Abstract
Benign prostatic hyperplasia (BPH) is linked to lower urinary tract symptoms (LUTS) such as incomplete bladder emptying, urinary frequency and urgency. Mechanisms responsible for BPH are not fully known. Here, we tested whether beta-catenin (CTNNB1) immunostaining intensity and distribution differ in human glandular BPH tissue specimens compared to normal prostate tissue. Multiplex immunostaining of CTNNB1, its putative transcriptional target gene lymphoid enhancer binding factor 1 (LEF1), and the epithelial marker E-cadherin were examined in clinical human prostate specimens with or without histological BPH (pure epithelial or mixed stromal-epithelial nodules). BPH specimens were obtained from 24 men who experienced LUTS and underwent transurethral resection of the prostate surgery. Control specimens were tumor-adjacent histologically normal prostate tissue from 48 patients who underwent radical prostatectomy. The resulting multispectral images were unmixed and optical densities recorded to quantify staining abundance, cellular (membranous, cytoplasmic, and nuclear) and tissue localization (stromal versus epithelial), and determination of percentage of CTNNB1-positive cells. The following CTNNB1 indices were significantly higher in BPH compared to normal prostate tissue: overall staining intensity, staining intensity in prostate stromal cell membranes, cytoplasm and nuclei, and prostate epithelial cell nuclei. The following LEF1 indices were significantly lower in BPH compared to tumor-adjacent normal prostate tissue: stromal LEF1 staining intensity, percentage of LEF1-positive stromal cells, and intensity of LEF1 staining in stromal cell membranes, cytoplasm, and nuclei. The percentage of stromal cells with CTNNB1+/LEF1- nuclei was higher and percentage of stromal cells with CTNNB1-/LEF1+ nuclei was lower in BPH compared to tumor-adjacent normal prostate tissues. These results support the hypothesis that CTNNB1 expression increases in specific BPH tissue compartments. Further, since nuclear LEF1 staining does not coincide with cytoplasmic or nuclear CTNNB1 staining, it does not appear to be a reliable index of CTNNB1 activity in adult human prostate.
Keywords: Prostate, beta-catenin, tissue microarray, multispectral, LEF1, stromal-epithelial, cellular localization
Introduction
Clinical benign prostatic hyperplasia (BPH) is an aging-dependent expansion of prostate tissue resulting from non-cancerous cell proliferation. Prostatic hyperplasia occurs in focal nodules consisting of prostatic stroma, glandular epithelium, or both. BPH is often accompanied by lower urinary tract symptoms (LUTS) including incomplete bladder emptying, increased frequency, and difficulty in starting and stopping urination. The underlying basis of BPH is not fully known, and current pharmacotherapies for LUTS associated with BPH only moderate patient symptoms rather than cure the disease. Thus, there is a need to identify the mechanistic basis of BPH and LUTS so that new and more effective therapies can be generated.
Several processes are hypothesized to participate in development of BPH and LUTS including: inflammation, fibrosis, hormones, proliferation, and an inappropriate reawakening of prostate developmental signaling pathways [1-9]. All of these involve beta-catenin (CTNNB1) signaling. CTNNB1 signaling is induced by inflammation, its activity correlates with fibrosis in a host of tissues, and it is required for prostate development [10-22].
The preponderance of studies focused on CTNNB1 in adult human prostate tissues has been directed towards prostate cancer. Many of these studies used CTNNB1 immunostaining to assess protein activity: the presence of CTNNB1 protein in cytoplasm and nuclei heralds its potential transformation into a transcriptional coactivator [23]. Although CTNNB1 participates in many processes suspected to contribute to BPH, few studies have specifically examined whether CTNNB1 signaling is elevated in BPH compared to histologically normal tissues. More often, BPH tissues are used as a control to examine whether CTNNB1 is activated in prostate cancer (reviewed by Whitaker et al. 2008).
Here, we used human prostate tissues and multiplex immunostaining to examine expression and distribution of CTNNB1 and its putative target gene, lymphoid enhancer binding factor 1 (LEF1), in human BPH and control tissues from tumor adjacent, histologically normal prostate tissue. We used multispectral imaging to objectively quantify immunostaining intensities within cellular compartments (membrane, cytoplasm, and nuclei) and across tissue populations (epithelium and stroma). We identified significant elevations in CTNNB1 staining intensity among BPH stromal cells and within BPH epithelial cell nuclei compared to histologically normal tissue. On the other hand, LEF1 staining decreased within BPH stroma and did not accompany nuclear CTNNB1 staining in most cell types, indicating it is not a reliable index of CTNNB1 signaling in adult human prostate. Together, our results are consistent with an increased CTNNB1 activity in BPH compared to histologically normal prostate tissue.
Materials and methods
Human tissues and immunohistochemistry
A prostate tissue microarray (TMA) containing duplicate cores was constructed using a Manual Tissue Arrayer (Beecher Instruments, Sun Prairie, WI; model MTA-1), as previously described [25,26]. The 0.6 mm cores were arranged 0.8 mm center to center. The TMA includes 96 cores (48 patients) of tumor-adjacent normal prostate (BPT) from prostatectomy and 48 cores (24 patients) of benign prostatic hyperplasia tissue (BPH) acquired from transurethral resection of the prostate (TURP) [27]. All BPH patients had history of lower urinary tract symptomology (LUTS) and surgical indications included history of urinary retention and failure of medical management of LUTS.
