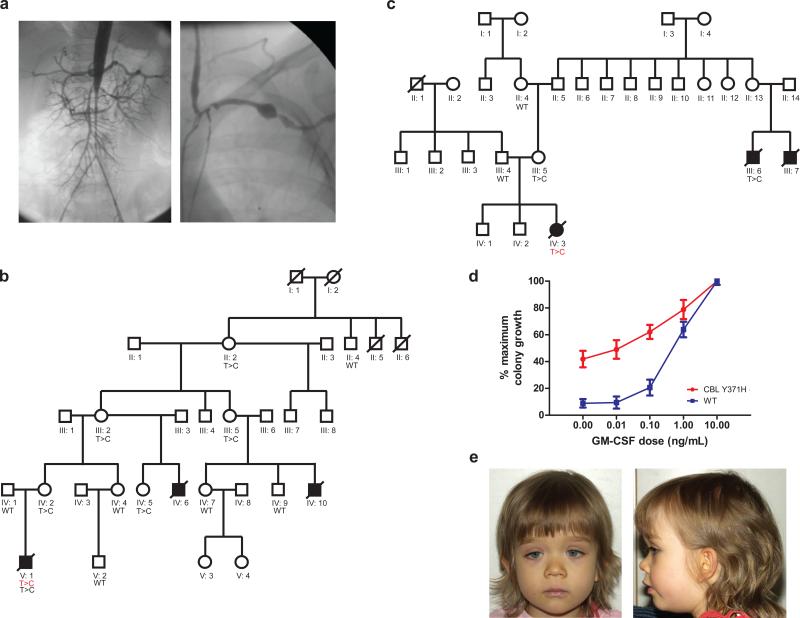
Figure 1. Germline mutations in CBL can be inherited in an autosomal dominant fashion and are associated with a phenotype, GM-CSF hypersensitivity and vasculitis.
Panel (a) demonstrates the angiograms from the aorta and left subclavian artery from patient D048 nine months after the diagnosis of Takayasu arteritis type III. Panel (b) The family tree of UPN1333 is shown in panel a, where the diseased bone marrow of UPN1333 displayed a homozygous CBL c.1111T>C (red) mutation as well as a heterozygous lesion from his buccal swab (black). Only women appear to be heterozygote carriers, and only boys appear to be affected by JMML in this family. Panel (c) The bone marrow of UPN1125 demonstrated a homozygous CBL mutation—her mother (III:5) is a known carrier, and she had two male cousins dying from JMML (III:6, III:7). Panel (d) demonstrates a classic GM-CSF hypersensitivity response on a colony assay for patients with CBL mutations (n=3) versus normal (n=13). Error bars represent standard error of the mean (s.e.m.) Panel (e) shows one toddler (D703) diagnosed with JMML and a homozygous mutation at p.C384R. She displays frontal bossing, downslanting palpebral fissues, hypertelorism, and a low nasal bridge. Photographs of her father, who harbors a heterozygous mutation at p.C384R, are included in Figures S1, panel d. Of note, both father and daughter also display bilateral ptosis.
We analyzed normal tissues from 17 of these children and detected a heterozygous CBL mutation in each. Mutational analysis of parental DNA was informative in 13 families and confirmed autosomal inheritance of a CBL mutation in seven (Figure 1b, 1c, Table 2, and Figure S1). Patients UPN1333 and UPN1125 were from large pedigrees in which several individuals had died of JMML (Figure 1b and c).
The proband in family 1 (UPN1333, V:1) (Figure 1a) was referred after transplantation for JMML. Initially diagnosed at 7 months of age, he received his first HSCT at age 13 months and then developed mixed chimerism 6 months later. A blood sample displayed a homozygous CBL mutation at c.1111T>C (Y371H). Importantly, analysis of buccal swab DNA revealed a heterozygous lesion. Both maternal relatives died from progressive JMML. Peripheral blood or buccal swabs from extended family members revealed multiple heterozygous individuals (Figure 1b). Interestingly, detailed medical history from the affected maternal great-grandmother revealed a history of infant leukemia characterized by a high white blood cell count and splenomegaly that resolved spontaneously. Sorted B (CD19) and T (CD3) cells, and granulocytes from her peripheral blood currently display a heterozygous c.1111T> C. Leukemia cells from UPN1333 displayed a classic pattern of GM-CSF hypersensitivity (Figure 1d) and increased phosphorylation of STAT5 in response to low doses of GM-CSF 28. Peripheral blood mononuclear cells from his heterozygous mother did not display either of these features, suggesting that homozygosity for the mutant CBL allele is essential for these cellular behaviors (data not shown).
The proband in family 2 (UPN1125, IV:3) was a girl diagnosed at 15 months of age with JMML. She also harbored a homozygous c.1111T>C CBL mutation in her bone marrow. Family history revealed that her mother had two male first cousins who were diagnosed with JMML and died before age 10. The first boy had archived frozen liver tissue available from autopsy and demonstrated a heterozygous c.1111T>C mutation. Interestingly, he also developed clinical signs and laboratory values consistent with small vessel vasculitis prior to his death. Her mother of UPN1125 was found to carry a heterozygous CBL mutation (Figure 1c).
