Abstract
The Epidermal Growth Factor Receptor (EGFR) is upregulated in numerous human cancers. Inhibition of EGFR signaling induces autophagy in tumor cells. Here we report an unanticipated role for the inactive EGFR in autophagy initiation. Inactive EGFR interacts with the oncoprotein LAPTM4B that is required for the endosomal accumulation of EGFR upon serum starvation. Inactive EGFR and LAPTM4B stabilize each other at endosomes and recruit the exocyst subcomplex containing Sec5. We show that inactive EGFR, LAPTM4B, and the Sec5 subcomplex are required for basal and starvation induced autophagy. LAPTM4B and Sec5 promote EGFR association with the autophagy inhibitor Rubicon, which in turn disassociates Beclin 1 from Rubicon to initiate autophagy. Thus, the oncoprotein LAPTM4B facilitates the role of inactive EGFR in autophagy initiation. This pathway is positioned to control tumor metabolism and promote tumor cell survival upon serum deprivation or metabolic stress.
Keywords: EGFR, LAPTM4B, Sec5, Autophagy
INTRODUCTION
Autophagy (macroautophagy) is a self-eating process where part of the cytoplasm, organelles and/or membrane are engulfed and eventually digested in lysosomes for cell homeostasis, development and/or nutrient recycling (Feng et al., 2014). Autophagy is inducible by nutrient starvation as a mechanism to protect cells (Klionsky et al., 2012). The core machinery required for autophagosome formation consists of autophagy-related (Atg) proteins that function in a specific order (Feng et al., 2014). The Beclin 1 (Atg6 in yeast) complex containing Beclin 1, VPS34, ATG14 and VPS15 plays essential roles in phagophore nucleation and autophagy initiation and is emerging as a regulatory complex by autophagy signals (He and Levine, 2010; Matsunaga et al., 2009; Sun et al., 2009; Zhong et al., 2009). LC3 (Atg8 in yeast) is an autophagy regulator downstream of Beclin 1, and a hallmark of autophagosome formation is the conversion of the cytosolic LC3-I into the lipidated LC3-II that associates with autophagosome membranes (Kabeya et al., 2000). The exocyst also plays a role in the Beclin 1 complex assembly during autophagy initiation (Bodemann et al., 2011), but how and where the exocyst and the autophagy complexes are regulated is not defined.
Epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase that plays fundamental roles in physiology and cancers (Schlessinger, 2000). Aberrant EGFR signaling due to EGFR mutations and/or protein overexpression is associated with most epithelial cancers (Mendelsohn and Baselga, 2006). Recent studies have shown a role for EGFR signaling in autophagy suppression by activation of the PI3K/AKT/mTOR pathway or by EGFR-mediated phosphorylation and inhibition of Beclin 1 (Wei et al., 2013). Autophagy is generally thought to prevent cancer initiation but is also positioned to promote cancer cell survival upon metabolic stress (Brech et al., 2009; White, 2012). Rapamycin, an autophagy inducer, exhibits cytotoxic effects in cancer cells treated with EGFR tyrosine kinase inhibitors (TKIs), erlotinib or gefitinib (Fung et al., 2012; Gorzalczany et al., 2011). However, autophagy induced by EGFR TKIs or neutralizing antibody shows cytoprotective roles (Choi et al., 2013; Dragowska et al., 2013; Eimer et al., 2011; Han et al., 2011; Li and Fan, 2010; Li et al., 2010b; Sobhakumari et al., 2013; Zou et al., 2013), suggesting that EGFR TKIs (or neutralizing antibody) may induce autophagy by a mechanism distinct from rapamycin. At least some EGFR overexpressing cells and tumors are dependent on autophagy for growth and survival (Jutten et al., 2013; Jutten and Rouschop, 2014). A gefitinib resistant subpopulation of lung adenocarcinoma cells depend on enhanced autophagic activity but not EGFR signaling for survival (Sakuma et al., 2013). The combined data suggest a kinase-independent role for EGFR in autophagy that regulates cancer cell survival.
The lysosomal-associated transmembrane protein 4B (LAPTM4B) is a tetra-transmembrane protein localized primarily to the late endosome and lysosome (Liu et al., 2004). LAPTM4B was originally identified in hepatocellular carcinoma patients as an abnormally overexpressed protein and was later found to be upregulated in many other human cancers including breast, lung, ovarian, and colon cancers (Kasper et al., 2005). Ectopic expression of LAPTM4B transforms normal cells and promotes proliferation, migration and invasion of cancer cells (Li et al., 2011a; Yang et al., 2010). LAPTM4B is proposed to promote cancer cell proliferation by upregulating PI3K/ATK signaling (Li et al., 2010a). We have shown that LAPTM4B blocks EGF-stimulated EGFR intraluminal sorting and lysosomal degradation, resulting in enhanced and prolonged EGFR signaling (unpublished data). A potential role for LAPTM4B in the promotion or inhibition of autophagosome maturation has been proposed (Li et al., 2011b; Vergarajauregui et al., 2011).
Here, we report an unexpected role for inactive EGFR in autophagy initiation. Upon serum starvation, inactive EGFR collaborates with LAPTM4B and Sec5 at endosomes to disassociate Rubicon from Beclin 1, which in turn initiates autophagy. EGFR TKIs also induce autophagy initiation, revealing that kinase-inactive EGFR functions in autophagy and this may be a mechanism for tumor cell resistance to EGFR TKIs.
RESULTS
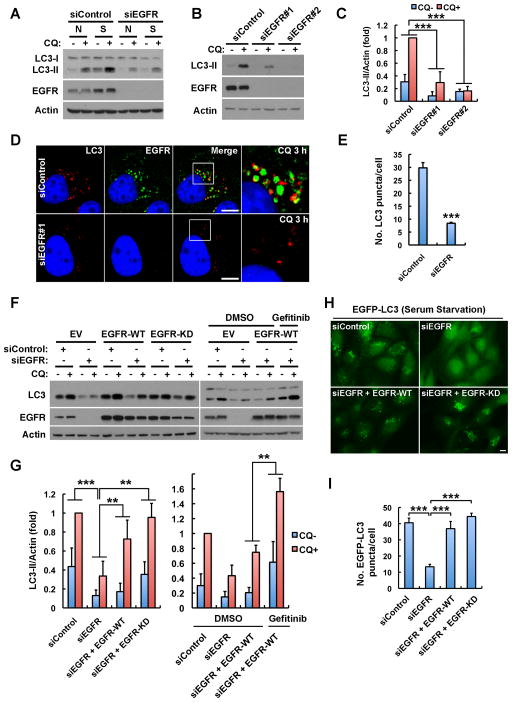
EGFR but not Its Kinase Activity Is Required for Autophagy
EGFR TKIs induce autophagy (Han et al., 2011; Jutten and Rouschop, 2014; Li et al., 2010b), suggesting a direct role for inactive EGFR in autophagy upregulation. To test this hypothesis, we examined if EGFR is required for autophagic flux. Knockdown of EGFR inhibited basal and serum starvation induced autophagic flux (Figure 1A), as shown by the LC3-II levels, before and after inhibition of its degradation by chloroquine. Two different EGFR siRNAs independently knocked down EGFR expression and inhibited autophagic flux (Figures 1B and 1C), and loss of EGFR markedly reduced endogenous LC3 puncta formation (Figures 1D and 1E), indicating a role for EGFR in autophagy initiation.
Figure 1. EGFR but Not Its Kinase Activity Is Required for Autophagy.
(A) Knockdown of EGFR inhibits basal and serum starvation induced LC3-II (autophagosome marker) turnover in MDA-MB-231 cells. Cells transfected with indicated siRNAs were cultured in normal medium (N) or serum free starvation medium (S) for 24 h, and then treated with 80 μM chloroquine (CQ) for 2 h to induce LC3-II accumulation. Whole cell lysates were harvested for Western blot analysis of LC3-II.
(B) Knockdown of EGFR by two distinct siRNAs inhibits LC3-II generation. MDA-MB-231 cells were cultured in normal medium. Unless otherwise indicated, the EGFR siRNA #1 was used in all the remaining experiments.
(C) Quantification of LC3-II levels in (B); mean + SD, n = 3, *** P < 0.001.
(D) CQ induces accumulation of LC3 puncta (autophagosomes) in control cells but not EGFR knockdown cells. Control or EGFR siRNA transfected cells in normal medium were pretreated with CQ for 3 h and fixed for immunostaining of endogenous LC3 (red) and EGFR (green). DAPI was used to stain the nuclei; Boxes indicate selected regions for magnified new; Bar, 10 μm.
(E) Quantification of LC3 puncta in (D); mean + SD, n = 3, *** P < 0.001.
(F) Left: Re-expression of wild type (WT) or the K721A kinase dead (KD) EGFR rescues LC3-II turnover in EGFR-knockdown cells. Right: Gefitinib (2 μM) treatment enhances autophagy rescue by EGFR-WT. SiRNA resistant EGFR-WT or -KD was expressed in MDA-MB-231 cells by lentivirus mediated infection. Endogenous EGFR was knocked down by siRNA #1, and cells were cultured in normal medium and treated with CQ for 2 h before whole cell lysate harvest for Western analysis. EV, empty vector.
(G) Quantification of LC3-II levels in (F); mean + SD, n = 3, ** P < 0.01, *** P < 0.001.
(H) Representative images of serum starvation induced EGFP-LC3 puncta in MDA-MB-231 cells treated with indicated siRNAs with or without re-expression of EGFR-WT or EGFR-KD. EGFP-LC3 was expressed by lentivirus infection and a monoclonal cell line stably expressing low levels of EGFP-LC3 was selected. Endogenous EGFR was knocked down by siRNA, and siRNA-resistant WT or KD EGFR was re-expressed using lentivirus. Cells were serum starved and fixed for fluorescence microscopy. Bar, 10 μm.
(I) Quantification of the number of EGFP-LC3 puncta in (H); mean + SD, n = 3, *** P < 0.001. See also Figure S1.
To determine whether the role for EGFR in autophagy is due to its overexpression, we tested the effects of EGFR knockdown in 7 cell lines, 4 of which do not overexpress EGFR (Figure S1A). Knockdown of EGFR inhibited autophagic flux in each cell line (Figure S1B), indicating a role in autophagy independent of receptor level. Importantly, the role for EGFR in autophagy initiation is receptor selective, as loss of PDGFRB, FGFR2, or c-Met did not suppress autophagy (Figures S1C–E).
Active EGFR blocks autophagy (Wei et al., 2013), and we confirmed that EGF stimulation inhibits autophagy initiation (Figure S1F); thus we hypothesized that inactive EGFR initiates autophagy. To test this hypothesis, siRNA resistant wild-type (WT) or K721A kinase dead (KD) EGFR was re-expressed in cells where endogenous EGFR was knocked down. Re-expressed EGFR-WT is auto-phosphorylated and activates AKT signaling when stimulated with EGF, but EGFR-KD does not auto-phosphorylate or activate AKT (Figure S1G). However, re-expression of either WT or KD EGFR rescued autophagic flux in EGFR knockdown cells (Figures 1F and 1G, left panels), and treatment of EGFR-WT re-expressing cells with the EGFR TKI gefitinib enhanced the rescue of autophagy initiation (Figures 1F and 1G, right panels), indicating the role for EGFR in autophagy initiation is kinase independent. Serum starvation induced an increase in GFP-LC3 puncta (autophagosomes) (Figure S1H), and loss of EGFR strongly inhibited autophagosome formation (Figures 1H and 1I). Re-expression of either WT or KD EGFR fully rescued the starvation-induced formation of GFP-LC3 puncta in EGFR knockdown cells (Figures 1H and 1I). These combined results reveal a kinase-independent role for EGFR in basal and serum starvation-induced autophagy.