Samples were processed and stained by immunohistochemistry (IHC) using antibodies for CTNNB1, LEF1, and E-cadherin, as previously described [26,28]. E-cadherin was used to aid in tissue segmentation and for identification of the plasma membrane. Tissues were stained using rabbit monoclonal anti-LEF1 (Cell Signaling Technology, Beverly, MA; 1:150 in Renoir Red [Biocare, Concord, CA]) followed by goat anti-rabbit Mach 2 HRP-Polymer (Biocare) as a secondary antibody. Mouse monoclonal anti-CTNNB1 (BD Transduction Laboratories; San Jose, CA; 1:200 in Biocare Renoir Red) was applied and goat anti-mouse Mach 2 HRP-Polymer (Biocare) was used as a secondary antibody. Monoclonal mouse anti-E-cadherin (Dako, Carpinteria, CA; 1:150 in Biocare Renoir Red) was applied and goat anti-mouse Mach 2 secondary antibody was used. Bajoran Purple chromogen (Biocare) was used to detect CTNNB1, DAB chromogen (Biocare) was used for LEF1, and Deep Space Black chromogen (Biocare) was used to detect E-cadherin.
Image analysis
Data acquisition and image analysis was performed as previously described [26,28,29]. Briefly, the TMA slide was loaded onto the Vectra slide scanner (PerkinElmer, Waltham, MA) and a scanning protocol was created. 8-bit Nuance multispectral image cubes were acquired using the 20x objective lens (0.5 µm/pixel). Using Nuance software (PerkinElmer), spectral curves from four control slides containing only hematoxylin, Deep Space Black, DAB, or Bajoran Purple were used to create a spectral library for separation of chromogens on multi-chromogenic slides. Tissue and cell segmentation was performed using inForm software (PerkinElmer), and an algorithm of differentiation was created using 18% of image cubes, assuring 97% segmentation accuracy [26]. The algorithm was applied to all image cubes and CTNNB1 and LEF1 was quantified in all tissue and cellular compartments. Cores with significant folding or <5% epithelium were eliminated from analysis.
Positivity and co-localization data was generated through thresholding of mean optical density (OD) values for each protein. A threshold of 0.1 was used for both CTNNB1 and LEF1 expression, similar to previous studies using the same multispectral imaging platform [30]. The following values were assessed for both proteins in all tissue and cellular compartments: total positivity, double positivity, double negativity, and single positivity (CTNNB1+/LEF1- and CTNNB1-/LEF1+).
Statistical analysis
Differences in protein expression between normal prostate and BPH were assessed using a two-tailed Student’s t-test. A P-value <0.05 was considered significant in all analyses, and GraphPad Prism (GraphPad Software, Inc., La Jolla, Ca) was used for all statistical analysis.
Results
Total protein expression of CTNNB1 and LEF1 in normal prostate and BPH
CTNNB1 and LEF1 protein was localized to the nucleus, cytoplasm, and membrane in both the prostatic epithelia and stroma (Figure 1A). The overall abundance of CTNNB1 staining was significantly higher in BPH (mean OD ± SEM: 0.093 ± 0.002) than normal prostate (0.085 ± 0.002; p = 0.02; Figure 1B). No difference in overall LEF1 staining was found between normal prostate (0.058 ± 0.006) and BPH (0.046 ± 0.004; p = 0.17); Figure 1C).
Figure 1.
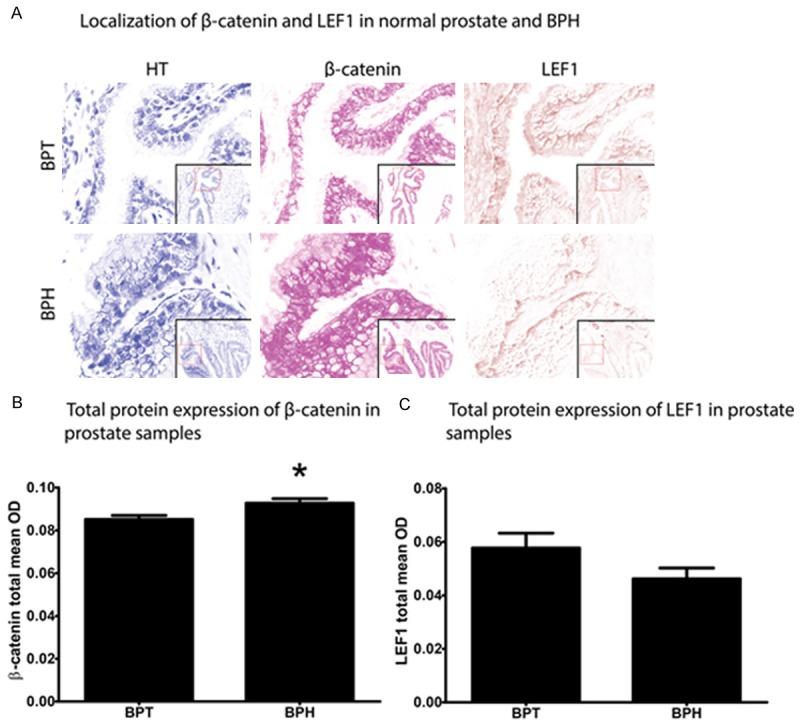
LEF1 and CTNNB1 localization and staining abundance in normal prostate and glandular benign prostatic hyperplasia (BPH). Nuance® software was used to build a spectral library to unmix multiplexed immunohistochemical staining, resulting in separation of hematoxylin (left column), Bajoran purple (middle column), and 3,3’-Diaminobenzidine (DAB; right column) chromogens (A). CTNNB1 and LEF1 staining intensity was quantified using inForm® software (B). Total CTNNB1 staining was significantly higher in BPH (mean = 0.093, SEM = 0.002) compared to benign prostatic tissue (BPT; 0.085 ± 0.002; p = 0.02) (C). Total LEF1 staining was not significantly different between BPT (0.058 ± 0.006) and BPH (0.046 ± 0.004; p = 0.17).