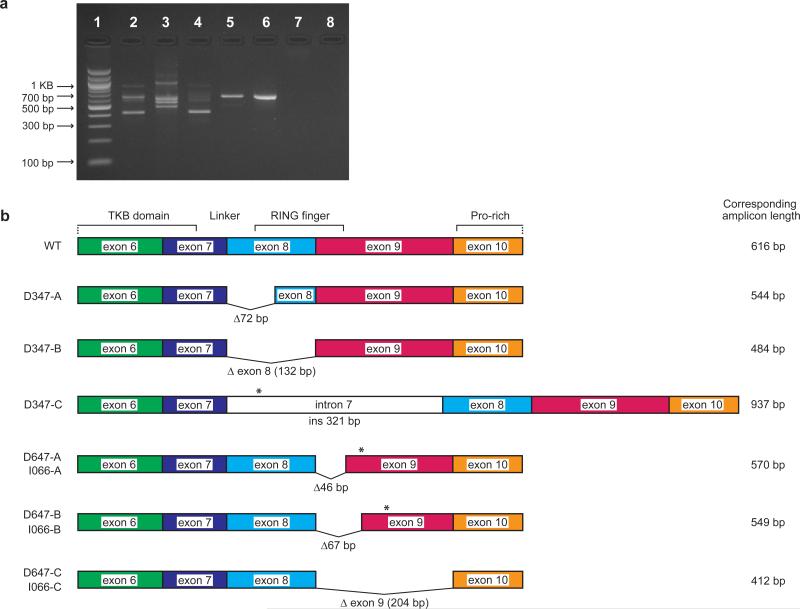
Three patients in our series displayed homozygous splice site mutations (Table 2, I066, D647, and D347). RT-PCR demonstrated several new splice products arising from these mutations (Figure 2); most notable are the 2 splice site variants that either delete the entirety of exon 8 (D347) or exon 9 (I066 and D647) (Figure S2) or retain an interstitial intron (e.g. intron 7 for D347). Each of the deletion splice variants encodes a protein that is predicted to lack critical regions of the linker and RING finger domains, while the intron 7 retention introduces a premature stop codon to abort translation upstream of the RING finger domain.
Figure 2. Consequences of splice site mutations in cDNA from individuals D347, 647, and I066.
Panel (a) RT-PCR using an exon 6 forward primer and an exon 10 reverse primer on cDNA generated from these patients. The wildtype amplicon is 616 base pairs long. Lane 1: MW ladder, Lane 2: I066, Lane 3: D347, Lane 4: D647, Lane 5: CBL point mutant, Lane 6: HM2833 CBL wildtype, Lane 7: Genomic DNA control, Lane 8: no template control. Panel (b) is a schematic representation of the splice site variants detected either recurrently (D347) or that were shared by I066 and D647. Sequences are shown in Figure S2. Deletions of base pairs are indicated by Δ# of base pairs and insertions by ins # base pairs. Premature stop codons are indicated by asterisks (*).
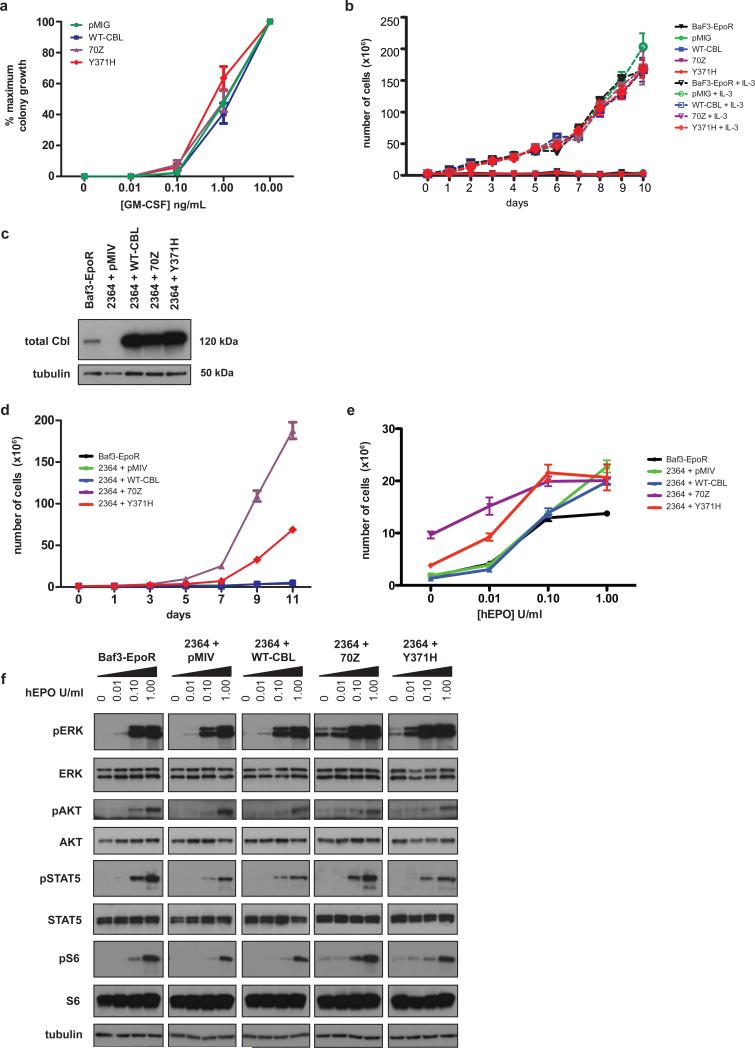
To investigate the functional properties of the variant mutant Cbl proteins encoded by homozygous point mutations, we first studied the effect of the common p.Y371H substitution on the growth of primary hematopoietic cells from murine fetal liver. This system reproduces the hypersensitivity to GM-CSF that is characteristic of JMML 16,29. Fetal liver cells transduced with retroviral vectors expressing wildtype or mutant Cbl proteins demonstrated no increased sensitivity to GM-CSF (Figure 3a). Similarly, expression of p.Y371H Cbl in a BaF3-EpoR cell line did not confer cytokine independence (Figure 3b). Based on the observation that JMML cells invariably lose the normal CBL allele and on recently published data21, we reasoned that reduction of normal Cbl expression might be mandatory to deregulate hematopoietic growth. Therefore, we introduced a short hairpin RNA, which markedly reduced the expression of murine Cbl in BaF3-EpoR cell lines (Figure 3c). We next transduced these cells with a series of wildtype and mutant human constructs and demonstrated strong expression of exogenous Cbl (Figure 3c). In this context, we observed cytokine independent proliferation (Figure 3d) upon expression of p.Y371H Cbl and a known murine oncogenic Cbl protein (70Z)30-32. The 70Z oncogenic protein deletes out 17 amino acids located from position 366-382 in the linker domain. Furthermore, the p.Y371H and 70Z transduced cells demonstrated hypersensitivity to increasing concentrations of human EPO (Figure 3e), mimicking the growth factor hypersensitivity seen in JMML (Figure 1d). In cells depleted of endogenous Cbl and expressing exogenous p.Y371H and 70Z Cbl proteins, we observed constitutive phosphorylation of ERK, AKT and S6 in cells deprived of cytokine. We also found heightened responses to low dose EPO (Figure 2f).
