Summary
BRAF and MEK inhibitors are effective in BRAF mutant melanoma, but most patients eventually relapse with acquired resistance, and others present intrinsic resistance to these drugs. Resistance is often mediated by pathway reactivation through receptor tyrosine kinase (RTK)/SRC-family kinase (SFK) signaling or mutant NRAS, which drive paradoxical reactivation of the pathway. We describe pan-RAF inhibitors (CCT196969, CCT241161) that also inhibit SFKs. These compounds do not drive paradoxical pathway activation and inhibit MEK/ERK in BRAF and NRAS mutant melanoma. They inhibit melanoma cells and patient-derived xenografts that are resistant to BRAF and BRAF/MEK inhibitors. Thus, paradox-breaking pan-RAF inhibitors that also inhibit SFKs could provide first-line treatment for BRAF and NRAS mutant melanomas and second-line treatment for patients who develop resistance.
Graphical Abstract
Highlights
-
•
pan-RAF inhibitors also inhibit SRC family kinases
-
•
The compounds do not induce paradoxical activation of ERK in RAS mutant cells
-
•
The compounds are active in BRAF and NRAS mutant melanomas
-
•
The compounds are active in PDXs resistant to BRAF or BRAF plus MEK inhibitors
Girotti et al. describe two pan-RAF inhibitors that also inhibit SRC-family kinases. These compounds do not drive paradoxical MEK/ERK activation and can inhibit MEK in NRAS mutant cells. Moreover, the agents can overcome resistance to clinical BRAF or combination BRAF/MEK inhibitors in patient-derived xenografts.
Significance
BRAF inhibitors are active in BRAF mutant melanoma patients, but the majority of patients will eventually develop resistance or present intrinsic resistance and so will not respond to BRAF inhibitors, despite the presence of a BRAF mutation. Here, we describe pan-RAF inhibitors that also target SRC and that are active in tumors from patients who developed resistance to BRAF-selective inhibitors and a BRAF plus MEK inhibitor combination. These compounds, therefore, provide vital second-line targeted therapies for relapsed patients, and a compound from the series is being developed to enter clinical trials.
Introduction
Malignant melanoma is the most deadly form of skin cancer. Current estimations are that each year there are >76,000 cases of melanoma with >9,000 deaths in the U.S. (www.cancer.org; American Cancer Society). In 2008, >100,000 cases with 22,000 deaths were estimated in Europe (Forsea et al., 2012), and >12,000 cases with ∼1,500 deaths were estimated in Australia (http://www.melanoma.org.au; Melanoma Institute Australia). Critically, 43%–50% of melanomas carry somatic mutations in BRAF, and those in another 20% carry mutations in NRAS (www.sanger.ac.uk/genetics/CGP/cosmic/). The mutant proteins are active and constitutively activate the RAS-RAF-MEK-ERK pathway, driving cancer cell proliferation and survival and, thereby, tumor progression.
Vemurafenib is an orally available and clinically active small-molecule inhibitor of BRAF that achieves increased progression-free and overall survival of patients with BRAF mutant melanoma, but not those with BRAF wild-type melanoma (Chapman et al., 2011; Flaherty et al., 2010; Sosman et al., 2012). However, despite initially impressive responses, most patients treated with vemurafenib develop acquired resistance after a relatively short period of disease control. Furthermore, ∼20% of patients having BRAF mutant melanoma present intrinsic resistance and do not respond to vemurafenib. Thus, resistance is a persistent clinical problem in the management of BRAF mutant melanoma, and second-line treatments are urgently required for patients with both intrinsic and acquired resistance to BRAF inhibitors.
Many mechanisms of resistance to BRAF inhibitors have been described, but in the majority of cases, it results from reactivation of the MEK/ERK pathway (Girotti et al., 2013; Johannessen et al., 2010; Nazarian et al., 2010; Shi et al., 2012; Straussman et al., 2012; Vergani et al., 2011; Villanueva et al., 2010; Wilson et al., 2012). Thus, amplification or upregulation of growth factors or receptor tyrosine kinases (RTKs), which signal through the SRC-family kinases (SFKs), can lead to pathway reactivation and resistance. Similarly, acquisition of secondary mutations in NRAS, which signals through CRAF (a close relative of BRAF), can also lead to resistance. In addition, amplification of mutant BRAF or alternative splicing of mutant BRAF mRNA, upregulation of the MEK kinase COT, or mutations in MEK can also drive resistance.
In addition to resistance, BRAF inhibitors mediate a curious paradox. Although they inhibit MEK/ERK signaling in BRAF mutant cells, they activate MEK/ERK signaling in RAS mutant cells. This is because, in the presence of oncogenic RAS, BRAF inhibitors drive the formation of BRAF-CRAF hetero- and homodimers containing one partner that is drug bound and one partner that is drug-free. The drug-bound partner drives activation of the drug-free partner through scaffolding or conformational functions, activating CRAF and, consequently, stimulating MEK and ERK hyperactivation (Hatzivassiliou et al., 2010; Heidorn et al., 2010; Poulikakos et al., 2010). In some contexts, paradoxical activation of the pathway can stimulate tumor growth and progression.
To overcome both resistance and paradoxical activation of the MEK/ERK pathway, strategies to achieve increased inhibition of the pathway by combined targeting of BRAF and MEK have been tested. The combination of dabrafenib, a BRAF inhibitor, with trametinib, a MEK inhibitor, was recently approved by the U.S. Food and Drug Administration for treating patients with mutant BRAF melanomas, based on phase II clinical trial data that show that the combination achieved higher response rates, longer median progression-free survival, and less cutaneous toxicity than dabrafenib alone (Flaherty et al., 2012; Long et al., 2014). However, despite these improved responses, patients on this drug combination still develop resistance, and most patients relapse after ∼9 months of treatment; furthermore, a recent study reported that, in these patients, resistance can be mediated by acquired mutations in MEK2 (Wagle et al., 2014). Independent of the mechanisms of resistance, there is an urgent need for second-line treatments for BRAF mutant melanoma patients who develop resistance to BRAF inhibitor mono- and combination therapies.
Results
CCT196969 and CCT241161 Are pan-RAF Inhibitors with Anti-SRC Activity
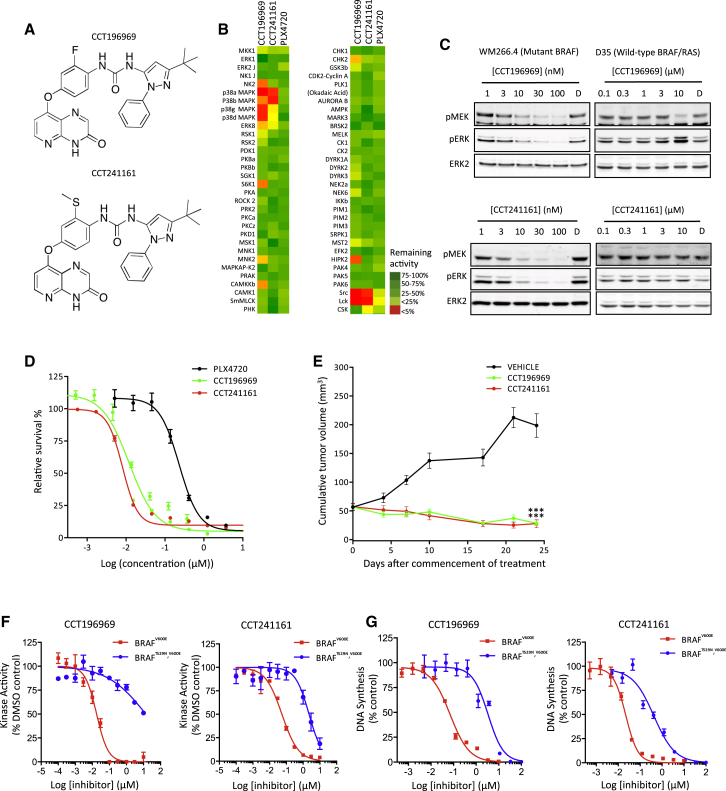
As previously described, we have pursued a drug discovery program in which we designed, synthesized, and characterized inhibitors of the inactive conformation of BRAFV600E (Ménard et al., 2009; Niculescu-Duvaz et al., 2009; Suijkerbuijk et al., 2010; Whittaker et al., 2010b; Zambon et al., 2010). Here, we describe two further inhibitors, CCT196969 and CCT241161 (Figure 1A; synthesis and characterization are described in the Supplemental Experimental Procedures available online). These compounds were found to inhibit BRAF, CRAF, and SFKs (Table 1). Since resistance to BRAF and BRAF/MEK inhibitors can be driven by RTKs signaling through SFKs, or mutant NRAS signaling through CRAF, we selected these compounds for further study. CCT196969 inhibits BRAF at 100 nM and BRAFV600E at 40 nM, while CCT241161 inhibits BRAF at 252 nM and BRAFV600E at 15 nM (Table 1). CCT196969 inhibits CRAF at 12 nM, SRC at 26 nM, and LCK at 14 nM, while CCT241161 inhibits CRAF at 6 nM, SRC at 15 nM, and LCK at 3 nM (Table 1). Neither compound inhibits MEK1 or the MEK1 kinase COT (Table 1), and, in a panel of protein kinases, they only inhibit SRC, LCK, and the p38 mitogen-activated protein kinases (MAPKs) (Figure 1B). Both inhibit MEK and ERK in WM266.4 cells (BRAF mutant), but not D35 cells (BRAF/RAS wild-type) (Figure 1C), and both inhibit growth of BRAF mutant melanoma cells more potently than PLX4720, an analog of the BRAF-selective inhibitor vemurafenib that has superior bioavailability in mice (Su et al., 2012a) (Figure 1D).
