Abstract
Optimal regulation of immune networks is essential for generation of effective immune responses, and defects in such networks can lead to immunodeficiency, while uncontrolled responses can result in autoimmune disorders. mTOR and STAT signaling cascades are key regulators of differentiation and function of cells of the immune system. Both pathways act as sensors and transducers of environmental stimuli, and recent evidence has revealed points of crosstalk between these pathways, highlighting synergistic regulation of immune cell differentiation and function. Here we review the current understanding of mTOR and STAT interactions in T cells and innate immune cells, and discuss potential mechanisms underlying these events. We further outline models for the intersection of these pathways in the regulation of immunity, and highlight important areas for future research.
Keywords: mTOR, STAT, immunity, signaling
The mammalian target of rapamycin (mTOR) and signal transducer and activator of transcription (STAT) signaling pathways have been identified as key regulators in the development, survival, and function of cells of the immune system, including CD8+ cytotoxic T (Tc) cells, CD4+ T helper (Th) cells, CD4+ T regulatory cells (Tregs), dendritic cells (DCs), and monocytes [1,2]. These pathways have been shown to play unique roles during innate and adaptive immunity, and deregulation of their activation has been associated with several diseases [1,2].
mTOR acts as a sensor of various environmental or cellular changes, such as nutrient availability, energy status, stress responses and signals generated by cytokine and hormone receptors [1,3]. This sensor capacity of mTOR integrates these signals in the immune microenvironment, ultimately controlling the fate of diverse immune cell types [1,3]. Two distinct mTOR complexes exist: mTORC1 and mTORC2 (Box 1) [4,5]. Although these complexes share mTOR as their common catalytic subunit, they also include unique components and have different upstream and downstream effectors (Box 2 and 3, Fig. 1) [4,5]. The regulatory effects of these complexes in the generation of immune responses have been examined in elegant studies involving selective in vitro and/or in vivo targeting of their components in immune cells [6,7]. In the case of mTORC1, extensive work has been done using the specific mTORC1 inhibitor rapamycin, a Streptomyces hygroscopicus derivative [6,7]. The mechanism of action of this drug involves structural inhibition of mTORC1 complexes via binding to the FK506 binding protein (FKBP12) [8]. The resulting rapamycin-FKBP12 complex binds the FRB (FKBP12/rapamycin-binding) domain of mTOR, resulting in selective inhibition of mTORC1 kinase activity [8]. As rapamycin works by direct binding to mTORC1 complexes, it was originally assumed that all mTORC1 functions are rapamycin-sensitive. However, the existence of rapamycin-insensitive mTORC1 functions has been documented [9]. More recently, catalytic mTOR inhibitors (e.g., Torin1, PP242, OSI-027) have been developed to target both mTORC1 and mTORC2 complexes [10,11]. These mTOR inhibitors, which act by binding to and inhibiting the catalytic site of mTOR, have shown more potent antineoplastic effects in vitro and in vivo against different types of malignant cells [11–13]. Surprisingly, catalytic mTOR inhibition has weaker effects on the growth and function of normal lymphocytes, and appears to be less immunosuppressive than rapamycin [14].
Box 1. mTOR complexes.
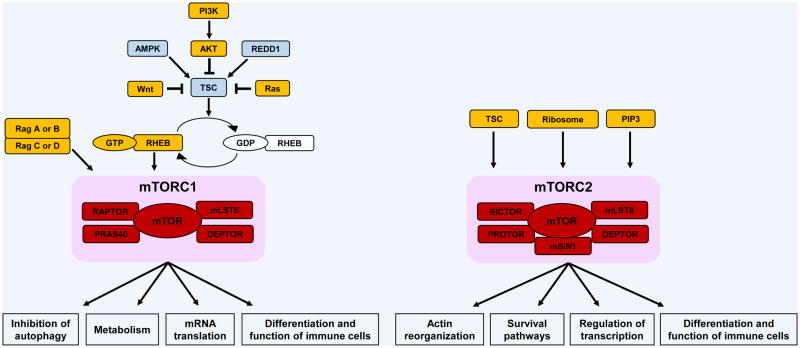
mTOR is an evolutionary conserved serine/threonine kinase that plays key regulatory roles in several biological processes, such as proliferation and survival of normal and malignant cells, cell differentiation, metabolism, and autophagy [5]. So far, two distinct mTOR complexes have been identified, mTORC1 and mTORC2. The core components of mTORC1 are the mTOR kinase, the regulatory-associated protein of mTOR (RAPTOR), the 40 kDa proline-rich AKT substrate (PRAS40), the DEP domain-containing mTOR-interacting protein (DEPTOR), and the mammalian lethal with SEC13 protein 8 (mLST8) [5,65]. mTORC2 is structurally different from mTORC1. It shares with mTORC1 mLST8 and DEPTOR, but it does not include RAPTOR and PRAS40 [5]. It contains other unique elements; the scaffolding protein RAPTOR-independent companion of TOR (RICTOR), the protein observed with RICTOR (PROTOR), and the mammalian stress-activated map kinase-interacting protein 1 (mSIN1) [5].
Box 2. Upstream regulation of mTOR.
In response to increased amino acid levels, activated Rag GTPases promote the translocation of mTORC1 into the lysosomal surface, where its activator, RHEB, is located [66,67]. Recently, it was reported that GAP activity toward Rags (GATOR) complex controls Rag A and B [68], whereas folliculin and folliculin interacting protein 1/2 control Rag C and D function [69]. Amino acid levels can also control mTORC1 activation through VPS34, a class III PI3K [5]. Similarly, increased levels of glucose recruit mTORC1 to the lysosomal surface through Rag GTPases [70]. Growth factors, cytokines, and immunological accessory molecules stimulate mTORC1 signaling via engagement of the PI3K pathway, which induces phosphorylation of AKT at threonine (Thr) 308. Active AKT decreases interaction between PRAS40 and mTORC1 and deactivates the tuberous sclerosis complex (TSC) formed by TSC1, TSC2 and TBC1D7 [71]. Inactive TSC promotes the conversion of GDP-bound RHEB to its GTP-bound state, which directly interacts and activates mTORC1 [72]. The activity of TSC can also be inhibited through the Ras and Wnt pathways, and it is activated by AMPK and REDD1 [3]. In contrast, mTORC2 activation is positively regulated by TSC [73], phosphatidylinositol 3,4,5-trisphosphate (PIP3) [74], and by association with the ribosome [75]. Additionally, the activation of both mTOR complexes can be mediated by phospholipase D, phosphatidic acid [76], and Rac1 [77].