Tissue compartment-specific expression of CTNNB1 and LEF1
To investigate differences in protein abundance within isolated prostate epithelial and stromal tissue compartments, CTNNB1 and LEF1 expression was quantified by mean OD and by percentage of positive cells in prostate epithelia and stroma (Figure 2). Epithelial CTNNB1 staining intensity was comparable in normal prostate and BPH when analyzed for mean OD (p = 0.15) and cellular positivity (p = 0.54). CTNNB1 protein staining abundance was significantly higher in BPH stroma (mean OD ± SEM: 0.028 ± 0.001) compared to normal prostate (0.021 ± 0.001; p<0.0001) when analyzed by mean OD. The percentage of stromal CTNNB1-positive cells did not change (p = 0.06).
Figure 2.
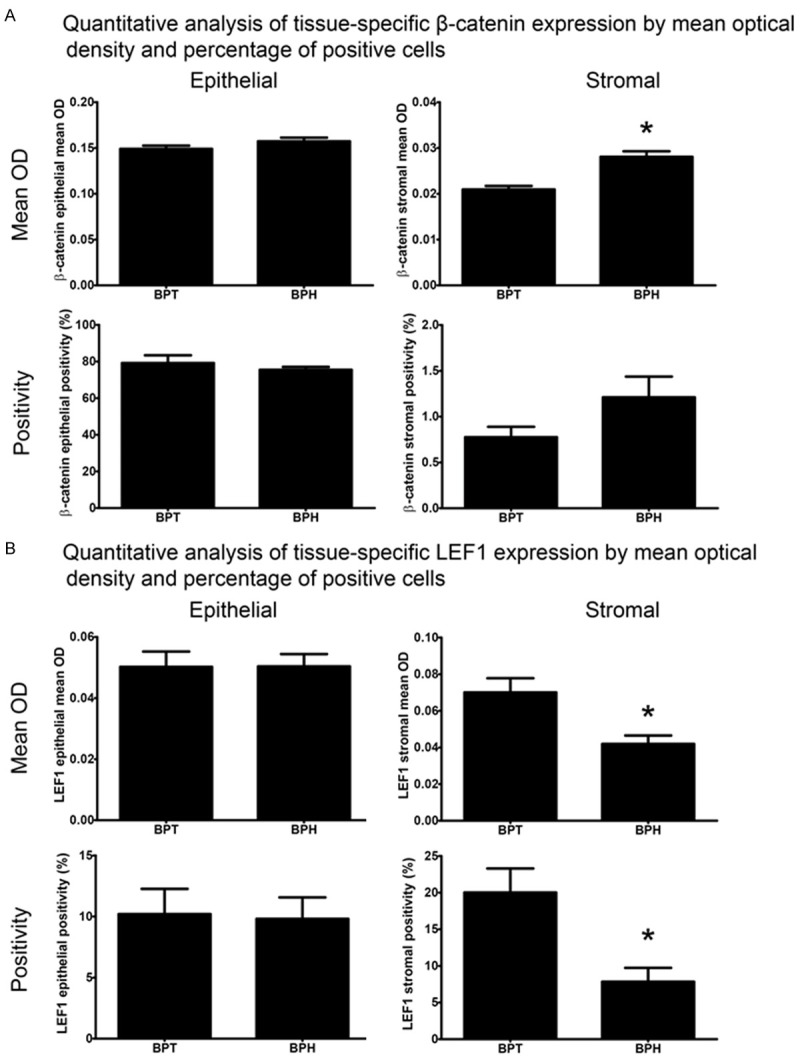
Quantitative analysis of tissue-specific CTNNB1 or LEF1 staining abundance by mean optical density (OD) and percentage of immunopositive cells within tissue. No differences in epithelial CTNNB1 staining were observed as assessed by mean OD (p = 0.15) or positivity (p = 0.54) analysis (A). Stromal CTNNB1 staining was significantly higher in BPH (mean = 0.028, SEM = 0.001) than BPT (0.021 ± 0.001; p<0.0001) in mean OD analysis. Stromal CTNNB1 positivity was higher in BPH than BPT but failed to reach significance (p = 0.06). Epithelial LEF1 staining was similar between BPT and BPH in both mean OD (p = 0.99) and positivity (p = 0.90) analysis (B). Analysis of stromal LEF1 staining by mean OD displayed significantly lower expression in BPH (0.042 ± 0.005) compared to BPT (0.070 ± 0.008; p = 0.02). LEF1 positivity in the stroma also showed a significantly lower amount of positive cells in BPH (mean = 7.84%, SEM = 1.89) compared to BPT (20.01 ± 3.28; p = 0.02).
