Summary
The universally conserved GTPase elongation factor G (EF-G) catalyzes the translocation of transfer RNA (tRNA) and messenger RNA (mRNA) on the ribosome after peptide bond formation. Despite numerous studies suggesting that EF-G undergoes extensive conformational rearrangements during translocation, high resolution structures exist for essentially only one conformation of EF-G in complex with the ribosome. Here, we report four atomic resolution crystal structures of EF-G bound to the ribosome programmed in the pre- and post-translocational states and to the ribosome trapped by the antibiotic dityromycin. We observe a previously unseen conformation of EF-G in the pretranslocation complex, which is independently captured by dityromycin on the ribosome. Our structures provide insights into the conformational space that EF-G samples on the ribosome and reveal that tRNA translocation on the ribosome is facilitated by a structural transition of EF-G from a compact to an elongated conformation, which can be prevented by the antibiotic dityromycin.
Introduction
Translation of the genetic code requires a codon-by-codon movement of mRNA and its associated tRNAs through the ribosome, a process which is catalyzed by the guanosine triphosphatase (GTPase) elongation factor G (EF-G) (Voorhees and Ramakrishnan, 2013). After each codon is decoded in the ribosome and peptide bond formation has occurred, the ribosome in the pre-translocational (PRE) state fluctuates between two conformations (Blanchard et al., 2004; Cornish et al., 2008; Fei et al., 2008; Munro et al., 2007) through a ratchet-like movement of the 30S ribosomal subunit with respect to the 50S subunit (Frank and Agrawal, 2000): a non-rotated form with tRNAs in the classical A/A and P/P states and a rotated form in which the CCA acceptor ends of tRNAs have moved from the A and P sites to the P and E sites on the 50S subunit, taking the hybrid A/P and P/E positions, respectively. EF-G in complex with GTP engages the PRE ribosome in both states, but binding to the non-rotated PRE ribosome is immediately followed by the ratchet-like movement of the ribosome into the rotated state (Chen et al., 2011; Chen et al., 2013a; Ermolenko and Noller, 2011; Holtkamp et al., 2014; Spiegel et al., 2007). In the next step, which is facilitated by the conformational changes of EF-G and concomitant GTP hydrolysis, the anticodon ends of tRNAs are translocated inside the ribosome via an intermediate step that involves swiveling of the 30S subunit head domain (Guo and Noller, 2012; Zhou et al., 2014), resulting in a post-translocational (POST) state of the ribosome in which the mRNA has been moved by one codon.
The structures of ribosome complexes with EF-G determined by cryo-electron microscopy (cryo-EM) (Agrawal et al., 1998; Connell et al., 2007; Frank and Agrawal, 2000; Ramrath et al., 2013; Ratje et al., 2010; Valle et al., 2003) and X-ray crystallography (Chen et al., 2013b; Gao et al., 2009; Pulk and Cate, 2013; Tourigny et al., 2013; Zhou et al., 2013, 2014) have shown that EF-G binds to the ribosome mainly through interactions between its G-domain (domain I) and the 50S subunit. These structures, however, represent either a POST state of the ribosome (Gao et al., 2009), or a state in transit during tRNA translocation (Chen et al., 2013b; Ramrath et al., 2013; Tourigny et al., 2013; Zhou et al., 2013, 2014), and exhibit an elongated form of EF-G with its domain IV projecting into the decoding center of the ribosome where the anticodon end of the A-site tRNA would be bound.
How EF-G binds a PRE ribosome and what position domain IV of EF-G takes to avoid collision with the A-site tRNA before translocation have remained an enigma in the field. Recently, a cryo-EM reconstruction of EF-G on the rotated PRE ribosome exhibited small conformational changes of EF-G compared to the one in the POST complex (Brilot et al., 2013), but the whole EF-G moves as a result of the G-domain rotating around the SRL such that domain IV is positioned next to the A-site tRNA. While the structure provides a snapshot of EF-G bound to the rotated PRE ribosome, the earlier event of EF-G sampling and binding to the non-rotated PRE ribosome remains to be determined. Meanwhile, several lines of evidence suggest that a large scale conformational change of EF-G occurs during translocation (Agrawal et al., 1999; Bulkley et al., 2014; Munro et al., 2010; Salsi et al., 2014; Stark et al., 2000; Wang et al., 2007), including a recent study of the antibiotic dityromycin (Bulkley et al., 2014). Dityromycin was shown to block EF-G-mediated tRNA translocation without affecting the binding of EF-G to the ribosome (Brandi et al., 2006; Brandi et al., 2012). The crystal structure of dityromycin in complex with the ribosome shows that dityromycin binds to ribosomal protein S12 (Bulkley et al., 2014), a position that would severely overlap with domain III of EF-G in the elongated form, thereby indicating the necessity for substantial domain rearrangements in EF-G on the PRE ribosome.
We have now determined the atomic resolution structures of EF-G bound to the ribosome in both the PRE and the POST states, as well as of an EF-G-ribosome complex trapped by the antibiotic dityromycin. We captured a new compact conformation of EF-G in the PRE complex which is also trapped by dityromycin. Together with the elongated form of EF-G in the POST complex, our structures reveal a conformational space that EF-G samples on the ribosome and suggest that tRNA translocation is accompanied by a structural transition of EF-G from a compact to an elongated conformation which can be blocked by the antibiotic dityromycin.