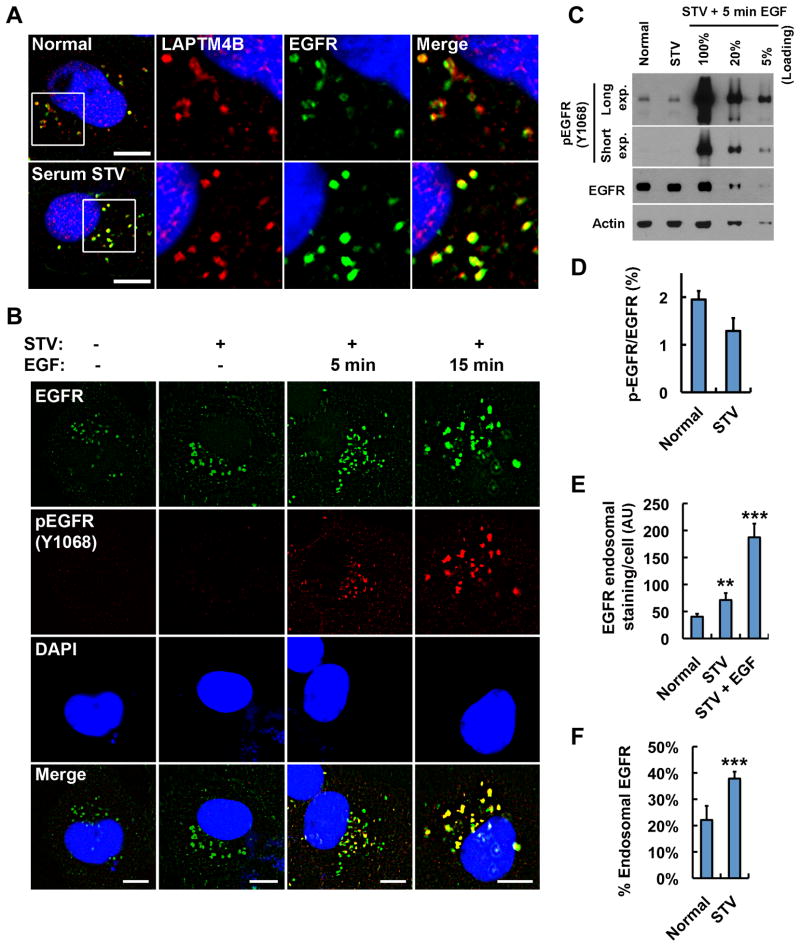
Starvation Induces Accumulation of Inactive EGFR at LAPTM4B Positive Endosomes
To explore how inactive EGFR controls autophagy, the subcellular localization of EGFR in normal and serum-starved cells was investigated. We previously showed that LAPTM4B inhibits EGF-stimulated EGFR lysosomal sorting (unpublished data). Here we show that EGFR partially colocalized with LAPTM4B at endosomes in cells grown with serum (Figure 2A, top). The specificity of EGFR immunostaining was confirmed by siRNA knockdown of EGFR (Figure S2A). Surprisingly, serum-starvation enhanced EGFR accumulation at LAPTM4B positive endosomes (Figure 2A, bottom), which include both late and early endosomes (Figure S2B). Starvation induced endosomal EGFR accumulation and EGFR-LAPTM4B colocalization were observed in all cells assayed (Figures S2C–F). Serum starvation also induced intracellular accumulation of c-Met and FGFR2, and c-Met colocalized well with LAPTM4B but FGFR2 did not (Figures S2G–J). However, neither FGFR2 nor c-Met was required for autophagy indicating that this is an EGFR selective function (Figure S1C–E).
Figure 2. Serum Starvation Induces Accumulation of Inactive EGFR at LAPTM4B Positive Endosomes.
(A) MDA-MB-231 cells were serum-starved or not for 24 h and fixed for co-staining of endogenous EGFR (green) and LAPTM4B (red). Boxes indicate regions for magnified new.
(B) MDA-MB-231 cells were serum-starved (STV) or not followed by EGF (100 ng/ml) stimulation for 5 or 15 min as indicated. Then cells were fixed for co-staining of EGFR (green) and phosphor-EGFR (Y1068, red).
(C) The whole cell lysates with or without serum starvation were analyzed by Western blot to detect phosphor-EGFR (Y1068) and total EGFR. Different loadings of the lysates from MDA-MB-231 cells with 5 min EGF (100 ng/ml) stimulation after starvation were used as a standard to quantify phosphor-EGFR levels in cells without EGF stimulation.
(D) Quantification of the phosphor-EGFR (Y1068) levels in (C); mean + SD, n = 3.
(E) Quantification of endosomal EGFR staining in cells cultured in normal medium, serum-free medium (STV), or serum-free medium plus EGF (100 ng/ml, 15 min), as shown in (B); mean + SD, n = 3, ** P < 0.01, *** P < 0.001.
(F) Quantification of the relative amount of endosomal EGFR staining in cells cultured in normal medium or serum-free medium (STV) normalized to the amount of endosomal EGFR staining in cells in serum-free medium plus EGF (100 ng/ml, 15 min); mean + SD, n = 3, *** P < 0.001.
DAPI was used to stain the nuclei; pEGFR, phosphor-EGFR (Y1068); Bar, 10 μm; See also Figure S2.
EGFR translocates to endosomes upon EGF stimulation and receptor autophosphorylation/activation (Wiley, 2003). We therefore examined whether the EGFR accumulated at LAPTM4B positive endosomes upon serum starvation was phosphorylated. EGF stimulated cells were used as positive controls for phospho-EGFR staining, as shown in Figure 2B. Interestingly, in either serum grown or the starved condition, phospho-EGFR was not detected by immunofluorescence, but there was clear endosomal EGFR accumulation (Figure 2B). The endosomal staining of EGFR appears stronger than the plasma membrane staining because of the point concentration at endosomes (Figures 2A, 2B, and S2). Quantitative Western blotting confirmed that less than 2% of the total EGFR was phosphorylated in cells cultured in either normal or serum starved medium, assuming that 5 min stimulation with 100 ng/ml EGF stimulated maximal EGFR phosphorylation (Figures 2C and 2D). Stimulation of cells with 100 ng/ml EGF for 15 min causes endocytosis and accumulation of over 95% of total EGFR at endosomes; we therefore calculated how much of EGFR was accumulated at endosomes in normal and starved cells by quantifying the endosomal EGFR staining in these conditions versus EGF (15 min) stimulated condition. This indicated that 20% or 40% of total EGFR is accumulated at endosomes in normal versus serum-starved conditions, respectively (Figures 2E and 2F). This demonstrates that in serum grown or starved cells the endosomal EGFR is primarily unphosphorylated. Importantly, upon serum starvation, the amount of endosomal EGFR increased from 20% to 40% (Figures 2B and 2F), but the total phospho-EGFR level decreased (Figures 2C and 2D), indicating that serum starvation induced endosomal accumulation of inactive EGFR.
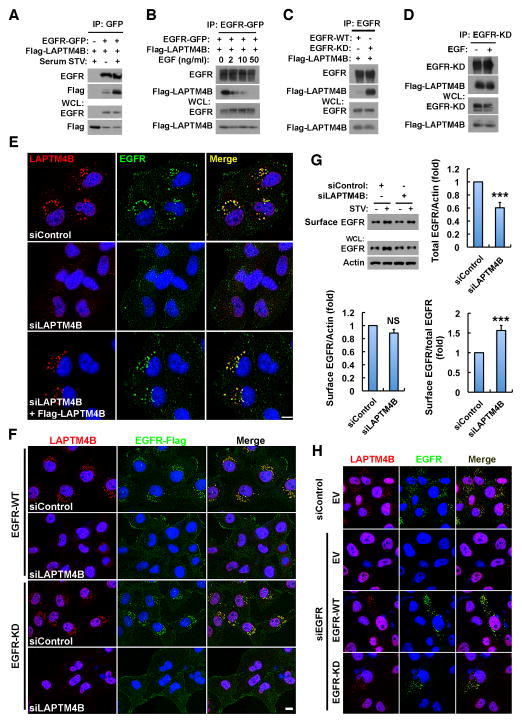
Inactive EGFR and LAPTM4B Interact and Stabilize Each Other at Endosomes
The interaction between EGFR and LAPTM4B was studied to explore the mechanism for EGFR endosomal localization. EGFR was co-immunoprecipitated (co-IP’ed) specifically with LAPTM4B but not other LAPTM members (Figure S3A). Serum starvation increased EGFR endosomal accumulation (Figure 2) and enhanced the LAPTM4B-EGFR association (Figure 3A), while EGF stimulation reduced the interaction (Figure 3B), indicating that LAPTM4B preferentially interacts with inactive EGFR. This was confirmed using the EGFR-KD mutant that has much stronger interaction with LAPTM4B compared with EGFR-WT (Figure 3C). EGF stimulation did not affect the LAPTM4B interaction with EGFR-KD (Figure 3D) that is not activated by EGF (Figure S1G). As serum starvation also induces endosomal accumulation of other receptors, LAPTM4B interaction with PDGFRB, FGFR2, and c-Met was tested. These receptors also co-IP’ed with LAPTM4B (Figure S3B), but none of these interactions were enhanced upon serum starvation (Figure S3C), indicating that the serum starvation promoted EGFR-LAPTM4B interaction is selective.
Figure 3. Inactive EGFR and LAPTM4B Interact and Stabilize Each Other at Endosomes.
(A) Co-immunoprecipitation (co-IP) of EGFR-GFP with Flag-LAPTM4B in HEK 293 cells cultured in normal or serum free medium.
(B) Co-IP of EGFR-GFP with Flag-LAPTM4B in HEK 293 cells cultured in serum free medium with 0, 2, 10, or 50 ng/ml EGF treatment (30 min).
(C) Co-IP of wild type (WT) or kinase dead (KD) EGFR with Flag-LAPTM4B in HEK 293 cells in normal medium.
(D) Co-IP of EGFR-KD with Flag-LAPTM4B in HEK 293 cells cultured in serum free medium with or without 50 ng/ml EGF treatment (30 min).
(E) LAPTM4B is required for the endosomal accumulation of endogenous EGFR. MDA-MB-231 cells were treated with control or LAPTM4B siRNA, and after 48 h the siRNA-resistant LAPTM4B was re-expressed via transient transfection. Cells were serum starved and fixed for co-staining of LAPTM4B (red) and EGFR (green).
(F) Ectopically expressed wild type (WT) and kinase dead (KD) EGFR depend on LAPTM4B for endosomal accumulation. MDA-MB-231 cells stably expressing C-terminally Flag-tagged EGFR-WT or -KD were treated with control or LAPTM4B siRNA, serum starved and fixed for co-staining of Flag (green) and endogenous LAPTM4B (red).
(G) Control and LAPTM4B knockdown cells were surface biotinylated, and surface and total EGFR levels were analyzed by western blot (top left). Quantification of total EGFR levels (top right), cell surface EGFR levels (bottom left), and the relative amounts of cell surface EGFR (bottom right) in serum starved control and LAPTM4B knockdown cells; mean + SD, n = 3; *** P < 0.001; NS, not significant.