Box 3. Effectors of mTOR pathways.
mTORC1 activation initiates mRNA translation by activating S6 kinase 1 (S6K1) and inhibiting 4E-binding protein 1 (4E-BP1) activities. S6K1 phosphorylates both S6 ribosomal protein (rpS6), which enhances the translation efficiency, and eukaryotic initiation factor 4B (eIF4B), which associates with eIF3 complex to form the translation-initiation complex. As part of this complex, eIF4-E dissociates from 4E-BP1, triggering the initiation of protein synthesis [5,65,78]. In contrast, active mTORC2 phosphorylates AGC kinases: serum and glucocorticoid-regulated kinase 1 (SGK1), protein kinase C (PKC), and AKT. mTORC2 activity is often evaluated by assessing the levels of AKT phosphorylation at serine (Ser) 473, as RICTOR, mSIN1, and mLST8 knockout cells present defective phosphorylation of AKT at this site in response to several stimuli [79]. Phosphorylation of AKT by mTORC2 primes it for further phosphorylation by 3-phosphoinositide-dependent protein kinase 1 (PDK1) at the Thr308 site, which in turn can induce mTORC1 signaling in the presence of several stimuli. Once activated by mTORC2, AKT inhibits the function of the transcription factors Forkhead box protein O1 (FOXO1) and FOXO3, as well as Kruppel-like factor 2 (KLF2). Similarly, SGK1 also inhibits the transcriptional activity of FOXO family members [3].
Figure 1.
mTOR complexes. mTORC1 and mTORC2 share mTOR as their common catalytic subunit, however, they also include unique elements in their structures. These two complexes have different upstream positive (represented in yellow) and negative (represented in blue) regulators and exert distinct biological functions.
Like mTOR, STAT proteins also act as environmental sensors, reporting on the presence of diverse cytokines, including interferons (IFNs) and interleukins (ILs) [15,16]. In mammals, there are seven STATs (STAT1, 2, 3, 4, 5A, 5B, and 6), which are linked to signaling downstream of over 30 receptors of the cytokine receptor superfamily [15]. STAT proteins are phosphorylated and activated by members of the Janus family of kinases, also known as JAK kinases [15,16]. There are 4 different known JAK kinases, JAK1, JAK2, JAK3, and TYK2, which associate to distinct cytokine receptor subunits in different receptor complexes [16]. The binding of cytokines to the respective receptors induces their oligomerization, bringing JAKs closer and priming them for auto- and/or trans-phosphorylation. Activated JAKs then phosphorylate intracellular tyrosine residues present in the receptor, creating docking sites for STAT proteins. Upon binding to cytokine receptors, STATs undergo JAK-dependent tyrosine phosphorylation and form homodimers and/or heterodimers that translocate to the nucleus where they recognize and bind to specific promoter regions of target genes to initiate transcription [16,17]. Thus, engagement of cytokine receptors in distinct immune cell types results in activation of different combinations of STAT proteins, establishing specificity of responses and ultimately determining the subsequent immunophenotype [15].
Although mTOR and STAT pathways exert unique roles during innate and adaptive immunity, recent studies have demonstrated that these pathways interact to control and optimize immune responses [18–21]. Here we review emerging evidence for cross-regulation between these pathways, and discuss how this cross-regulation suggests an orchestrated dynamic regulation of immune responses by various stimuli with important implications for the development of future therapies against immune disorders.
Coordinated functions of mTOR and STAT pathways in T cells
In past years, strong evidence has associated mTOR signaling with T cell activation and lineage specification, and with the development of T cell memory [22,23]. STAT proteins have been associated with T cell development, differentiation, and survival [24–26]. Recent evidence suggests that STAT transcription factors transduce environmental stimuli in part by controlling activation of lineage-specific enhancers and, at the same time, suppressing enhancers associated with alternative cell fates [27]. As both STAT and mTOR pathways are involved in key processes of T cell development and maintenance, cross-talk between these two pathways is likely to play a role in signal integration and interpretation within the cell, and to be important in the determination of T cell fate and function. Indeed, although many of the specific biochemical events accounting for mTOR-STAT cross-regulation in T cells remain to be precisely determined, such coordination appears to be essential and critically important.