Results
Crystallization of the L9-GTPase fusion protein with the ribosome
To crystalize EF-G with the ribosome in different stages of translocation, we used a newly developed strategy that entails a fusion of the N-terminal domain (NTD) of ribosomal protein L9 to the N-terminus of EF-G (Figure S1). By varying the length of the linker between the L9-NTD and EF-G, one construct of L9-EF-G crystallized with the ribosome lacking the endogenous L9 under the same condition and in the same space group as the wild-type ribosome (Blaha et al., 2009). Using this strategy, we first determined the structure of EF-G bound to the ribosome in the POST state. The structure of the POST complex shows that EF-G binds to the ribosome in the same manner as seen in the previously determined structure (Figure 1B and 1D, Figure 2A) (Gao et al., 2009), while the NTD of L9, that is fused to the N-terminus of EF-G, binds to its canonical site on the neighboring ribosome (Figure S1). This indicates that the chimeric fusion does not interfere with the conformation of EF-G. Importantly, this method allows us to crystalize EF-G with the ribosome in the PRE state revealing drastic conformational changes. We have applied this strategy successfully in a recent study of elongation factor 4 bound to a clockwise ratcheted ribosome (Gagnon et al., 2014), showing this crystallization strategy to be an excellent tool to study the structures of GTPases on the ribosome in different functional states, especially when only weak or transient interactions are involved.
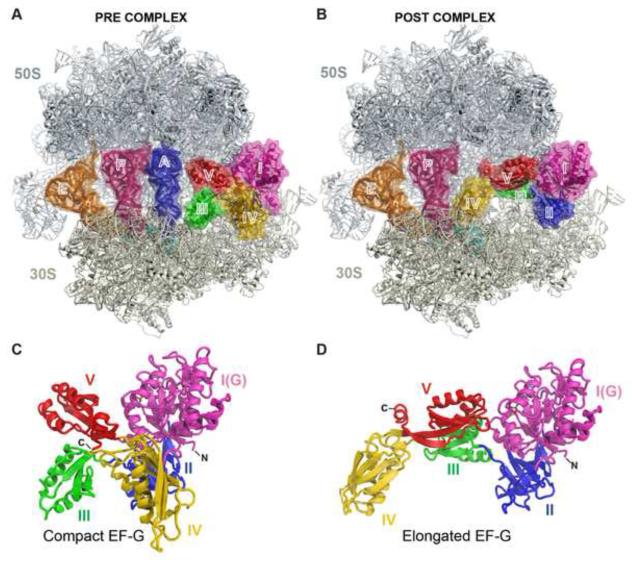
Figure 1. The structures of EF-G bound to the pre- and post-translocation ribosome.
(A and B) Overview of EF-G bound to the PRE (A) and the POST (B) ribosome. Shown are the 50S (gray) and the 30S (ivory) subunits, the A-site (blue), P-site (pink) and E-site (orange) tRNAs, mRNA (cyan) and EF-G with its five domains colored differently.
(C and D) Cartoon representations of EF-G shown in the compact conformation (C) from the PRE complex and the elongated conformation (D) from the POST complex. Domains of EF-G are colored and labeled as in panels A and B.
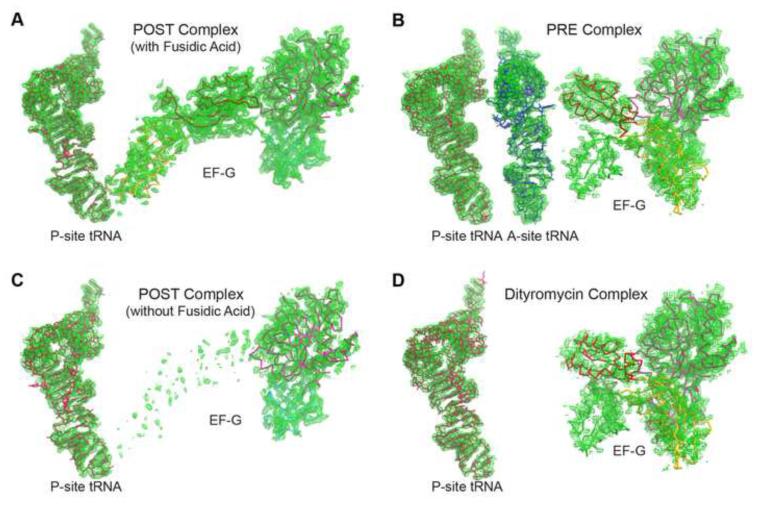
Figure 2. Partial electron density for the POST and PRE complexes.
(A-D) Unbiased Fobs – Fcalc difference Fourier map of EF-G and the P-site tRNA in the POST complex in the presence of fusidic acid (A), EF-G and the A- and P-site tRNAs in the PRE complex (B), EF-G and the P-site tRNA in the POST complex in the absence of fusidic acid (C), and EF-G and the P-site tRNA in the dityromycin complex (D). All maps are contoured at 2.5σ obtained after initial refinement with an empty ribosome as a starting model.
See also Table S1.
Structure of a PRE ribosome in complex with EF-G
When regular aminoacyl-tRNAs were placed in the ribosomal P and A sites to prepare a PRE ribosome, the resultant structure of the complex always displayed a post-translocated ribosome, suggesting that the system is capable of active translocation. In order to lock the ribosome in the PRE state, we have taken advantage of non-hydrolysable aminoacyl-tRNA analogs (Voorhees et al., 2009) to prevent de-aminoacylation of the P-site-bound tRNA and its subsequent movement into the E site. Accordingly, we prepared a PRE ribosome with non-hydrolysable fMet-NH-tRNAfMet in the P site and Phe-NH-tRNAPhe in the A site. This complex was then co-crystallized with EF-G.