Figure 3. p.Y371H does not confer cytokine sensitivity or cytokine independent growth until silencing of murine Cbl.
Panel (a) Transduction of p.Y371H or the known murine oncogeneic mutant 70Z in wildtype hematopoietic cells from fetal liver, did not confer hypersensitivity to GM-CSF, nor did expression of these mutants in BaF3-EpoR cells result in cytokine independent growth (b). Panel (c) An shRNA to murine Cbl (cbl.2364) demonstrated near complete shutdown of expression in BaF3-EpoR cells by Western blot with re-expression upon introduction of the human WT, 70Z, or p.Y371H in these same cell lines. Panel (d) Both the p.Y371H and 70Z Cbl conferred cytokine independent growth in the presence of cbl.2364. Controls included the Venus vector pMIV and BaF3-EpoR. Error bars for triplicate replicates (s.e.m.) are shown and when not visible, indicate tight clustering. Using a paired t-test: day 7 comparing 2364+ WT-Cbl versus 2364+p.Y371H, p-value= 0.017, and at day 9: p-value <0.001. Panel (e) Serial transduction of the hairpin (2364) and p.Y371H or 70Z constructs also conferred hypersensitive growth after assessing cell proliferation on day 5 in increasing concentrations of Epo. Using a paired t-test at each concentration of Epo when comparing 2364+WT-Cbl versus 2364+p.Y371H: Epo 0 unit/ml: p = 0.036, Epo 0.01 units/ml: p= 0.0015, Epo 0.1 units/ml: p= 0.029, Epo 1 unit/ml (saturating dose) P= 0.697. Panel (f) Both the p.Y371H and 70Z containing cells demonstrated activation of pERK, pAKT, and pS6 in the absence of Epo or in low dose 0.01 unit/mL of Epo in comparison to negative controls. All cell proliferation work was done in triplicate.
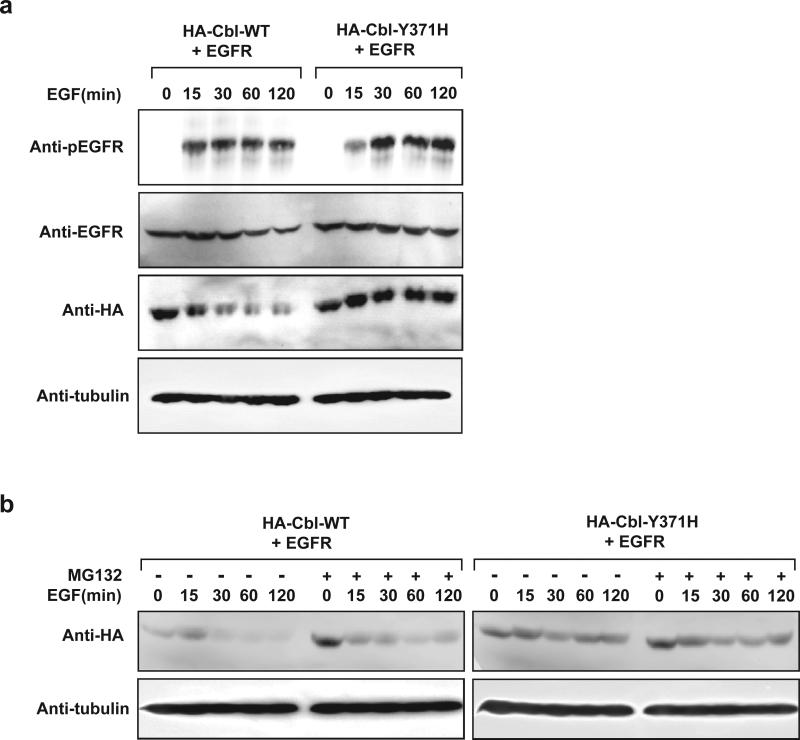
In order to determine if mutant Cbl proteins retain E3 ligase activity, we assessed their ability to promote ubiquitylation of a known Cbl substrate. Soon after activation, the epidermal growth factor receptor (EGFR) undergoes Cbl-dependent polyubiquitylation and proteosomal degradation. HA-Cbl(p.Y371H)-expressing HEK293 cells exhibited markedly elevated levels of phosphorylated EGFR upon EGF stimulation in comparison to HA-Cbl(WT)-expressing counterpart (Figure 4a), which indicates a possible defect in clearance of pEGFR by Cbl(p.Y371H). Cbl(p.C384R) mutant exhibited a similar defect in pEGFR clearance (data not shown). Upon polyubiquitylation of targets, Cbl promotes its own polyubiquitylation and subsequent auto-degradation. Consistent with this model, levels of Cbl(WT), but not Cbl(p.Y371H or p.C384R), rapidly diminished following EGF treatment in the absence of proteasome inhibitor MG132 (Figure 4a, b and data not shown,). However, Cbl(WT) levels were markedly stabilized in the presence of MG132 while Cbl(p.Y371H) levels remained relatively high irrespective of MG132 (Figure 4b), which suggest that Cbl mutants p.Y371H and p.C384R have an inherent defect in E3 function. Consistent with this notion, Cbl(WT) promoted robust polyubiquitylation of pEGFR while p.Y371H and p.C384R showed diminished capacity to polyubiquitylate pEGFR in an in vitro ubiquitylation assay (data not shown), similar to that of Cbl(70Z), which is known to be defective in E3 ligase activity, and in concordance with recently published data by Sanada et al 21, who demonstrated that p.Y371S and p.Q367P mutants detected in human patients also have diminished ubiquitylation activity. The Y371 residue most commonly affected in germline CBL mutations has been the focus of extensive biochemical analysis 21. This literature supports a key role for Y371 in maintaining the integrity of the alpha-helical structure of the linker region, which plays a critical role in substrate specificity. Interestingly, Y371 has been found to be phosphorylated despite its predicted location away from the protein surface. Substitution of this residue with phenylalanine results in a nononcogenic form of the protein, which lacks E3 ligase activity. Conversely substitution with a glutamate constitutively activates E3 ligase activity 33,34.