Cooperation between mTOR and STAT signals in the determination of effector versus memory Tc cell fate
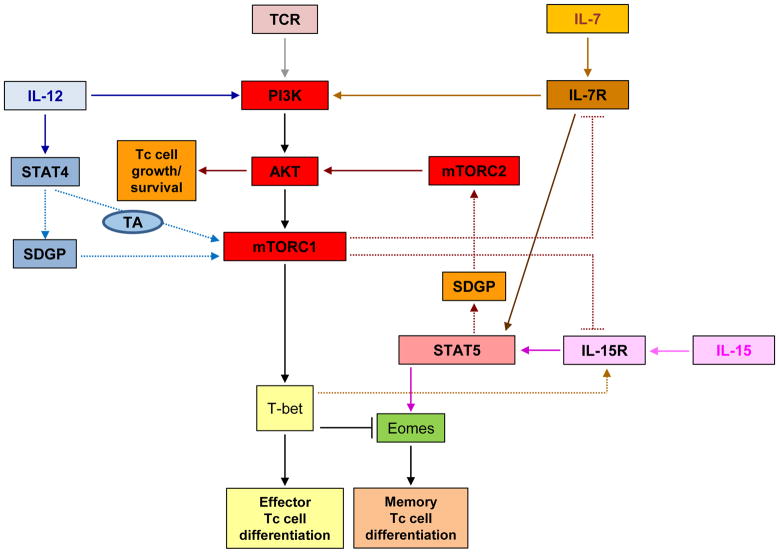
Rao and colleagues [19] reported that IL-12 drives type I effector maturation by enhancing and sustaining mTOR phosphorylation in naïve Tc (OT-1) cells stimulated with antigen and the co-stimulatory molecule B7 (Ag+B7.1). The functions of both phosphoinositide 3-kinase (PI3K) and STAT4 were required for the effects of IL-12 on mTOR activity, indicating synergistic co-regulation of PI3K- and STAT4-dependent cellular events in this process. Notably, inhibition of PI3K blocked IL-12 enhanced, mTOR-mediated, S6K phosphorylation at early and late time points. The absence of STAT4 did not affect the IL-12-induced phosphorylation of S6K at early time points, but resulted in failure of IL-12 to sustain S6K phosphorylation at 48 hours. Thus, coordinated engagement of both PI3K- and STAT4-dependent pathways is necessary for optimal induction of mTOR activity by IL-12 in Tc cells (Fig. 2). Notably, inhibition of mTOR activity with rapamycin during IL-12 stimulation blocked the expression of T-bet, a transcription factor critical for type I effector differentiation. In contrast, rapamycin treatment enhanced eomesodermin (Eomes) expression, favoring differentiation into memory T cells. These findings are intriguing, as they establish a requirement for STAT4 for mTOR activity in this system, and provide the basis for future work to define the mechanisms by which STAT4 impacts mTOR activity, and the CD8+ T cell maturation and memory formation. One possibility is that STAT4 regulates transcriptional activation of the mTOR gene [19] (Fig. 2). An alternative scenario would involve transcriptional activation of another upstream effector of the mTOR pathway that is required for mTOR/S6K activity (Fig. 2). The requirement of STAT4 in the process also raises the possibility of involvement of other STATs that may form heterodimers with STAT4; studies in that direction may be warranted.
Figure 2.
Cross-regulation of effector/memory Tc cell fate by mTOR- and STAT-pathways. Proposed model for cross-talk of mTOR and STAT signals in response to TCR-engagement or treatment with different interleukins. Dashed lines represent possible intermediate pathways accounting for regulation between mTOR complexes and STATs. Abbreviations: SDGP, STAT-dependent gene product; TA, transcriptional activation; Eomes, Eomesodermin.
Studies examining the mechanisms underlying homeostatic proliferation (HP) [28] of naïve T cells under conditions of lymphopenia have also revealed important roles for mTOR in Tc cell memory formation. Li et al. [29] examined HP of OVA-specific Tc cells (OT-1) transferred into intact or irradiated syngeneic recipient mice, and found a requirement for mTOR activation downstream of IL-7 (Fig. 2). Similar to the findings of Rao et al. [19], the authors found that rapamycin treatment inhibited T-bet expression downstream of mTOR, which in turn enhanced the transition to a memory phenotype by relieving T-bet-mediated repression of Eomes expression. In this system, mTOR induced T-bet expression was required for Tc cell memory formation in response to IL-15, which in turn induces phosphorylation of STAT5. These findings suggest an indirect interaction between mTOR and STAT5 activation in CD8+ cells during HP (Fig. 2), and together with Rao et al [19], highlight the relevance of cross-talk by TCR and cytokine signals transduced by the mTOR and STAT pathways in memory formation. STAT5 activation is also critical for sustaining effector Tc cell responses [30,31]. Whether mTOR-induced T-bet expression downstream of TCR and IL-12 stimulation [19] indirectly promotes STAT5 activity in effector Tc cells remains to be determined.
STAT5 and mTOR signaling events differentially regulate effector and memory Tc cell survival, and a balance between the functions of these pathways may be required for optimal immune responses [20]. During viral infection, the majority of Tc cells differentiate into short-lived effectors, but a minority contract to produce memory cells that are sustained by IL-7 and IL-15 [32]. AKT is activated in memory cells in response to both cytokine and TCR stimulation, [20,33–35], suggesting that downstream survival pathways account for the longer life span of memory cells (see Box 3, and Fig. 1). However, following viral infection, constitutive activation of AKT (at Ser473 and Thr308) does not increase effector Tc cell survival or memory cell generation [20]. Instead, it elicits a negative feedback mechanism that results in the inhibition of expression of the IL-7 and IL-15 receptors, and impaired STAT5 signaling. It is important to note that mTORC2 activation induces phosphorylation of AKT at Ser473, whereas mTORC1 signaling is activated downstream of AKT phosphorylation on Thr308. Since mTORC1 inhibition correlates with enhanced memory Tc cell formation [19, 22, 29], it is plausible that mTORC1 activation induces a negative feedback regulatory loop that represses IL-7 and IL-15 receptor-signals (Fig. 2). On the other hand, constitutive activation of STAT5 in T cells enhances baseline phosphorylation of AKT at Ser473 in response to IL-15, and increases survival of effector Tc cells and formation of more memory Tc cells [20]. Thus, it is possible that following viral infection, IL-7- or IL-15-induced STAT5-dependent targets activate mTORC2 complexes, resulting in enhanced effector and memory Tc cell growth and survival, while downstream mTORC1 engagement prevents uncontrolled IL-7- or IL-15-induced STAT5 activity (Fig. 2). Thus, complex coordination of mTOR- and STAT5-mediated signals may be required for effector and memory Tc-cell longevity. It is likely that in effector T cells, mTORC1 indirectly controls STAT5 signaling through regulation of expression of T-bet [29] and IL-7 and IL-15 receptors [20], while STAT5 activation promotes transcription of activators of mTORC2, enhancing effector T cell survival and memory T cell formation [20] (Fig. 2).