(H) Inactive EGFR stabilizes endosomal LAPTM4B. MDA-MB-231 cells were transfected with control or EGFR siRNA, and after 48 h cells were transfected with empty vector (EV) or siRNA-resistant EGFR-WT or -KD and starved overnight, followed by fixing and co-staining of EGFR (green) and LAPTM4B (red).
WCL, whole cell lysate; STV, serum starvation; Bar, 10 μm; DAPI was used to stain the nuclei. See also Figure S3.
As LAPTM4B is an endosomal protein (Shao et al., 2003; Vergarajauregui et al., 2011), we determined whether LAPTM4B could modulate EGFR endosomal accumulation. MDA-MB-231 cells express variable amounts of LAPTM4B, and in cells with low LAPTM4B there was less endosomal EGFR (Figure S3D, arrow head), suggesting LAPTM4B modulation of EGFR localization. Strikingly, upon knockdown of LAPTM4B, EGFR endosomal accumulation was lost, but EGFR targeting was fully rescued by re-expression of siRNA-resistant LAPTM4B (Figure 3E). Loss of LAPTM4B also resulted in a loss of endosomal EGFR in A431 and HeLa cells indicating that this was independent of cell type (Figures S3E and S3F). To confirm that LAPTM4B mediates endosomal accumulation of inactive EGFR, cells stably expressing C-terminally Flag tagged WT or KD EGFR were generated. Both WT and KD EGFR had endosomal localization, but this was disrupted by loss of LAPTM4B (Figure 3F), demonstrating that LAPTM4B is required for the endosomal accumulation of inactive EGFR. Though c-Met and FGFR2 also accumulate at endosomes upon serum starvation, LAPTM4B knockdown reduced the endosomal localization of c-Met but not FGFR2 (Figures S3G and S3H). Interestingly, while c-Met localization appears to be impacted by LAPTM4B, c-Met was not required for autophagy initiation (Figures S1C–E).
To further explore LAPTM4B regulation of EGFR endosome accumulation, a biotinylation assay was used to quantify the amount of cell surface EGFR. This demonstrated that while the total EGFR level was reduced in LAPTM4B knockdown cells, the level of EGFR at the cell surface was not affected (Figure 3G), consistent with a specific loss of endosomal EGFR in LAPTM4B knockdown cells. This indicates that LAPTM4B mediates endosomal EGFR accumulation by stabilizing inactive EGFR at endosomes. Likewise, inactive EGFR also stabilizes LAPTM4B at endosomes, as knockdown of EGFR caused a loss of endosomal LAPTM4B staining, and re-expression of either WT or KD EGFR rescued LAPTM4B staining at endosomes (Figure 3H).
Importantly, the endosomal localization of EGFR or LAPTM4B is not dependent on Hrs or TSG101 (Figure S3I), both of which are essential for the ESCRT-mediated EGFR intraluminal sorting upon ligand stimulation (Sorkin and Goh, 2008). This indicates that the pathway mediating accumulation of inactive EGFR at LAPTM4B positive endosomes is different from that for ligand stimulated EGFR endosomal sorting.
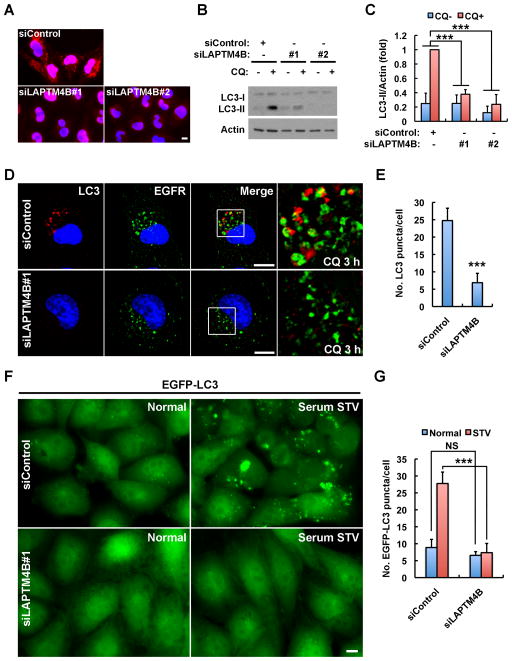
LAPTM4B Is Required for Basal and Serum Starvation-Induced Autophagy
The data suggest that EGFR dependent autophagy initiation may be linked to LAPTM4B-mediated EGFR endosomal localization. To determine if LAPTM4B is required for autophagy, different siRNAs were used to knockdown LAPTM4B and LC3-II turnover was quantified. Both LAPTM4B siRNAs knocked down LAPTM4B expression and inhibited basal autophagic flux (Figures 4A–C). The chloroquine induced accumulation of endogenous LC3 puncta was also strongly inhibited in LAPTM4B siRNA-treated cells (Figures 4D and 4E), phenocopying that of EGFR knockdown. Similarly, starvation-induced formation of the GFP-LC3 puncta was markedly decreased upon loss of LAPTM4B (Figures 4F and 4G). Consistently, ectopic expression of LAPTM4B increased LC3-II turnover (Figures S4A and S4B) as well as basal GFP-LC3 puncta formation (Figure S4C), and re-expression of siRNA-resistant LAPTM4B rescued starvation-induced GFP-LC3 puncta formation in LAPTM4B knockdown cells (Figure S4D). Significantly, knockdown of EGFR and LAPTM4B together did not further inhibit LC3-II turnover compared to the single knockdown of either protein (Figures S4E and S4F), suggesting that LAPTM4B and EGFR function in the same pathway.
Figure 4. LAPTM4B Is Required for Basal and Serum Starvation Induced Autophagy.
(A) Immunofluorescence images showing the knockdown efficiency of two distinct LAPTM4B siRNAs. Nuclear staining is nonspecific.
(B) Knockdown of LAPTM4B by two distinct siRNAs suppresses LC3-II turnover in MDA-MB-231 cells cultured in normal medium. CQ, chloroquine.
(C) Quantification of LC3-II in (B); mean + SD, n = 3, *** P < 0.001.
(D) Knockdown of LAPTM4B results in a loss of chloroquine (CQ) induced accumulation of endogenous LC3 puncta. Note: different from Figure 3E, some EGFR intracellular puncta were observed here in LAPTM4B knockdown cells because of chloroquine treatment that blocked basal EGFR lysosomal degradation. Boxes are selected regions for magnified view.
(E) Quantification of the number of LC3 puncta in (D); mean + SD, n = 3, *** P < 0.001.
(F) Knockdown of LAPTM4B causes a loss of serum starvation induced EGFP-LC3 puncta formation. MDA-MB-231 cells stably expressing low levels of EGFP-LC3 as described in Figure 1H were treated with control or LAPTM4B siRNA and serum starved or not before fixing for microscopy.
(G) Quantification of the number of EGFP-LC3 puncta in (F); mean + SD, n = 3; NS, not significant; *** P < 0.001.
Bar: 10 μm. DAPI was used to stain the nuclei. See also Figure S4.
To explore if EGFR and LAPTM4B have mechanisms independent of each other for autophagy initiation, we examined autophagic turnover in cells with knockdown of one protein and overexpression of the other. Interestingly, overexpression of EGFR largely rescued autophagy in LAPTM4B knockdown cells (Figure S4G and S4H), whereas overexpression of LAPTM4B weakly, if at all, rescued autophagy in EGFR knockdown cells (Figures S4I and S4J). This suggests that LAPTM4B is a cofactor for EGFR driven autophagy initiation.
Inactive EGFR and LAPTM4B Interact with the Exocyst for Autophagy Initiation
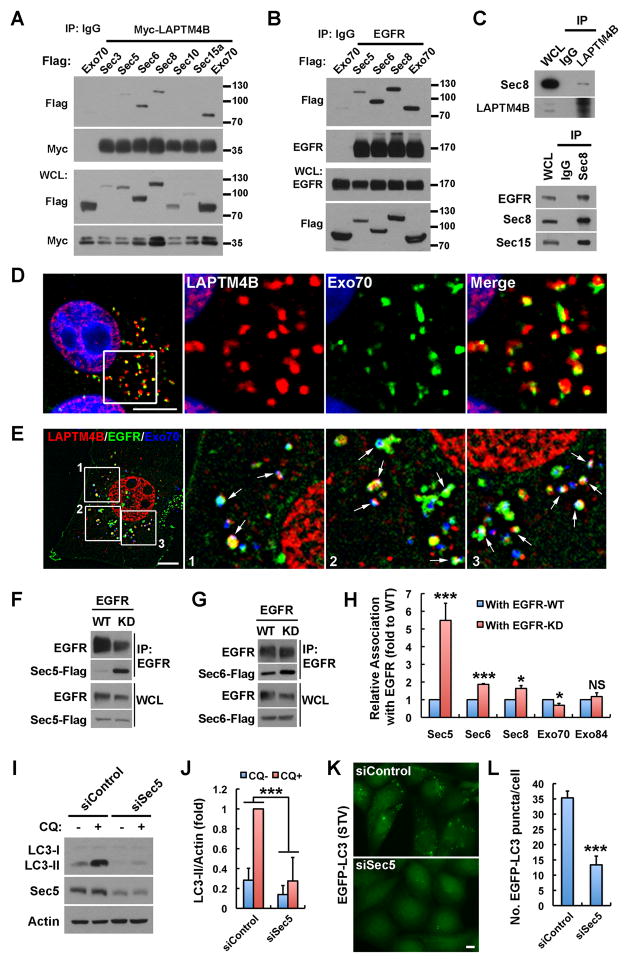
The data indicate that serum starvation promotes the assembly of an endosomal EGFR-LAPTM4B complex that regulates autophagy initiation. Recently, exocyst complexes have been implicated in starvation induced autophagy (Bodemann et al., 2011). A proteomic screen demonstrated that exocyst subunits interact with the EGFR complex (Foerster et al., 2013); we therefore explored if LAPTM4B and inactive EGFR interact with exocyst complex to modulate autophagy. As shown in Figures 5A and 5B, LAPTM4B and EGFR interact with multiple subunits of the exocyst complex. Sec8 is a core subunit of the exocyst and in endogenous co-IP Sec8 associated with LAPTM4B and EGFR (Figure 5C). Exo70 also interacted with LAPTM4B and EGFR and localized to subdomains of LAPTM4B positive endosomes (Figure 5D), and a fraction of endosomes were positive for EGFR, LAPTM4B and Exo70 (Figure 5E).
Figure 5. Inactive EGFR and LAPTM4B Interact with Exocyst for Autophagy Initiation.
(A and B) Co-immunoprecipitation (co-IP) of LAPTM4B or EGFR with different exocyst subunits in HEK 293 cells co-transfected with indicated proteins.
(C) Endogenous co-IP of Sec8 with LAPTM4B or EGFR in MDA-MB-231 cells.
(D) Co-localization of LAPTM4B and Exo70-Flag. MDA-MB-231 cells were fixed and stained with anti-LAPTM4B (red) and anti-Flag (green). DAPI was used to stain the nuclei.
(E) Colocalization of LAPTM4B (red), EGFR-GFP (green), and Exo70-Flag (blue) in MDA-MB-231 cells.
(F and G) Co-IP of EGFR-WT or -KD with Sec5 or Sec6 in HEK 293 cells co-transfected with indicated proteins.
(H) Quantification of the relative amounts of different exocyst subunits co-IP’ed with EGFR-WT or -KD; mean + SD, n ≥ 3; NS, not significant; * P < 0.05; *** P < 0.001.