Figure 4. Cbl mutant proteins exhibit prolonged protein turnover and are associated with increased phosphorylated EGFR levels upon EGF stimulation.
Panel (a) HEK293 cells transfected with plasmids encoding EGFR in combination with HA-Cbl(WT) or HA Cbl(p.Y371H) were serum starved for 18 h followed by 15 min EGF (50 ng/ml) stimulation. Cells were then washed and maintained in serum-free media for the indicated periods of time. Equal amounts of whole cell extracts were resolved on SDS-PAGE and immunoblotted with the indicated antibodies. Panel (b) HEK293 cells transfected with plasmids encoding EGFR in combination with HA-Cbl(WT) or HA-Cbl(p.Y371H) were serum starved for 18 h followed by 15 min EGF (50 ng/ml) stimulation. Cells were then washed and maintained in serum-free media with (+) or without (-) MG132 for the indicated periods of time. Equal amounts of whole cell extracts were resolved on SDS-PAGE and immunoblotted with the indicated antibodies.
In most ways, CBL appears to function as a classic tumor suppressor gene in this cohort, with germline heterozygosity predisposing to neoplasia upon reduction to homozygosity in target tissues. However, the predominance of specific missense mutations makes CBL distinct from most tumor suppressor genes such as RB and NF1, which typically demonstrate more severe loss of function mutations such as deletion or protein truncation. This suggests that the mutant Cbl proteins retain an essential biochemical function 20,21. It is further supported by our data showing that exclusive expression of a mutant CBL allele has positive effects on cytokine signaling and proliferation. The specific disruption of E3 ligase activity may leave intact adapter functions, resulting in a relative imbalance of CBL's positive and negative roles in signal transduction. Sanada et al hypothesized that this may result in inhibitory effects on Cbl-b, a related family member21. This is similar to what has been reported for p53, a classic tumor suppressor gene with specific gain of function mutations in the context of loss of heterozygosity 35,36. Beyond missense mutations, truncated Cbl proteins are also known to confer transforming effects by countering the negative action of full-length Cbl on RTK signaling. The Drosophila analogue of the mammalian v-Cbl oncogene (i.e. Dv-Cbl), for example, functions as oncogenic dominant-negative variant whose expression results in vivo in enhanced signaling of the EGFR cascade and cooperates with activating mutations in the Ras pathway to ultimately produce melanotic tumors 37.
In addition, clinical data reveals that some patients may experience spontaneous resolution of JMML but go on to develop clinical features consistent with vasculitis and other autoimmune phenomena later in life. Interestingly, autoimmunity has also characterized genetically engineered mouse strains with Cbl mutations. Lck-Cre+ c-Cblflox/flox Cbl-b−/− mice, which delete c-Cbl in T cells, develop severe vascular lesions with massive infiltration of T-cells and high concentrations of anti-double stranded DNA antibodies 38. Furthermore, these T cells are hypersensitive to T cell receptor signaling and display prolonged ERK phosphorylation. Similarly, Cd19-Cre+ c-Cblflox/flox Cbl-b−/− mice that delete c-Cbl in B cells develop a lupus-like syndrome associated with perivascular infiltration and hyperactivation of B cell receptor signaling 39. In mice, loss of Cbl-b is required for these phenotypes. Oncogenic c-Cbl proteins thus may inhibit Cbl-b in vivo, resulting in a functional loss of both Cbl and Cbl-b, which in turn contributes to dysregulated lymphocyte signaling and subsequent vasculitis. Indeed, the redundant role of c-Cbl and Cbl-b was further explored by Sanada, et al, who demonstrated the inhibitory effects of c-Cbl p.Y371S on wildtype Cbl-b 21. It is of clinical interest that patients with JMML and homozygous CBL mutations who undergo HSCT are not known to develop vasculitis later in life, implying that a normal immune system is critical to preventing this late manifestation.