Epithelial LEF1 expression was similar in normal prostate and BPH for both the number of LEF1-positive cells (p = 0.90) and average staining intensity across epithelial cells (p = 0.99). Stromal LEF1 staining intensity was significantly decreased in BPH (mean OD ± SEM: 0.042 ± 0.005) compared to normal prostate (0.070 ± 0.008; p = 0.02). The percentage of LEF1-positive stromal cells in BPH (7.84 ± 1.86) was significantly lower than in normal prostate (20.01 ± 3.28; p = 0.02).
Cellular compartment-specific expression of CTNNB1 and LEF1
In preliminary analysis of stained samples, CTNNB1 expression was found in all three subcellular compartments, with higher expression observed in the nucleus than the plasma membrane (p<0.0001). Expression of CTNNB1 in the cytoplasm was lower than the nucleus and membrane (p<0.0001). LEF1 was also localized to all three subcellular compartments, but no differences in expression were observed between nucleus, cytoplasm, and plasma membrane (p = 0.14). The appearance of detectable CTNNB1 staining in cytosol or nuclei indicates its potential activation into a transcriptional coactivator. We therefore quantified CTNNB1 and LEF1 protein expression within isolated cellular compartments including nucleus, cytoplasm, and cell membranes (Table 1). Within the epithelium, no significant differences in CTNNB1 or LEF1 expression were observed in any cellular compartment (p>0.05), with the exception of nuclear CTNNB1 staining, which was significantly higher in BPH compared to normal prostate (p = 0.02). In the stroma, CTNNB1 expression was significantly higher in all BPH cellular compartments (total, nuclear, cytoplasmic, and membrane) compared to normal prostate (p<0.0001). Stromal LEF1 expression was significantly decreased in the nucleus (p = 0.01), cytoplasm (p = 0.02), and membrane (p = 0.02).
Table 1.
Cellular expression of β-catenin and LEF1 in the epithelium and stroma of normal prostate tissue and benign prostatic hyperplasia specimens, mean OD (± SEM)
| Epithelial | Stromal | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Total | Nucleus | Cytoplasm | Membrane | Total | Nucleus | Cytoplasm | Membrane | |
| β-catenin | ||||||||
| BPT | 0.149 (±0.004) | 0.173 (± 0.004) | 0.126 (± 0.003) | 0.149 (± 0.004) | 0.021 (± 0.001) | 0.034 (± 0.001) | 0.011 (± 0.001) | 0.018 (± 0.001) |
| BPH | 0.157 (± 0.004) | 0.188 (± 0.004) | 0.131 (± 0.004) | 0.153 (± 0.005) | 0.028 (± 0.001) | 0.044 (± 0.002) | 0.015 (± 0.001) | 0.024 (± 0.001) |
| P-value | 0.15 | 0.02 | 0.39 | 0.45 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| LEF1 | ||||||||
| BPT | 0.050 (± 0.005) | 0.056 (± 0.006) | 0.050 (± 0.005) | 0.046 (± 0.005) | 0.070 (± 0.008) | 0.074 (± 0.007) | 0.066 (± 0.007) | 0.070 (± 0.008) |
| BPH | 0.050 (± 0.004) | 0.057 (± 0.005) | 0.048 (± 0.003) | 0.046 (± 0.004) | 0.042 (± 0.005) | 0.045 (± 0.005) | 0.040 (± 0.004) | 0.041 (± 0.005) |
| P-value | 0.99 | 0.81 | 0.81 | 1.00 | 0.02 | 0.01 | 0.02 | 0.02 |
Abbreviations: benign prostatic tissue (BPT), benign prostatic hyperplasia (BPH).
Co-localization of CTNNB1 and LEF1 in prostate tissues
Using positivity thresholds and inForm software (PerkinElmer), we then investigated the co-localization of CTNNB1 and LEF1 in different tissue and cellular compartments within prostate tissues (Table 2). No significant differences in the percentage of double positive (CTNNB1+/LEF1+), double negative (CTNNB1-/LEF1-), or single positive (CTNNB1+/LEF1- or CTNNB1-/LEF1+) cells were observed between BPH and normal prostate in the epithelia (p>0.05). However, within cellular compartments, the percentage of double negative stromal cells in BPH was higher in the nucleus (p = 0.03), cytoplasm (p = 0.01), membrane (p = 0.02), and overall (p = 0.02). No changes in stromal double positive cells were observed between BPH and normal prostate (p>0.05). The percentage of stromal CTNNB1+/LEF1- single-positive cells was higher in BPH in the nucleus (p = 0.0009), membrane (p = 0.02), and overall (p = 0.04), but not in the cytoplasm (p = 0.21). The percentage of stromal CTNNB1-/LEF1+ single-positive cells was lower in BPH compared to normal prostate in all compartments (p<0.05).
Table 2.
