Abstract
Therapeutic delivery of cardiomyocytes derived from human pluripotent stem cells (hPSC-CMs) represents a novel clinical approach to regenerate the injured myocardium. However, poor survival and contractility of these cells are a significant bottleneck to their clinical use. To better understand the role of cell-cell communication in enhancing the phenotype and contractile properties of hPSC-CMs, we developed a three-dimensional (3D) hydrogel composed of hPSC-CMs, human pluripotent stem cell-derived endothelial cells (hPSC-ECs), and/or human amniotic mesenchymal stem cells (hAMSCs). The objective of this study was to examine the role of multi-cellular interactions among hPSC-ECs and hAMSCs on the survival and long-term contractile phenotype of hPSC-CMs in a 3D hydrogel. Quantification of spontaneous contractility of hPSC-CMs in tri-culture demonstrated a 6-fold increase in the area of contractile motion after 6 weeks with characteristic rhythmic contraction frequency, when compared to hPSC-CMs alone (P < 0.05). This finding was supported by a statistically significant increase in cardiac troponin T protein expression in the tri-culture hydrogel construct at 6 weeks, when compared to hPSC-CMs alone (P < 0.001). The sustained hPSC-CM survival and contractility in tri-culture was associated with a significant upregulation in the gene expression of L-type Ca2+ ion channel, Cav1.2, and the inward-rectifier potassium channel, Kir2.1 (P < 0.05), suggesting a role of ion channels in mediating these processes. These findings demonstrate that multi-cellular interactions modulate hPSC-CM phenotype, function, and survival, and they will have important implications in engineering cardiac tissues for treatment of cardiovascular diseases.
Keywords: Induced pluripotent stem cell, differentiation, cardiomyocyte, endothelial cell, mesenchymal stem cell, cardiac patch
Introduction
Cardiovascular disease is the leading cause of death in the US [1]. Current stem cell-based clinical trials have shown only moderate benefit in improving cardiac function [2]. Contributing factors to the limited therapeutic effect of implanted cells include ineffective electromechanical coupling and poor transplant cell survival. The native myocardium consists of multiple cell populations, including cardiomyocytes, endothelial cells, and stromal cells, which together exhibit an integral role in the synchronous behavior of the heart [3]. Accordingly, engineered human heart tissue that promotes multi-cellular interactions among these cell populations may simulate a cardiac syncytium that mimics the native myocardium [4,5] and lead to improved cardiac function.
Recent advances in cardiovascular differentiation of human pluripotent stem cells (hPSCs) including human embryonic stem cells (hESCs) and human induced pluripotent stem cell (hiPSCs) make them an extremely attractive and seemingly limitless cell source for the generation of engineered human heart tissue [6-12]. Towards the goal of developing a hPSC-based hydrogel construct to promote cardiac regeneration, the objective of this study was to examine the role of multi-cellular interactions among hPSC-derived cardiomyocytes (hPSC-CMs), hPSC-derived endothelial cells (hPSC-ECs), and human amniotic mesenchymal stem cells (hAMSCs) on the survival and long-term contractile phenotype of hPSC-CMs in a three-dimensional (3D) hydrogel platform. We demonstrate that multi-cellular interactions with hPSC-ECs and hAMSCs enhance hPSC-CM survival, electromechanical force generation, and contractility in vitro for at least 6 weeks.
Material and methods
Cardiac differentiation of hPSCs
The hPSC-CMs were derived from the human embryonic stem cell line H7 [13] (WA07, WiCell Research Institute) according to our previously described differentiation methods [9]. Briefly, the cells were grown in custom-made E8 media on Matrigel-coated dishes (Corning) and passaged with Ethylenediaminetetraacetic acid (EDTA) in the presence of thiazovivin (2 μM, Selleck Chemicals) for 24 hours [14-16]. The hPSC-CMs were derived using a modified small-molecule monolayer method using 6 μM CHIR99021 and 5 μM IWR-1-endo (both Selleck Chemicals) and Roswell Park Memorial Institute (RPMI) 1640 basal medium supplemented with 2% B27 without insulin (RPMI+B27-ins; Life Technologies) [6].
Endothelial differentiation of hPSCs
The hPSC-ECs were generated and characterized as described previously using hiPSCs reprogrammed from adult human dermal fibroblasts [10-12]. Briefly, confluent cultures of hiPSCs (HUF5) were incubated with type IV collagenase (Life Technologies) and transferred to ultra-low attachment dishes (Corning) containing differentiation media for 4 days to form embryoid bodies (EBs). The differentiation media consisted of α-Minimum Eagle’s Medium, 20% fetal bovine serum (FBS), 2-mercaptoethanol (50 μM), 1% non-essential amino acids (all from Life Technologies), bone morphogenetic protein 4 (BMP4; 50 ng/mL, Peprotech) and vascular endothelial growth factor A (VEGFA, 50 ng/mL, Peprotech). After 4 days, the EBs were then seeded on 0.2% gelatin-coated dishes and cultured for another 10 days in differentiation media in the absence of BMP4. Differentiation medium was changed every 48 hours for 14 days and then purified by fluorescence activated cell sorting (FACS) as described below.