Crystals obtained using this approach diffracted to 2.8 Å resolution, and after molecular replacement using an empty ribosome as the search model, we observed well-resolved electron density for mRNA, tRNAs in the A, P and E sites, and domains I, II and V of EF-G. Unexpectedly, the orientation for domain V of EF-G had to be inverted to fit into the electron density compared with the POST complex (Figure 2B). Additional density was confined to regions near domains I and II of EF-G, which was unambiguously assigned to domains III and IV after refinement. As expected for a ribosome with an aminoacyl-tRNA in the P site (Valle et al., 2003), the ribosome is in the non-rotated state with tRNAs in the classical P/P and A/A positions. While domains I and II of EF-G bind to the ribosome in the same manner as they do in previously determined complexes (Gao et al., 2009; Tourigny et al., 2013), domains III-V undergo dramatic conformational changes, resulting in a structure of EF-G bound to the ribosome that has not been previously observed (Figure 1A and 1C and movie S1).
EF-G in a compact conformation
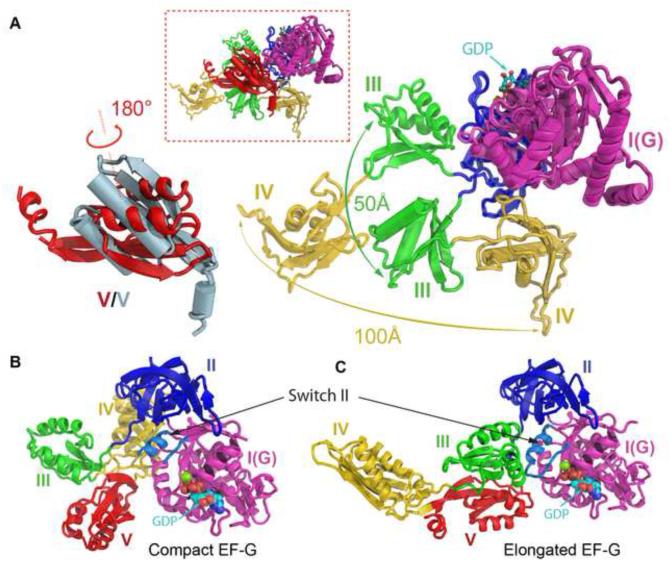
Instead of exhibiting the usual elongated shape seen in previous structures, EF-G in the PRE complex adopts a compact conformation in which domain IV is in close proximity to domains I and II. A superimposition of domain I from the PRE and the POST complexes reveals that domains III-V move as a relatively rigid entity, swiveling around the center of domain V, such that domain V remains in the same position but flips by ~180° with a simultaneous ~90° self-rotation (Figure 3A and movies S1 and S2). As a result, the tip of domain IV swivels by ~100 Å between the two conformations with a 90° self-rotation, a possibility that was not previously anticipated for EF-G. These movements are concomitant with a swing of domain III which disengages from its interactions with domain I near the catalytic site and moves outwards by ~50 Å (Figure 3A). It appears that the rearrangement of domains III-V relies on the loop connecting domains II and III, which is able to turn 90° between the two conformations of EF-G (Figure 3A). Overall, the two relatively rigid entities in the EF-G structure, domains I-II and domains IIIV, are loosely connected through a flexible loop without apparent interaction in the compact conformation, and as a result, the catalytic site is fully exposed where switches I and II are both disordered (Figure 3B and 3C).
Figure 3. Comparison of the EF-G structures in the elongated and the compact conformations.
(A) Inset is a superimposition of the structures of the compact and the elongated EF-G through domain I. Helices in the compact EF-G are displayed as cylinders. Lower right is a close-up view of the movements of domains III and IV. For clarity, conformational change of domain V is displayed separately in the lower left where the one from the compact EF-G is colored in lightblue. The GDP nucleotide is shown as spheres.
(B and C) Structures of the compact (B) and the elongated (C) EF-G viewed from the catalytic site. The switch II is colored in lightblue as indicated. The switch I loop is disordered and not shown in both complexes.
Interactions between the compact EF-G and the ribosome
While domains I and II of EF-G interact with the ribosome in essentially the same manner in both the PRE and the POST complexes, the interfaces between domains III-V and the ribosome are significantly different. Instead of occupying its usual binding position in the inter-subunit cleft of the ribosome between proteins S12 and L14, domain III moves to the opposite side of ribosomal protein S12 in the PRE complex, being positioned close to the A-site tRNA and h34 in the 30S head (Figure 4B). Domain IV hangs over the 30S shoulder with its tip pointing toward protein S4 in the PRE complex, in contrast with the POST complex where domain IV reaches into the A site (Figure 4C). Strikingly, the position of domain IV is similar to the interpretation made of a previous low-resolution cryo-EM reconstruction (Stark et al., 2000), in which the antibiotic thiostrepton was used to trap EF-G on the PRE ribosome. Domain V is positioned in the vicinity of the same elements of the ribosome in both the PRE and the POST complexes (Figure 4D), but the backward folding of EF-G in the PRE complex makes domain V more distant from these elements, such that helices H43/44 in the stalk base become mobile and the nucleotides at the tips of H43/44 and the sarcin-ricin loop (SRL) are more exposed in the PRE complex (Figure 5). Interestingly, the new interface between domain V and the stalk base in the PRE complex may allow the binding of the antibiotic thiostrepton to the stalk base (Figure S2), suggesting the prior cryo-EM structure (Stark et al., 2000) may have captured a similar compact conformation of EF-G.