Co-localization of β-catenin and LEF1 in the epithelium and stroma of normal prostate tissue and benign prostatic hyperplasia specimens, % (SEM)
| Epithelial | Stromal | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| β-cat-/LEF1- | β-cat+/LEF1+ | β-cat+/LEF1- | β-cat-/LEF1+ | β-cat-/LEF1- | β-cat+/LEF1+ | β-cat+/LEF1- | β-cat-/LEF1+ | |
| TOTAL | ||||||||
| BPT | 20.92 (± 1.49) | 5.96 (± 1.29) | 68.88 (± 2.23) | 4.25 (± 1.19) | 79.30 (± 3.27) | 0.09 (± 0.03) | 0.69 (± 0.11) | 19.93 (± 3.26) |
| BPH | 21.03 (± 1.29) | 6.30 (± 1.17) | 69.17 (± 2.32) | 3.51 (± 0.76) | 91.00 (± 1.79) | 0.05 (± 0.02) | 1.16 (± 0.23) | 7.79 (± 1.88) |
| P-value | 0.96 | 0.86 | 0.94 | 0.68 | 0.02 | 0.44 | 0.04 | 0.02 |
| NUCLEUS | ||||||||
| BPT | 14.19 (± 0.94) | 8.69 (± 1.58) | 72.92 (± 2.15) | 4.22 (± 1.02) | 76.95 (± 2.97) | 0.58 (± 0.15) | 1.48 (± 0.21) | 21.00 (± 2.89) |
| BPH | 13.70 (± 0.76) | 9.92 (± 1.53) | 72.47 (± 2.33) | 3.92 (± 0.76) | 86.72 (± 1.94) | 0.26 (± 0.07) | 3.39 (± 0.66) | 9.65 (± 2.14) |
| P-value | 0.73 | 0.62 | 0.90 | 0.85 | 0.03 | 0.16 | 0.0009 | 0.01 |
| CYTOPLASM | ||||||||
| BPT | 34.95 (± 1.77) | 5.12 (± 1.04) | 54.72 (± 2.07) | 5.22 (± 1.31) | 81.43 (± 3.13) | 0.04 (± 0.02) | 0.30 (± 0.06) | 18.24 (± 3.12) |
| BPH | 35.83 (± 1.87) | 5.49 (± 0.96) | 54.51 (± 2.31) | 4.17 (± 0.82) | 93.05 (± 1.50) | 0.02 (± 0.01) | 0.42 (± 0.07) | 6.51 (± 1.54) |
| P-value | 0.76 | 0.82 | 0.95 | 0.59 | 0.01 | 0.38 | 0.21 | 0.01 |
| MEMBRANE | ||||||||
| BPT | 19.77 (± 1.63) | 4.32 (± 0.94) | 71.36 (± 2.27) | 4.55 (± 1.22) | 79.10 (± 3.41) | 0.13 (± 0.05) | 1.11 (± 0.14) | 19.66 (± 3.36) |
| BPH | 21.60 (± 1.76) | 4.34 (± 0.95) | 70.27 (± 2.59) | 3.80 (± 0.93) | 90.79 (± 1.61) | 0.07 (± 0.02) | 1.84 (± 0.35) | 7.30 (± 1.74) |
| P-value | 0.49 | 0.99 | 0.77 | 0.68 | 0.02 | 0.46 | 0.02 | 0.02 |
Abbreviations: benign prostatic tissue (BPT), benign prostatic hyperplasia (BPH).
Discussion
The role of CTNNB1, and it’s downstream target, LEF1, in the development of BPH, requires elucidation. We discovered that CTNNB1 protein staining is elevated in BPH compared to normal prostate tissue. CTNNB1 expression is elevated in BPH stromal cell cytoplasm and nuclei and in BPH epithelial cell nuclei. These staining patterns are consistent with potential increases in CTNNB1-mediated transcriptional coactivation in both epithelium and stroma of human BPH tissue. Our current results substantiate the need for more investigations assessing CTNNB1 localization and expression as we have performed here, in other states including cancer. A specific need is to test whether CTNNB1 abundance, and the presence of cytosolic or nuclear CTNNB1, associates with LUTS in general, and determine the specific manifestations of LUTS in which CTNNB1 may be involved.
Our results are among the first to describe immunostaining differences consistent with CTNNB1 activation in human BPH. Most other studies of prostatic CTNNB1 expression in humans and dogs (which are also susceptible to BPH and prostate cancer), have reported CTNNB1 immunostaining in BPH specimens only in relation to prostate cancer. In most cases, CTNNB1 abundance and/or cytoplasmic and nuclear localization is greater in prostate cancer compared to BPH [31-33]. A few studies evaluated CTNNB1 in BPH. Whitaker, et al. [24] reported the presence of nuclear CTNNB1 staining in human BPH prostate tissues, and Lean, et al. [34] observed a significant increase in cytoplasmic and nuclear CTNNB1 staining in canine BPH specimens compared to histologically normal canine prostate tissues. These results are consistent with our current observation that the number of prostate epithelial cells with detectable CTNNB1 staining is elevated in BPH compared to histologically normal tissue.
CTNNB1 is constitutively localized to cell membranes in most benign epithelial cells, where it mediates cell adhesion by functioning as a member of the adherens junction complex. While membranous CTNBB1 expression is readily evident in benign prostatic epithelium, cytoplasmic and nuclear CTNNB1 is more difficult to visualize and assessments of CTNNB1 staining in these cell compartments are subject to reviewer bias. One potential reason that we were able to reveal elevated CTNNB1 staining in BPH is our use of the ultra-sensitive method of multispectral imaging. The Vectra system accurately and reproducibly unmixes overlapping chromogens and can quantify, while limiting reviewer bias, the abundance and positivity of immunostain across tissues and within compartments [29,30,35].