Figure 1.
CCT196969 and CCT241161 Are BRAF Inhibitors
(A) Chemical structures of CCT196969 and CCT241161.
(B) Efficacy of PLX4720, CCT196969, and CCT241161 (1 μM) against a panel of 63 protein kinases. Color bar shows percent activity compared to DMSO.
(C) Phospho-MEK (pMEK), phospho-ERK (pERK), and ERK2 in WM266.4 and D35 cells treated for 24 hr with DMSO (D), CCT196969, or CCT241161.
(D) BRAFV600E A375 cell proliferation assay (CellTiter Glo) with PLX4720, CCT196969, or CCT241161.
(E) BRAFV600E A375 xenograft growth in nude mice treated with vehicle, CCT196969, or CCT241161 14 days after cell injection. ∗∗∗p ≤ 0.001 (t test, two-tailed).
(F) BRAFV600E and BRAFT529N,V600E kinase inhibition by CCT196969 and CCT241161.
(G) Growth of Ba/F3 cells ([3H]-thymidine incorporation) expressing BRAFV600E or BRAFT529N,V600E treated with CCT196969 or CCT241161.
Bars represent SEM. See also Figure S1 and Tables S1–S3.
Table 1.
IC50 Values of CCT196969 and CCT241161
| Kinase | CCT196969 (μM ) | CCT241161 (μM) |
|---|---|---|
| BRAF | 0.1 | 0.03 |
| V600E-BRAF | 0.04 | 0.015 |
| CRAF | 0.01 | 0.006 |
| MEK1 | >10 | >10 |
| COT | >10 | >10 |
| SRC | 0.03 | 0.01 |
| LCK | 0.02 | 0.003 |
CCT196969 and CCT241161 Are Well Tolerated pan-RAF Inhibitors
A comprehensive safety profile analysis on CCT196969 shows that the compound is extremely well tolerated at the doses assessed and does not produce any significant adverse effects in vivo. A single dose at 20 mg/kg does not produce any clinical signs and produces no observed adverse effects in CD-1 mice. When administered at 40 mg/kg, we observed slight, transient tachypnoea 1 hr after dosing, but no effect on body weight, so 40 mg/kg is defined as the maximum tolerated dose (single dose). At 20 mg/kg daily × 24 days, we observed no clinical signs or body weight loss, and at 25 mg/kg daily for 19 days, we did not observe any mortality, although tachypnoea with decreased activity and excitation were seen. However, as mentioned earlier, the treated group did not show any body weight loss or reduction in food intake. In addition, at the end of the study, microscopical examination of tissues did not identify any treatment-related changes.
Oral dosing at 10 mg/kg/day results in plasma concentrations ∼1 μM at 24 hr and 14 hr, for CCT196969 and CCT241161, respectively (Figure S1A; Table S1), with areas under the curve of ∼416,000 and ∼275,000 nM.hr, respectively. These compounds are equally orally bioavailable at ∼55%, and even at 10 mg/kg/day, we achieve plasma levels well above the half-maximal inhibition of cell proliferation (GI50) values for BRAF-selective-inhibitor-resistant cells (GI50 = 0.4 μM, mean value from Figure 2A) and NRAS mutant melanoma cells (GI50 = 0.6 μM, mean value from Figure 2A). We confirm that doses of 30 mg/kg for 4 days do not cause significant weight loss (Figure S1B), so we selected 20 mg/kg/day (7 days/week; no weekend break) based on efficacy and tolerability. Critically, at these doses, we achieve tumor regression with BRAF mutant A375 tumor xenografts in nude mice (Figure 1E), although CCT196969 is also effective at 10 mg/kg/day (data not shown). CCT196969 and CCT241161 achieve plasma exposures of ∼40 μM and ∼50 μM, respectively (Table S2), which are similar to those seen for vemurafenib in humans (Flaherty et al., 2010). Note, also, that 1 hr after the last dose was administered at the end of the therapy experiments, the concentration of drug in the tumors was ∼7 μM for CCT196969 and 6.5–10 μM for CCT241161 (Table S3), levels that are well above the GI50 values for growth inhibition of cancer cells.
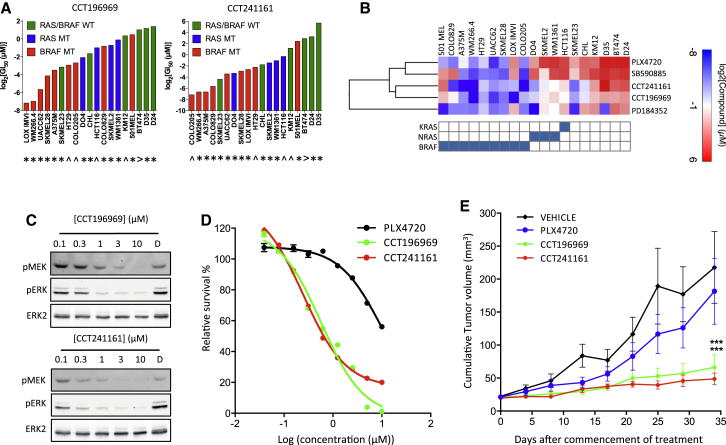
Figure 2.
CCT196969 and CCT241161 Inhibit RAS Mutant Cells
(A) Cell growth inhibition by CCT196969 or CCT241161 (expressed as log2 GI50 in micromolar) in cells carrying BRAF (red), RAS (blue), or neither (green) mutation. ∗, melanoma cell line; ˆ, colorectal carcinoma cell line; >, breast cancer cell line. WT, wild-type.
(B) Heat map showing sensitivity of cancer cell lines bearing mutations in BRAF, NRAS, or KRAS (shown in the grid below the heat map) presented as GI50 values determined after a 5-day exposure to each compound (BRAF inhibitors PLX4720 and SB590885, MEK inhibitor PD184352, and our compounds CCT241161 and CCT196969) and analysis by sulphorhodamine B staining. Values were log2-transformed, and hierarchical clustering was performed with “one minus Pearson correlation” using Gene E (www.broadinstitute.org/cancer/software/GENE-E/).
(C) Phospho-MEK (pMEK), phospho-ERK (pERK), and ERK2 in D04 cells treated for 24 hr with DMSO (D), CCT196969, or CCT241161.