Isolation of hAMSCs from human placenta
The hAMSCs were isolated from fresh human placenta as described in our previous studies [17]. Briefly, human placentas were obtained from healthy donors at the Stanford University Medical Center and placed in Hank’s Balanced Salt Solution (HBSS, Life Technologies) for transport. The amniotic membrane was separated from the chorion and washed 3 to 5 times in 0.9% NaCl. Membranes were dissected into 2 cm2 pieces and digested in trypsin-EDTA (Life Technologies) for 30 min at 37°C, 5% CO2. Digested tissue was centrifuged, the supernatant removed, and digested in HBSS with type 1 collagenase (1:1 weight to volume ratio, Life Technologies), 0.01% papain (Sigma-Aldrich) and 10% fetal bovine serum (FBS, Life Technologies) for 2 hours at 37°C, 5% CO2. Cells were filtered through a 70 μm sterile filter (BD Biosciences). Cells were centrifuged at 200 × g for 5 min. The collected hAMSCs were cultured in DMEM with 100 mg/L sodium pyruvate, 29.2 mg/ml L-glutamine in 0.85% NaCl, 10% FBS, 1% pen-strep and 10 ng/mL epidermal growth factor (R&D Systems).
Fluorescence activated cell sorting and flow cytometry
For flow cytometric analysis of hPSC-CM differentiation efficacy at day 15 of differentiation, cells were dissociated with TrypLE Express for 10 min at 37°C and transferred to flow cytometry tubes (BD Biosciences). Cells were then fixed with 1% paraformaldehyde, permeabilized with 90% methanol, and then incubated with TNNT2 (cardiac troponin T, Thermo Scientific), followed by secondary antibody incubation with Alexa Fluor-conjugated antibody (Life Technologies). Isotype-matched antibody served as a negative control. Cells were analyzed using a FACSAria II (BD Biosciences). Data were analyzed using FlowJo 8.7 (Tree Star).
The hPSC-ECs were purified at day 14 of endothelial differentiation according to our previous methods [12]. Briefly, differentiating cells were dissociated using accutase (Sigma-Aldrich, blocked with 5% bovine serum albumin (BSA), and then incubated with phycoerythrin-conjugated anti-human CD31 antibody (eBioscience). Isotype-matched antibody served as a negative control. Cells were sorted using a BD Digital Vantage cell sorter (BD Biosciences) and then expanded in culture in EGM-2MV (Lonza).
Immunofluorescence staining
The identity of hPSC-CMs, hPSC-ECs, and hMSCs was verified by immunofluorescence staining of phenotypic markers. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton-X (Sigma-Aldrich), and blocked in 10% goat serum (Sigma-Aldrich) or 1% BSA. For hPSC-CMs, the primary antibodies consisted of cardiac troponin T (Thermo Scientific) and α-actinin (Santa Cruz Biotechnology). For hPSC-ECs, the primary antibody consisted of VE-cadherin (CD144; Santa Cruz Biotechnology). For hAMSCs, Thy-1 (Biolegend) antibody was used. Following incubation in primary antibodies, the cells were then incubated with Alexa Fluor-conjugated secondary antibodies (Life Technologies). Cell nuclei were labeled with DAPI (Life Technologies).
Generation of hydrogel constructs
Growth factor-reduced Matrigel was placed on top of glass coverslips to create a hydrogel (200-500 μm thick) to allow for cell adhesion and migration. Matrigel was chosen to allow for proper cell growth for all cell types used. The cell ratio utilized was 5:1:1 hPSC-CM:hPSC-EC:hAMSC, based on preliminary studies showing that this ratio improved cell survival and contractility (data not included). Each hydrogel was seeded with 2.5 × 105 hPSC-CMs with the addition of 5 × 104 hPSC-ECs and/or 5 × 104 hAMSCs in RPMI+B27-insulin culture medium. The media was changed every 2 days.
Contractility analysis
At time points of 2, 4, and 6 weeks, movies of cell contractility within the engineered hydrogel constructs were captured at 640x480 resolution with a VistaVision inverted microscope (VWR) with a 10x objective at ~12-13 frames per second (n = 4). Video analysis of deformation and contractility was performed as described by Navarrete et al. [18]. Briefly, captured image stacks were analyzed frame-by-frame using a Fourier-based cross-correlation algorithm to identify movement of cell-seeded constructs. Average movement for each region over time was plotted to quantify contractile motion over time. Maximum contractile motion was calculated as the mean peak values for each trace. Percent beating was quantified as movement vectors that exceeded the detection limit of the algorithm (0.2 pixels) divided by the number of movement vectors calculated for the entire region.