Our observation of increased nuclear CTNNB1 staining in BPH specimens suggest its potential role in pathogenesis and raises several testable hypotheses about how it may function in BPH. BPH has been hypothesized to involve a reactivation of developmental signaling pathways and epithelial CTNNB1 signaling plays a critical role in prostatic ductal development by organizing proliferative growth processes that establish appropriate prostatic bud patterns. We previously determined that CTNNB1 increases mRNA expression of several TGF beta family members, including bone morphogenetic proteins (BMPs)-2, -4 and -7 [36]. While there is little information about the role of BMPs in BPH, BMP 2 and 4 proteins were previously detected in a fraction of human BPH specimens [37], and BMP2 was shown to interact in vitro with the profibrogenic mediator periostin [38]. This provides a new testable mechanism for the etiology of fibrogenic changes that have been reported in BPH [9].
A surprising finding of this study was the observation of elevated CTNNB1 staining in BPH prostate stroma. Though the consequences of CTNNB1 gain-of-function and loss-of-function have been carefully examined in prostatic epithelium, the function of beta-catenin in prostatic stroma is less well understood. Potential roles of CTNNB1 in prostatic stroma are suggested from studies of CTNNB1 function in kidney stroma. CTNNB1 is activated in kidney stroma (interstitial fibroblasts) during reparative processes, and CTNNB1 overexpression in this cell type is sufficient for fibroblast-to-myofibroblast transition, and the onset of fibrosis without injury [39]. CTNNB1 is also activated in kidney pericytes during injury, and is required therein for damage-induced inflammation and fibrosis [40]. Whether CTNNB1 activation in prostate stroma elicits actions similar to that of CTNNB1 activation in kidney stroma, remains to be determined.
Though cytosolic and nuclear accumulation of CTNNB1 have historically been used as biomarkers for its potential engagement as a transcriptional coactivator, additional molecular evidence is needed to more accurately assess CTNNB1 activity. Though many CTNNB1 target genes are known across mammalian tissues, they are expressed in a context- and tissue-dependent manner. Several CTNNB1 target genes have been elucidated in developing mouse prostate including mRNAs for Axin2, Lef1, Wnt inhibitory factor 1, Bmps 2, 4, and 7, T cell factor 7, and Myc [10,12,36]. In the current study, LEF1 protein was present in about 10% of the epithelial cells and 18% of stroma cells. However, LEF1 did not appear to mark cells that also showed potential CTNNB1 activation, based on its nuclear localization in tumor adjacent normal prostate tissue. CTNNB1 localized to the nucleus of 72.92% of prostate epithelial cells that lacked detectable LEF1 staining, but only 8.69% of cells with detectable LEF1 staining. Further, CTNNB1 localized to the nucleus of 1.48% of prostate stromal cells without detectable LEF1, but only 0.58% of cells with detectable LEF1. This pattern was similar in BPH tissues. These results indicate that LEF1 is present in histologically normal and BPH prostate tissue, but is unlikely to be an accurate readout of CTNNB1 signaling in these tissues. Future validation of other CTNNB1 target genes in human BPH prostate specimens will reveal its potential mechanisms of action in this disease process.
Acknowledgements
This work was supported by National Institutes of Health grants R01DK093690 and R01CA123199 (WAR), U54DK104310A (WAR, PCM, CMV), K01DK083425 (CMV), and R01ES001332 (REP), DK091193 and CA140217 (PCM). The authors thank the University of Wisconsin Translational Research Initiatives in Pathology laboratory (UWCCC grant P30 CA014520) and the University of Wisconsin Laboratory for Optical and Computational Instrumentation for the use of their facilities and services. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.St Sauver JL, Jacobsen SJ. Inflammatory Mechanisms Associated with Prostatic Inflammation and Lower Urinary Tract Symptoms. Current Prostate Reports. 2008;6:67–73. doi: 10.1007/s11918-008-0011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishra VC, Allen DJ, Nicolaou C, Sharif H, Hudd C, Karim OM, Motiwala HG, Laniado ME. Does intraprostatic inflammation have a role in the pathogenesis and progression of benign prostatic hyperplasia? BJU Int. 2007;100:327–331. doi: 10.1111/j.1464-410X.2007.06910.x. [DOI] [PubMed] [Google Scholar]
- 3.Di Silverio F, Gentile V, De Matteis A, Mariotti G, Giuseppe V, Luigi PA, Sciarra A. Distribution of inflammation, pre-malignant lesions, incidental carcinoma in histologically confirmed benign prostatic hyperplasia: a retrospective analysis. Eur Urol. 2003;43:164–175. doi: 10.1016/s0302-2838(02)00548-1. [DOI] [PubMed] [Google Scholar]
- 4.Roehrborn CG, Kaplan SA, Noble WD, Lucia MS, Slawin KM, McVary KT. The impact of acute or chronic inflammation in basaeline biopsy on the risk of clinical progression of BPE: results from the MTOPS study. 2005 May 21-25; San Antonio, TX. [Google Scholar]