(D) NRAS mutant D04 cell proliferation assay (CellTiter Glo) with PLX4720, CCT196969, or CCT241161.
(E) NRAS mutant D04 xenograft growth in nude mice treated with vehicle, PLX4720, CCT196969, or CCT241161 10 days after cell injection. ∗∗∗p ≤ 0.001 (t test, two-tailed).
Bars represent SEM. See also Figure S2.
To directly test if CCT196969 and CCT241161 are BRAF inhibitors, we replaced the “gatekeeper” threonine 529 (T529) in BRAF with asparagine to block drug binding without compromising kinase activity (Whittaker et al., 2010a). We saw that CCT196969 is 753-fold and CCT241161 is 42-fold less active against BRAFT529N,V600E than BRAFV600E (Figure 1F), demonstrating that the T529N substitution impairs binding of these drugs to BRAFV600E. To test the effects of these mutations in cells, we used Ba/F3 cells. As we have shown previously, Ba/F3 cells grow in an interleukin-3 (IL-3)-dependent manner, but when transformed with BRAFV600E and BRAFT529N,V600E, their growth becomes IL-3 independent but dependent on oncogenic BRAF (Whittaker et al., 2010a). Critically, we show that the growth of Ba/F3 cells transformed with BRAFT529N,V600E is 48-fold and 19-fold less sensitive to CCT196969 and CCT241161, respectively, than cells transformed with BRAFV600E (Figure 1G), demonstrating directly that these drugs inhibit BRAFV600E in cells.
CCT196969 and CCT241161 Are Paradox Breakers
Taken together, the aforementioned data confirm that CCT196969 and CCT241161 are orally available, well-tolerated BRAF inhibitors that directly inhibit BRAFV600E in cells. We show that CCT196969 and CCT241161 are active against melanoma and colorectal cancer cell lines that are mutant for BRAF (Figures 2A and 2B). In addition, unlike the BRAF-selective inhibitors PLX4720 and SB590885, but in common with the MEK inhibitor PD184352, CCT196969 and CCT241161 are also active against RAS mutant melanoma and colorectal cancer cells (Figures 2A and 2B). In general, CCT196969 and CCT241161 are not active against cancer cells that are wild-type for BRAF and NRAS, but curiously, SK-Mel 23 cells are sensitive to these compounds (Figure 2A). The reasons for this are unclear, but ERK activity is elevated in these cells and sensitive to CCT196969 and CCT241161 (Figure S2), suggesting that their growth depends on this pathway, presumably due to events upstream of RAS. Notably, in contrast to previously described BRAF inhibitors (Hatzivassiliou et al., 2010; Heidorn et al., 2010; Poulikakos et al., 2010), CCT196969 and CCT241161 inhibit rather than activate MEK in NRAS mutant cells (Figure 2C), and they inhibit NRAS mutant cell growth more efficiently than does PLX4720 (Figure 2D). Furthermore, in contrast to the BRAF inhibitor PLX4720, CCT196969 and CCT241161 inhibit the growth of NRAS mutant DO4 tumor xenografts in nude mice (Figure 2E). Thus, CCT196969 and CCT241161 are paradox-breaking pan-RAF inhibitors that are active against both BRAF mutant and NRAS mutant melanomas.
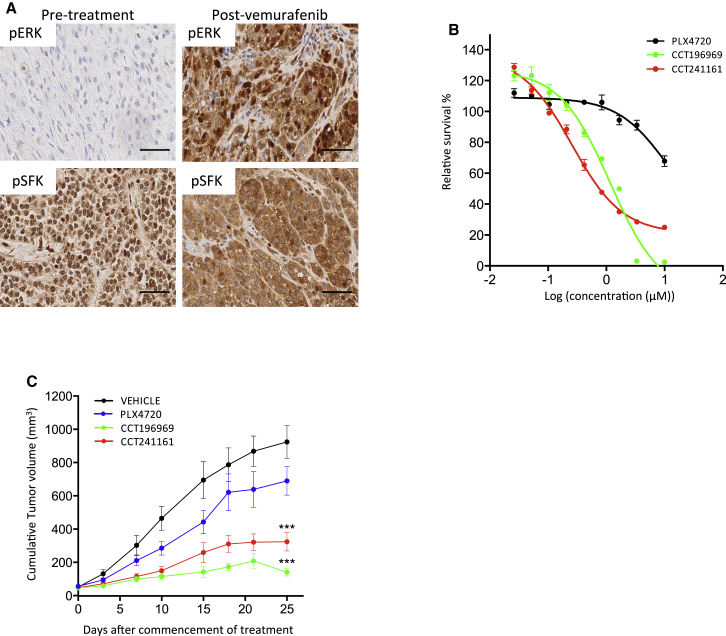
CCT196969 and CCT241161 Inhibit BRAF-Inhibitor-Resistant Melanoma Cell Lines
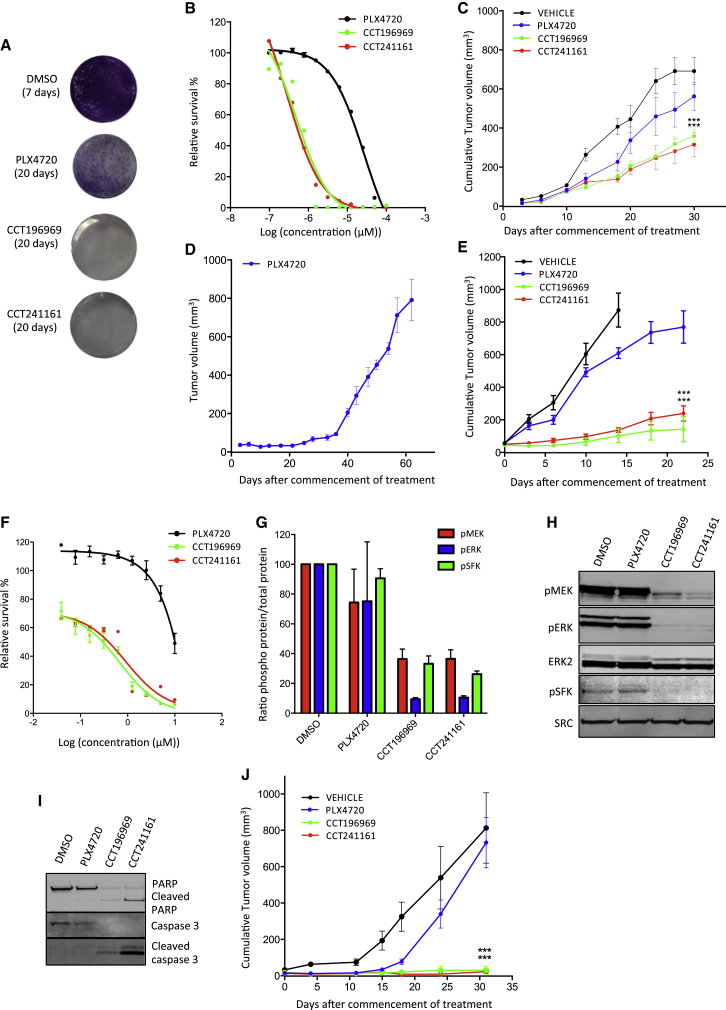
We tested whether our compounds are active in melanomas that are resistant to BRAF inhibitors. A375 cells that are continually exposed to PLX4720 developed resistance as demonstrated by the regrowth of cells after 20 days, but no cells are able to grow in parallel cultures exposed to CCT196969 or CCT241161 (Figure 3A). Note that A375 cells that have developed resistance to PLX4720 following continual exposure to the drug are still sensitive to CCT196969 and CCT241161 (Figure 3B), and more important, CCT196969 and CCT241161 inhibit the growth of PLX4720-resistant A375 xenografts (A375/R) in mice (Figure 3C), without causing any body weight loss to the mice (Figure S3A). Next, we induced resistance to PLX4720 in a patient-derived xenograft (PDX) from a patient (patient #1; Table S4) who presented stage III BRAF mutant melanoma and had a tumor removed for palliation. The tumor was propagated in immunocompromised mice, and the mice were then treated with PLX4720 until the tumor developed resistance (Figures 3D and S3A). Note that despite its resistance to PLX4720, this tumor remains sensitive to CCT196969 and CCT241161 (Figure 3E).