Quantification of cardiac troponin t expression
For quantification of cardiac phenotype, each hydrogel construct composed of hPSC-CMs alone or in co-culture with hPSC-ECs and/or hAMSCs was immunofluorescently stained at 2, 4, and 6 weeks for cardiac troponin T and DAPI (n = 4). Images captured on a laser scanner confocal microscope. Cardiac troponin T expression was analyzed by Image J and expressed as the area of troponin T expression normalized to the area of total nuclei.
Atomic force microscopy (AFM) and calcium transients quantification
AFM was performed on a cluster of cells from hPSC-CM+hPSC-EC co-culture on Matrigel after 3 days. The constructs were cultured in 50 mm Fluorodish cell culture dishes (World Precision Instruments). Thirty minutes before simultaneous imaging by AFM and fluorescence microscopy, the cell media was replaced with Fluo-4 NW (Life Technologies) in assay buffer and incubated at 37°C on the AFM stage. Force measurements were acquired with a MFP3D-Bio AFM (Asylum Research) on an Eclipse Ti microscope base (Nikon) with a SHOCONG cantilever (Applied NanoStructures). The spring constant and of the cantilever was determined by the thermal method. The 488 laser used for epifluorescent illumination was controlled by Micro-Manager. Fluo-4 fluorescence images were acquired on an XR/Mega-10EX camera (Stanford Photonics) at 55 frames per second (2 × 2 binning). After engaging the cells with 200 pN of force, corresponding to less than 50 nm of indentation, force exerted by the cells was measured. The raw deflection voltage signal from the AFM controller was collected at 5 kHz in LabVIEW (National Instruments) along with digital pulses from camera acquisition acquired at 500 kHz to allow precise synchronization of force with calcium flux. Deflection voltage was multiplied by the spring constant and optical sensitivity of the cantilever to find the exerted force. The force data was de-noised in Matlab using wavelet filtering with the coif3 wavelet at level 5, yielding a pseudo-frequency of 706 Hz. To find the beating rate, peak-to-peak distances were measured with a peak finding algorithm in Matlab (http://www.mathworks.com/matlabcentral/fileexchange/25500-peakfinder).
Quantitative real-time polymerase chain reaction
To analyze gene expression, cells were dissociated from hydrogel constructs with TrypLE Express for 15 min at 37°C, triturated and diluted in RPMI+B27-ins, and centrifuged at 200xg for 4 min. Media was aspirated and pellets of cells were snap frozen in liquid nitrogen and stored at -80°C. RNA was isolated using an RNeasy Plus kit (Qiagen), and cDNA was produced using a High Capacity RNA-to-cDNA kit (Life Technologies). Quantitative PCR was performed using TaqMan Gene Expression Assays (Life Technologies, Table 1) and TaqMan Gene Expression Master Mix using a 7900HT Real-Time PCR System (all Life Technologies). All PCR reactions were normalized to the 18S endogenous control gene, and assessed using the comparative Ct method (n = 3).
Table 1.
PCR Primers
| Category | Gene symbol | TaqMan Assay Primer/Probe |
|---|---|---|
| Housekeeping | 18S | Hs99999901_s1 |
| Pluripotency | POU5F1 | Hs00999632_g1 |
| NANOG | Hs04260366_g1 | |
| Endothelial | PECAM1 | Hs00169777_m1 |
| VEGFA | Hs00900055_m1 | |
| CDH5 | Hs00901463_m1 | |
| Gap junction | GJC1 | Hs00271416_s1 |
| GJA1 | Hs00748445_s1 | |
| Cardiac development | NKX2-5 | Hs00231763_m1 |
| ISL1 | Hs00158126_m1 | |
| TNNT2 | Hs00943911_m1 | |
| Ion channels | CACNA1C | Hs00167681_m1 |
| KCNJ2 | Hs01876357_s1 |
Statistical analysis
Data are presented as mean ± standard deviation. Gene and protein expression between groups were analyzed using one way analysis of variance, with significant differences defined by P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***). Contractile property at various time points was analyzed using a two-way ANOVA with Tukey-Kramer honestly significantly different (HSD) test.
Results
Characterization of cellular phenotype
The hPSC-CMs, hPSC-ECs, and hAMSCs were previously characterized according to our previous studies [9,12,17]. Confirmation of the identity of the three cell populations was performed by conducting cellular morphology and immunocytochemical analysis (Figure 1A-D). The hPSC-CMs expressed phenotypic markers for cardiac troponin T and α-actinin, and flow cytometry demonstrated that populations were > 90% positive for troponin T (Figure 1A, 1B). The hPSC-ECs expressed phenotypic marker VE-Cadherin (CD144) (Figure 1C). The hAMSCs were previously purified by the positive expression of SSEA3, SSEA4, TRA-1-81, Thy-1, and c-kit [17]. Consistent with the purification markers used, the hAMSCs were morphologically elongated and expressed mesenchymal marker Thy1 (CD90) (Figure 1D). Using these highly purified populations of hPSC-CMs, hPSC-ECs, and hAMSCs, we quantified the role of multi-cellular interactions using a 5:1:1 ratio of hPSC-CM:hPSC-EC:hAMSCs, along with control constructs containing hPSC-CMs only or in co-culture with hPSC-ECs or hAMSCs. Each construct was assayed for cardiac function and survival based on cardiac troponin T expression, contractile area using time lapse imaging, gene expression of pluripotency, gap junctions, and cardiac and endothelial phenotype, and force generation and calcium transients using AFM (Figure 1E).