Other work also supports cross-talk between STAT5 and mTOR/AKT activation in T cells. IL-7 was shown to stimulate Glut1, the primary glucose transporter in immune cells, trafficking and glucose uptake in a STAT5-dependent manner, leading to sustained mTOR and AKT activation [36]. Remarkably, IL-7-stimulated phosphorylation of AKT and translocation of Glut1 to the cell surface were blocked by STAT5A/B RNAi in early lymphoid cells. Expression of a DNA-binding mutant form of STAT5A in these cells impaired the sustained phosphorylation of mTOR and AKT (Ser 473 and Thr 308). Altogether, these results suggest that activation of mTOR/AKT by IL-7 involves a mechanism dependent on the transcriptional activity of STAT5, further supporting the notion that STAT5 may induce or repress transcription of genes encoding proteins that regulate mTORC2/AKT activity in response to IL-7. It will be important to discern whether this putative role for STAT5 is specific to T cells, or it is also observed in other immune cell types, and whether other cytokines utilize similar STAT5-dependent mechanisms to regulate mTOR activation.
mTOR-dependent regulation of STATs during naïve CD4+ T cell differentiation
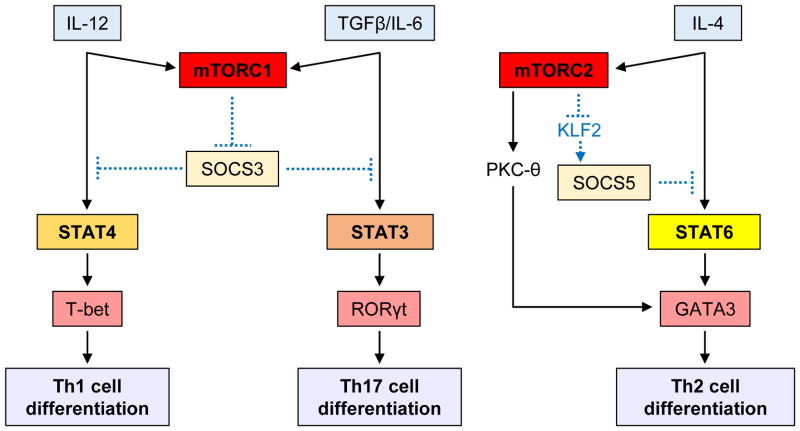
Naïve CD4+ T cells become activated upon TCR engagement and differentiate into specific effector lineages depending on the function and activity of cytokines present within the inflammatory microenvironment. Different cytokines activate specific STAT proteins, which subsequently upregulate lineage-specific transcription factors. For instance, IL-12 promotes differentiation of Th1 cells through activation of STAT4 and T-bet, while IL-4 induces development of Th2 cells through activation of STAT6 and GATA3. IL-6 and TGF-β elicit Th17 cell differentiation via activation of STAT3 and RORγt, while IL-2 and TGFβ promote generation of Tregs through activation of STAT5 and Foxp3. Various lines of evidence have shown a role for mTOR in CD4+ effector lineage differentiation (reviewed in [23]), and recent studies have revealed cross-talk between the STAT and mTOR pathways in these processes. Using in vitro and in vivo models, Delgoffe et al. [37] demonstrated that mTOR deficiency in T cells correlates with diminished cytokine-dependent activation of STAT4, STAT3, and STAT6. Consequently, mTOR-deficient naïve CD4+ T cells were unable to differentiate into Th1, Th17, or Th2 effector cells under specific stimulatory conditions, and skewed towards T regulatory (Treg) cell differentiation. In this process, the activities of both mTORC1 and mTORC2 are required for suppression of generation of Treg cells [37]. Thus, it appears that the mTOR pathway integrates diverse environmental signals in T cells and controls T cell lineage commitment via modulating activation of specific STATs. However, questions remain regarding the mechanisms by which mTOR complexes regulate STAT activation. One potential mechanism may involve direct or indirect mTOR-dependent phosphorylation of STAT-proteins. In such scenario, mTOR complexes may phosphorylate STATs in residues required for their activation or may engage downstream effector kinases that phosphorylate STATs. Alternatively, mTOR-dependent signals may control activation of STAT-proteins by phosphorylation-independent mechanisms. Consistent with the latter hypothesis, a recent study demonstrated that in RHEB-deficient T cells activation of STAT4 and STAT3 in response to IL-12 and IL-6 is decreased, resulting in inability to differentiate into Th1 and Th17 cells [38]. Such reduced STAT4 and STAT6 activation was associated with increased expression levels of suppressor of cytokine signaling 3 (SOCS3). On the other hand, RICTOR-deficient T cells showed reduced IL-4-induced activation of STAT6 and failed to differentiate into Th2 cells [38]. In this case, RICTOR deficiency was associated with increased expression levels of SOCS5. The expression of SOCS proteins is cytokine-dependent and interferes with signaling from the inducing cytokine, either by competing with STAT proteins for binding to phosphorylated tyrosine residues within the receptor cytoplasmic domains or by inducing ubiquitination of SH2 and N-terminal bound substrates, resulting in their proteasomal degradation [39]. Thus, it is possible that during its engagement by the IL-12 receptor, mTORC1 negatively regulates SOCS3 expression and indirectly promotes differentiation of Th1 cells through STAT4 activation (Fig. 3). As the major role of SOCS3 is suppression of signaling by the IL-6 family of cytokines via inhibition of STAT3 activation [40], a proposed model for the regulatory effects of mTOR in IL-6 and TGFβ stimulation, involves mTORC1 repression of SOCS3 expression, indirectly inducing differentiation of Th17 cells via STAT3 activation (Fig. 3). On the other hand, under IL-4 stimulation, mTORC2 represses SOCS5 expression, indirectly controlling STAT6-induced differentiation of Th2 cells (Fig. 3).
Figure 3.