Figure 3.
CCT196969 and CCT241161 Inhibit SFK in Patient-Derived Resistant Cells
(A) A375 cell colony formation with DMSO, PLX4720, CCT196969, or CCT241161 (0.5 μM) after 7 or 20 days.
(B) A375/R cell proliferation assay (CellTiter Glo) with PLX4720, CCT196969, and CCT241161.
(C) A375/R xenograft growth in nude mice treated with vehicle, PLX4720, CCT196969, or CCT241161. ∗∗∗p ≤ 0.001 (t test, two-tailed).
(D) Patient #1 PDX growth in NSG mice treated with PLX4720.
(E) Growth of PLX4720-resistant PDX from patient #1, from (D), in mice treated with PLX4720, CCT196969, or CCT241161 7 days after cell injection. ∗∗∗p ≤ 0.001 (t test, two-tailed).
(F) Patient #2 cell proliferation assay (CellTiter Glo) with PLX4720, CCT196969, or CCT241161.
(G) RPPA quantification for pMEK, pERK, and pSFK in three vemurafenib-resistant patient-derived cell lines treated with DMSO (vehicle), PLX4720, CCT196969, and CCT241161 (1 μM; 4 hr).
(H) pMEK, pERK, ERK2, pSFK, and SRC in patient #2 cells treated with DMSO, PLX4720, CCT196969, or CCT241161 (1 μM; 4 hr).
(I) PARP and caspase 3 in patient #2 cells treated with DMSO, PLX4720, CCT196969, or CCT241161 (1 μM; 4 hr).
(J) Patient #2 cell xenograft growth in nude mice treated with vehicle, PLX4720, CCT196969, or CCT241161 16 days after tumor implant. ∗∗∗p ≤ 0.001 (t test, two-tailed).
Bars represent SEM. See also Figure S3 and Tables S4 and S5.
We also examined our inhibitors in samples from a second patient (patient #2; Table S4), who presented stage IV BRAF mutant metastatic melanoma and achieved a partial response to vemurafenib but relapsed after only 3 months. A cell line derived from a vemurafenib-resistant melanoma is resistant to PLX4720 but sensitive to CCT196969 and CCT241161 (Figure 3F), so we treated this cell line and two other cell lines (biological replicates)—derived from two patients who developed resistance to vemurafenib—with PLX4720, CCT196969, or CCT241161 and performed reverse phase protein arrays (RPPAs) to examine the phosphorylation of 25 proteins. For most of the proteins, we did not observe significant differences following treatment with any of the compounds, but MEK, ERK and SRC phosphorylation were strongly suppressed by CCT196969 and CCT241161, but not by PLX4720 (Figure 3G; Table S5). We confirm that CCT196969 and CCT241161 inhibit MEK, ERK, and SRC in the cells from patient #2, whereas PLX4720 does not (Figure 3H).
Next, we compared the inhibition of SRC in these resistant cells treated with CCT196969 and CCT241161 or three other pan-RAF inhibitors (RAF265, TAK632, and MLN2480) (K. Galvin, 2012, Am. Assoc. Cancer Res., conference; Nakamura et al., 2013; Su et al., 2012b), or another BRAF inhibitor (ARQ736) (Y.Z. Yu et al., 2010, Am. Assoc. Cancer Res., conference), which have entered clinical trials. Notably, all six compounds inhibit ERK, but only CCT196969 and CCT241161 also inhibit SFKs (Figure S3B), and CCT196969 inhibits the growth of the cells more potently than any of the other inhibitors (Figure S3C). We also assessed SRC inhibitor 1, a selective SFK inhibitor (Bain et al., 2007) in these resistant cells. Although SRC inhibitor 1 is inactive alone, it increases the activity of the pan-RAF inhibitor TAK632 against these cells (Figure S3D). This confirms that concurrent inhibition of RAF and SFKs cooperates to inhibit the growth of cells that are resistant to BRAF-selective inhibitors.
We also tested whether inhibition of p38 MAPK by CCT196969 and CCT241161 contributed to the inhibition of growth of the cells. We show that the cells are highly resistant to the p38 MAPK inhibitor SB203580 (50% inhibitory concentration [IC50] = 29.4 μM) and that SB203580 does not cooperate with the pan-RAF inhibitors RAF265 or MLN2480 to inhibit the growth of the cells (Figure S3E). Thus, p38 MAPK does not appear to play a role in regulating the growth of these cells, so inhibition of p38 MAPK by CCT196969 and CCT241161 does not contribute to the inhibition of their growth.
Notably, CCT196969 and CCT241161 induce caspase 3 and PARP cleavage, demonstrating that they induce apoptosis, whereas PLX4720 does not (Figure 3I). Finally, we show that xenografts grown from the cells of patient #2’s tumor are resistant to PLX4720, whereas CCT196969 and CCT241161 achieve complete inhibition of these xenografts (Figure 3J) without causing any body weight loss to the mice (Figure S3A).
CCT196969 and CCT241161 Inhibit the Growth of PDXs from Patients with Acquired and Intrinsic Resistance to BRAF Inhibitors
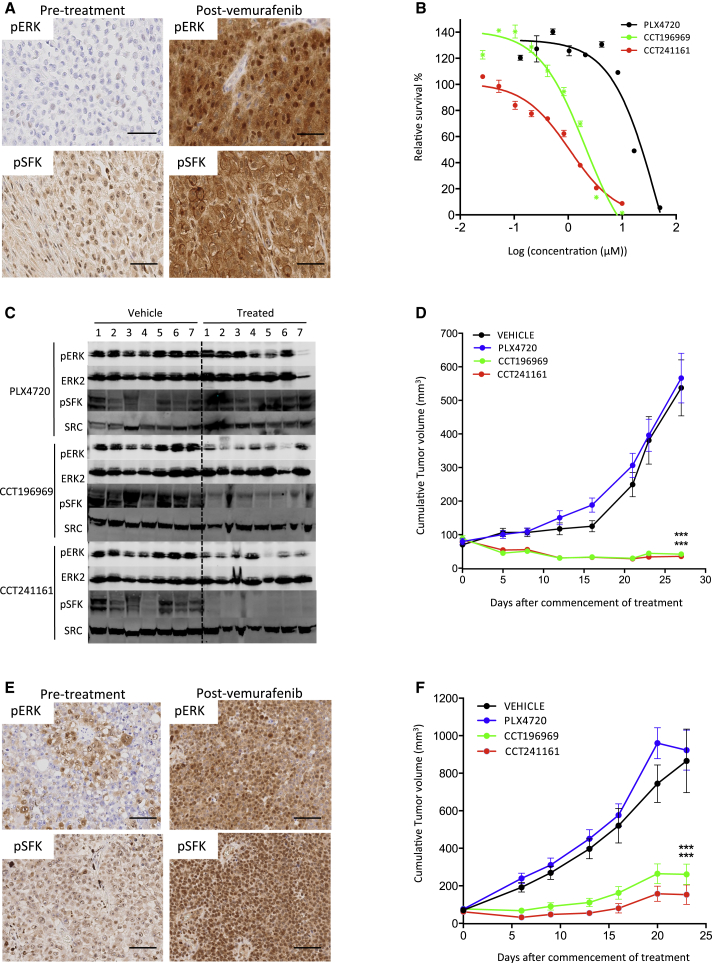
Next, we tested CCT196969 and CCT241161 in a tumor from a patient with stage IV BRAF mutant melanoma who achieved a complete response to vemurafenib but relapsed after 15 months with acquired resistance (patient #3; Table S4). We show that tumors from this patient express the melanoma markers HMB45 and S100 before and after treatment (Figure S4A). Note that, compared to the pretreatment tumor, ERK and SFK phosphorylation is elevated in the resistant tumor (Figure 4A), and cells from the resistant tumor are not inhibited by PLX4720, whereas they are sensitive to CCT196969 and CCT241161 (Figure 4B). Furthermore, CCT196969 and CCT241161 inhibit ERK and SRC and induce tumor regression in a PDX from the resistant tumor (Figures 4C and 4D), again without causing body weight loss in the mice (Figure S4B). Note that PLX4720 does not inhibit ERK or SRC in this PDX (Figure 4C), and accordingly, neither does it inhibit the growth of this PDX (Figure 4D).