- 5.Nickel JC, Roehrborn CG, O’Leary MP, Bostwick DG, Somerville MC, Rittmaster RS. The relationship between prostate inflammation and lower urinary tract symptoms: examination of baseline data from the REDUCE trial. Eur Urol. 2008;54:1379–1384. doi: 10.1016/j.eururo.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robert G, Descazeaud A, Nicolaiew N, Terry S, Sirab N, Vacherot F, Maille P, Allory Y, de la Taille A. Inflammation in benign prostatic hyperplasia: a 282 patients’ immunohistochemical analysis. Prostate. 2009;69:1774–1780. doi: 10.1002/pros.21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNeal JE. Origin and evolution of benign prostatic enlargement. Invest Urol. 1978;15:340–345. [PubMed] [Google Scholar]
- 8.Bercovich E, Barabino G, Pirozzi-Farina F, Deriu M. A multivariate analysis of lower urinary tract ageing and urinary symptoms: the role of fibrosis. Archivio italiano di urologia, andrologia: organo ufficiale [di] Societa italiana di ecografia urologica e nefrologica/Associazione ricerche in urologia. 1999;71:287–292. [PubMed] [Google Scholar]
- 9.Ma J, Gharaee-Kermani M, Kunju L, Hollingsworth JM, Adler J, Arruda EM, Macoska JA. Prostatic Fibrosis is Associated with Lower Urinary Tract Symptoms. J Urol. 2012;188:1375–1381. doi: 10.1016/j.juro.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simons BW, Hurley PJ, Huang Z, Ross AE, Miller R, Marchionni L, Berman DM, Schaeffer EM. Wnt signaling though beta-catenin is required for prostate lineage specification. Dev Biol. 2012;371:246–55. doi: 10.1016/j.ydbio.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehta V, Schmitz CT, Keil KP, Joshi PS, Abler LL, Lin TM, Taketo MM, Sun X, Vezina CM. Beta-catenin (CTNNB1) induces Bmp expression in urogenital sinus epithelium and participates in prostatic bud initiation and patterning. Dev Biol. 2013;376:125–35. doi: 10.1016/j.ydbio.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis JC, Thomsen MK, Taketo MM, Swain A. beta-Catenin Is Required for Prostate Development and Cooperates with Pten Loss to Drive Invasive Carcinoma. PLoS Genet. 2013;9:e1003180. doi: 10.1371/journal.pgen.1003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu HS, Liu CC, Lin JH, Hsu TW, Su K, Hung SC. Repair of naphthalene-induced acute tracheal injury by basal cells depends on beta-catenin. J Thorac Cardiovasc Surg. 2014;148:322–32. doi: 10.1016/j.jtcvs.2013.10.039. [DOI] [PubMed] [Google Scholar]
- 14.Zemans RL, McClendon J, Aschner Y, Briones N, Young SK, Lau LF, Kahn M, Downey GP. Role of beta-catenin-regulated CCN matricellular proteins in epithelial repair after inflammatory lung injury. Am J Physiol Lung Cell Mol Physiol. 2013;304:L415–427. doi: 10.1152/ajplung.00180.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L, Rao JN, Zou T, Xiao L, Smith A, Zhuang R, Turner DJ, Wang JY. Activation of Wnt3a signaling stimulates intestinal epithelial repair by promoting c-Myc-regulated gene expression. Am J Physiol Cell Physiol. 2012;302:C277–285. doi: 10.1152/ajpcell.00341.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan J, Gherghe C, Liu D, Hamlett E, Srikantha L, Rodgers L, Regan JN, Rojas M, Willis M, Leask A, Majesky M, Deb A. Wnt1/betacatenin injury response activates the epicardium and cardiac fibroblasts to promote cardiac repair. EMBO J. 2012;31:429–442. doi: 10.1038/emboj.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim X, Tan SH, Koh WL, Chau RM, Yan KS, Kuo CJ, van Amerongen R, Klein AM, Nusse R. Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science. 2013;342:1226–1230. doi: 10.1126/science.1239730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson MD, Wickline ED, Bowen WB, Lu A, Singh S, Misse A, Monga SP. Spontaneous repopulation of beta-catenin null livers with beta-catenin-positive hepatocytes after chronic murine liver injury. Hepatology. 2011;54:1333–1343. doi: 10.1002/hep.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akhmetshina A, Palumbo K, Dees C, Bergmann C, Venalis P, Zerr P, Horn A, Kireva T, Beyer C, Zwerina J, Schneider H, Sadowski A, Riener MO, MacDougald OA, Distler O, Schett G, Distler JH. Activation of canonical Wnt signalling is required for TGF-beta-mediated fibrosis. Nat Commun. 2012;3:735. doi: 10.1038/ncomms1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chilosi M, Poletti V, Zamo A, Lestani M, Montagna L, Piccoli P, Pedron S, Bertaso M, Scarpa A, Murer B, Cancellieri A, Maestro R, Semenzato G, Doglioni C. Aberrant Wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol. 2003;162:1495–1502. doi: 10.1016/s0002-9440(10)64282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng JH, She H, Han YP, Wang J, Xiong S, Asahina K, Tsukamoto H. Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. Am J Physiol Gastrointest Liver Physiol. 2008;294:G39–49. doi: 10.1152/ajpgi.00263.2007. [DOI] [PubMed] [Google Scholar]