We describe a new syndrome in which affected children display several congenital anomalies that overlap with NF1, NS, and Legius, suggesting that the affected proteins converge on the Ras/MAPK pathway. Indeed, several Ras/MAPK pathway proteins regulate developmental programs in multiple species - for instance, the Drosophila homologs of each of these genes--CBL (D-cbl), PTPN11 (csw), NF1, and SPRED perform critical functions for growth and patterning 40-43.
Patients with germline CBL mutations are at increased risk of developing JMML, which may follow an aggressive clinical course or resolve without treatment. Some affected individuals develop vasculitis later in life. The CBL mutations found in JMML can arise de novo or can be transmitted through the germline, and human leukemia samples invariably show loss of the normal CBL allele. Consistent with this tumor suppressor function, JMML-associated Cbl proteins confer cytokine hypersensitivity in transduced BaF3-EpoR cells in the absence of wildtype Cbl, have defective E3 ligase activity, and constitutively activate key Ras effector pathways. The role of aberrant Cbl signaling in vasculitis remains to be determined, and it will be particularly interesting to investigate if patients with germline CBL mutations who have been cured of JMML after HSCT remain at risk of developing vasculitis. It is also of great interest that some of these patients continue to display homozygous CBL mutations in their peripheral blood despite having improved their blood counts. Finally, our data provide strong evidence that Cbl is a key negative regulator of Ras signaling networks in hematopoietic cells and it will be important to identify key targets of the Cbl ubiquitin ligase and to uncover other biochemical mechanisms involved in growth control.
METHODS
Subjects
Patients were diagnosed and treated either in Europe under the auspices of the European Working Group of Myelodysplastic Syndromes in Childhood (EWOG-MDS) or enrolled as research subjects at the University of California, San Francisco. The Committees on Human Research at each of the institutions in EWOG-MDS, as well as UCSF approved these studies. Informed consent was obtained from parents or guardians, and in the case of pedigree analysis, all screened relatives. Family and clinical histories were reviewed, as were physical exams at diagnosis.
Mutation Screening
Bone marrow or peripheral blood samples at diagnosis were obtained. Mononuclear cells were isolated using standard Histopaque 1111. Buccal swabs, fibroblasts, or tissues unaffected by tumor were also obtained when available. Genomic DNA was extracted using PureGene reagents (Qiagen, Foster City, CA). Patients were screened for mutations in CBL, NRAS, KRAS, and PTPN11 as previously described6,7,20.
RNA was prepared according to usual methods. cDNA was generated using the SuperScript® III First-Strand Synthesis System for RT-PCR (Invitrogen). Splice variants were identified through PCR using an annealing temperature of 58 °C and the following primers: 5’-TTGAGGGAACACATACTCGCT-3’ and 5’-TATGTTACTGCTGATGGGAACA-3’. Splice variants were gel extracted using the QIAquick Gel Extraction Kit (Qiagen). The resulting fragments were subcloned using the TOPO® TA Cloning Kit (Invitrogen) according to the manufacturer's instructions. Positive colonies were picked, mini prepped using the QIAprep Spin Miniprep Kit (Qiagen), and sequenced with standard M13 Forward (-20) and M13 Reverse primers.
Cbl and shRNA expression constructs
For the CFU-GM experiments, Gateway technology (Invitrogen) was used to clone WT and mutant Cbl cDNAs into the murine stem cell virus (MSCV) backbone containing a green fluorescent protein (GFP) cassette driven by an internal ribosome entry site (IRES) downstream of the Cbl sequence (pMIG). The human WT and 70Z Cbl plasmids were a kind gift from Hamid Band 44. For the subsequent experiments using Ba/F3 cells, the same Cbl cDNAs were cloned into an MSCV-IRES-Venus (pMIV) backbone to allow for co-transduction with the GFP-tagged shRNA. MiR30 based shRNA sequences targeting murine Cbl were graciously designed by Johannes Zuber and Scott Lowe. We selected one of these putative sequences (cbl.2364) and custom ordered a single 110-bp oligonucleotide (Bioneer) to serve as a template for PCR amplification. PCR products were digested with XhoI and EcoRI and ligated into the LTR-driven MiR30 SV40-GFP (LMS) MSCV-based vector (also graciously provided by the Lowe lab) to produce the LMS-2364 retrovirus encoding the Cbl shRNA. The sequence for cbl.2364 is TCGAGAAGGTATATTGCTGTTGACAGTGAGCGATACCTATGAAGCGATGTAT AATAGTGAAGCCACAGATGTATTATACATCGCTTCATAGGTACTGCCTACTG CCTCGG.