Proposed models for cross-regulation between mTOR and STAT pathways in CD4+ T cells. Naïve CD4+ T cells become activated upon TCR engagement and differentiate into specific effector lineages depending on the function and activity of cytokines present within the inflammatory microenvironment. Dashed lines represent possible intermediates accounting for regulation between mTOR complexes and STATs.
It should be also noted that conditional deletion of RICTOR in T cells results in a substantial decrease in Th2 cell differentiation, and impaired generation of Th1 cells [41]. Transduction of RICTOR-deficient T cells with constitutively activated AKT plasmid rescues T-bet expression and generation of Th1 cells, whereas transduction of RICTOR-deficient T cells with constitutively active PKC-θ plasmid restores GATA-3 expression and differentiation of Th2 cells [41]. These results suggest that mTORC2 is critical both for generation of Th1 cells via AKT phosphorylation at Ser473 and for development of Th2 cells via PKC-θ signaling (Fig. 3). However, it should be noted that in the study of Delgoffe et al. [38], RICTOR-deficient T cells exhibited increased phosphorylation of AKT at Thr308 and S6K1 activation and, consequently, differentiated into Th1 cells under IL-12 stimulation. In contrast, in the study of Lee et al. [41], RICTOR-deficient T cells exhibited reduced AKT activity and defective differentiation into Th1 cells. The discrepancy between the two studies can be explained, at least in part, by some differences in the experimental designs. Delgoffe et. al. [38] bred mice with loxP-flanked Rictor alleles with mice expressing Cre recombinase from the Cd4 promoter and enhancer regions, while Lee et. al. [41] used mice expressing Cre from the distal promoter of the gene encoding the kinase Lck. Independently of the experimental differences, the different outcomes of these two studies could reflect different levels of AKT activation and raise the possibility that mTORC1 is selectively involved in differentiation of Th1 cells (Fig. 3).
Prior evidence suggests that mTOR activity controls the maintenance and function of the CD4+ Treg cell population, which is crucial for the development of immune tolerance [42, 43]. There is also evidence for STAT5 involvement, as RNAi of STAT5B in human CD4+ T cells results in decreased Treg cell numbers upon stimulation under Treg skewing conditions, and diminished suppressive function [44]. These findings suggest interactions and coordinated actions between STAT and mTOR pathways in Treg differentiation, and potentially function and maintenance in the periphery. Consistent with this, a recent study showed that T-cell-specific TSC1-deficient mice and RICTOR-deficient mice have increased numbers of Treg cells, which correlated with higher STAT5 activation and proliferation rates in response to IL-2 [45]. Rapamycin treatment was found to further enhance the number of Treg cells in these mice. Thus, generation of Treg cells in response to IL-2 stimulation appears to be controlled by TSC via rapamycin-insensitive (RI) mTORC1 signals and/or mTORC2-mediated signals, while STAT5 activity is required for their proliferation [45]. As mTORC1 inhibits expression of SOCS3 [38], which in turn inhibits IL-2 induced STAT5 activation [46], it is possible that rapamycin-resistant mTORC1 integrates IL-2 signals to suppress SOCS3 expression and enhance STAT5 phosphorylation. In contrast, mTORC2 integrates IL-2 signals to induce SOCS3 transcription, suppressing STAT5 activation. Thus, a balance between RI-mTORC1 and mTORC2 pathways and their effects on STAT-pathways appears to be required for optimal differentiation of Treg cells.
mTOR-dependent regulation of STAT activation in dendritic cells and macrophages
Monocytes and macrophages are important effectors and regulators of innate immunity, whereas dendritic cells (DCs) initiate and control adaptive immune responses [47]. There is accumulating evidence that mTOR and STAT3 signaling are required for proper development and function of DCs [48–51], suggesting potential cross-talk between these pathways. Two independent studies [52,53] demonstrated that selective mTORC1 inhibition using rapamycin inhibits IL-10 expression and STAT3 phosphorylation in LPS-stimulated DCs. Additionally, STAT3 activation is also blocked by rapamycin in LPS-stimulated monocytes and macrophages [54]. More recently, it was shown that treatment with an anti–IL-10 antibody strongly reduced phosphorylation of STAT3 in LPS-stimulated monocytes, and that mTORC1 activity was not essential for IL-10 mediated STAT3 phosphorylation, as rapamycin did not affect the IL-10/IL-10 receptor-mediated activation of STAT3 [55]. Thus, mTORC1 activity seems to indirectly control STAT3 activation by regulating IL-10 production [55].
Other work has shown that treatment with rapamycin decreases the expression of B7-homolog 1 (B7-H1) in LPS-stimulated DCs, while extended exposure to catalytic mTOR inhibitors enhanced it [56]. Unexpectedly, LPS-stimulated RICTOR - deficient DCs presented reduced B7-H1 expression. These observations support the notion that a rapamycin-resistant and mTORC2-independent mTOR mechanism negatively regulates B7-H1 expression. DCs conditioned with the catalytic mTOR inhibitor Torin 1 were found to express lower levels of SOCS3, resulting in increased activation of STAT3, increased B7-H1 expression, and enhanced Tregs induction. On the other hand, there was decreased STAT3 phosphorylation in LPS-stimulated RICTOR - deficient DCs, and no change in SOCS3 expression [56]. Thus, rapamycin-resistant mTORC1 appears to integrate Toll like receptor 4 (TLR4) signals to control SOCS3 expression and STAT3 activation, whereas mTORC2-depedent effector(s) seem to be required for STAT3 phosphorylation in response to LPS stimulation.