Figure 4.
CCT196969 and CCT241161 Inhibit Tumors with Acquired Resistance to Vemurafenib
(A) pERK and pSFK in pre- and postvemurafenib treatment tumors from patient #3. Scale bars, 50 μm.
(B) Patient #3 tumor cell proliferation assay (CellTiter Glo) with PLX4720, CCT196969, and CCT241161.
(C) pERK, ERK2, and pSFK in patient #3 PDX in mice treated with PLX4720, CCT196969, or CCT241161.
(D) Growth of patient #3 PDX in NSG mice treated with PLX4720, CCT196969, or CCT241161.
(E) pERK and pSFK in pre- and post-vemurafenib treatment tumors from patient #4. Scale bars, 100 μm.
(F) Growth of patient #4 PDX in NSG mice treated with PLX4720, CCT196969, or CCT241161 21 days after tumor implant. ∗∗∗p ≤ 0.001 (t test, two-tailed).
Bars represent SEM. See also Figure S4.
We also tested CCT196969 and CCT241161 in a PDX from a patient with stage IV BRAF mutant melanoma who had achieved a partial response to vemurafenib but who then relapsed with acquired resistance after only 5 months (patient #4; Table S4). Again, we confirm that the tumors from this patient express melanoma markers before and after vemurafenib treatment (Figure S4A), that ERK and SFK phosphorylation is elevated in the resistant tumor (Figure 4E), and that a PDX from the resistant tumor is resistant to PLX4720 but sensitive to CCT196969 and CCT241161 (Figure 4F). Note that also here, CCT196969 and CCT241161 do not cause body weight loss in the mice (Figure S4B).
Subsequently, we tested CCT196969 and CCT241161 in a PDX from a patient with stage IV BRAF mutant melanoma who did not respond to vemurafenib and was diagnosed with progressive disease due to intrinsic resistance (patient # 5; Table S4). As before, the tumors from this patient expressed melanoma markers before and after treatment (Figure S5A), and ERK and SFK phosphorylation was elevated in the tumors following vemurafenib treatment (Figure 5A). Note that cells from this patient’s resistant tumor are more sensitive to CCT196969 and CCT241161 than to PLX4720 (Figure 5B), and critically, a PDX from the resistant tumor was more sensitive to CCT196969 and CCT241161 than to PLX4720 (Figure 5C). Also in this experiment, we did not observe any loss in body weight in the mice (Figure S5B).
Figure 5.
CCT196969 and CCT241161 Inhibit PDX from a Patient with Intrinsic Resistance to Vemurafenib
(A) pERK and pSFK in pre- and post-vemurafenib treatment tumors from patient #5. Scale bars, 50 μm.
(B) Growth of cell lines from patient #5 tumor (CellTiter Glo) with PLX4720, CCT196969, and CCT241161.
(C) Growth of PDX from patient #5 in NSG mice treated with PLX4720, CCT196969, or CCT241161 21 days after tumor implant. ∗∗∗p ≤ 0.001 (t test, two-tailed).
Bars represent SEM. See also Figure S5.
Thus, CCT196969 and CCT241161 inhibit both BRAF and SRC. Moreover, they inhibit the growth of PDXs from tumors that are resistant to BRAF inhibitors and have increased pSFK. Critically, we find that SFK phosphorylation is increased, particularly in the plasma membrane, in six of another seven melanomas from patients who presented acquired or intrinsic resistance to vemurafenib (Figure S5C; data not shown). Thus, we show that SFK phosphorylation is increased in nine of the ten tumors we examined, confirming the critical role of SRC signaling in resistance.
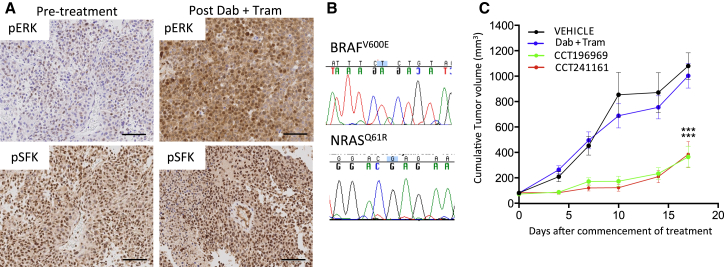
CCT196969 and CCT241161 Inhibit the Growth of a Melanoma with Acquired Resistance to Dabrafenib and Trametinib
The aforementioned data show that SFK signaling is increased in the majority of BRAF-inhibitor-resistant tumors, and furthermore, that tumors with increased SFK phosphorylation are sensitive to CCT196969 and CCT241161. However, not all resistant tumors show increased SFK phosphorylation, so we tested CCT196969 and CCT241161 in a PDX from a patient with stage IV BRAF mutant melanoma who achieved a partial response to dabrafenib plus trametinib but relapsed after only 5 months (patient #13; Table S4). Again, this patient’s tumors expressed melanoma markers before and after treatment (Figure S6A), and critically, although ERK phosphorylation is elevated in this resistant tumor, SFK phosphorylation is not (Figure 6A), suggesting that resistance is mediated by events downstream of SFKs. We confirm that the BRAFV600E mutation persists in the resistant tumor, but additionally, we observed an acquired NRASQ61R mutation that is not present in the pretreatment tumor (Figure 6B). Critically, a PDX from this patient is resistant to dabrafenib plus trametinib but sensitive to CCT196969 and CCT241161 (Figure 6C), and no body weight loss was observed in the mice (Figure S6B).
Figure 6.
CCT196969 and CCT241161 Inhibit Tumors with Acquired Resistance to Dabrafenib plus Trametinib
(A) pERK and pSFK in PDX from patient #13 before and after treatment with dabrafenib plus trametinib. Scale bars, 100 μm.
(B) Sequencing electropherograms confirming BRAFV600E and NRASQ61R mutations in PDX from patient #13.
(C) Growth of PDX from patient #13 in NSG mice treated with dabrafenib plus trametinib (dab + tram), CCT196969, or CCT241161 21 days after tumor implant. ∗∗∗p ≤ 0.001 (t test, two-tailed).
Bars represent SEM. See also Figure S6.
Discussion
Acquired resistance and intrinsic resistance to BRAF inhibitors are persistent problems in the treatment of BRAF mutant melanomas (Chapman et al., 2011; Flaherty et al., 2010, 2012; Sosman et al., 2012), even when BRAF and MEK inhibitors are combined (Flaherty et al., 2012). The advent of immunotherapies based on anti-CTLA-4 (e.g., ipilimumab) or anti-PD-1 (e.g., nivolumab and pembrolizumab) has recently revolutionized the treatment of melanoma, with excellent clinical results (A. Ribas et al., 2014, ASCO, conference; Weber et al., 2013; Wolchok et al., 2013), suggesting that patients who develop resistance to BRAF inhibitors should be considered for immunotherapy as a second line of treatment. However, recent evidence (Ackerman et al., 2014) shows that outcomes with ipilimumab following BRAF inhibitor discontinuation are poor, indicating that immunotherapies may provide better efficacy as first-line rather than second-line treatments. Consistent with this hypothesis, patients #3, #4, #5, and #13, described earlier, all eventually failed on BRAF inhibitor or BRAF plus MEK inhibitor combinations and were subsequently treated with ipilimumab, but none responded to this second-line treatment. Thus, there is a critical lack of second-line treatment options for patients who develop resistance to currently approved targeted therapies.