(I) Knockdown of Sec5 suppresses LC3-II turnover in MDA-MB-231 cells cultured in normal medium.
(J) Quantification of LC3-II levels in (I); mean + SD, n = 3; *** P < 0.001.
(K) Knockdown of Sec5 suppresses serum starvation induced EGFP-LC3 puncta formation in MDA-MB-231 cells.
(L) Quantification of the number of EGFP-LC3 puncta in (K); mean + SD, n = 3; *** P < 0.001. Boxes are selected regions for magnified new; Bar: 10 μm; See also Figure S5.
As LAPTM4B selectively interacts with inactive EGFR, we determined if exocyst subunits interact similarly. Strikingly, Sec5 interacted with EGFR-KD much more strongly than with EGFR-WT (Figures 5F and 5H). Sec6 and Sec8 also associated with EGFR-KD more strongly (Figures 5G, 5H and S5A), but Exo70 and Exo84 did not (Figures 5H, S5B and S5C) demonstrating that inactive EGFR selectively associates with the Sec5 exocyst subcomplex.
To determine if the Sec5 subcomplex modulates autophagy, Sec5 was knocked down, and this strongly inhibited basal LC3 turnover (Figures 5I and 5J) and serum starvation induced GFP-LC3 puncta formation (Figures 5K and 5L), indicating that Sec5 is required for basal and serum starvation induced autophagy initiation. This was unexpected as Sec5 has inhibitory roles in amino acid starvation induced autophagy (Bodemann et al., 2011). Sec6, Sec8 and Exo70 also interacted with LAPTM4B and EGFR (Figures 5A and 5B), and all were required for autophagic turnover in MDA-MB-231 cells (Figures S5D–F).
EGFR, LAPTM4B and Sec5 Regulate Rubicon Disassociation from the Beclin 1 Complex
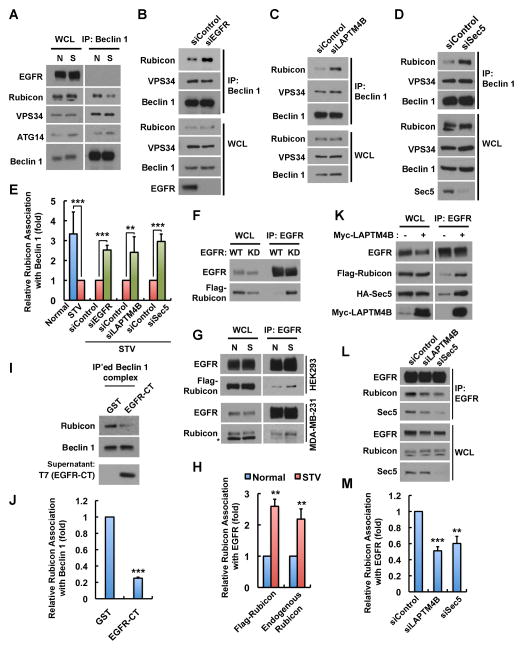
Beclin 1 is a core subunit of the autophagy initiation complex involved in phagophore formation (He and Levine, 2010). To investigate how inactive EGFR may modulate the Beclin 1 complex, changes in the Beclin 1 interactome (He and Levine, 2010) upon serum starvation were explored. Although serum starvation induced strong autophagosome formation and autophagic flux in cells (Figures 1A and S1H), only the association between Beclin 1 and the autophagy inhibitor Rubicon was reduced whereas the Beclin 1 association with VPS34 or ATG14 was not affected by serum starvation (Figures 6A and 6E). Importantly, serum starvation strongly stimulated autophagy in control cells, but in Rubicon knockdown cells basal autophagy is dramatically enhanced, and serum starvation does not further induce autophagy (Figure S6), signifying that the disassociation of Rubicon from Beclin 1 is a major mechanism for serum starvation induced autophagy. EGFR, LAPTM4B and Sec5 appear to have a collaborative role in autophagy initiation, as loss of any of these components blocked autophagy (Figures 1, 4 and 5) and inhibited Rubicon disassociation from Beclin 1 (Figures 6B–E).
Figure 6. EGFR, LAPTM4B and Sec5 Regulate Rubicon Disassociation from the Beclin 1 Complex.
(A) Co-immunoprecipitation (co-IP) of Beclin 1 with ATG14, VPS34, and Rubicon in normal (N) and serum starved (S) MDA-MB-231 cells.
(B–D) Co-IP of Beclin 1 with VPS34 and Rubicon in serum starved MDA-MB-231 cells pretreated with indicated siRNAs.
(E) Quantification of relative Rubicon association with Beclin 1 in (A–D); mean + SD, n = 3; ** P < 0.01; *** P < 0.001.
(F) Co-IP of Rubicon with EGFR-WT or -KD in HEK 293 cells co-transfected with indicated proteins.
(G) Co-IP of EGFR with Flag-Rubicon or endogenous Rubicon in normal (N) and serum starved (S) conditions. Asterisk indicates a nonspecific band.
(H) Quantification of relative Rubicon association with EGFR in (G); mean + SD, n = 3; ** P < 0.01.
(I) Endogenous Beclin 1 complex was IP’ed from MDA-MB-231 cells using anti-Beclin 1. The beads with immunocomplex were washed, divided into equal volumes and then incubated with purified GST or EGFR-C tail (EGFR-CT, amino acids 959-1186) for 20 min at room temperature. Changes of Rubicon association with Beclin 1 after the incubation were analyzed by Western blot.
(J) Quantification of relative Rubicon association with Beclin 1 in (I); mean + SD, n = 5; *** P < 0.001.
(K) LAPTM4B overexpression facilitates EGFR co-IP with Rubicon and Sec5 in HEK 293 cells.
(L) Knockdown of LAPTM4B or Sec5 diminishes Rubicon co-IP with EGFR in serum starved MDA-MB-231 cells.
(M) Quantification of relative Rubicon association with EGFR in (L); mean + SD, n = 3; ** P < 0.01; *** P < 0.001.
N, normal medium with serum; S, STV, serum free starvation medium; WCL, whole cell lysate. See also Figure S6.
Rubicon strongly and selectively associates with EGFR-KD (Figure 6F), and consistently, serum starvation significantly enhanced the co-IP of EGFR with overexpressed or endogenous Rubicon (Figure 6G and 6H), suggesting that inactive EGFR regulates autophagy by interacting with Rubicon. To determine if the EGFR C-tail (EGFR-CT) could modulate the Rubicon-Beclin 1 interaction in vitro, the Beclin 1 complex was purified by IP and incubated with purified GST or EGFR-CT. As shown in Figures 6I and 6J, incubation with EGFR-CT efficiently disassociated Rubicon from Beclin 1 in vitro. The EGFR-CT lacks the kinase domain and is not phosphorylated, consistent with the concept that the inactive EGFR functions as a nonphosphorylated receptor in autophagy initiation and removes Rubicon from the Beclin 1 complex.
LAPTM4B is overexpressed in most epithelial cancers, and increased LAPTM4B expression enhanced the EGFR interaction with Rubicon and Sec5 (Figure 6K). Sec5 has been shown to associate with Rubicon (Bodemann et al., 2011), and the knockdown of either LAPTM4B or Sec5 diminished the Rubicon interaction with EGFR (Figures 6L and 6M), indicating that LAPTM4B and Sec5 facilitate the Rubicon interaction with inactive EGFR. Thus, the endosomal inactive EGFR, with LAPTM4B and Sec5, promotes disassociation of Rubicon from the Beclin 1 complex, which in turn initiates autophagy.
Erlotinib and Gefitinib Induce a Sec5-Dependent Role for Inactive EGFR in Autophagy Initiation
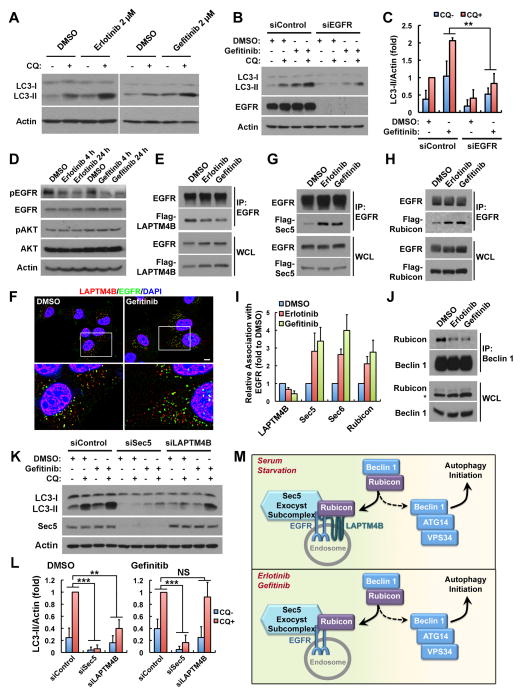
The EGFR TKIs, erlotinib and gefitinib, induce autophagy in multiple cancer cell lines and the underlying mechanisms have been linked to the inhibition of the downstream PI3K/AKT/mTOR pathway and/or the inhibition of EGFR-mediated Beclin 1 phosphorylation (Dragowska et al., 2013; Jutten and Rouschop, 2014; Sobhakumari et al., 2013; Wei et al., 2013). The role for inactive EGFR in autophagy initiation suggests that EGFR TKIs may induce a direct role for EGFR in autophagy. Erlotinib or gefitinib at clinically relevant concentrations (Baselga et al., 2002; Soulieres et al., 2004) stimulated autophagic activity comparable to that induced by serum starvation or rapamycin (Figures 7A, S7A and S7B). Further, TKI-induced autophagy is EGFR dependent as loss of EGFR strongly suppressed gefitinib-induced autophagy (Figures 7B and 7C). TKIs downregulated phosphorylation of EGFR but not Akt (Figure 7D), consistent with a direct role for inactive EGFR but not Akt/mTOR inactivation in the EGFR TKI-induced autophagy.
Figure 7. Erlotinib and Gefitinib Induce a Sec5-Dependent Role for Inactive EGFR in Autophagy Initiation.
(A) The EGFR TKIs, erlotinib and gefitinib, induce autophagy in MDA-MB-231 cells. Cells were treated with DMSO or 2 μM of erlotinib or gefitinib for 24 h, followed by 2 h of 80 μM chloroquine (CQ) treatment as indicated, and whole cell lysates were analyzed for LC3 levels.
(B) Knockdown of EGFR blocks gefitinib-induced autophagy in MDA-MB-231 cells. Control or EGFR knockdown cells were treated with DMSO or 2 μM gefitinib for 24 h, followed by 2 h of CQ treatment as indicated.
(C) Quantification of LC3-II levels in (B); mean + SD, n = 3; ** P < 0.01.
(D) Erlotinib/gefitinib inhibits EGFR but not Akt phosphorylation in MDA-MB-231 cells. Cells were treated with DMSO or 2 μM of erlotinib or gefitinib for 4 h or 24 h, followed by whole cell lysate harvest and Western blot analysis of indicated proteins. Specific antibodies recognizing pEGFR (Y1068) and pAkt (S473) were used.
(E) Erlotinib/gefitinib inhibits EGFR association with LAPTM4B. HEK293 cells co-transfected with EGFR and Flag-LAPTM4B were treated with MDSO or 2 μM erlotinib or gefitinib for 2 h and whole cell lysates were harvested for co-immunoprecipitation (co-IP) assay.
(F) Gefitinib treatment stimulates EGFR accumulation at endosomes. MDA-MB-231 cells treated with DMSO or 2 μM of gefitinib for 24 h were fixed for co-staining of endogenous LAPTM4B (red) and EGFR (green). Bar, 10 μm.