Figure 1.
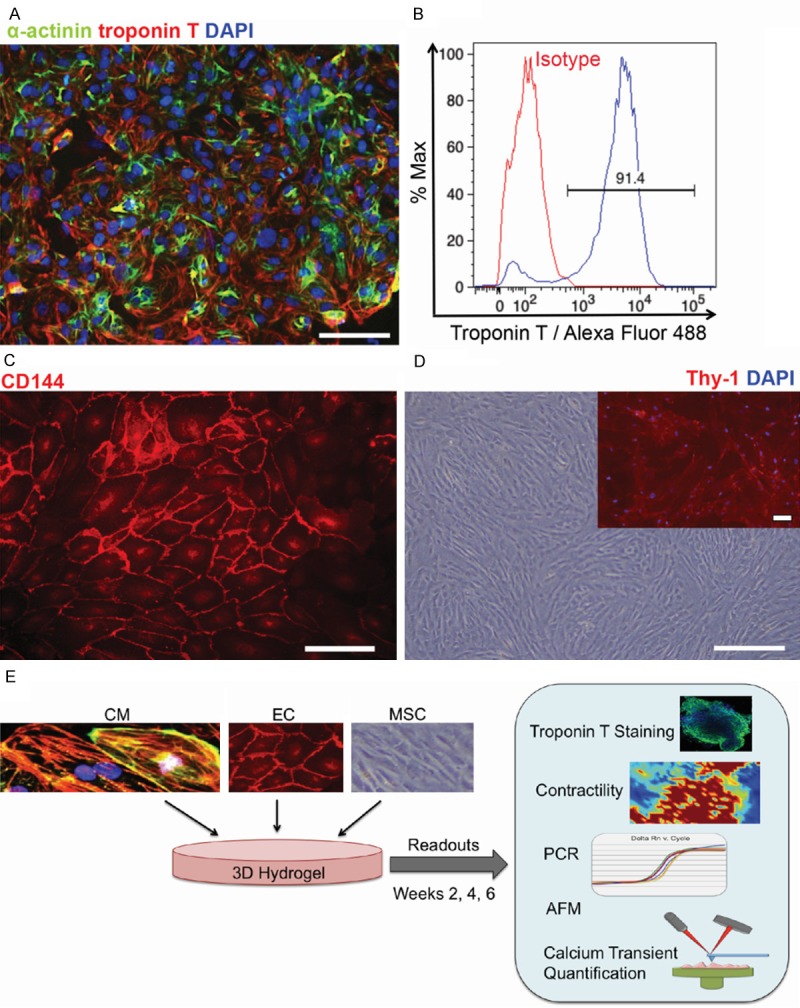
Phenotypic expression of of hPSC-derived lineages and hAMSCs. A: Immunofluorescence staining of hPSC-CMs at day 15 of differentiation for cardiac troponin T and α-actinin. B: Flow cytometric analysis of hPSC-CMs for Troponin T expression. C: Immunofluorescence staining of hPSC-ECs for endothelial marker VE-Cadherin (CD144). D: Phase contrast images of hAMSCs and immunofluorescence staining of Thy-1 (inset). E: Schematic of experimental design in which hPSC-CMs (CM), hPSC-ECs (EC), and hAMSCs (MSC) are cultured in 3D hydrogel for analysis of contractile function, calcium transients, AFM, and gene expression. Scale bar, 100 μm.
Multi-cellular interactions sustain cardiac troponin t expression
Cardiac troponin T expression within the various constructs was assessed by quantitative immunofluorescence staining. Representative confocal images of troponin T in hydrogels consisting of hPSC-CMs only, co-cultures (hPSC-CMs + hPSC-ECs or hPSC-CMs + hAMSCs), and tri-culture (hPSC-CMs + hPSC-ECs + hAMSCs) are demonstrated in Figure 2A-L. Whereas there was a progressive decrease in the area of troponin T in hPSC-CMs alone over the course of 6 weeks, there was a relative increase in cardiac troponin-T expression in the tri-culture hydrogel. Moreover, troponin T expression in the co-culture with hPSC-ECs + hPSC-CMs (P < 0.05) and the tri-culture (P < 0.001) demonstrated statistically higher troponin T expression, when compared to hPSC-CMs alone at 6 weeks in culture. However, the co-culture with hAMSCs + hPSC-CMs at 6 weeks was not statistically significant compared to hPSC-CM culture, suggesting that an increase in troponin expression was dependent on the interaction of hPSC-ECs with hPSC-CMs, and that the expression was further enhanced with interactions with hAMSCs.