Hematopoietic progenitor assays
All experimental procedures involving mice were reviewed and approved by the UCSF Committee on Animal Research. These assays were performed as described previously using murine fetal liver cells transduced with MSCV-Cbl-IRES-GFP retroviruses engineered to express WT or mutant Cbl proteins. For the human CFU-GM assays, mononuclear cells from peripheral blood or bone marrow were plated in MethoCult H4230 (StemCell Technologies), supplemented with recombinant human GM-CSF (Peprotech) and counted 14 days later as described previously 28. For the CFU-GM assays, GFP-positive cells were sorted using a FACS Aria (BD Biosciences) and then seeded in methylcellulose medium (M3231; StemCell Technologies) 45, supplemented with recombinant murine GM-CSF (Peprotech). Colonies were counted by indirect microscopy after 8 days.
Cell Viability and Proliferation assays and Western blots
Murine pro-B Ba/F3 cells were transduced with MSCV-EpoR-IRES-puro as described previously 46. These cells were maintained in RPMI-1640 with 10% FCS (HyClone), penicillin, streptomycin, L-glutamine, 10 ng/ml puromycin (Calbiochem), and 10 µg/ml murine IL-3 (Peprotech).
The Ba/F3-EpoR cells were transduced with the LMS-2364 construct or the LMS vector alone. GFP-positive cells were sorted on a FACS Aria (BD Biosciences) and then transduced with the MSCV-Cbl-IRES-Venus retroviruses expressing WT or mutant Cbl proteins. Cells positive for both GFP and YFP expression were sorted on the FACS Aria and studied in proliferation and Western blot assays.
For the proliferation assays, cells were washed 3x and then cultured for 6 hours in cytokine-free media before being plated in 6-well plates at a density of 500,000 cells/ml at increasing doses of hEPO (R&D Systems). Growth was monitored every other day using a ViCell cell counter (Beckman Coulter).
For Western blot analysis, cells were washed 3x and cultured for 6 hours in cytokine-free media before being stimulated for 15 minutes with increasing doses of hEPO (R&D Systems). Whole-cell lysates were blotted and probed with the following antibodies: anti-phospho-p44/42 mitogen-activated protein kinase/extracellular signal-regulated kinase 1/2 (ERK1/2) (Thr202/Tyr204, cat. 9101), anti-phospho-AKT (S473, cat. 4060), anti-phospho-S6 (Ser235/236, cat. 2211) (all from Cell Signaling Technology); anti-phospho-STAT5 (cat. 44-390G) (Invitrogen) and anti-alpha-tubulin (Abnova). ERK1/2 (cat. 9102), AKT (cat. 9272), S6 (2217), and STAT5 (9363) antibodies were from Cell Signaling Technology.
HEKCells
HEK293 cells were obtained from the American Type Culture Collection (Rockville, MD) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Wisent, St. Bruno, QC, Canada) at 37°C in a humidified 5% CO2 atmosphere.
Antibodies
Mouse monoclonal antibodies against HA (12CA5) and α-tubulin were obtained from Boehringer Ingelheim (Laval, QC, Canada) and Sigma (Milwaukee, WI), respectively. Rabbit polyclonal antibodies against pEGFR, EGFR and ubiquitin were obtained from Upstate (Temecula, CA), Santa Cruz Biotechnology (Santa Cruz, CA) and DAKO Canada (Mississauga, ON, Canada), respectively. EGF ligand was obtained from Sigma (Milwaukee, WI). MG132 proteasome inhibitor was obtained from Boston Biochem (Cambridge, MA).
Plasmids
Plasmids encoding HA-CBL(WT, 70Z, Y371H, C384R) were subcloned into the pcDNA-DEST4.0 vector via Gateway Cloning technology (Invitrogen, Carlsbad, CA) and confirmed by DNA sequencing. Plasmid encoding EGFR was generated as previously described 47.
Immunoblotting and immunoprecipitation
Immunoprecipitation and immunoblotting were performed as described previously 48. Cells were lysed in EBC buffer (50mM Tris pH 8.0, 120mM NaCl and 0.5% NP-40) supplemented with protease and phosphatase inhibitors (Roche, Laval, QC, Canada). Cell lysates were immunoprecipitated with indicated antibodies in the presence of Protein-A agarose beads (Repligen, Waltham, MA). Bound proteins were washed five times with NETN buffer (20mM Tris pH 8.0, 120mM NaCl, 1mM EDTA, and 0.5% NP-40), eluted by boiling in sodium dodecyl sulfate (SDS)-containing sample buffer, and resolved by SDS-polyacrylamide gel electrophoresis (PAGE).
In vitro ubiquitylation assay
In vitro ubiquitylation assay was performed as described previously 49.
Supplementary Material
Acknowledgements
The authors would gratefully like to acknowledge the generous and selfless participation of the families included in this report.
Supported in part by the National Institutes of Health Grants CA113557 (M.L.L), the V-Foundation for Cancer Research (MLL and BSB), K08 CA103868 (NIH/NCI) (BSB) The Leukemia Lymphoma Society 6059-09, 2157-08 (MLL), the Frank A. Campini Foundation (MLL and BSB), The Concern Foundation (BSB) R01 CA104282 (NIH/NCI) (MLL and BSB), Deutsche Forschungsgemeinschaft (Grant No. KR3473/1-1 to CF), Deutsche Krebshilfe (Grant No. 108220 to CN, CF), Deutsche José Carreras Leukämiestiftung (Grant No. R08/19 to CF), and the Canadian Cancer Society (Grant No. 16056 to MO).