(G and H) Erlotinib/gefitinib (2 μM) treatment enhances EGFR association with Sec5 and Rubicon in HEK 293 cells co-transfected with EGFR and Sec5 or Rubicon.
(I) Quantification of relative EGFR association with indicated proteins in (E), (G) and (H); mean + SD, n = 3.
(J) Erlotinib/gefitinib (2 μM) induces disassociation of Rubicon from Beclin 1 in MDA-MB-231 cells. Asterisk indicates a nonspecific band.
(K) Gefitinib induced autophagy requires Sec5 but not LAPTM4B. MDA-MB-231 cells were transfected with indicated siRNAs, treated with DMSO or 2 μM gefitinib for 24 h, followed with or without 2 h of CQ treatment. Whole cell lysate were harvested for Western blot analysis of LC3-II.
(L) Quantification of LC3-II levels in (K); mean + SD, n = 3; ** P < 0.01; *** P < 0.001; NS, not significant.
(M) A model for the role of inactive EGFR in autophagy initiation. Serum starvation enhances EGFR interaction with LAPTM4B, resulting in increased accumulation of inactive EGFR at endosomes where EGFR also gains stronger interaction with Sec5. LAPTM4B and Sec5 facilitate Rubicon association with EGFR, which promotes the release of Beclin 1 from Rubicon for autophagy initiation. The EGFR TKIs, erlotinib and gefitinib, stimulate LAPTM4B-independent EGFR accumulation at endosomes where EGFR still gains enhanced Sec5 and Rubicon interaction so as to release Rubicon-free Beclin 1 for autophagy initiation.
WCL, whole cell lysate; See also Figure S7.
As erlotinib and gefitinib inhibit EGFR kinase activity, whether the EGFR-LAPTM4B-Sec5 pathway is stimulated for autophagy initiation was investigated. Erlotinib and gefitinib inhibited the interaction between EGFR and LAPTM4B (Figures 7E and 7I), but still stimulated accumulation of EGFR at endosomes (Figures 7F and S7C), similar to serum starvation (Figure 2). The EGFR interactions with Sec5, Sec6, and Rubicon were all enhanced by erlotinib or gefitinib (Figures 7G–I and S7D). As with serum starvation, erlotinib or gefitinib also promoted Rubicon disassociation from the Beclin 1 complex (Figure 7J), indicating that erlotinib and gefitinib induce a LAPTM4B-independent role for EGFR in Rubicon regulation. Finally and consistently, while Sec5 and LAPTM4B were required for basal autophagy (Figures 7K and 7L, left), loss of Sec5 but not LAPTM4B inhibited the gefitinib (or erlotinib, not shown) stimulated LC3-II turnover (Figures 7K and 7L, right). Together, these results reveal that the EGFR TKIs, erlotinib and gefitinib, stimulate a direct role for inactive EGFR in autophagy initiation that is dependent on Sec5 but independent of LAPTM4B.
Together, the data suggest a model that serum starvation induces LAPTM4B dependent EGFR accumulation at endosomes, where inactive EGFR synergizes with LAPTM4B and Sec5 in stimulating autophagy initiation independently of its tyrosine kinase activity by facilitating the disassociation of Rubicon from Beclin 1 (Figure 7M, top). Similarly, EGFR TKIs induce LAPTM4B-independent accumulation of EGFR at endosomes where EGFR associates with Sec5 and Rubicon to release Beclin 1 for autophagy initiation (Figure 7M, bottom).
DISCUSSION
Autophagy is activated in many cancers and facilitates cancer cell survival in stressed conditions (Brech et al., 2009; White, 2012). EGFR plays crucial roles in many epithelial cancers, and its signaling has been linked to pro-survival, proliferative, and pro-metastatic functions of cancer cells (Mendelsohn and Baselga, 2006). There are multiple links of EGFR to autophagy: (1) EGFR signaling activates the PI3K/AKT/mTOR pathway that inhibits autophagy; (2) activated EGFR directly phosphorylates and inhibits Beclin 1, a key component in autophagy initiation; (3) EGFR TKIs upregulate autophagy in many cancer cells (Jutten and Rouschop, 2014; Li and Fan, 2010; Wei et al., 2013). These studies all support a role for EGFR signaling in autophagy suppression.
Our data reveal a direct and unexpected role for inactive EGFR in autophagy initiation. Upon serum starvation, inactive EGFR forms a complex with LAPTM4B at endosomes where it recruits the Sec5 exocyst subcomplex. This EGFR complex binds the autophagy inhibitor Rubicon resulting in its disassociation from Beclin 1, releasing the Rubicon-free Beclin 1 complex to initiate autophagy (Figure 7M, top). These results establish the concept that while in nutrient rich conditions activated EGFR suppresses autophagy and promotes cancer cell proliferation and survival through agonist stimulated EGFR signaling, in metabolically stressed conditions inactive EGFR alternatively facilitates cancer cell survival by activating autophagy. Consistent with our findings, kinase independent roles for EGFR in cancer cell survival have been reported (Coker et al., 1994; Ewald et al., 2003; Weihua et al., 2008), and cancer cells expressing EGFR have been shown to depend on autophagy for survival and growth (Jutten et al., 2013; Jutten and Rouschop, 2014; Sakuma et al., 2013).
In clinical treatments, the EGFR TKIs, erlotinib and gefitinib, are effective in non-small cell lung cancers (NSCLCs) that have activating mutations in EGFR, but show no effects in most solid tumors with WT EGFR (Mendelsohn and Baselga, 2006; Paez et al., 2004). Recent studies have shown that treatment with EGFR TKIs increased autophagy with cytoprotective effects in many cancer cells (Dragowska et al., 2013; Eimer et al., 2011; Gorzalczany et al., 2011; Han et al., 2011; Jutten and Rouschop, 2014; Li et al., 2013; Sakuma et al., 2013; Sobhakumari et al., 2013; Zou et al., 2013). EGFR TKIs activate autophagy by blocking the role for active EGFR in autophagy suppression in lung cancer cells with activating EGFR mutations (Wei et al., 2013), but the mechanism by which the EGFR TKIs induce autophagy in cancer cells with WT EGFR was not defined. We show that EGFR TKIs induce a direct role for inactivated EGFR in autophagy initiation. EGFR TKIs mimic serum starvation and trigger endosomal accumulation of inactive EGFR that gains enhanced interaction with the Sec5 exocyst subcomplex and Rubicon, releasing Beclin 1 to initiate autophagy (Figure 7M, bottom). Thus, though EGFR TKIs block the cellular functions mediated by EGFR kinase signaling, they activate a role for inactive EGFR in autophagy that could potentially provide a survival advantage and TKI resistance in WT EGFR expressing cancers. In fact, there is in vitro and in vivo evidence that co-targeting EGFR and autophagy is a promising strategy to overcome TKI resistance of cancer cells with WT EGFR, even when these cancer cells carry a different oncogenic factor like K-Ras or p53 mutation (Choi et al., 2013; Dragowska et al., 2013; Jutten and Rouschop, 2014; Zou et al., 2013). For example, while erlotinib or hydroxychloroquine (HCQ, a clinically available autophagy inhibitor) alone show no anti-tumor activity in xenografts derived from H460 NSCLC cells expressing WT EGFR but mutant K-Ras, combination of the two drugs resulted in dramatic inhibition of tumor growth (Zou et al., 2013). A recent clinical trial has confirmed the safety and tolerance for co-treatment of erlotinib with HCQ (Goldberg et al., 2012), and additional trials combining HCQ with erlotinib or gefitinib are ongoing for the treatment of NSCLC.
EGFR and/or LAPTM4B are overexpressed in the majority of human cancers and contribute to cancer cell survival and proliferation (Kasper et al., 2005; Shao et al., 2003). The results presented here highlight the roles for the oncoprotein LAPTM4B in EGFR mediated cell survival functions. We have previously shown that LAPTM4B promotes active EGFR signaling by blocking EGF stimulated EGFR intraluminal sorting and lysosomal degradation (unpublished data). In the EGF stimulated condition, LAPTM4B has a weak interaction with EGFR but it inhibits the function of Hrs, a key subunit of the Endosomal Sorting Complex Required for Transport-0 (ESCRT-0), through enhancing the ubiquitination of Hrs by the E3 ubiquitin ligase Nedd4 (unpublished data). Here, we show that upon serum starvation, LAPTM4B senses EGFR inactivation at endosomes and selectively forms a complex with inactive EGFR to initiate autophagy. In both conditions, LAPTM4B facilitates the pro-survival functions of EGFR in cancer cells. The lack of a requirement for LAPTM4B in EGFR-TKIs induced autophagy is not surprising, as LAPTM4B appears to be a cofactor for EGFR driven autophagy and the effects of LAPTM4B knockdown on autophagy can be largely compensated by EGFR overexpression (Figures S4G and S4H). Importantly, in chemotherapies, LAPTM4B appears to increase drug resistance by multiple mechanisms (Li et al., 2010a; Li et al., 2010c). Thus, although LAPTM4B is not required for the erlotinib/gefitinib-induced EGFR functions in autophagy initiation, it may enhance EGFR-TKI resistance through other pathways. This supports LAPTM4B as a therapeutic target for EGFR positive cancers or a combined target for anti-EGFR therapies.
Multivesicular endosomes (MVEs) and autophagosomes are closely related as in mammalian cells autophagosomes often fuse with endosomes to form amphisomes before the final formation of autolysosomes (Fader and Colombo, 2009; Lamb et al., 2013). Therefore, endosomes have an established role in autophagosome maturation. Our results demonstrate that the endosome-localized inactive EGFR and LAPTM4B play pivotal roles in autophagy initiation. This emphasizes the importance of endosomes not only as canonical degradative compartments of the autophagosome content but also as signaling organelles that activate the autophagy pathway. The EGFR mediated Rubicon-Beclin 1 disassociation may occur at the endosomal surface, but the Beclin 1 complex functions in autophagosomal membrane nucleation at the endoplasmic reticulum (ER) (Hamasaki et al., 2013; Lamb et al., 2013). This suggests that the phagophore initiation sites at ER are in close proximity to endosomal Beclin 1 releasing sites. In support of this, LC3 puncta were observed adjacent to EGFR positive endosomes (Figures 1D and 4D, top panels), suggesting that autophagosomes are formed at the ER close to EGFR positive endosomes. Consistently, there are tight associations between the ER network and endosomes through ER-endosome contacts in mammalian cells (Eden et al., 2010; Friedman et al., 2013; Helle et al., 2013). Although most studies have focused on the roles for ER, mitochondria and Golgi in phagophore initiation, the ER-endosome contact sites are positioned to play key roles. In this case, autophagosome maturation could be coupled, as autophagosomes would be formed close to endosomes.
In summary, this study identifies a key role for inactive EGFR in autophagy initiation in cells with WT EGFR gene in both serum starved and EGFR TKIs treated cells. It provides new insights into the mechanistic regulation of cancer cell survival by EGFR expression in metabolically stressed tumors and suggests a basis for EGFR TKI resistance in the treatments of tumors without EGFR mutations.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, Plasmids, siRNA, Antibodies and Reagents
MDA-MB-231, HeLa, A431 and HEK293 cells were cultured in DMEM (Corning) with 10% FBS (Invitrogen). Transfection of plasmids and siRNA oligonucleotides was carried out using Lipofectamine 2000 and Lipofectamine RNAiMax (Invitrogen), respectively, following manufacturer’s instructions. For serum starvation, cells were washed with DMEM twice and cultured in DMEM without FBS for indicated time periods. Detailed description of reagents, DNA plasmids, antibody origins, siRNA sequences, and cell treatments is included in the Extended Experimental Procedures.