Figure 2.
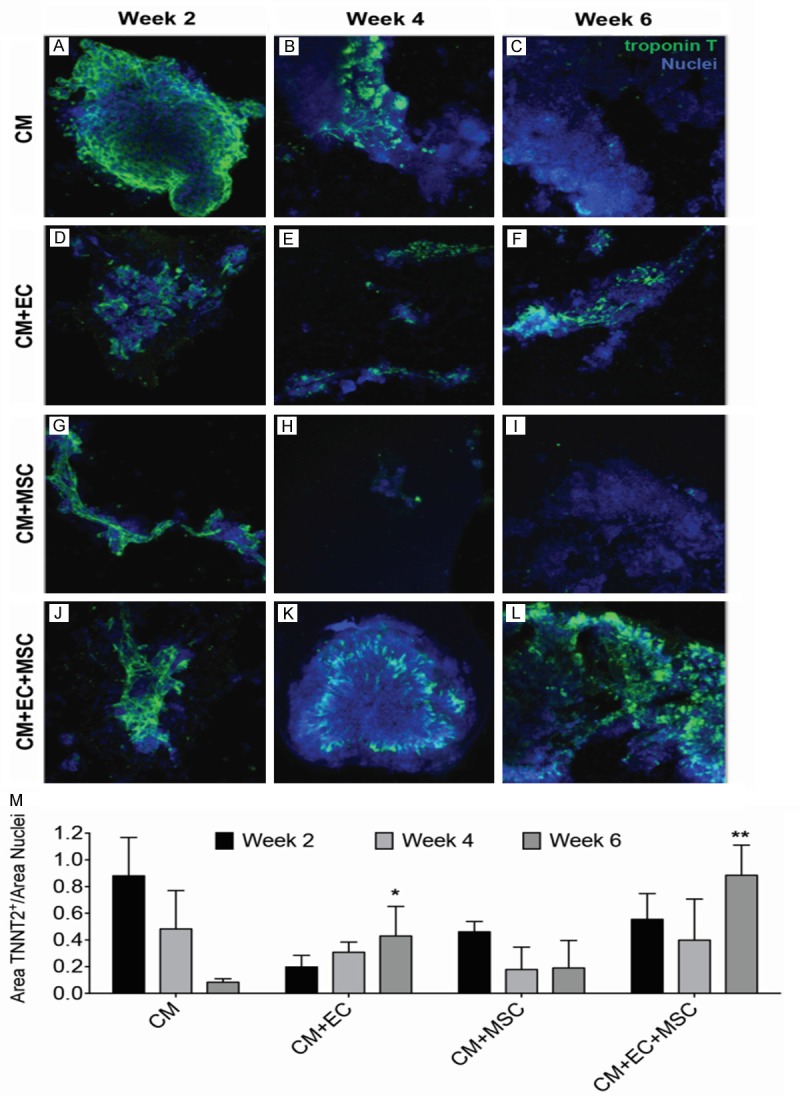
Effect of multi-cellular culture conditions on cardiac troponin T expression in 3D hydrogels. A-L. Representative images of cardiac troponin T expression after 2, 4, or 6 weeks of culture showing hPSC-CMs (CM) alone, CM and hPSC-ECs (CM+EC), CM and hAMSCs (CM+MSC), and CM+EC+MSC. M. Effects of long-term (2, 4, 6 weeks) multi-cellular culture on troponin T protein expression (n = 4). Statistically significant comparisons denoted by *P < 0.05, **P < 0.01, when compared to CM at week 6.
Multi-cellular interactions sustain hPSC-CM contractility
Although hPSC-CMs alone had the highest percentage (> 90%) of contractile regions at 2 weeks, by 6 weeks only 20% of the cells remained contractile (Figure 3A). In contrast, the mean magnitude of contractility for tri-culture constructs at week 6 was nearly nine times higher than that of hPSC-CMs alone (P < 0.05, Figure 3B). Strikingly, the tri-culture constructs at week 6 still displayed rhythmic beating while other cell combinations exhibited minimal contractility (Figure 3C). Furthermore, two-way ANOVA analysis demonstrated a statistical difference for cell type(s) (P < 0.001) as well as duration in culture (P < 0.001). There was a significant and synergistic interaction effect (P < 0.05) between the cell type(s) and time in culture, suggesting that both cell type(s) and incubation period in culture impact contractility. This data suggested that multi-cellular interactions with hPSC-ECs and hAMSCs were required for sustained hPSC-CM contractility for up to 6 weeks.
Figure 3.