DHS is supported by T32GM007618 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
MLL is a Clinical Scholar of the Leukemia Lymphoma Society
Footnotes
Author Contributions:
CMN coordinated and collected clinical data from the EWOG-MDS patients and wrote the manuscript; MWK collected clinical data, performed laboratory assays including sequencing, proliferation assays, and prepared figures; DHS performed laboratory assays including the shRNA experiments, the proliferation assays and western blots; IF collected clinical data; ME collected patients samples and performed mutational analysis on highly purified populations of blood cells; NJB contributed patient samples; SB performed ubiquitin assays; JZF contributed patient samples; TAG performed RNA isolation and cDNA sequencing; PM contributed patient samples, IS, GH, SC, PJL, CK and PGS contributed patient samples, AH provided age matched control samples from children with asthma; MS performed mutational analysis; JS, MMvH, HH, FL contributed patient samples and collected clinical data; DS collected clinical data; SA performed colony assays, performed cDNA sequencing; LC collected clinical data; RCR performed ubiquitylation assays; SSS performed ubiquitylation assays; MO supervised the ubiquitylation assays and wrote the manuscript; BSB oversaw the shRNA and cell proliferative experiments and wrote the manuscript; CF collected clinical data and performed sequencing; MLL coordinated and collected clinical and laboratory data from the U.S., oversaw all of the laboratory work, coordinated the data, and wrote the manuscript.
Gene Bank accession number used for sequencing: nm_005188.2
References
- 1.Niemeyer CM, Kratz CP. Paediatric myelodysplastic syndromes and juvenile myelomonocytic leukaemia: molecular classification and treatment options. Br J Haematol. 2008;140:610–24. doi: 10.1111/j.1365-2141.2007.06958.x. [DOI] [PubMed] [Google Scholar]
- 2.Locatelli F, et al. Hematopoietic stem cell transplantation (HSCT) in children with juvenile myelomonocytic leukemia (JMML): results of the EWOG-MDS/EBMT trial. Blood. 2005;105:410–9. doi: 10.1182/blood-2004-05-1944. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda K, et al. Spontaneous improvement of hematologic abnormalities in patients having juvenile myelomonocytic leukemia with specific RAS mutations. Blood. 2007;109:5477–80. doi: 10.1182/blood-2006-09-046649. [DOI] [PubMed] [Google Scholar]
- 4.Flotho C, et al. Genotype-phenotype correlation in cases of juvenile myelomonocytic leukemia with clonal RAS mutations. Blood. 2008;111:966–7. doi: 10.1182/blood-2007-09-111831. author reply 967-8. [DOI] [PubMed] [Google Scholar]
- 5.Niemeyer CM, et al. Chronic myelomonocytic leukemia in childhood: a retrospective analysis of 110 cases. European Working Group on Myelodysplastic Syndromes in Childhood (EWOG-MDS). Blood. 1997;89:3534–43. [PubMed] [Google Scholar]
- 6.Kalra R, Paderanga D, Olson K, Shannon KM. Genetic analysis is consistent with the hypothesis that NF1 limits myeloid cell growth through p21ras. Blood. 1994;84:3435–3439. [PubMed] [Google Scholar]
- 7.Loh ML, et al. Somatic mutations in PTPN11 implicate the protein tyrosine phosphatase SHP-2 in leukemogenesis. Blood. 2003 doi: 10.1182/blood-2003-09-3287. [DOI] [PubMed] [Google Scholar]
- 8.Shannon KM, et al. Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. N Engl J Med. 1994;330:597–601. doi: 10.1056/NEJM199403033300903. [DOI] [PubMed] [Google Scholar]
- 9.Tartaglia M, et al. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34:148–50. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- 10.Levine RL, et al. The JAK2V617F activating mutation occurs in chronic myelomonocytic leukemia and acute myeloid leukemia, but not in acute lymphoblastic leukemia or chronic lymphocytic leukemia. Blood. 2005;106:3377–3379. doi: 10.1182/blood-2005-05-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onida F, et al. Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients. Blood. 2002;99:840–9. doi: 10.1182/blood.v99.3.840. [DOI] [PubMed] [Google Scholar]
- 12.Braun BS, et al. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004;101:597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan IT, et al. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest. 2004;113:528–38. doi: 10.1172/JCI20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le DT, et al. Somatic Inactivation of Nf1 in Hematopoietic Cells Results in a Progressive Myeloproliferative Disorder. Blood. 2004 doi: 10.1182/blood-2003-08-2650. [DOI] [PubMed] [Google Scholar]
- 15.Mohi MG, et al. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer Cell. 2005;7:179–91. doi: 10.1016/j.ccr.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Emanuel PD, Bates LJ, Castleberry RP, Gualtieri RJ, Zuckerman KS. Seletive hypersensitivity to granulocyte-macrophage colony stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood. 1991;77:925–929. [PubMed] [Google Scholar]
- 17.Ramshaw HS, Bardy PG, Lee MA, Lopez AF. Chronic myelomonocytic leukemia requires granulocyte-macrophage colony- stimulating factor for growth in vitro and in vivo. Exp Hematol. 2002;30:1124–31. doi: 10.1016/s0301-472x(02)00903-7. [DOI] [PubMed] [Google Scholar]
- 18.Dunbar AJ, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008;68:10349–57. doi: 10.1158/0008-5472.CAN-08-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grand FH, et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood. 2009 doi: 10.1182/blood-2008-12-194548. [DOI] [PubMed] [Google Scholar]