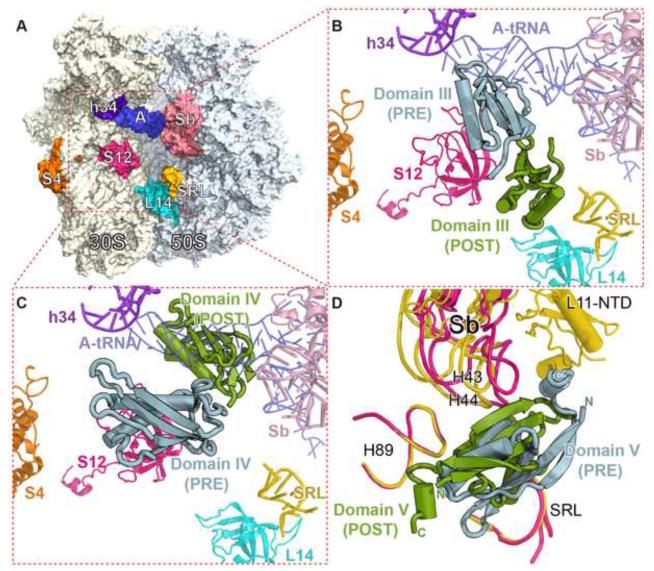
Figure 4. Interfaces between domains III, IV and V of EF-G and the ribosome in the PRE and the POST complexes.
(A) Overview of the ribosome showing the orientation of the insets B and C. Elements of the ribosome are indicated and colored differently.
(B and C) Positions of domains III (B) and IV (C) on the ribosome in the two complex structures.
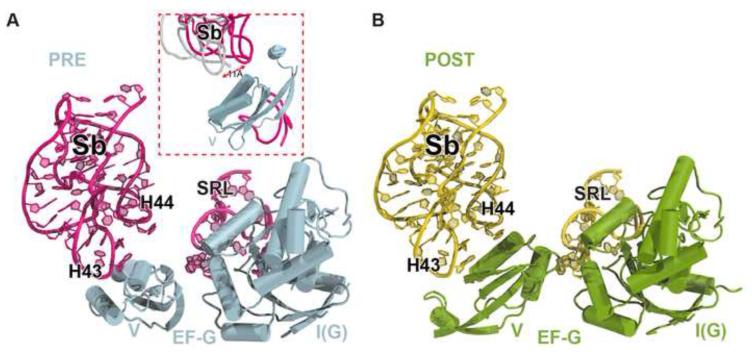
(D) Surroundings of domain V in the two complex structures. Helix 43/44, L11-NTD in the stalk base (Sb), Helix 89 and the sarcin-ricin loop (SRL) are shown in yellow in the POST complex and pink in the PRE complex. The L11-NTD in the PRE complex is not visible.
Figure 5. Contacts between domain V of EF-G, the stalk base (Sb) and the sarcin-ricin loop (SRL).
(A and B) Nucleotides at the tips of H43, H44 and the SRL are exposed in the PRE complex and are in close contacts with domain V in the POST complex. Positions of Sb from the two copies in the crystal of the PRE complex shown in the inset of panel A demonstrate that Sb becomes flexible in the PRE state.
See also Figure S2.
EF-G has two super-domains that are loosely connected
The compact EF-G reveals a hinge joint that divides EF-G into two super-domains: domains I-II and domains III-V. To explore the inter-domain flexibility of EF-G, we re-determined the structure of the POST complex in the absence of fusidic acid. We observed well resolved density for domains I, II and GDP, which bind tightly to the ribosome. However, domains III-V become flexible as shown by their residual density, which is only strong enough to indicate that EF-G is in the elongated conformation (Figure 2C). Soaking fusidic acid into the crystal restores density for all domains of EF-G (Figure 2A). These observations confirm that EF-G favors the elongated conformation (Czworkowski and Moore, 1997) while harboring an intrinsic flexibility between the two relatively independent super-domains that are connected through a hinge. In the compact EF-G conformation, the absence of interaction between the super-domains (Figure 3C) strongly suggests that its binding partner, the PRE ribosome, stabilizes EF-G in a compact form.