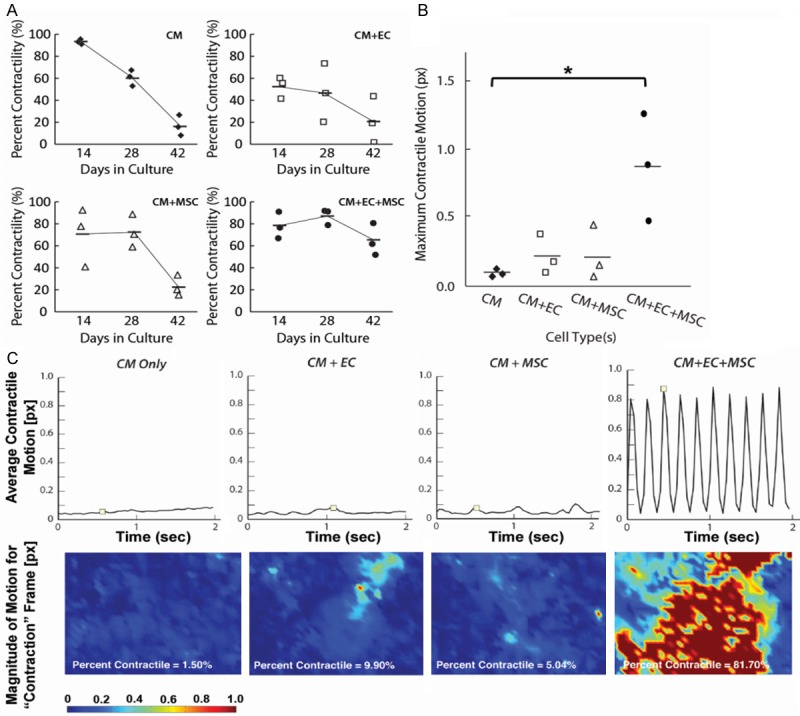
Effect of multi-cellular culture conditions on contractility in 3D hydrogels. A: Percent hPSC-CM contractility as a function of time in multi-cellular culture. B: Maximum contractile motion after 6 weeks in multi-cellular hydrogel constructs (*P < 0.05). C: Average contractile motion trace shows average motion of cells for entire field of view (top), and magnitude panels (below) show spatial distribution of contractile activity for frames marked above (n = 3). Abbreviations: hPSC-CM (CM), hPSC-EC (EC), hAMSC (MSC).
Multi-cellular interactions modulate gene expression
To better understand the mechanism by which multi-cellular interactions among hPSC-CMS, hPSC-ECs, and hAMSCs supported stained contractility of hPSC-CMs, we examined the expression of genes associated with cell phenotype or function, including genes that are related to cardiac development, gap junctional proteins, and ion channels (Figure 4). As expected, there was a significant (P < 0.05) increase in the expression of endothelial markers PECAM1 and VE-cadherin in the multi-cellular conditions containing hPSC-ECs (Figure 4A). However, we did not observe significant differences in gene expression for CM structural genes troponin T (TNNT2) and β-myosin heavy chain (MYH7), or for mature CM subtype-specific markers myosin light chain-2 (MYL2) and myosin light chain-7 (MYL7). Notably, there was a significant increase in the expression of the voltage dependent L-type Ca2+ ion channel Cav1.2 (CACNA1C) and the inward-rectifier potassium channel Kir2.1 (KCNJ2) in tri-culture (P < 0.05), suggesting that down-regulation of ion channels might partially account for the reduced contraction seen in the hPSC-CM alone hydrogels (Figures 3 and 4B).
Figure 4.
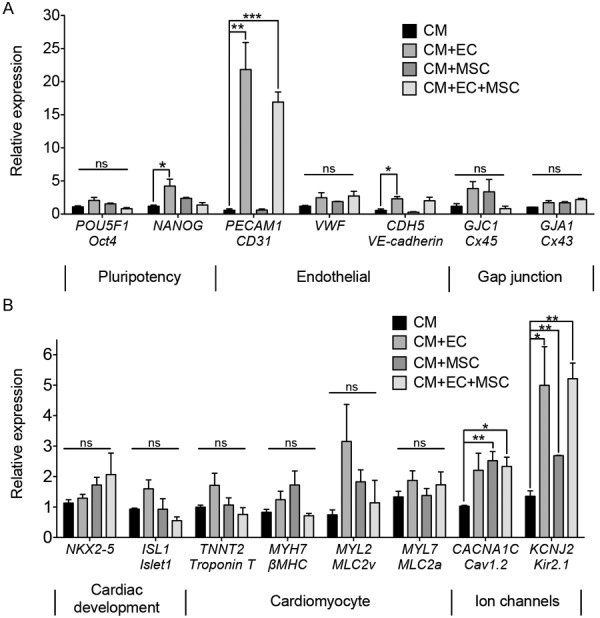
Effect of multi-cellular culture conditions on gene expression in 3D hydrogels after 2 weeks in culture. A: Expression of genes for pluripotency markers, endothelial cells, and gap junctions. B: Expression of genes for cardiac markers (n = 3). Statistically significant comparisons denoted by * = P < 0.05, ** = P < 0.01, *** = P < 0.005. Abbreviations: hPSC-CM (CM), hPSC-EC (EC), hAMSC (MSC).