Regulation of STAT activation by mTORC2 complexes during engagement of the Type I IFN receptor
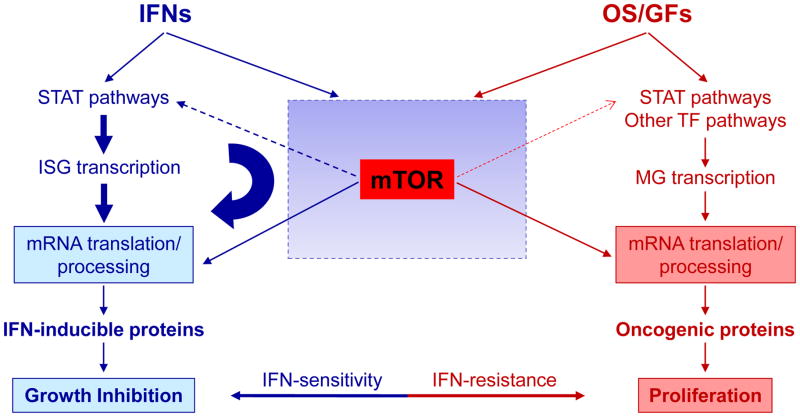
There has been extensive evidence that STAT signaling pathways play key and essential roles in the generation of Type I IFN responses [57–59]. Recent studies have also provided evidence that mTORC2 activity is required for transcriptional regulation of IFN-stimulated genes (ISGs) [18,60]. In addition, mTORC1-mediated signaling events are essential for mRNA translation of ISGs [61,62], suggesting cooperation between mTOR and STAT pathways for optimal expression of IFN-inducible proteins. Recent findings suggest a mechanism by which mTORC2-generated signals may regulate transcriptional activation of ISGs, via regulation of STAT1 and STAT2 activities [18]. Specifically, RICTOR and SIN1 were found to be required for Type I IFN-induced phosphorylation of STAT1 on serine 727, and of STAT2 on tyrosine 689 [18]. Consequently, formation of specific STAT-DNA-binding complexes required for expression of IFN-inducible genes is impaired in the absence of RICTOR or SIN. These findings can be brought together in a model for cross-regulation of STAT-pathways by either control of mRNA translation by mTORC1 complexes and indirect regulation of STAT-phosphorylation and transcriptional activation of ISGs by mTORC2 complexes (Fig. 4). An implication of this model is that competition for specific use of mTOR pathways between IFNs and oncogenic signals/growth factors may impact signaling specificity and differential biological outcomes [59] (Fig. 4).
Figure 4.
Proposed model for potential competition for the use of the mTOR pathway between interferons (IFNs) and oncogenic signals/growth factors (OS/GFs) for transcription of genes and mRNA translation/protein expression of products that account for distinct and opposing biological responses. In the case of the Type I IFN receptor there is also documented cross-regulation of STAT-pathways by mTORC2-dependent phosphorylation of STAT1 and STAT2, although the intermediate kinases and specific pathways involved remain to be identified. Abbreviations: ISG, IFN-stimulated genes; MG, mitogenic genes; TF, transcription factors.
Concluding remarks
In recent years, there has been accumulating evidence that mTOR and STAT pathways are critical for the control and generation of immune responses. Although the effectors of each pathway are required for expression of gene products that modulate key immune functions, there is also evidence that these pathways cross-regulate each other (Box 4). The evidence for a crosstalk between mTOR and JAK/STAT pathways lends support for the targeting of these two pathways as new treatment strategies for several disorders [63,64]. Notably, there is already evidence that treatment of myeloproliferative neoplastic cells with JAK2 inhibitors (AZD1480 and ruxolitinib) in combination with mTOR inhibitors (RAD001 and PP242) results in synergistic anti-proliferative effects when compared to each drug used alone [64]. The continuation of this work, including studies to precisely define elements accounting for cross-regulation between the two pathways, may ultimately lead to the development of more selective and potent therapies for the treatment of autoimmune disorders and/or certain malignancies.
Box 4. Proposed mechanisms underlying the interaction between mTOR and STAT pathways in immune cells.
There is evidence that mTOR and STAT pathways cross-regulate each other by either inducing or suppressing their activation in several immune cell types. However, several of the proposed mechanisms remain to be confirmed and the sequence of events precisely defined. Below is a list of suggested mechanisms/hypotheses regarding potential mTOR and STAT interactions in immune cells:
mTOR complexes regulate expression of SOCS genes and/or proteins and other mTOR-dependent targets, which directly interfere with cytokine-induced STAT phosphorylation [18, 29, 38, 45, 55, 56, 80–82].
mTOR phosphorylates serine residues within the STATs transactivation domain, promoting STAT-induced gene transcription [83].
mTORC2 complexes control activation of other kinases that activate/phosphorylate STATs and modulate their activities [18].
STAT-dependent gene products are required for cytokine-induced activation of mTOR [19, 20, 36].
STAT3 controls mTOR activity in response to different cytokines. Cytokines activate TYK2 and JAK1, which phosphorylate intracellular receptor domains creating a binding site for PI3K. STAT3 can act as an adapter protein linking PI3K to cytokine receptors, resulting in activation of AKT/mTOR pathways downstream of PI3K [reviewed in 84].
Highlights.