Dityromycin traps a compact form of EF-G on the ribosome
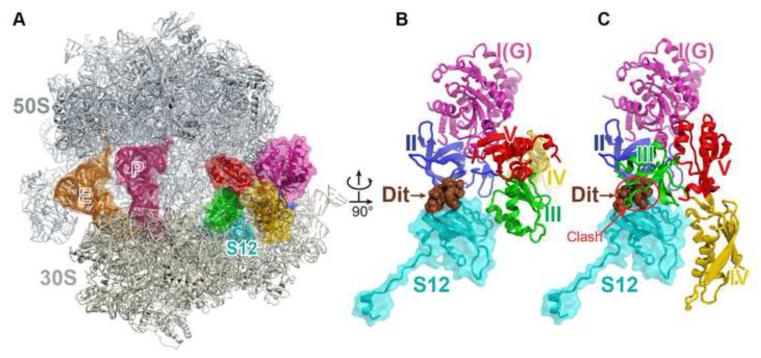
The newly characterized antibiotic dityromycin/GE82832 targets ribosomal protein S12 and traps EF-G in a pre-translocational state (Bulkley et al., 2014). To investigate the conformation of EF-G on the ribosome trapped by dityromycin, we determined a structure of EF-G bound to the ribosome in the presence of dityromycin (Figure 6A). The ribosome was programmed in the POST state to remove the structural constraints imposed by the A-site tRNA on the conformation of EF-G. Remarkably, EF-G adopts the same compact form on the ribosome in response to the binding of dityromycin instead of the one observed in the POST complex (Figure 6B and 6C and movie S2). Our structure shows that the binding position of dityromycin on protein S12 does not interfere with domain III of EF-G in the compact conformation while it would clash with domain III when EF-G is in the elongated conformation.
Figure 6. The structure of EF-G bound to the ribosome trapped by the antibiotic dityromycin.
(A) Overview of the structure of the complex. The components are colored in the same scheme as in Figure 1, except that here ribosomal protein S12, instead of mRNA, is shown in cyan. Dityromycin (Dit) binds to protein S12 located behind EF-G.
(B) Close up of EF-G, S12 and dityromycin (brown). (C) Model of EF-G from the POST complex on the ribosome showing the steric collision between domain III and dityromycin.
See also Movie S2.
Discussion
EF-G exhibits a great degree of conformational flexibility on the ribosome
This study unveils a strikingly new compact conformation of EF-G on the ribosome, suggesting that EF-G is far more flexible than previously thought. The transient nature of this conformation may account for the fact that it has eluded direct observation for decades despite extensive studies involving various techniques. EF-G favors the elongated conformation in solution regardless of the identity of the bound nucleotide (Czworkowski and Moore, 1997; Czworkowski et al., 1994) through weak association between two super-domains. Our structures show that this weak association breaks upon binding of EF-G to the PRE ribosome. It is notable that the interface between the two super-domains in EF-G is not strengthened by the binding of GTP when EF-G is off the ribosome, since switches I and II remain disordered in both states (Hansson et al., 2005; Ticu et al., 2009). This suggests that the flexibility of EF-G is minimally influenced by the bound nucleotide.
A pretranslocation complex of the ribosome with EF-G
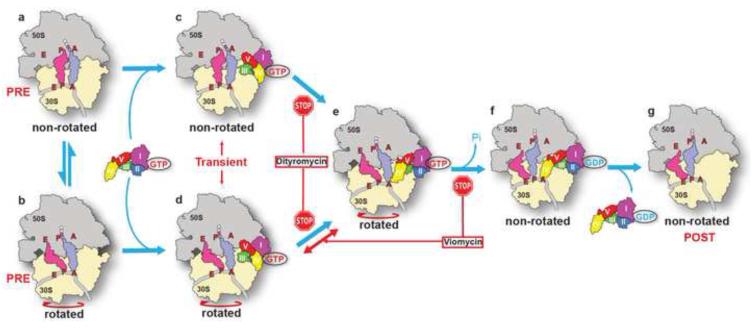
It has been a challenge to obtain the structural conformation of EF-G-GTP on a pre-translocation ribosome due to its transient existence and the difficulty in preparing a homogenous sample of the PRE ribosome that is deficient in tRNA translocation upon EF-G binding (Figure 7, steps a and b). Although the antibiotic viomycin can prevent translocation by keeping the PRE ribosome in the rotated state, considerable conformational heterogeneity still exists (Brilot et al., 2013; Stanley et al., 2010). In this study, we found the placement of non-hydrolysable aminoacyl-tRNA in the P site to be effective in stalling the ribosome in the PRE state albeit before peptidyl transfer. Such ribosomes are essentially identical to the authentic PRE ribosome (after peptidyl transfer) in the non-rotated state. However, we found only in the presence of the GDP nucleotide, and not GTP, does EF-G crystalize with the non-rotated ribosome under our experimental conditions. This is likely because of the unstable binding of domains I and II of EF-G on the non-rotated ribosome in the presence of GTP as confirmed by a recent crystal structure (Pulk and Cate, 2013). It has been established that EF-G bound with GTP engages the PRE ribosome. Therefore, in the bona fide PRE complex, GTP instead of GDP associates with EF-G (Figure 7, steps c and d), although minor positional adjustments of domains I and II of EF-G should be anticipated (Gao et al., 2009; Pulk and Cate, 2013; Tourigny et al., 2013).
Figure 7. Conformational changes of EF-G on the ribosome.
EF-G in complex with GTP transiently folds into a compact conformation, from an elongated conformation in the ribosome-free state, after engaging the ribosome in the pre-translocational state to avoid a collision with the A-site tRNA (steps a-d). Rotation of the 30S subunit enables domain IV moving next to the A-site tRNA, a step that can be blocked by the antibiotic dityromycin (steps c-e). Further conformational changes of EF-G with concomitant GTP hydrolysis facilitate tRNA translocation (steps e-f). This process involves swiveling of the 30S head domain (Ramrath et al., 2013), which is not shown here. The antibiotic viomycin prevents translocation by locking the ribosome in the rotated state, while not affecting the initial conformational changes of EF-G (Munro et al., 2010). tRNA translocation is completed by dissociation of EF-G in complex with GDP from the ribosome (steps f-g).