Force generation and calcium transients
To determine whether the hPSC-CMs resembled primary CMs in function, we quantified force generation and calcium transients using AFM. Since hPSC-CM contractility was similar in all treatment groups at early time points, we selected only one of the groups (hPSC-CM+hPSC-EC) for assessment of force generation and calcium transients. After 3 days in co-culture with hPSC-ECs in the hydrogel construct, the hPSC-CMs were found to have a well-coordinated contractile period of 1.62 ± 0.17 s, corresponding to a 0.624 ± 0.64 Hz beat frequency (Figure 5). To compare the contractility rate measured by AFM to optical microscopy, calcium flux was measured by fluo-4 fluorescence simultaneously with the AFM force measurements. At an imaging rate of 55 fps, the contractile frequency of 0.596 ± 0.059 Hz (1.69 ± 0.16 s period) was measured. The peak force exerted by the cell cluster as measured by AFM was almost 3 nN. Notably, this force and contractile rate was highly regular and corresponds well to previous results with hPSC-CM clusters [19], suggesting that interactions of hPSC-CMs with other cell types does not interfere with their contractile function.
Figure 5.
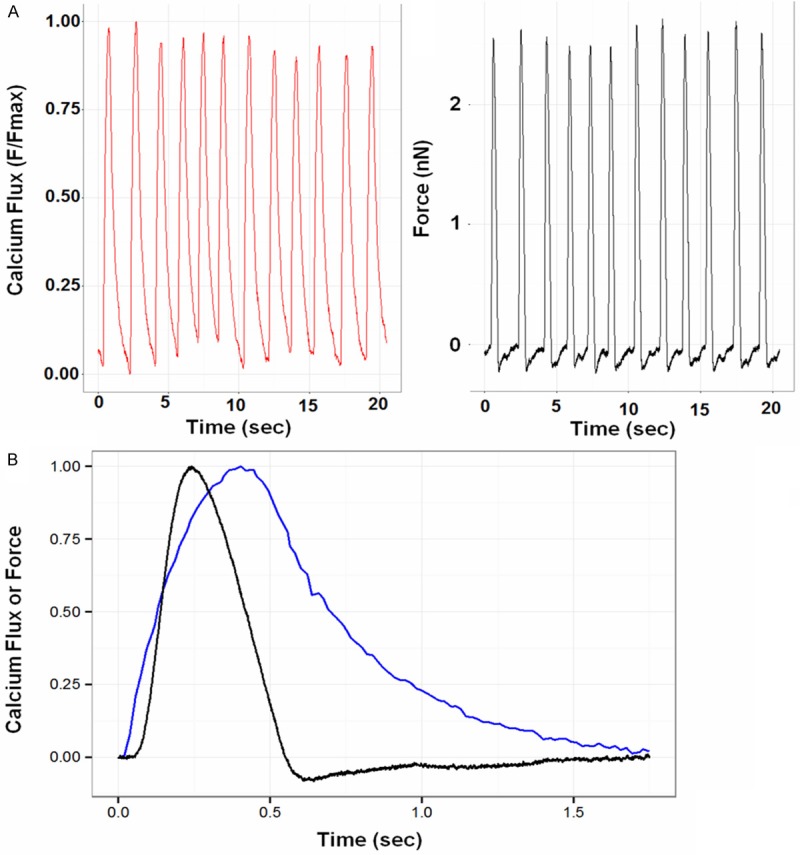
Atomic force microscopy (AFM) and calcium transients measurement. A: AFM of hPSC-CMs co-cultured with hPSC-ECs on day 3 in 3D hydrogel. B: Corresponding AFM force displacement curves with calcium transients (Flux = blue, force = black).
The AFM and fluorescence microscopy data were directly aligned to measure the temporal lag between force generation and calcium flux. Calcium flux was found to precede the force generation by 53 ± 6 ms. However, the maximum exerted force preceded maximum calcium flux by 170 ± 20 ms. At maximum force, calcium flux reached an average of 76 ± 3% of its maximum. At maximum calcium flux, force had dropped to 56 ± 9% of its maximum. At minimum force, calcium flux fell to 56 ± 7% of its maximum. These results indicate that calcium flux precedes force generation, and the force generation cycle is largely completed before the calcium flux cycle is completed.