Critical roles for mTOR and STAT signaling pathways in the control of immune cell fate
Emerging evidence for crosstalk between mTOR and STAT pathways in immune regulation
Cooperation between these pathways enables optimal induction of immune responses
Acknowledgments
The research of Dr. Platanias is supported by grants CA77816, CA155566, CA161196 and CA121192 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yang H, et al. Modulation of TSC-mTOR signaling on immune cells in immunity and autoimmunity. J Cell Physiol. 2014;229:17–26. doi: 10.1002/jcp.24426. [DOI] [PubMed] [Google Scholar]
- 2.Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene. 2013;32:2601–2613. doi: 10.1038/onc.2012.347. [DOI] [PubMed] [Google Scholar]
- 3.Powell JD, et al. Regulation of immune responses by mTOR. Annu Rev Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beauchamp EM, Platanias LC. The evolution of the TOR pathway and its role in cancer. Oncogene. 2013;32:3923–3932. doi: 10.1038/onc.2012.567. [DOI] [PubMed] [Google Scholar]
- 6.Xu X, et al. mTOR, linking metabolism and immunity. Semin Immunol. 2012;24:429–435. doi: 10.1016/j.smim.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang K, Chi H. Tuning mTOR activity for immune balance. J Clin Invest. 2013;123:5001–5004. doi: 10.1172/JCI73202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benjamin D, et al. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10:868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 9.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson V, et al. Next generation of mammalian target of rapamycin inhibitors for the treatment of cancer. Expert Opin Investig Drugs. 2013;22:715–722. doi: 10.1517/13543784.2013.787066. [DOI] [PubMed] [Google Scholar]
- 12.Altman J, et al. Dual mTORC2/mTORC1 targeting results in potent suppressive effects on acute myeloid leukemia (AML) progenitors. Clin Cancer Res. 2011;17:4378–4388. doi: 10.1158/1078-0432.CCR-10-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carayol N, et al. Critical roles for mTORC2- and rapamycin-insensitive mTORC1-complexes in growth and survival of BCR-ABL-expressing leukemic cells. Proc Natl Acad Sci U S A. 2010;107:12469–12474. doi: 10.1073/pnas.1005114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janes MR, et al. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat Med. 2010;16:205–213. doi: 10.1038/nm.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Shea JJ, et al. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368:161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity. 2012;36:542–550. doi: 10.1016/j.immuni.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaur S, et al. Critical roles for Rictor/Sin1 complexes in IFN-dependent gene transcription and generation of antiproliferative responses. J Biol Chem. 2014;289:6581–6591. doi: 10.1074/jbc.M113.537852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao RR, et al. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hand TW, et al. Differential effects of STAT5 and PI3K/AKT signaling on effector and memory CD8 T-cell survival. Proc Natl Acad Sci U S A. 2010;107:16601–16606. doi: 10.1073/pnas.1003457107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada O, et al. JAK-STAT and JAK-PI3K-mTORC1 pathways regulate telomerase transcriptionally and posttranslationally in ATL cells. Mol Cancer Ther. 2012;11:1112–1121. doi: 10.1158/1535-7163.MCT-11-0850. [DOI] [PubMed] [Google Scholar]
- 22.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi H. Regulation and function of mTOR signaling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Shea JJ, et al. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol. 2011;11:239–250. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui W, et al. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 2011;35:792–805. doi: 10.1016/j.immuni.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegel AM, et al. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity. 2011;35:806–818. doi: 10.1016/j.immuni.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vahedi G, et al. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takada K, Jameson SC. Naive T cell homeostasis: from awareness of space to a sense of place. Nat Rev Immunol. 2009;9:823–832. doi: 10.1038/nri2657. [DOI] [PubMed] [Google Scholar]
- 29.Li Q, et al. A central role for mTOR kinase in homeostatic proliferation induced CD8+ T cell memory and tumor immunity. Immunity. 2011;34:541–553. doi: 10.1016/j.immuni.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly J, et al. A role for Stat5 in CD8+ T cell homeostasis. J Immunol. 2003;170:210–217. doi: 10.4049/jimmunol.170.1.210. [DOI] [PubMed] [Google Scholar]
- 31.Tripathi P, et al. STAT5 is critical to maintain effector CD8+ T cell responses. J Immunol. 2010;185:2116–2124. doi: 10.4049/jimmunol.1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hand TW, Kaech SM. Intrinsic and extrinsic control of effector T cell survival and memory T cell development. Immunol Res. 2009;45:46–61. doi: 10.1007/s12026-008-8027-z. [DOI] [PubMed] [Google Scholar]
- 33.Kim HR, et al. Dual roles of IL-15 in maintaining IL-7Ralpha low CCR7- memory CD8+ T cells in humans via recovering the phosphatidylinositol 3-kinase/AKT pathway. J Immunol. 2007;179:6734–6740. doi: 10.4049/jimmunol.179.10.6734. [DOI] [PubMed] [Google Scholar]
- 34.Riou C, et al. Convergence of TCR and cytokine signaling leads to FOXO3a phosphorylation and drives the survival of CD4+ central memory T cells. J Exp Med. 2007;204:79–91. doi: 10.1084/jem.20061681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henson SM, et al. KLRG1 signaling induces defective Akt (ser473) phosphorylation and proliferative dysfunction of highly differentiated CD8+ T cells. Blood. 2009;113:6619–6628. doi: 10.1182/blood-2009-01-199588. [DOI] [PubMed] [Google Scholar]
- 36.Wofford JA, et al. IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T-cell survival. Blood. 2008;111:2101–2111. doi: 10.1182/blood-2007-06-096297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delgoffe GM, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delgoffe GM, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linossi EM, et al. Suppression of cytokine signaling: the SOCS perspective. Cytokine Growth Factor Rev. 2013;24:241–248. doi: 10.1016/j.cytogfr.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshimura A, et al. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 41.Lee K, et al. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powell JD, Delgoffe GM. The mammalian target of rapamycin: linking T cell differentiation, function, and metabolism. Immunity. 2010;33:301–311. doi: 10.1016/j.immuni.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park Y, et al. TSC1 regulates the balance between effector and regulatory T cells. J Clin Invest. 2013;123:5165–5178. doi: 10.1172/JCI69751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenks JA, et al. Differentiating the roles of STAT5B and STAT5A in human CD4+ T cells. Clin Immunol. 2013;148:227–236. doi: 10.1016/j.clim.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H, et al. Disruption of TSC1/2 signaling complex reveals a checkpoint governing thymic CD4+ CD25+ Foxp3+ regulatory T-cell development in mice. FASEB J. 2013;27:3979–3990. doi: 10.1096/fj.13-235408. [DOI] [PubMed] [Google Scholar]