Immunofluorescence Microscopy
Immunostaining of tagged and endogenous proteins were performed using standard procedures. Cells on glass coverslips were washed, fixed, permeabilized, blocked and stained with indicated antibodies. Cells were mounted using Vectashield and visualized with a Nikon TE2000-U microscope. Co-localization quantification was performed using the Coloc 2 plugin for Fiji (ImageJ). See the Extended Experimental Procedures for details.
Co-immunoprecipitation and Western Blotting Assays
Immunoprecipitation of indicated proteins was performed using nondenaturing cell extracts. Immunocomplex and whole cell lysates were analyzed by SDS polyacrylamide gel electrophoresis (SDS PAGE), followed by immunoblotting with indicated antibodies. See the Extended Experimental Procedures for details.
Autophagy Assays
Autophagy was analyzed by (1) western blotting detection of LC3-II turnover in the presence and absence of autophagy inhibitor chloroquine, by (2) quantifying chloroquine-induced accumulation of endogenous LC3 puncta and by (3) quantifying of the number of EGFP-LC3 puncta in cells stably expressing low levels of EGFP-LC3 using fluorescence microscopy. See the Extended Experimental Procedures for details.
Biotinylation Assay
Cells were surface biotinylated and harvested for purification of biotinylated proteins using streptavidin affinity gel. Whole cell lysates and purified biotinylated proteins were analyzed by immunoblotting. See the Extended Experimental Procedures for details.
Supplementary Material
(A) Western blot analysis comparing the EGFR expression levels in 7 different cell lines. Among them, 4 cell lines (HeLa, SUM159, SUM1315, and Hs578T) show low levels of EGFR expression, while the other 3 cell lines (A431, MDA-MB-231, and MDA-MB-468) express high levels of EGFR.
(B) Knockdown of EGFR inhibits LC3-II turnover in all the cell lines tested. The data establish that EGFR has a role in autophagy initiation in various cell lines independent of the EGFR expression level. Cells were cultured in normal medium with serum. CQ, chloroquine (80 μM) treatment for 2 h.
(C) Western blot data showing the siRNA knockdown efficiency of PDGFRB, c-Met, and FGFR2 in MDA-MB-231 cells.
(D) LC3-II turnover specifically requires EGFR but not PDGFRB, c-Met, or FGFR2 in MDA-MB-231 cells. Cells were transfected with indicated siRNA and 72 h after siRNA transfection, cells cultured in normal medium were treated or not with 80 μM chloroquine (CQ) for 2 h, followed by whole cell lysate harvest for Western blot analysis of LC3 levels.
(E) Quantification of LC3-II levels in conditions shown in (D); mean + SD; n = 3.
(F) EGF stimulation inhibits LC3-II turnover in serum starved MDA-MB-231 cells. Cells were serum starved over night and then treated with or without 50 ng/ml EGF for 2 h. Chloroquine (80 μM) was added to the medium as indicated at the same time with EGF. After the 2 h of incubation, cells were harvested for Western blot analysis of LC3 levels.
(G) Evaluation of EGFR and AKT signaling in EGFR knockdown cells re-expressing C-terminally Flag-tagged EGFR-WT or EGFR-KD. MDA-MB-231 cells stably expressing siRNA resistant EGFR-WT or EGFR-KD was transfected with siRNA to knock down endogenous EGFR. Cells were then cultured in normal (N) or serum free (S) medium for 24 h, followed by EGF treatment as indicated. Cells were then harvested for Western blot analysis of p-EGFR, EGFR-Flag, p-AKT, and AKT levels.
(H) Serum starvation induces the formation of EGFP-LC3 puncta in MDA-MB-231 cells stably expressing EGFP-LC3. MDA-MB-231 cells were infected with lentivirus to induce stable expression of EGFP-LC3. A monoclonal cell line expressing low levels of EGFP-LC3 was selected for all the serum starvation induced EGFP-LC3 puncta formation experiments in this manuscript. Note: without starvation almost no EGFP-LC3 aggregates/puncta was observed in this cell line, indicating low expression level of EGFP-LC3. Bar: 10 μm.
(A) Knockdown of EGFR causes a loss of EGFR staining by the Clone LA22 EGFR antibody in MDA-MB-231 cells.
(B) EGFR-GFP colocalizes well with LAPTMB and partially with EEA1. MDA-MB-231 cells transfected with EGFR-GFP were starved and stained for EEA1 (top) or LAPTM4B (bottom).
(C and E) Serum starvation induces endosomal accumulation of EGFR and enhances the colocalization between EGFR and LAPTM4B in A431 (C) and HeLa (E) cells. Cells were starved (bottom) or not (top) and then fixed for co-staining of endogenous EGFR (green) and LAPTM4B (red).
(D and F) Quantification of the relative intensities of EGFR endosomal staining (left) and the colocalization between EGFR and LAPTM4B (right) in conditions shown in (C) and (E), respectively. For colocalization, the threshoulded Manders M1 and M2 coefficients were expressed as percentages to show the fraction of intensities in one channel above threshold that was colocalized with intensities in the other channel above threshold. In each condition, 100–200 cells were analyzed for quantification. Mean + SD; n = 3.
(G and I) Serum starvation induces endosomal accumulation of c-Met (G) and FGFR2 (I) in MDA-MB-231 cells. Cells were starved (bottom) or not (top) and then fixed for co-staining of endogenous c-Met or FGFR2 (green) with LAPTM4B (red).
(H and J) Quantification of the intensities of endosomal staining of c-Met or FGFR2 (left) and the colocalization of c-Met or FGFR2 with LAPTM4B (right) in conditions shown in (G) and (I), respectively. In each condition, 100–200 cells were analyzed for quantification. Mean + SD; n = 3.
DAPI was used to stain the nuclei. Bar: 10 μm.
(A) EGFR specifically coimmunoprecipitates (co-IP) with LAPTM4B but not LAPTM4A or LAPTM5 in HEK293 cells co-transfected with indicated proteins.
(B) LAPTM4B is co-IP’ed with not only EGFR but also PDGFRB, FGFR2, and c-Met. Myc-LAPTM4B was co-transfected with indicated receptor constructs into HEK293 cells. Each receptor expresses a C-terminal GFP tag and was IP’ed by anti-GFP.
(C) Serum starvation does not affect LAPTM4B co-IP with PDGFRB, FGFR2, or c-Met. HEK293 cells co-transfected with indicated constructs were serum starved (S) or not (N) over night before harvested for the co-IP assay.
(D) Cells with lower endogenous LAPTM4B levels (arrowhead) have less endosomal EGFR staining. Parental MDA-MB-231 were starved and fixed for immuno-staining of LAPTM4B (red) and EGFR (green).
(E and F) Knockdown of LAPTM4B results in loss of endosomal EGFR accumulation in A431 (E) and HeLa (F) cells. Control or LAPTM4B knockdown cells were starved and fixed, followed by immuno-staining of LAPTM4B (red) and EGFR (green).
(G and H) Knockdown of LAPTM4B results in loss of endosomal accumulation of c-Met (G) but not FGFR2 (H) in MDA-MB-231 cells. Control or LAPTM4B knockdown cells were starved and fixed, followed by immuno-staining of LAPTM4B (red) with c-Met or FGFR2 (green).
(I) Knockdown of Hrs or TSG101 does not affect EGFR accumulation at LAPTM4B positive endosomes.
DAPI was used to stain the nuclei; Boxes are selected regions for magnified view; Bar, 10 μm.
(A) LAPTM4B overexpression enhances LC3-II turnover in MDA-MB-231 cells. LAPTM4B overexpression was mediated by lentivirus infection. A pool of the infected cells was used for this experiment.
(B) Quantification of LC3-II levels in conditions shown in (A); mean + SD; n = 3.
(C) LAPTM4B overexpression enhances basal EGFP-LC3 puncta formation. EGFP-LC3 (green) cells were transfected with Flag-LAPTM4B and after 48 h cells were seeded on coverslips and cultured in normal medium with serum. Then cells were fixed for staining of Flag-LAPTM4B (red). Bar, 10 μm.
(D) Re-expression of siRNA-resistant LAPTM4B rescues starvation-induced EGFP-LC3 puncta formation in LAPTM4B-knockdown cells. EGFP-LC3 (green) cells were transfected with siRNA-resistant Flag-LAPTM4B DNA and after 48 h cells were seeded on coverslips and reverse transfected with LAPTM4B siRNA. And 48 h after siRNA transfection, cells were serum starved over night and then fixed for staining of Flag-LAPTM4B (red). Bar, 10 μm.
(E) Double knockdown of EGFR and LAPTM4B has similar effects to single knockdown of either EGFR or LAPTM4B on LC3-II turnover in MDA-MB-231 cells.
(F) Quantification of LC3-II levels in conditions shown in (E); mean + SD; n = 3.
(G) EGFR overexpression largely rescues LC3-II turnover in LAPTM4B knockdown MDA-MB-231 cells. EGFR overexpression was mediated by lentivirus infection, and endogenous LAPTM4B was knocked down by siRNA.
(H) Quantification of LC3-II levels in conditions shown in (G); mean + SD; n = 3.
(I) LAPTM4B overexpression could not rescues LC3-II turnover in EGFR knockdown MDA-MB-231 cells. LAPTM4B overexpression was mediated by lentivirus infection, and endogenous EGFR was knocked down by siRNA.
(J) Quantification of LC3-II levels in conditions shown in (I); mean + SD; n = 3. CQ, chloroquine (80 μM) treatment for 2 h.
(A–C) Co-immunoprecipitation of WT or KD EGFR with Sec8-Flag (A), Exo70-Flag (B) or Exo84-Flag (C) in HEK 293 cells co-transfected with indicated EGFR and exocyst subunit constructs.
(D) Representative Western blot images showing efficient knockdown of Sec6, Sec8, and Exo70 in MDA-MB-231 cells.
(E) Knockdown of Sec5, Sec6, Sec8 or Exo70 inhibits LC3-II turnover in MDA-MB-231 cells.
(F) Quantification of LC3-II levels in conditions shown in (E). Mean + SD; n = 3.
(A) Representative Western blot images showing efficient knockdown of Rubicon in MDA-MB-231 cells. Asterisk indicates a nonspecific band.
(B) Knockdown of Rubicon enhances EGFP-LC3 puncta formation. Control or Rubicon knockdown MDA-MB-231 cells stably expressing EGFP-LC3 as described in Figure S1H were serum starved or not and then fixed for microscopy.
(C) Quantification of the number of EGFP-LC3 puncta in (B); mean + SD, n = 3.
(A) Serum starvation (STV) and Rapamycin (Rapa) induce autophagic turnover in MDA-MB-231 cells. For Rapamycin treatment, cells were incubated with indicated concentrations of Rapamycin for 2 h and then treated with or without chloroquine (80 μM) for another 2 h without medium change, followed by cell harvest for Western blot analysis.
(B) Quantification of LC3-II levels in conditions shown in Figures 7A and S7A.
Mean + SD; n ≥ 4.
(C) Immunofluorescence images showing that erlotinib (2 μM) treatment for 24 h induces EGFR accumulation at endosomes distinct from LAPTM4B positive endosomes in MDA-MB-231 cells. After erlotinib treatment, cells were fixed and co-stained for endogenous LAPTM4B (red) and EGFR (green). DAPI was used to stain the nuclei. Bar, 10 μm.