Our PRE complex addresses the long-standing puzzle—how EF-G avoids a collision with the A-site tRNA before promoting its translocation: domain IV simply folds backwards through an inter-domain joint. Such a conformational change is consistent with the movement of domain IV with respect to ribosomal protein S12 revealed by a recent single-molecule fluorescence resonance energy transfer (smFRET) study (Salsi et al., 2014). Evidently, the compact conformation of EF-G on the non-rotated ribosome displays the extreme end of the conformational space where domain IV reaches its closest proximity to domain II (Figure 7, step c). It is likely that domain IV could freely move next to the anticodon stem loop of the A-site tRNA following the rotating motion of the 30S subunit, thereby adopting a conformation similar to that seen in a recent cryo-EM structure of EF-G bound to the rotated PRE ribosome (Brilot et al., 2013) (Figure 7, steps c-e). Therefore, EF-G may not necessarily adopt the fully compact form when engaging the rotated PRE ribosome unless the ribosome is locked in the PRE state (Figure 7, step d). Interestingly, a smFRET study observed fluctuations of FRET between the C-terminus of EF-G and the A-site tRNA to zero after EF-G-GTP binds to the viomycin-trapped PRE ribosome (Munro et al., 2010); part of the fluctuations was attributed to the conformational changes of EF-G on the ribosome. We reason that EF-G-GTP can sporadically sample the fully compact conformation on the rotated PRE ribosome. Our dityromycin complex further confirms the flexibility of EF-G and the existence of its compact form on the ribosome.
Although it is debatable whether the antibiotic thiostrepton inhibits EF-G turnover on the ribosome (Rodnina et al., 1999) or binding to the ribosome (Walter et al., 2012), our data suggest that it may be able to coexist with a compact EF-G on the ribosome, thereby supporting the previous cryo-EM study (Stark et al., 2000). However, the close proximity of thiostrepton to domain V of EF-G and the SRL may interfere with stable binding of EF-G to the ribosome.
Insights into EF-G-mediated tRNA translocation
A comparison of the structures of our PRE and POST complexes immediately suggests that tRNA translocation is associated with a structural transition of EF-G from the compact to the previously observed elongated form (Figure 7 and movie S2). The path of domain IV movement merges with that of the anticodon stem loop of the A-site tRNA, in which the A-site tRNA travels into the P site while domain IV enters the vacated A site. Single-molecule studies have suggested that conformational changes of EF-G are coordinated with the ribosome during translocation (Chen et al., 2013b; Munro et al., 2010). Our work offers clues into the communication between EF-G and the ribosome during translocation. The structural transition of EF-G involves domain III passing over ribosomal protein S12 to approach the GTP-binding pocket in domain I (Figure 4B). Protein S12 seems to pose a steric restriction on the path of domain III in the non-rotated PRE complex, which could presumably be lifted by the rotation of the 30S subunit. On the other hand, dityromycin binding on top of protein S12 completely blocks the path taken by domain III, thereby preventing a structural transition of EF-G and inhibiting tRNA translocation (Figure 7, steps c-e and movie S2). Interestingly, removal of S12 from the ribosome stimulates spontaneous translocation (Cukras et al., 2003), underlining the significance of the interaction between S12 and EF-G during translocation. Domain V flips underneath the stalk base (H43 and H44) during the conformational change of EF-G. Consistent with chemical probing data (Bowen et al., 2005; Wilson and Nechifor, 2004), our structures show that the stalk base, which is mobile in the PRE complex, is stabilized by domain V in the POST complex. This may account for the different FRET states observed between the G’ domain of EF-G and protein L11 in the stalk base (Wang et al., 2007). As part of the GTPase-associated center on the ribosome, the stalk (through its bound protein L12) may stimulate GTP hydrolysis (Mohr et al., 2002) or control the timing of phosphate release (Savelsbergh et al., 2005) by sensing the orientation of domain V. Considering that the swivel of the 30S head domain effectively translocates the anticodon ends of the tRNAs (Guo and Noller, 2012; Ramrath et al., 2013; Zhou et al., 2014) and the critical role of domain IV in translocation (Rodnina et al., 1997), we speculate that an interaction between domain IV and the 30S head may be essential for translocation. In support of this proposal, h34 in the 30S head, which slightly overlaps with the path taken by domain IV during the transformation of EF-G (Figure 4C), was found to be protected during translocation (Matassova et al., 2001). It is generally accepted that GTP hydrolysis in EF-G precedes tRNA translocation (Pan et al., 2007; Savelsbergh et al., 2003), but how exactly these two processes are coupled remains less understood. To activate the catalytic center, domain III of EF-G needs to be positioned next to domain I to close the GTP binding pocket (Chen et al., 2013c; Martemyanov and Gudkov, 2000; Pulk and Cate, 2013), which would require EF-G to form at least a partially extended conformation (Figure 7, step e). This suggests that the earlier stage of conformational change of EF-G is independent of GTP hydrolysis (Salsi et al., 2014). Energy from GTP hydrolysis could be harnessed to promote further conformational changes of EF-G to the fully elongated conformation with domain IV reaching into the A site (Figure 7, step f). Such a process, coupled with a swivel of the 30S head, would result in translocation of the anticodon ends of tRNAs in the ribosome.
EXPERIMENTAL PROCEDURES
Additional details can be found online in Supplemental Information.