Here, we describe CCT196969 and CCT241161, BRAF/CRAF inhibitors that are also active against SFKs. These agents block BRAF mutant and NRAS mutant melanoma cell growth in vitro and in vivo. They are active against treatment-naive BRAF mutant tumors, against melanomas that are resistant to BRAF-selective drugs, and against a sample from a patient who was resistant to a BRAF/MEK inhibitor combination. The inhibitors are active in tumors from patients with acquired or intrinsic resistance. Critically, pERK was increased in all of the resistant patient tumors, consistent with resistance being mediated by MEK/ERK pathway activation. SFK phosphorylation was also increased in nine of 11 resistant tumors, but in the patient whose resistance was associated with an acquired mutation in NRAS, SFK phosphorylation was not increased.
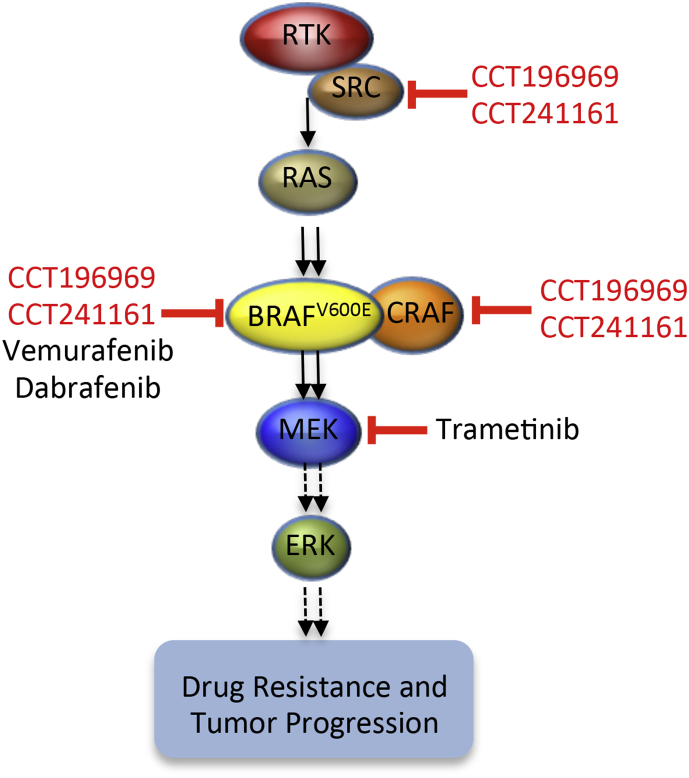
In many patients, BRAF-inhibitor resistance is mediated by MEK/ERK pathway reactivation driven by upregulation of RTK signaling or acquisition of mutations in NRAS (Fedorenko et al., 2011; Girotti et al., 2013; Johannessen et al., 2010; Nazarian et al., 2010; Van Allen et al., 2014; Vergani et al., 2011; Villanueva et al., 2010; Wagle et al., 2011, 2014). RTKs signal through SFKs, RAS signals through CRAF, and CCT196969 and CCT241161 are equipotent against BRAFV600E, CRAF, and SFKs. Accordingly, we posit that our inhibitors are active against tumors when resistance is mediated by upregulation of RTKs because they signal through SFKs, which are inhibited by our compounds (Figure 7). Conversely, they are active against tumors when resistance is mediated by mutant NRAS because it signals through CRAF, which is also a target of our compounds (Figure 7). Furthermore, because our compounds inhibit both BRAF and CRAF (Figure 7), they do not induce paradoxical activation of the MEK/ERK pathway and so are also active against NRAS mutant tumors.
Figure 7.
Model Showing Targets of Vemurafenib, Dabrafenib, Trametinib, CCT196969, and CCT241161
Thus, we have developed pan-RAF/SFK inhibitors that are orally available and well tolerated at therapeutic doses. They are active against treatment-naive BRAF and NRAS mutant tumors and against a range of tumors that are resistant to BRAF and BRAF plus MEK inhibitors, critically achieving regressions in a range of tumors. We also note that they were active in PDXs from patients who subsequently failed ipilimumab treatment. For treatment-naive patients, the presence of a BRAF or NRAS mutation may serve as a predictive biomarker to select patients who could benefit from treatment with these inhibitors. For patients whose tumors are resistant to current BRAF and MEK inhibitors, upregulated RTK signaling evidenced by increased SFK phosphorylation or pathway reactivation evidenced by increased ERK phosphorylation may provide predictive biomarkers to select patients for treatment. The presence of an NRAS mutation may also serve as a predictive biomarker for patient selection in the resistant setting. Thus, we posit that these inhibitors could provide first-line therapy for treatment-naive patients and second-line therapy for a range of patients with relapsed melanoma. We aim to conduct phase I clinical trials with this series of inhibitors starting in 2015.
Experimental Procedures
Cell Culture
Cell lines were cultured under standard conditions and routinely monitored for mycoplasma contamination by PCR and were not used if found to be positive. A375/R was derived from A375 cells (Girotti et al., 2013). All of the cell lines were cultured in Dulbecco’s modified Eagle’s medium or RPMI medium, both supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cell lines from patient samples were established from fresh tumor biopsies.
Kinase Assays
A DELFIA-based (PerkinElmer) 96-well assay was used to measure BRAF kinase activity as described elsewhere (Niculescu-Duvaz et al., 2006). Assays were performed in duplicate with an 11-point concentration response, and IC50 values were determined using GraphPad Prism software (GraphPad Software). IC50 values are the mean of three independent assays. The inhibition of BRAF, BRAFV600E, CRAF, SRC, LCK, MEK1, and COT were determined using Z9-LYTE Kinase Assay Kits, and the IC50 values (concentration in micromolar) for CCT196969 and CCT241161 against each kinase are presented in Table 1.
Short-Term Growth Inhibition Assays
Cultured cells were seeded into 96-well plates (2,000 cells per well). At 24 hr later, serial dilutions of the BRAF inhibitors PLX4720 and SB590885, the MEK inhibitor PD184352, or our compounds CCT241161 and CCT196969 were added. Cells were incubated for a further 72 hr, and viability was measured by CellTiter-Glo assays (Roche). Relative survival in the presence of drugs was normalized to the untreated controls after background subtraction.
Tritium-Labeled Thymidine Incorporation
Cell proliferation was assessed by measuring tritium-labeled thymidine incorporation. A total of 10,000 Ba/F3 cells were seeded into the wells of 96-well plates, and compounds were added to the desired concentration as described by Whittaker et al. (2010a). Assays were performed in quadruplicate with 10-point dilution series, and IC50 values were calculated using GraphPad Prism software. Values reported are the mean of three independent assays.
Long-Term Cell Proliferation Assays
Cells were seeded into six-well plates (5 × 104 cells per well) and cultured both in the absence and presence of drugs as indicated. Assays were performed independently at least three times, and average results are represented.
RPPA
The three cell lines derived from tumors displaying resistance to vemurafenib were incubated with DMSO (control), PLX4720, CCT196969, or CCT241161 (1 μM; 4 hr). Protein extracts were prepared in CLB1 lysis buffer (Zeptosens-Bayer Technology Services), and samples were analyzed by Zeptosens RPPA as described elsewhere (van Oostrum et al., 2009).
Histology and Immunohistochemistry
Tumors were formalin-fixed and prepared as described elsewhere (Dhomen et al., 2009) for staining with hematoxylin and eosin, rabbit pSRC (Invitrogen 44660G) and pERK (Cell Signaling 20G11). Positive and negative controls were included in each experiment. The scoring of the pattern and intensity of staining was performed in a blinded manner.