(D) Co-immunoprecipitation images showing the effects of erlotinib and gefitinib treatment (2 μM) on the interaction between EGFR and Sec6 in HEK 293 cells co-transfected with EGFR and Sec6-Flag.
Acknowledgments
We thank MA White (UT Southwestern) for sharing reagents, SJ Parsons (University of Virginia) for the EGFR-K721A construct, R Zhou (Peking University) for the LAPTM4B construct, R Puertollano (National Institutes of Health) for the LAPTM4A construct, Paul Bertics (University of Wisconsin-Madison) for the EGFR-GFP construct, JE Ladbury (University of Texas MD Anderson Cancer Center) for FGFR2-GFP construct, H Band (University of Nebraska Medical Center) for PDGFRB-GFP construct, and D Wheeler (University of Wisconsin-Madison) for c-Met-GFP construct. We thank Drs. Randall Kimple, Alan Rapraeger, and Paul Lambert from University of Wisconsin-Madison and Drs. Andrew C Hedman, Weimin Li and Suyong Choi in the laboratory for extensive comments on the manuscript, research discussions and reagents. This work is supported by National Institute of Health grants CA104708 and GM057549 to R.A.A., Howard Hughes Medical Institute International Student Research Fellowship (59107631) to X.T., and American Heart Association (AHA) Scientist Development Grant 12SDG11950022 to Y.S.
Footnotes
AUTHOR CONTRIBUTIONS
XT and RAA conceived the study. XT, NT, YS and RAA designed experiments. XT, NT and YS performed experiments. XT and RAA wrote the manuscript.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baselga J, Rischin D, Ranson M, Calvert H, Raymond E, Kieback DG, Kaye SB, Gianni L, Harris A, Bjork T, et al. Phase I Safety, Pharmacokinetic, and Pharmacodynamic Trial of ZD1839, a Selective Oral Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor, in Patients With Five Selected Solid Tumor Types. Journal of Clinical Oncology. 2002;20:4292–4302. doi: 10.1200/JCO.2002.03.100. [DOI] [PubMed] [Google Scholar]
- Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, Maiti M, Hazelett CC, Wauson EC, Balakireva M, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011;144:253–267. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brech A, Ahlquist T, Lothe RA, Stenmark H. Autophagy in tumour suppression and promotion. Mol Oncol. 2009;3:366–375. doi: 10.1016/j.molonc.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Jeong EH, Lee TG, Kim SY, Kim HR, Kim CH. Autophagy Inhibition with Monensin Enhances Cell Cycle Arrest and Apoptosis Induced by mTOR or Epidermal Growth Factor Receptor Inhibitors in Lung Cancer Cells. Tuberc Respir Dis (Seoul) 2013;75:9–17. doi: 10.4046/trd.2013.75.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker KJ, Staros JV, Guyer CA. A kinase-negative epidermal growth factor receptor that retains the capacity to stimulate DNA synthesis. Proc Natl Acad Sci U S A. 1994;91:6967–6971. doi: 10.1073/pnas.91.15.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragowska WH, Weppler SA, Wang JC, Wong LY, Kapanen AI, Rawji JS, Warburton C, Qadir MA, Donohue E, Roberge M, et al. Induction of autophagy is an early response to gefitinib and a potential therapeutic target in breast cancer. PLoS One. 2013;8:e76503. doi: 10.1371/journal.pone.0076503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden ER, White IJ, Tsapara A, Futter CE. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol. 2010;12:267–272. doi: 10.1038/ncb2026. [DOI] [PubMed] [Google Scholar]
- Eimer S, Belaud-Rotureau MA, Airiau K, Jeanneteau M, Laharanne E, Véron N, Vital A, Loiseau H, Merlio JP, Belloc F. Autophagy inhibition cooperates with erlotinib to induce glioblastoma cell death. Cancer Biol Ther. 2011;11:1017–1027. doi: 10.4161/cbt.11.12.15693. [DOI] [PubMed] [Google Scholar]
- Ewald JA, Wilkinson JC, Guyer CA, Staros JV. Ligand- and kinase activity-independent cell survival mediated by the epidermal growth factor receptor expressed in 32D cells. Exp Cell Res. 2003;282:121–131. doi: 10.1016/s0014-4827(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 2009;16:70–78. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
- Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster S, Kacprowski T, Dhople VM, Hammer E, Herzog S, Saafan H, Bien-Möller S, Albrecht M, Völker U, Ritter CA. Characterization of the EGFR interactome reveals associated protein complex networks and intracellular receptor dynamics. PROTEOMICS. 2013;13:3131–3144. doi: 10.1002/pmic.201300154. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Dibenedetto JR, West M, Rowland AA, Voeltz GK. Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol Biol Cell. 2013;24:1030–1040. doi: 10.1091/mbc.E12-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung C, Chen X, Grandis JR, Duvvuri U. EGFR tyrosine kinase inhibition induces autophagy in cancer cells. Cancer Biol Ther. 2012;13:1417–1424. doi: 10.4161/cbt.22002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg SB, Supko JG, Neal JW, Muzikansky A, Digumarthy S, Fidias P, Temel JS, Heist RS, Shaw AT, McCarthy PO, et al. A Phase I Study of Erlotinib and Hydroxychloroquine in Advanced Non AiSmall-Cell Lung Cancer. Journal of Thoracic Oncology. 2012;7:1602–1608. doi: 10.1097/JTO.0b013e318262de4a. doi:1610.1097/JTO.1600b1013e318262de318264a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzalczany Y, Gilad Y, Amihai D, Hammel I, Sagi-Eisenberg R, Merimsky O. Combining an EGFR directed tyrosine kinase inhibitor with autophagy-inducing drugs: a beneficial strategy to combat non-small cell lung cancer. Cancer Lett. 2011;310:207–215. doi: 10.1016/j.canlet.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Han W, Pan H, Chen Y, Sun J, Wang Y, Li J, Ge W, Feng L, Lin X, Wang X, et al. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS One. 2011;6:e18691. doi: 10.1371/journal.pone.0018691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle SC, Kanfer G, Kolar K, Lang A, Michel AH, Kornmann B. Organization and function of membrane contact sites. Biochim Biophys Acta. 2013;1833:2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- Jutten B, Keulers TG, Schaaf MB, Savelkouls K, Theys J, Span PN, Vooijs MA, Bussink J, Rouschop KM, Rouschop KM. EGFR overexpressing cells and tumors are dependent on autophagy for growth and survival. Radiother Oncol. 2013;108:479–483. doi: 10.1016/j.radonc.2013.06.033. [DOI] [PubMed] [Google Scholar]
- Jutten B, Rouschop KM. EGFR signaling and autophagy dependence for growth, survival, and therapy resistance. Cell Cycle. 2014;13:42–51. doi: 10.4161/cc.27518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper G, Vogel A, Klaman I, Gröne J, Petersen I, Weber B, Castaños-Vélez E, Staub E, Mennerich D. The human LAPTM4b transcript is upregulated in various types of solid tumours and seems to play a dual functional role during tumour progression. Cancer Letters. 2005;224:93–103. doi: 10.1016/j.canlet.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nature reviews Molecular cell biology. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- Li L, Shan Y, Yang H, Zhang S, Lin M, Zhu P, Chen XY, Yi J, McNutt MA, Shao GZ, et al. Upregulation of LAPTM4B-35 promotes malignant transformation and tumorigenesis in L02 human liver cell line. Anat Rec (Hoboken) 2011a;294:1135–1142. doi: 10.1002/ar.21421. [DOI] [PubMed] [Google Scholar]
- Li L, Wei XH, Pan YP, Li HC, Yang H, He QH, Pang Y, Shan Y, Xiong FX, Shao GZ, et al. LAPTM4B: a novel cancer-associated gene motivates multidrug resistance through efflux and activating PI3K/AKT signaling. Oncogene. 2010a;29:5785–5795. doi: 10.1038/onc.2010.303. [DOI] [PubMed] [Google Scholar]
- Li X, Fan Z. The epidermal growth factor receptor antibody cetuximab induces autophagy in cancer cells by downregulating HIF-1alpha and Bcl-2 and activating the beclin 1/hVps34 complex. Cancer Res. 2010;70:5942–5952. doi: 10.1158/0008-5472.CAN-10-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lu Y, Pan T, Fan Z. Roles of autophagy in cetuximab-mediated cancer therapy against EGFR. Autophagy. 2010b;6:1066–1077. doi: 10.4161/auto.6.8.13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Tian R, Wang Q, Zhao JJ, Iglehart JD, Wang ZC, Richardson AL. Lysosomal transmembrane protein LAPTM4B promotes autophagy and tolerance to metabolic stress in cancer cells. Cancer Res. 2011b;71:7481–7489. doi: 10.1158/0008-5472.CAN-11-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zou L, Li Q, Haibe-Kains B, Tian R, Li Y, Desmedt C, Sotiriou C, Szallasi Z, Iglehart JD, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med. 2010c;16:214–218. doi: 10.1038/nm.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Lam SK, Mak JC, Zheng CY, Ho JC. Erlotinib-induced autophagy in epidermal growth factor receptor mutated non-small cell lung cancer. Lung Cancer. 2013;81:354–361. doi: 10.1016/j.lungcan.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhou R, Zhang Q, Zhang Y, Jin Y, Lin M, Rui J, Ye D. Structure analysis and expressions of a novel tetratransmembrane protein, lysosoma-associated protein transmembrane 4 beta associated with hepatocellular carcinoma. World J Gastroenterol. 2004;10:1555–1559. doi: 10.3748/wjg.v10.i11.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Mendelsohn J, Baselga J. Epidermal growth factor receptor targeting in cancer. Semin Oncol. 2006;33:369–385. doi: 10.1053/j.seminoncol.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Matsukuma S, Nakamura Y, Yoshihara M, Koizume S, Sekiguchi H, Saito H, Nakayama H, Kameda Y, Yokose T, et al. Enhanced autophagy is required for survival in EGFR-independent EGFR-mutant lung adenocarcinoma cells. Lab Invest. 2013;93:1137–1146. doi: 10.1038/labinvest.2013.102. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Shao G, Zhou R, Zhang Q, Zhang Y, Liu J, Rui J, Wei X, Ye D. Molecular cloning and characterization of LAPTM4B, a novel gene upregulated in hepatocellular carcinoma. Oncogene. 2003;22:5060–5069. doi: 10.1038/sj.onc.1206832. [DOI] [PubMed] [Google Scholar]
- Sobhakumari A, Schickling BM, Love-Homan L, Raeburn A, Fletcher EV, Case AJ, Domann FE, Miller FJJ, Simons AL. NOX4 mediates cytoprotective autophagy induced by the EGFR inhibitor erlotinib in head and neck cancer cells. Toxicol Appl Pharmacol. 2013;272:736–745. doi: 10.1016/j.taap.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2008;314:3093–3106. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulieres D, Senzer NN, Vokes EE, Hidalgo M, Agarwala SS, Siu LL. Multicenter Phase II Study of Erlotinib, an Oral Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor, in Patients With Recurrent or Metastatic Squamous Cell Cancer of the Head and Neck. Journal of Clinical Oncology. 2004;22:77–85. doi: 10.1200/JCO.2004.06.075. [DOI] [PubMed] [Google Scholar]
- Sun Q, Fan W, Zhong Q. Regulation of Beclin 1 in autophagy. Autophagy. 2009;5:713–716. doi: 10.4161/auto.5.5.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergarajauregui S, Martina JA, Puertollano R. LAPTMs regulate lysosomal function and interact with mucolipin 1: new clues for understanding mucolipidosis type IV. J Cell Sci. 2011;124:459–468. doi: 10.1242/jcs.076240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, Kinch L, Koduru P, Christudass CS, Veltri RW, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ, Hung MC. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–393. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley HS. Trafficking of the ErbB receptors and its influence on signaling. Exp Cell Res. 2003;284:78–88. doi: 10.1016/s0014-4827(03)00002-8. [DOI] [PubMed] [Google Scholar]
- Yang H, Xiong F, Wei X, Yang Y, McNutt M, Zhou R. Overexpression of LAPTM4B-35 promotes growth and metastasis of hepatocellular carcinoma in vitro and in vivo. Cancer Letters. 2010;294:236–244. doi: 10.1016/j.canlet.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Wang Q, Li X, Yan Y, Backer J, Chait B, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Ling YH, Sironi J, Schwartz EL, Perez-Soler R, Piperdi B. The autophagy inhibitor chloroquine overcomes the innate resistance of wild-type EGFR non-small-cell lung cancer cells to erlotinib. J Thorac Oncol. 2013;8:693–702. doi: 10.1097/JTO.0b013e31828c7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Western blot analysis comparing the EGFR expression levels in 7 different cell lines. Among them, 4 cell lines (HeLa, SUM159, SUM1315, and Hs578T) show low levels of EGFR expression, while the other 3 cell lines (A431, MDA-MB-231, and MDA-MB-468) express high levels of EGFR.