Preparation of the L9–EF-G protein fusions and the Thermus thermophilus mutant ribosomes lacking L9
The N-terminal domain (74 residues) of ribosomal protein L9 was fused to the N-terminus of EF-G with varying lengths of the linker between L9 and EF-G, as previously described (Gagnon et al., 2014). The Escherichia coli BL21 (DE3) Star (Invitrogen, Carlsbad, CA) cells were transformed with each construct and expression of the protein fusion was induced when the absorbance reached ~2.0 at 600 nm.
Before lysis, cells with the over-expressed protein fusion were re-suspended at 4 °C in a buffer containing 20 mM Tris–HCl pH 8.0, 50 mM NaCl, 1 mM β-mercaptoethanol. Pure L9–EF-G fusion protein was obtained after multiple purification steps, including hydrophobic interaction, anion exchange and size exclusion chromatography. The purified fractions containing the L9– EF-G fusion were concentrated to ~100 μM in a buffer containing 10 mM Tris–HCl pH 7.5, 200 mM KCl, 10 mM Mg(CH3COO)2 and 1 mM β-mercaptoethanol.
The T. thermophilus 70S ribosomes lacking L9 were prepared from the same mutant T. thermophilus 70S:L91-58 strain used in the previous study (Gagnon et al., 2014).
Complex formation and crystallization
The complexes were formed as previously described with some modifications (Gagnon et al., 2012). The POST complex was formed by incubating ribosomes with mRNA and a P-site fMettRNAfMet. Thereafter, the L9–EF-G fusion protein was added together with the guanosine 5′-diphosphate (GDP) nucleotide. The PRE complex was formed as described above, except that fMet-NH-tRNAfMet and Phe-NH-tRNAPhe were used. For the dityromycin-containing complex, a P-site Phe-tRNAPhe and dityromycin were used during complex formation.
Ribosome crystals were grown at room temperature in sitting drop trays. All complexes grew in the presence of the L9–EF-G fusion connecting residues 1-74 of ribosomal protein L9 and residues 8-691 of EF-G. The crystals were cryo-protected and frozen in a liquid nitrogen stream before plunging in liquid nitrogen.
Data collection
X-ray diffraction data were collected at the beamlines X25 at Brookhaven National Laboratory (Upton, NY), and 24-ID-C at the Advanced Photon Source at Argonne National Laboratory (Argonne, IL) using 0.2° or 0.3° oscillations. Data were integrated and scaled with the XDS program package (Kabsch, 1993).
Molecular replacement, model building, and structure refinement
Molecular replacement was performed using PHASER (McCoy et al., 2007). Two 70S ribosomes were found per asymmetric unit of the crystal. EF-G, mRNA and tRNAs in the A, P or E sites were built into the Fobs – Fcalc electron density map using Coot (Emsley and Cowtan, 2004) and the structures were refined in PHENIX (Adams et al., 2002). The E-site tRNA in all complexes comes from the excess of tRNAs in the sample preparation. In the POST complex, domains III, IV and V of EF-G were only resolved when the crystals were soaked with fusidic acid. The final refinement statistics for all the complexes are provided in Table S1. Figures 1-6 were generated using PyMOL (DeLano, 2006).
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Moore, S. Seetharaman, Y. Polikanov, I. Lomakin and A. Innis for valuable discussions and critical reading of the manuscript. We thank R. Grodzicki and Y. Polikanov for preparing tRNAs. The antibiotic dityromycin was kindly provided by the Gualerzi laboratory at the University of Camerino, Italy. We also thank the staffs of the Advanced Photon Source beamline 24-ID and the National Synchrotron Light Source beamline X25 for help during data collection and the Center for Structural Biology facility at Yale University for computational support. This work was supported by NIH grant GM022778.
Footnotes
ACCESSION NUMBERS Atomic coordinates and structure factors have been deposited in the Protein Data Bank with the accession codes 4WPO (PRE complex), 4WQF (POST complex with fusidic acid), 4WQY (POST complex without fusidic acid) and 4WQU (Dityromycin complex).
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures, two figures, one table and two movies and can be found with this article online.