- 46.Cohney SJ, et al. SOCS-3 is tyrosine phosphorylated in response to interleukin-2 and suppresses STAT5 phosphorylation and lymphocyte proliferation. Mol Cell Biol. 1999;19:4980–4988. doi: 10.1128/mcb.19.7.4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geissmann F, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Laar L, et al. Human CD34-derived myeloid dendritic cell development requires intact phosphatidylinositol 3-kinase-protein kinase B-mammalian target of rapamycin signaling. J Immunol. 2010;184:6600–6611. doi: 10.4049/jimmunol.0903089. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, et al. Tuberous sclerosis 1 (Tsc1)-dependent metabolic checkpoint controls development of dendritic cells. Proc Natl Acad Sci U S A. 2013;110:E4894–E4903. doi: 10.1073/pnas.1308905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wölfle SJ, et al. PD-L1 expression on tolerogenic APCs is controlled by STAT-3. Eur J Immunol. 2011;41:413–424. doi: 10.1002/eji.201040979. [DOI] [PubMed] [Google Scholar]
- 51.Sun Y, et al. Cutting edge: Negative regulation of dendritic cells through acetylation of the nonhistone protein STAT-3. J Immunol. 2009;182:5899–5903. doi: 10.4049/jimmunol.0804388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohtani M, et al. Mammalian target of rapamycin and glycogen synthase kinase 3 differentially regulate lipopolysaccharide-induced interleukin-12 production in dendritic cells. Blood. 2008;112:635–643. doi: 10.1182/blood-2008-02-137430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haidinger M, et al. A versatile role of mammalian target of rapamycin in human dendritic cell function and differentiation. J Immunol. 2010;185:3919–3931. doi: 10.4049/jimmunol.1000296. [DOI] [PubMed] [Google Scholar]
- 54.Weichhart T, et al. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 2008;29:565–577. doi: 10.1016/j.immuni.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 55.Wang H, et al. Convergence of the mammalian target of rapamycin complex 1- and glycogen synthase kinase 3-β-signaling pathways regulates the innate inflammatory response. J Immunol. 2011;186:5217–26. doi: 10.4049/jimmunol.1002513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosborough BR, et al. Murine dendritic cell rapamycin-resistant and rictor-independent mTOR controls IL-10, B7-H1, and regulatory T-cell induction. Blood. 2013;121:3619–3630. doi: 10.1182/blood-2012-08-448290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 58.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fish EN, Platanias LC. Interferon Receptor Signaling in Malignancy: a network of cellular pathways defining biological outcomes. Mol Cancer Res. 2014 doi: 10.1158/1541-7786.MCR-14-0450. pii: molcanres.0450.2014. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaur S, et al. Regulatory effects of mTORC2 complexes in type I IFN signaling and in the generation of IFN responses. Proc Natl Acad Sci U S A. 2012;109:7723–7728. doi: 10.1073/pnas.1118122109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaur S, et al. Regulatory effects of mammalian target of rapamycin-activated pathways in type I and II interferon signaling. J Biol Chem. 2007;282:1757–1768. doi: 10.1074/jbc.M607365200. [DOI] [PubMed] [Google Scholar]
- 62.Kaur S, et al. Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc Natl Acad Sci U S A. 2008;105:4808–4813. doi: 10.1073/pnas.0710907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tasian SK, et al. Aberrant STAT5 and PI3K/mTOR pathway signaling occurs in human CRLF2-rearranged B-precursor acute lymphoblastic leukemia. Blood. 2012;120:833–842. doi: 10.1182/blood-2011-12-389932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bogani C, et al. mTOR inhibitors alone and in combination with JAK2 inhibitors effectively inhibit cells of myeloproliferative neoplasms. PLoS One. 2013;8:e54826. doi: 10.1371/journal.pone.0054826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gentzler RD, et al. An overview of the mTOR pathway as a target in cancer therapy. Expert Opin Ther Targets. 2012;16:481–489. doi: 10.1517/14728222.2012.677439. [DOI] [PubMed] [Google Scholar]
- 66.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bar-Peled L, et al. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bar-Peled L, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsun ZY, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Efeyan A, et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dibble CC, et al. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126:1713–1719. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu L, Parent CA. Review series: TOR kinase complexes and cell migration. J Cell Biol. 2011;94:815–824. doi: 10.1083/jcb.201102090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gan X, et al. Evidence for direct activation of mTORC2 kinase activity by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 2011;286:10998–11002. doi: 10.1074/jbc.M110.195016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zinzalla V, et al. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 76.Toschi A, et al. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol Cell Biol. 2009;29:1411–1420. doi: 10.1128/MCB.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saci A, et al. Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol Cell. 2011;42:50–61. doi: 10.1016/j.molcel.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 79.Chen CH, et al. ER stress inhibits mTORC2 and Akt signaling through GSK-3β-mediated phosphorylation of rictor. Sci Signal. 2011;4:ra10. doi: 10.1126/scisignal.2001731. [DOI] [PubMed] [Google Scholar]
- 80.Busch S, et al. mTOR mediates human trophoblast invasion through regulation of matrix-remodeling enzymes and is associated with serine phosphorylation of STAT3. Exp Cell Res. 2009;315:1724–1733. doi: 10.1016/j.yexcr.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 81.Kim JH, et al. Signal transducer and activator of transcription 3 (STAT3) mediates amino acid inhibition of insulin signaling through serine 727 phosphorylation. J Biol Chem. 2009;284:35425–35432. doi: 10.1074/jbc.M109.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fielhaber JA, et al. Inhibition of mammalian target of rapamycin augments lipopolysaccharide-induced lung injury and apoptosis. J Immunol. 2012;188:4535–4542. doi: 10.4049/jimmunol.1003655. [DOI] [PubMed] [Google Scholar]
- 83.Kiu H, Nicholson SE. Biology and significance of the JAK/STAT signaling pathways. Growth Factors. 2012;30:88–106. doi: 10.3109/08977194.2012.660936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hervas-Stubbs S, et al. Direct effects of type I interferons on cells of the immune system. Clin Cancer Res. 2011;17:2619–2627. doi: 10.1158/1078-0432.CCR-10-1114. [DOI] [PubMed] [Google Scholar]