(B) Knockdown of EGFR inhibits LC3-II turnover in all the cell lines tested. The data establish that EGFR has a role in autophagy initiation in various cell lines independent of the EGFR expression level. Cells were cultured in normal medium with serum. CQ, chloroquine (80 μM) treatment for 2 h.
(C) Western blot data showing the siRNA knockdown efficiency of PDGFRB, c-Met, and FGFR2 in MDA-MB-231 cells.
(D) LC3-II turnover specifically requires EGFR but not PDGFRB, c-Met, or FGFR2 in MDA-MB-231 cells. Cells were transfected with indicated siRNA and 72 h after siRNA transfection, cells cultured in normal medium were treated or not with 80 μM chloroquine (CQ) for 2 h, followed by whole cell lysate harvest for Western blot analysis of LC3 levels.
(E) Quantification of LC3-II levels in conditions shown in (D); mean + SD; n = 3.
(F) EGF stimulation inhibits LC3-II turnover in serum starved MDA-MB-231 cells. Cells were serum starved over night and then treated with or without 50 ng/ml EGF for 2 h. Chloroquine (80 μM) was added to the medium as indicated at the same time with EGF. After the 2 h of incubation, cells were harvested for Western blot analysis of LC3 levels.
(G) Evaluation of EGFR and AKT signaling in EGFR knockdown cells re-expressing C-terminally Flag-tagged EGFR-WT or EGFR-KD. MDA-MB-231 cells stably expressing siRNA resistant EGFR-WT or EGFR-KD was transfected with siRNA to knock down endogenous EGFR. Cells were then cultured in normal (N) or serum free (S) medium for 24 h, followed by EGF treatment as indicated. Cells were then harvested for Western blot analysis of p-EGFR, EGFR-Flag, p-AKT, and AKT levels.
(H) Serum starvation induces the formation of EGFP-LC3 puncta in MDA-MB-231 cells stably expressing EGFP-LC3. MDA-MB-231 cells were infected with lentivirus to induce stable expression of EGFP-LC3. A monoclonal cell line expressing low levels of EGFP-LC3 was selected for all the serum starvation induced EGFP-LC3 puncta formation experiments in this manuscript. Note: without starvation almost no EGFP-LC3 aggregates/puncta was observed in this cell line, indicating low expression level of EGFP-LC3. Bar: 10 μm.
(A) Knockdown of EGFR causes a loss of EGFR staining by the Clone LA22 EGFR antibody in MDA-MB-231 cells.
(B) EGFR-GFP colocalizes well with LAPTMB and partially with EEA1. MDA-MB-231 cells transfected with EGFR-GFP were starved and stained for EEA1 (top) or LAPTM4B (bottom).
(C and E) Serum starvation induces endosomal accumulation of EGFR and enhances the colocalization between EGFR and LAPTM4B in A431 (C) and HeLa (E) cells. Cells were starved (bottom) or not (top) and then fixed for co-staining of endogenous EGFR (green) and LAPTM4B (red).
(D and F) Quantification of the relative intensities of EGFR endosomal staining (left) and the colocalization between EGFR and LAPTM4B (right) in conditions shown in (C) and (E), respectively. For colocalization, the threshoulded Manders M1 and M2 coefficients were expressed as percentages to show the fraction of intensities in one channel above threshold that was colocalized with intensities in the other channel above threshold. In each condition, 100–200 cells were analyzed for quantification. Mean + SD; n = 3.
(G and I) Serum starvation induces endosomal accumulation of c-Met (G) and FGFR2 (I) in MDA-MB-231 cells. Cells were starved (bottom) or not (top) and then fixed for co-staining of endogenous c-Met or FGFR2 (green) with LAPTM4B (red).
(H and J) Quantification of the intensities of endosomal staining of c-Met or FGFR2 (left) and the colocalization of c-Met or FGFR2 with LAPTM4B (right) in conditions shown in (G) and (I), respectively. In each condition, 100–200 cells were analyzed for quantification. Mean + SD; n = 3.
DAPI was used to stain the nuclei. Bar: 10 μm.
(A) EGFR specifically coimmunoprecipitates (co-IP) with LAPTM4B but not LAPTM4A or LAPTM5 in HEK293 cells co-transfected with indicated proteins.
(B) LAPTM4B is co-IP’ed with not only EGFR but also PDGFRB, FGFR2, and c-Met. Myc-LAPTM4B was co-transfected with indicated receptor constructs into HEK293 cells. Each receptor expresses a C-terminal GFP tag and was IP’ed by anti-GFP.
(C) Serum starvation does not affect LAPTM4B co-IP with PDGFRB, FGFR2, or c-Met. HEK293 cells co-transfected with indicated constructs were serum starved (S) or not (N) over night before harvested for the co-IP assay.
(D) Cells with lower endogenous LAPTM4B levels (arrowhead) have less endosomal EGFR staining. Parental MDA-MB-231 were starved and fixed for immuno-staining of LAPTM4B (red) and EGFR (green).
(E and F) Knockdown of LAPTM4B results in loss of endosomal EGFR accumulation in A431 (E) and HeLa (F) cells. Control or LAPTM4B knockdown cells were starved and fixed, followed by immuno-staining of LAPTM4B (red) and EGFR (green).
(G and H) Knockdown of LAPTM4B results in loss of endosomal accumulation of c-Met (G) but not FGFR2 (H) in MDA-MB-231 cells. Control or LAPTM4B knockdown cells were starved and fixed, followed by immuno-staining of LAPTM4B (red) with c-Met or FGFR2 (green).
(I) Knockdown of Hrs or TSG101 does not affect EGFR accumulation at LAPTM4B positive endosomes.
DAPI was used to stain the nuclei; Boxes are selected regions for magnified view; Bar, 10 μm.
(A) LAPTM4B overexpression enhances LC3-II turnover in MDA-MB-231 cells. LAPTM4B overexpression was mediated by lentivirus infection. A pool of the infected cells was used for this experiment.
(B) Quantification of LC3-II levels in conditions shown in (A); mean + SD; n = 3.
(C) LAPTM4B overexpression enhances basal EGFP-LC3 puncta formation. EGFP-LC3 (green) cells were transfected with Flag-LAPTM4B and after 48 h cells were seeded on coverslips and cultured in normal medium with serum. Then cells were fixed for staining of Flag-LAPTM4B (red). Bar, 10 μm.
(D) Re-expression of siRNA-resistant LAPTM4B rescues starvation-induced EGFP-LC3 puncta formation in LAPTM4B-knockdown cells. EGFP-LC3 (green) cells were transfected with siRNA-resistant Flag-LAPTM4B DNA and after 48 h cells were seeded on coverslips and reverse transfected with LAPTM4B siRNA. And 48 h after siRNA transfection, cells were serum starved over night and then fixed for staining of Flag-LAPTM4B (red). Bar, 10 μm.
(E) Double knockdown of EGFR and LAPTM4B has similar effects to single knockdown of either EGFR or LAPTM4B on LC3-II turnover in MDA-MB-231 cells.
(F) Quantification of LC3-II levels in conditions shown in (E); mean + SD; n = 3.
(G) EGFR overexpression largely rescues LC3-II turnover in LAPTM4B knockdown MDA-MB-231 cells. EGFR overexpression was mediated by lentivirus infection, and endogenous LAPTM4B was knocked down by siRNA.
(H) Quantification of LC3-II levels in conditions shown in (G); mean + SD; n = 3.
(I) LAPTM4B overexpression could not rescues LC3-II turnover in EGFR knockdown MDA-MB-231 cells. LAPTM4B overexpression was mediated by lentivirus infection, and endogenous EGFR was knocked down by siRNA.
(J) Quantification of LC3-II levels in conditions shown in (I); mean + SD; n = 3. CQ, chloroquine (80 μM) treatment for 2 h.
(A–C) Co-immunoprecipitation of WT or KD EGFR with Sec8-Flag (A), Exo70-Flag (B) or Exo84-Flag (C) in HEK 293 cells co-transfected with indicated EGFR and exocyst subunit constructs.
(D) Representative Western blot images showing efficient knockdown of Sec6, Sec8, and Exo70 in MDA-MB-231 cells.
(E) Knockdown of Sec5, Sec6, Sec8 or Exo70 inhibits LC3-II turnover in MDA-MB-231 cells.
(F) Quantification of LC3-II levels in conditions shown in (E). Mean + SD; n = 3.
(A) Representative Western blot images showing efficient knockdown of Rubicon in MDA-MB-231 cells. Asterisk indicates a nonspecific band.
(B) Knockdown of Rubicon enhances EGFP-LC3 puncta formation. Control or Rubicon knockdown MDA-MB-231 cells stably expressing EGFP-LC3 as described in Figure S1H were serum starved or not and then fixed for microscopy.
(C) Quantification of the number of EGFP-LC3 puncta in (B); mean + SD, n = 3.
(A) Serum starvation (STV) and Rapamycin (Rapa) induce autophagic turnover in MDA-MB-231 cells. For Rapamycin treatment, cells were incubated with indicated concentrations of Rapamycin for 2 h and then treated with or without chloroquine (80 μM) for another 2 h without medium change, followed by cell harvest for Western blot analysis.
(B) Quantification of LC3-II levels in conditions shown in Figures 7A and S7A.
Mean + SD; n ≥ 4.
(C) Immunofluorescence images showing that erlotinib (2 μM) treatment for 24 h induces EGFR accumulation at endosomes distinct from LAPTM4B positive endosomes in MDA-MB-231 cells. After erlotinib treatment, cells were fixed and co-stained for endogenous LAPTM4B (red) and EGFR (green). DAPI was used to stain the nuclei. Bar, 10 μm.
(D) Co-immunoprecipitation images showing the effects of erlotinib and gefitinib treatment (2 μM) on the interaction between EGFR and Sec6 in HEK 293 cells co-transfected with EGFR and Sec6-Flag.