AUTHOR CONTRIBUTIONS J.L., M.G.G., D.B and T.A.S conceived the project. J.L. and M.G.G. designed and performed all experiments. D.B. contributed to the purification of ribosomes. J.L., M.G.G. and T.A.S. analyzed the data and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol. 1999;6:643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- Agrawal RK, Penczek P, Grassucci RA, Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc Natl Acad Sci U S A. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL, Jr., Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WS, Van Dyke N, Murgola EJ, Lodmell JS, Hill WE. Interaction of thiostrepton and elongation factor-G with the ribosomal protein L11-binding domain. J Biol Chem. 2005;280:2934–2943. doi: 10.1074/jbc.M407008200. [DOI] [PubMed] [Google Scholar]
- Brandi L, Fabbretti A, Di Stefano M, Lazzarini A, Abbondi M, Gualerzi CO. Characterization of GE82832, a peptide inhibitor of translocation interacting with bacterial 30S ribosomal subunits. Rna. 2006;12:1262–1270. doi: 10.1261/rna.61206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi L, Maffioli S, Donadio S, Quaglia F, Sette M, Milon P, Gualerzi CO, Fabbretti A. Structural and functional characterization of the bacterial translocation inhibitor GE82832. FEBS Lett. 2012;586:3373–3378. doi: 10.1016/j.febslet.2012.07.040. [DOI] [PubMed] [Google Scholar]
- Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci U S A. 2013;110:20994–20999. doi: 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulkley D, Brandi L, Polikanov YS, Fabbretti A, O’Connor M, Gualerzi CO, Steitz TA. The Antibiotics Dityromycin and GE82832 Bind Protein S12 and Block EF-GCatalyzed Translocation. Cell Rep. 2014;6:357–365. doi: 10.1016/j.celrep.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Stevens B, Kaur J, Cabral D, Liu H, Wang Y, Zhang H, Rosenblum G, Smilansky Z, Goldman YE, et al. Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol Cell. 2011;42:367–377. doi: 10.1016/j.molcel.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Petrov A, Tsai A, O’Leary SE, Puglisi JD. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat Struct Mol Biol. 2013a;20:718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Feng S, Kumar V, Ero R, Gao YG. Structure of EF-G-ribosome complex in a pretranslocation state. Nat Struct Mol Biol. 2013b;20:1077–1084. doi: 10.1038/nsmb.2645. [DOI] [PubMed] [Google Scholar]
- Chen Y, Feng S, Kumar V, Ero R, Gao YG. Structure of EF-G-ribosome complex in a pretranslocation state. Nature structural & molecular biology. 2013c;20:1077–1084. doi: 10.1038/nsmb.2645. [DOI] [PubMed] [Google Scholar]
- Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25:751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol Cell. 2003;12:321–328. doi: 10.1016/s1097-2765(03)00275-2. [DOI] [PubMed] [Google Scholar]
- Czworkowski J, Moore PB. The conformational properties of elongation factor G and the mechanism of translocation. Biochemistry. 1997;36:10327–10334. doi: 10.1021/bi970610k. [DOI] [PubMed] [Google Scholar]
- Czworkowski J, Wang J, Steitz TA, Moore PB. The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J. 1994;13:3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2006 http://wwwpymolorg.
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol. 2011;18:457–462. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Gagnon MG, Lin J, Bulkley D, Steitz TA. Crystal structure of elongation factor 4 bound to a clockwise ratcheted ribosome. Science. 2014;345:684–687. doi: 10.1126/science.1253525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon MG, Seetharaman SV, Bulkley D, Steitz TA. Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science. 2012;335:1370–1372. doi: 10.1126/science.1217443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci U S A. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson S, Singh R, Gudkov AT, Liljas A, Logan DT. Crystal structure of a mutant elongation factor G trapped with a GTP analogue. FEBS Lett. 2005;579:4492–4497. doi: 10.1016/j.febslet.2005.07.016. [DOI] [PubMed] [Google Scholar]
- Holtkamp W, Cunha CE, Peske F, Konevega AL, Wintermeyer W, Rodnina MV. GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J. 2014;33:1073–1085. doi: 10.1002/embj.201387465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- Martemyanov KA, Gudkov AT. Domain III of elongation factor G from Thermus thermophilus is essential for induction of GTP hydrolysis on the ribosome. The Journal of biological chemistry. 2000;275:35820–35824. doi: 10.1074/jbc.M002656200. [DOI] [PubMed] [Google Scholar]
- Matassova AB, Rodnina MV, Wintermeyer W. Elongation factor G-induced structural change in helix 34 of 16S rRNA related to translocation on the ribosome. Rna. 2001;7:1879–1885. [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr D, Wintermeyer W, Rodnina MV. GTPase activation of elongation factors Tu and G on the ribosome. Biochemistry. 2002;41:12520–12528. doi: 10.1021/bi026301y. [DOI] [PubMed] [Google Scholar]
- Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat Struct Mol Biol. 2010;17:1470–1477. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Molecular cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340:1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrath DJ, Lancaster L, Sprink T, Mielke T, Loerke J, Noller HF, Spahn CM. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci U S A. 2013;110:20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Matassova NB, Katunin VI, Semenkov YP, Wintermeyer W. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9586–9590. doi: 10.1073/pnas.96.17.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsi E, Farah E, Dann J, Ermolenko DN. Following movement of domain IV of elongation factor G during ribosomal translocation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:15060–15065. doi: 10.1073/pnas.1410873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Molecular cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. Rna. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat Struct Mol Biol. 2010;17:289–293. doi: 10.1038/nsmb.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- Ticu C, Nechifor R, Nguyen B, Desrosiers M, Wilson KS. Conformational changes in switch I of EF-G drive its directional cycling on and off the ribosome. EMBO J. 2009;28:2053–2065. doi: 10.1038/emboj.2009.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annu Rev Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter JD, Hunter M, Cobb M, Traeger G, Spiegel PC. Thiostrepton inhibits stable 70S ribosome binding and ribosome-dependent GTPase activation of elongation factor G and elongation factor 4. Nucleic Acids Res. 2012;40:360–370. doi: 10.1093/nar/gkr623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qin H, Kudaravalli RD, Kirillov SV, Dempsey GT, Pan D, Cooperman BS, Goldman YE. Single-molecule structural dynamics of EF-G-ribosome interaction during translocation. Biochemistry. 2007;46:10767–10775. doi: 10.1021/bi700657d. [DOI] [PubMed] [Google Scholar]
- Wilson KS, Nechifor R. Interactions of translational factor EF-G with the bacterial ribosome before and after mRNA translocation. J Mol Biol. 2004;337:15–30. doi: 10.1016/j.jmb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science. 2014;345:1188–1191. doi: 10.1126/science.1255030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.