Tissue, Cell Lysates, and Immunoblots
Tissue and cell lysates were prepared with NP-40 buffer containing 5% β-mercaptoethanol, 150 mM NaCl, 50 mM Tris, pH 7.5, 2 mM EDTA, pH 8, 25 mM NaF, 1% NP-40, protease inhibitors (Complete, Roche), and Phosphatase Inhibitor Cocktails 2 and 3 (Sigma-Aldrich). All lysates were freshly prepared and resolved by SDS gel electrophoresis for western blotting. Primary antibodies were: ERK2 from Santa Cruz Biotechnology (clone C-14) and p-ERK1/2 T202/Y204 (clone D13.14.4E), p-SFK Y416 (clone D49G4), cleaved caspase 3 (clone 5A1E), caspase 3 (clone 3G2), cleaved PARP (clone D64E10), PARP (clone 46D11), and pMEK S217/221 (clone 41G9), all from Cell Signaling. Specific bands were detected using fluorescent-labeled secondary antibodies (Invitrogen; Li-COR Biosciences) and analyzed using an Odyssey Infrared Scanner (Li-COR Biosciences).
Mouse Xenografts and In Vivo Drug Studies
All procedures involving animals were approved by Cancer Research UK Manchester Institute's or the Institute of Cancer Research's Animal Welfare and Ethical Review bodies in accordance with the Animals (Scientific Procedures) Act 1986 and according to the guidelines of the Committee of the National Cancer Research Institute (Workman et al., 2010). Five- to 6-week-old female nude mice were injected subcutaneously with either 1 × 106 A375/R, or A375, or D04 or 5 × 106 patient #2 cells. Tumors were allowed to establish for 7 days, sizes were matched, and then the mice were randomly allocated to groups of eight to ten animals. No blinding was used in the treatment schedules for these studies. Based on literature precedents, groups of eight to ten animals were used to provide sufficient animals per cohort to provide statistically significant data while keeping the number of animals used to a minimum. Treatment was by oral gavage daily with vehicle (5% DMSO, 95% water), 90 mg/kg PLX4720, 20 mg/kg CCT196969, or 20 mg/kg CCT241161. All the inhibitors were administered 7 days/week, with no weekend break. Tumor size was determined by caliper measurements of tumor length, width, and depth; volume was calculated as volume = 0.5236 × length × width × depth (in millimeters). In accordance with our license to perform animal experiments, animals were excluded from the experiments if they displayed signs of distress, excessive body weight loss (>20%), or illness.
Patient Samples and Patient-Derived Xenografts
Tumor samples were collected under the Manchester Cancer Research Centre (MCRC) Biobank ethics application #07/H1003/161+5, with full informed consent from the patients. The work presented in this article was approved by MCRC Biobank Access Committee applications 12_JOBR_01 and 13_RIMA_01. Metastatic melanoma tumor samples from patients #1, #2, #3, #4, #5, and #13 were obtained immediately after surgery. For patients #1, #3, #4, #5, and #13, necrotic parts of the tumor were removed, and 5 × 5 × 5 mm pieces were implanted subcutaneously in the right flank of 5- to 6-week-old female IL-2 NSG mice. When the PDXs reached ∼1,400 mm3 volume, they were excised, and viable tissue was dissected into 5 mm cubes and transplanted into additional mice using the same procedure. Genomic and histological analyses had confirmed that the tumors at each point were derived from the starting material. Following transplantation, tumors were allowed to grow to ∼60–80 mm3, which was generally 14 to 21 days after the trocars had been implanted. Animals were randomized before initiation of treatment, for 23–30 days, by daily orogastric gavage of the following groups: PLX4720 (45 mg/kg); dabrafenib (25 mg/kg) plus trametinib (0.15 mg/kg); CCT196969 (20 mg/kg); CCT2141161 (20 mg/kg); vehicle (5% DMSO, 95% water). All the drugs were administered 7 days/week and without weekends off.
Kinase Assays
The IC50 assays for kinases were performed at Invitrogen using the Z’-LYTE kinase assay platform (10 points with 3-fold dilutions, or 5 points with 10-fold dilutions, in duplicate). The IC50 values were calculated using nonlinear regression with XL-Fit data analysis software, version 4.1 (IDBS).
Sanger Sequencing
Tumor gDNA was prepared using the QIAGEN DNA extraction kit. gDNA was subsequently amplified by PCR, and the products were sequenced using dye-terminator chemistry as described elsewhere (Turajlic et al., 2012). Sequences were visualized using Sequencher software. Oligonucleotide primer sequences are available on request.
Author Contributions
M.R.G., C.S., and R. Marais designed experiments and research aims and wrote the manuscript. M.R.G., F.L., N.P., D.N.D., A.Z., L.D., S.W., and G.S. performed experiments, analyzed the data, and wrote the manuscript. M.R.G., A.V., G.S., M.P., B.M.J.M.S., D.M., R. McLeary, L.J., L.F., S.E., B.S.-L., K.M., G.A., A.A.M., J.B., A.F., P.L., M.F., N.C., and J.H. contributed to acquisition of data and manuscript preparation. A.F. and P.L. provided clinical samples. M.R.G., F.L., R. McLeary, and S.E. performed in vivo experiments and contributed to data analysis.
Acknowledgments
We thank the patients and their families who contributed tissues to these studies. This work was supported by the Cancer Research UK Manchester Institute (grant C15759/A12328), Cancer Research UK (grant A17240), the Wellcome Trust (grants WT1005X and 100282/Z/12/Z), and the Division of Cancer Therapeutics at The Institute of Cancer Research (grant C309/A11566). Development of RPPA (Reverse Phase Protein Array) in Edinburgh was funded jointly by a Cancer Research UK Programme grant to M.F. (C157/A15703) and an European Research Council Advanced Investigator grant to M.F. (No. 294440 Cancer Innovation). Employees and former employees of the Institute of Cancer Research may benefit financially from any drug discovery programs that are commercialized.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Contributor Information
Richard Marais, Email: richard.marais@cruk.manchester.ac.uk.
Caroline Springer, Email: caroline.springer@icr.ac.uk.