- 20.Loh ML, et al. Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood. 2009;114:1859–63. doi: 10.1182/blood-2009-01-198416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanada M, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460:904–8. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 22.Makishima H, et al. Mutations of e3 ubiquitin ligase cbl family members constitute a novel common pathogenic lesion in myeloid malignancies. J Clin Oncol. 2009;27:6109–16. doi: 10.1200/JCO.2009.23.7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bader-Meunier B, et al. Occurrence of myeloproliferative disorder in patients with the Noonan syndrome. J Pediatr. 1997;130:885–889. doi: 10.1016/s0022-3476(97)70273-7. [DOI] [PubMed] [Google Scholar]
- 24.Tartaglia M, et al. PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. Am J Hum Genet. 2002;70:1555–63. doi: 10.1086/340847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rauen KA, et al. Proceedings from the 2009 genetic syndromes of the Ras/MAPK pathway: From bedside to bench and back. Am J Med Genet A. 152A:4–24. doi: 10.1002/ajmg.a.33183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan RJ, Cooper T, Kratz CP, Weiss B, Loh ML. Juvenile myelomonocytic leukemia: A report from the 2nd International JMML Symposium. Leuk Res. 2008 doi: 10.1016/j.leukres.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasle H, et al. A pediatric approach to the WHO classification of myelodysplastic and myeloproliferative diseases. Leukemia. 2003;17:277–82. doi: 10.1038/sj.leu.2402765. [DOI] [PubMed] [Google Scholar]
- 28.Kotecha N, et al. Single-Cell Profiling Identifies Aberrant STAT5 Activation in Myeloid Malignancies with Specific Clinical and Biologic Correlates. Cancer Cell. 2008;14:335–43. doi: 10.1016/j.ccr.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schubbert S, et al. Functional analysis of leukemia-associated PTPN11 mutations in primary hematopoietic cells. Blood. 2005;106:311–7. doi: 10.1182/blood-2004-11-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langdon WY, Hyland CD, Grumont RJ, Morse HC., 3rd The c-cbl proto-oncogene is preferentially expressed in thymus and testis tissue and encodes a nuclear protein. J Virol. 1989;63:5420–4. doi: 10.1128/jvi.63.12.5420-5424.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andoniou CE, Thien CB, Langdon WY. Tumour induction by activated abl involves tyrosine phosphorylation of the product of the cbl oncogene. Embo J. 1994;13:4515–23. doi: 10.1002/j.1460-2075.1994.tb06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blake TJ, Shapiro M, Morse HC, 3rd, Langdon WY. The sequences of the human and mouse c-cbl proto-oncogenes show v-cbl was generated by a large truncation encompassing a proline-rich domain and a leucine zipper-like motif. Oncogene. 1991;6:653–7. [PubMed] [Google Scholar]
- 33.Kassenbrock CK, Anderson SM. Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. J Biol Chem. 2004;279:28017–27. doi: 10.1074/jbc.M404114200. [DOI] [PubMed] [Google Scholar]
- 34.Thien CB, Langdon WY. Cbl: many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 35.Dittmer D, et al. Gain of function mutations in p53. Nat Genet. 1993;4:42–6. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- 36.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–72. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Robertson H, Hime GR, Lada H, Bowtell DD. A Drosophila analogue of v-Cbl is a dominant-negative oncoprotein in vivo. Oncogene. 2000;19:3299–308. doi: 10.1038/sj.onc.1203624. [DOI] [PubMed] [Google Scholar]
- 38.Naramura M, et al. c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR down-modulation. Nat Immunol. 2002;3:1192–9. doi: 10.1038/ni855. [DOI] [PubMed] [Google Scholar]
- 39.Kitaura Y, et al. Control of the B cell-intrinsic tolerance programs by ubiquitin ligases Cbl and Cbl-b. Immunity. 2007;26:567–78. doi: 10.1016/j.immuni.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hashimoto S, Nakano H, Singh G, Katyal S. Expression of Spred and Sprouty in developing rat lung. Mech Dev. 2002;119(Suppl 1):S303–9. doi: 10.1016/s0925-4773(03)00132-1. [DOI] [PubMed] [Google Scholar]
- 41.Oishi K, et al. Transgenic Drosophila models of Noonan syndrome causing PTPN11 gain-of-function mutations. Hum Mol Genet. 2006;15:543–53. doi: 10.1093/hmg/ddi471. [DOI] [PubMed] [Google Scholar]
- 42.Pai LM, Barcelo G, Schupbach T. D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell. 2000;103:51–61. doi: 10.1016/s0092-8674(00)00104-5. [DOI] [PubMed] [Google Scholar]
- 43.The I, et al. Rescue of a Drosophila NF1 mutant phenotype by protein kinase A. Science. 1997;276:791–4. doi: 10.1126/science.276.5313.791. [DOI] [PubMed] [Google Scholar]
- 44.Levkowitz G, et al. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–74. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schubbert S, et al. Biochemical and functional characterization of germ line KRAS mutations. Mol Cell Biol. 2007;27:7765–70. doi: 10.1128/MCB.00965-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mullighan CG, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2009;106:9414–8. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Huang DY, Huong SM, Huang ES. Integrin alphavbeta3 is a coreceptor for human cytomegalovirus. Nat Med. 2005;11:515–21. doi: 10.1038/nm1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohh M, et al. The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol Cell. 1998;1:959–68. doi: 10.1016/s1097-2765(00)80096-9. [DOI] [PubMed] [Google Scholar]
- 49.Ohh M, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–7. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.