- 22.Xu-Dubois YC, Baugey E, Peltier J, Colombat M, Ouali N, Jouanneau C, Rondeau E, Hertig A. Epithelial phenotypic changes are associated with a tubular active fibrogenic process in human renal grafts. Hum Pathol. 2013;44:1251–1261. doi: 10.1016/j.humpath.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Funayama N, Fagotto F, McCrea P, Gumbiner BM. Embryonic axis induction by the armadillo repeat domain of beta-catenin: evidence for intracellular signaling. J Cell Biol. 1995;128:959–968. doi: 10.1083/jcb.128.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitaker HC, Girling J, Warren AY, Leung H, Mills IG, Neal DE. Alterations in beta-catenin expression and localization in prostate cancer. Prostate. 2008;68:1196–1205. doi: 10.1002/pros.20780. [DOI] [PubMed] [Google Scholar]
- 25.Slezak J, Truong M, Huang W, Jarrard D. HP1gamma expression is elevated in prostate cancer and is superior to Gleason score as a predictor of biochemical recurrence after radical prostatectomy. BMC cancer. 2013;13:148. doi: 10.1186/1471-2407-13-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W, Hennrick K, Drew S. A colorful future of quantitative pathology: validation of Vectra technology using chromogenic multiplexed immunohistochemistry and prostate tissue microarrays. Hum Pathol. 2013;44:29–38. doi: 10.1016/j.humpath.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Bauman TM, Nicholson TM, Abler LL, Eliceiri KW, Huang W, Vezina CM, Ricke WA. Characterization of fibrillar collagens and extracellular matrix of glandular benign prostatic hyperplasia nodules. PLoS one. 2014;9:e109102. doi: 10.1371/journal.pone.0109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholson TM, Ricke EA, Marker PC, Miano JM, Mayer RD, Timms BG, vom Saal FS, Wood RW, Ricke WA. Testosterone and 17beta-estradiol induce glandular prostatic growth, bladder outlet obstruction, and voiding dysfunction in male mice. Endocrinology. 2012;153:5556–5565. doi: 10.1210/en.2012-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauman TM, Sehgal PD, Johnson KA, Pier T, Bruskewitz RC, Ricke WA, Huang W. Finasteride treatment alters tissue specific androgen receptor expression in prostate tissues. Prostate. 2014;74:923–932. doi: 10.1002/pros.22810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abel EJ, Bauman TM, Weiker M, Shi F, Downs TM, Jarrard DF, Huang W. Analysis and validation of tissue biomarkers for renal cell carcinoma using automated high-throughput evaluation of protein expression. Hum Pathol. 2014;45:1092–1099. doi: 10.1016/j.humpath.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita N, Uemura H, Tsumatani K, Cho M, Hirao Y, Okajima E, Konishi N, Hiasa Y. E-cadherin and alpha-, beta- and gamma-catenin expression in prostate cancers: correlation with tumour invasion. Br J Cancer. 1999;79:1879–1883. doi: 10.1038/sj.bjc.6690299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arenas MI, Romo E, Royuela M, Fraile B, Paniagua R. E-, N- and P-cadherin, and alpha-, beta- and gamma-catenin protein expression in normal, hyperplastic and carcinomatous human prostate. Histochem J. 2000;32:659–667. doi: 10.1023/a:1004111331752. [DOI] [PubMed] [Google Scholar]
- 33.Rodrigues MM, Rema A, Gartner MF, Laufer-Amorim R. Role of adhesion molecules and proliferation hyperplasic, pre neoplastic and neoplastic lesions in canine prostate. Pak J Biol Sci. 2013;16:1324–9. doi: 10.3923/pjbs.2013.1324.1329. [DOI] [PubMed] [Google Scholar]
- 34.Lean FZ, Kontos S, Palmieri C. Expression of beta-catenin and mesenchymal markers in canine prostatic hyperplasia and carcinoma. J Comp Pathol. 2014;150:373–81. doi: 10.1016/j.jcpa.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Huang W, Hennrick K, Drew S. A colorful future of quantitative pathology: validation of Vectra technology using chromogenic multiplexed immunohistochemistry and prostate tissue microarrays. Hum Pathol. 2013;44:29–38. doi: 10.1016/j.humpath.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Mehta V, Schmitz CT, Keil KP, Joshi PS, Abler LL, Lin TM, Taketo MM, Sun X, Vezina CM. Beta- catenin (CTNNB1) induces Bmp expression in urogenital sinus epithelium and participates in prostatic bud initiation and patterning. Dev Biol. 2013;376:125–135. doi: 10.1016/j.ydbio.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bentley H, Hamdy FC, Hart KA, Seid JM, Williams JL, Johnstone D, Russell RG. Expression of bone morphogenetic proteins in human prostatic adenocarcinoma and benign prostatic hyperplasia. Br J Cancer. 1992;66:1159–1163. doi: 10.1038/bjc.1992.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang EY, Jeong MS, Park EK, Kim JH, Jang SB. Structural characterization and interaction of periostin and bone morphogenetic protein for regulation of collagen cross-linking. Biochem Biophys Res Commun. 2014;449:425–431. doi: 10.1016/j.bbrc.2014.05.055. [DOI] [PubMed] [Google Scholar]
- 39.DiRocco DP, Kobayashi A, Taketo MM, McMahon AP, Humphreys BD. Wnt4/beta-catenin signaling in medullary kidney myofibroblasts. J Am Soc Nephrol. 2013;24:1399–1412. doi: 10.1681/ASN.2012050512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren S, Johnson BG, Kida Y, Ip C, Davidson KC, Lin SL, Kobayashi A, Lang RA, Hadjantonakis AK, Moon RT, Duffield JS. LRP-6 is a coreceptor for multiple fibrogenic signaling pathways in pericytes and myofibroblasts that are inhibited by DKK-1. Proc Natl Acad Sci U S A. 2013;110:1440–1445. doi: 10.1073/pnas.1211179110. [DOI] [PMC free article] [PubMed] [Google Scholar]