Supplemental Information
References
- Ackerman A., Klein O., McDermott D.F., Wang W., Ibrahim N., Lawrence D.P., Gunturi A., Flaherty K.T., Hodi F.S., Kefford R. Outcomes of patients with metastatic melanoma treated with immunotherapy prior to or after BRAF inhibitors. Cancer. 2014;120:1695–1701. doi: 10.1002/cncr.28620. [DOI] [PubMed] [Google Scholar]
- Bain J., Plater L., Elliott M., Shpiro N., Hastie C.J., McLauchlan H., Klevernic I., Arthur J.S., Alessi D.R., Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhomen N., Reis-Filho J.S., da Rocha Dias S., Hayward R., Savage K., Delmas V., Larue L., Pritchard C., Marais R. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- Fedorenko I.V., Paraiso K.H., Smalley K.S. Acquired and intrinsic BRAF inhibitor resistance in BRAF V600E mutant melanoma. Biochem. Pharmacol. 2011;82:201–209. doi: 10.1016/j.bcp.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty K.T., Puzanov I., Kim K.B., Ribas A., McArthur G.A., Sosman J.A., O’Dwyer P.J., Lee R.J., Grippo J.F., Nolop K., Chapman P.B. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty K.T., Infante J.R., Daud A., Gonzalez R., Kefford R.F., Sosman J., Hamid O., Schuchter L., Cebon J., Ibrahim N. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsea A.M., Del Marmol V., de Vries E., Bailey E.E., Geller A.C. Melanoma incidence and mortality in Europe: new estimates, persistent disparities. Br. J. Dermatol. 2012;167:1124–1130. doi: 10.1111/j.1365-2133.2012.11125.x. [DOI] [PubMed] [Google Scholar]
- Girotti M.R., Pedersen M., Sanchez-Laorden B., Viros A., Turajlic S., Niculescu-Duvaz D., Zambon A., Sinclair J., Hayes A., Gore M. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer discovery. 2013;3:158–167. doi: 10.1158/2159-8290.CD-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzivassiliou G., Song K., Yen I., Brandhuber B.J., Anderson D.J., Alvarado R., Ludlam M.J., Stokoe D., Gloor S.L., Vigers G. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- Heidorn S.J., Milagre C., Whittaker S., Nourry A., Niculescu-Duvas I., Dhomen N., Hussain J., Reis-Filho J.S., Springer C.J., Pritchard C., Marais R. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen C.M., Boehm J.S., Kim S.Y., Thomas S.R., Wardwell L., Johnson L.A., Emery C.M., Stransky N., Cogdill A.P., Barretina J. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G.V., Stroyakovskiy D., Gogas H., Levchenko E., de Braud F., Larkin J., Garbe C., Jouary T., Hauschild A., Grob J.J. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 2014;371:1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- Ménard D., Niculescu-Duvaz I., Dijkstra H.P., Niculescu-Duvaz D., Suijkerbuijk B.M., Zambon A., Nourry A., Roman E., Davies L., Manne H.A. Novel potent BRAF inhibitors: toward 1 nM compounds through optimization of the central phenyl ring. J. Med. Chem. 2009;52:3881–3891. doi: 10.1021/jm900242c. [DOI] [PubMed] [Google Scholar]
- Nakamura A., Arita T., Tsuchiya S., Donelan J., Chouitar J., Carideo E., Galvin K., Okaniwa M., Ishikawa T., Yoshida S. Antitumor activity of the selective pan-RAF inhibitor TAK-632 in BRAF inhibitor-resistant melanoma. Cancer Res. 2013;73:7043–7055. doi: 10.1158/0008-5472.CAN-13-1825. [DOI] [PubMed] [Google Scholar]
- Nazarian R., Shi H., Wang Q., Kong X., Koya R.C., Lee H., Chen Z., Lee M.K., Attar N., Sazegar H. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu-Duvaz I., Roman E., Whittaker S.R., Friedlos F., Kirk R., Scanlon I.J., Davies L.C., Niculescu-Duvaz D., Marais R., Springer C.J. Novel inhibitors of B-RAF based on a disubstituted pyrazine scaffold. Generation of a nanomolar lead. J. Med. Chem. 2006;49:407–416. doi: 10.1021/jm050983g. [DOI] [PubMed] [Google Scholar]
- Niculescu-Duvaz D., Gaulon C., Dijkstra H.P., Niculescu-Duvaz I., Zambon A., Ménard D., Suijkerbuijk B.M., Nourry A., Davies L., Manne H. Pyridoimidazolones as novel potent inhibitors of v-Raf murine sarcoma viral oncogene homologue B1 (BRAF) J. Med. Chem. 2009;52:2255–2264. doi: 10.1021/jm801509w. [DOI] [PubMed] [Google Scholar]
- Poulikakos P.I., Zhang C., Bollag G., Shokat K.M., Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Moriceau G., Kong X., Lee M.K., Lee H., Koya R.C., Ng C., Chodon T., Scolyer R.A., Dahlman K.B. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosman J.A., Kim K.B., Schuchter L., Gonzalez R., Pavlick A.C., Weber J.S., McArthur G.A., Hutson T.E., Moschos S.J., Flaherty K.T. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straussman R., Morikawa T., Shee K., Barzily-Rokni M., Qian Z.R., Du J., Davis A., Mongare M.M., Gould J., Frederick D.T. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F., Viros A., Milagre C., Trunzer K., Bollag G., Spleiss O., Reis-Filho J.S., Kong X., Koya R.C., Flaherty K.T. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N. Engl. J. Med. 2012;366:207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Vilgelm A.E., Kelley M.C., Hawkins O.E., Liu Y., Boyd K.L., Kantrow S., Splittgerber R.C., Short S.P., Sobolik T. RAF265 inhibits the growth of advanced human melanoma tumors. Clinical cancer research. 2012;18:2184–2198. doi: 10.1158/1078-0432.CCR-11-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk B.M., Niculescu-Duvaz I., Gaulon C., Dijkstra H.P., Niculescu-Duvaz D., Ménard D., Zambon A., Nourry A., Davies L., Manne H.A. Development of novel, highly potent inhibitors of V-RAF murine sarcoma viral oncogene homologue B1 (BRAF): increasing cellular potency through optimization of a distal heteroaromatic group. J. Med. Chem. 2010;53:2741–2756. doi: 10.1021/jm900607f. [DOI] [PubMed] [Google Scholar]
- Turajlic S., Furney S.J., Lambros M.B., Mitsopoulos C., Kozarewa I., Geyer F.C., Mackay A., Hakas J., Zvelebil M., Lord C.J. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res. 2012;22:196–207. doi: 10.1101/gr.125591.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen E.M., Wagle N., Sucker A., Treacy D.J., Johannessen C.M., Goetz E.M., Place C.S., Taylor-Weiner A., Whittaker S., Kryukov G.V. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oostrum J., Calonder C., Rechsteiner D., Ehrat M., Mestan J., Fabbro D., Voshol H. Tracing pathway activities with kinase inhibitors and reverse phase protein arrays. Proteomics Clin. Appl. 2009;3:412–422. doi: 10.1002/prca.200800070. [DOI] [PubMed] [Google Scholar]
- Vergani E., Vallacchi V., Frigerio S., Deho P., Mondellini P., Perego P., Cassinelli G., Lanzi C., Testi M.A., Rivoltini L. Identification of MET and SRC activation in melanoma cell lines showing primary resistance to PLX4032. Neoplasia. 2011;13:1132–1142. doi: 10.1593/neo.111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J., Vultur A., Lee J.T., Somasundaram R., Fukunaga-Kalabis M., Cipolla A.K., Wubbenhorst B., Xu X., Gimotty P.A., Kee D. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N., Emery C., Berger M.F., Davis M.J., Sawyer A., Pochanard P., Kehoe S.M., Johannessen C.M., Macconaill L.E., Hahn W.C. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N., Van Allen E.M., Treacy D.J., Frederick D.T., Cooper Z.A., Taylor-Weiner A., Rosenberg M., Goetz E.M., Sullivan R.J., Farlow D.N. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61–68. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.S., Kudchadkar R.R., Yu B., Gallenstein D., Horak C.E., Inzunza H.D., Zhao X., Martinez A.J., Wang W., Gibney G. Safety, efficacy, and biomarkers of nivolumab with vaccine in ipilimumab-refractory or -naive melanoma. J. Clin. Oncol. 2013;31:4311–4318. doi: 10.1200/JCO.2013.51.4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker S., Kirk R., Hayward R., Zambon A., Viros A., Cantarino N., Affolter A., Nourry A., Niculescu-Duvaz D., Springer C., Marais R. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci. Transl. Med. 2010;2:35ra41. doi: 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- Whittaker S., Ménard D., Kirk R., Ogilvie L., Hedley D., Zambon A., Lopes F., Preece N., Manne H., Rana S. A novel, selective, and efficacious nanomolar pyridopyrazinone inhibitor of V600EBRAF. Cancer Res. 2010;70:8036–8044. doi: 10.1158/0008-5472.CAN-10-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T.R., Fridlyand J., Yan Y., Penuel E., Burton L., Chan E., Peng J., Lin E., Wang Y., Sosman J. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolchok J.D., Kluger H., Callahan M.K., Postow M.A., Rizvi N.A., Lesokhin A.M., Segal N.H., Ariyan C.E., Gordon R.A., Reed K. Nivolumab plus ipilimumab in advanced melanoma. N. Engl. J. Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman P., Aboagye E.O., Balkwill F., Balmain A., Bruder G., Chaplin D.J., Double J.A., Everitt J., Farningham D.A., Glennie M.J., Committee of the National Cancer Research Institute Guidelines for the welfare and use of animals in cancer research. Br. J. Cancer. 2010;102:1555–1577. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon A., Ménard D., Suijkerbuijk B.M., Niculescu-Duvaz I., Whittaker S., Niculescu-Duvaz D., Nourry A., Davies L., Manne H.A., Lopes F. Novel hinge binder improves activity and pharmacokinetic properties of BRAF inhibitors. J. Med. Chem. 2010;53:5639–5655. doi: 10.1021/jm100383b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.