Discussion
The salient findings of this study are that multi-cellular interactions of hPSC-CMs with hPSC-ECs and hAMSCs significantly enhance troponin T expression (Figure 2) and sustain synchronous contractile motion (Figure 3), which could be in part due to a significant increased gene expression of the voltage dependent L-type Ca2+ ion channel Cav1.2 and the inward-rectifier potassium channel Kir2.1 (Figure 4). In co-culture with hPSC-ECs, the force generation and calcium transients of hPSC-CMs are consistent with values previously reported from hPSC-CM clusters (Figure 5), suggesting that cellular interactions of hPSC-CMs with other cell types do not affect cell function. A persistent challenge in engineering a cardiac tissue is the combinatorial complexity in recapitulating the physiological composition and interactions of the multiple cell populations involved. The data from this study demonstrate the benefits of multi-cellular interactions of hPSC-CMs with hPSC-ECs and hAMSCs. A prior study reported that the tri-culture of neonatal cardiomyocytes cultured onto micropatterned substrates pre-seeded with fibroblasts and endothelial cells generated morphologically and functionally superior cardiac organoids, when compared to primary cardiomyocytes co-cultured with either fibroblasts or endothelial cells [20]. Caspi et al. demonstrated that multicellular interactions of hPSC-CMs with hPSC-ECs and murine embryonic fibroblasts in a porous scaffold for 2 weeks significantly induced the expression of cardiac-specific genes and enhanced action potential propagation, and these findings concur with those from this current study [21]. Our investigation further evaluated the maintenance of the electromechanical, contractile, and physiologic properties of hPSC-CMs for up to 6 weeks. Although the mechanism by which multi-cellular interactions with hPSC-ECs and hAMSCs modulate cardiac phenotype is not well-known, it is likely due to cell-cell interactions that may promote electromechanical property or paracrine factors released by vascular or stromal cells [4,22].
It is well-recognized that cardiac cells physiologically exist in a complex 3D environment that is not adequately mimicked in two-dimensional (2D) Petri dishes [23]. Our 3D hydrogel system enables the hPSC-CMs to exert cell-cell and cell-ECM interactions that better represent the physiological myocardial environment, which may account for the improvement in cardiac function. Our results are in agreement with other studies that demonstrate that 3D microenvironment can enhance cardiac phenotype by increasing cell contractility and expression of phenotypic markers, when compared to 2D environments [24,25].
Conclusion
In summary, this study demonstrates the role of multi-cellular interactions of hPSC-CMs with hPSC-ECs and hAMSCs in a 3D hydrogel construct. In tri-culture, the constructs demonstrated improved contractile behavior and survival. These findings will facilitate the translation of the hPSC-CMs in multi-cellular culture platform for potential novel therapeutic approach in patients with advanced heart failure.
Acknowledgements
This study was supported in part by grants from the National Institutes of Health, Cardiovascular Cell Therapy Research Network 5 UM 1HL087318-08 (PCY), the National Institutes of Health 5R01HL097516-02 (PCY) and R00HL098688 (NFH), the National Science Foundation 1249008 (NFH), the Department of Defense W81XWH-12-C-0111 (NFH), Merit Review Award 1I01BX002310 from the Department of Veterans Affairs Biomedical Laboratory Research and Development (NFH), and the Stanford Cardiovascular Institute (PCY and NFH), and the American Heart Association 12POST12050254 (PWB).
Disclosure of conflict of interest
None to declare.
References
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Magid D, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, Moy CS, Mussolino ME, Nichol G, Paynter NP, Schreiner PJ, Sorlie PD, Stein J, Turan TN, Virani SS, Wong ND, Woo D, Turner MB American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics--2013 update: A report from the american heart association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Telukuntla KS, Suncion VY, Schulman IH, Hare JM. The advancing field of cell-based therapy: Insights and lessons from clinical trials. J Am Heart Assoc. 2013;2:e000338. doi: 10.1161/JAHA.113.000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee I, Fuseler JW, Price RL, Borg TK, Baudino TA. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am J Physiol Heart Circ Physiol. 2007;293:H1883–1891. doi: 10.1152/ajpheart.00514.2007. [DOI] [PubMed] [Google Scholar]
- 4.Narmoneva DA, Vukmirovic R, Davis ME, Kamm RD, Lee RT. Endothelial cells promote cardiac myocyte survival and spatial reorganization: Implications for cardiac regeneration. Circulation. 2004;110:962–968. doi: 10.1161/01.CIR.0000140667.37070.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsieh PC, Davis ME, Lisowski LK, Lee RT. Endothelial-cardiomyocyte interactions in cardiac development and repair. Annu Rev Physiol. 2006;68:51–66. doi: 10.1146/annurev.physiol.68.040104.124629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical wnt signaling. Proc Natl Acad Sci U S A. 2012;109:E1848–1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, Barron MR, Hou L, Soerens AG, Yu J, Palecek SP, Lyons GE, Thomson JA, Herron TJ, Jalife J, Kamp TJ. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: The matrix sandwich method. Circ Res. 2012;111:1125–1136. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: Human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang NF, Dewi RE, Okogbaa J, Lee JC, Jalilrufaihah A, Heilshorn SC, Cooke JP. Chemotaxis of human induced pluripotent stem cell-derived endothelial cells. Am J Transl Res. 2013;5:510–520. [PMC free article] [PubMed] [Google Scholar]
- 11.Rufaihah AJ, Huang NF, Kim J, Herold J, Volz KS, Park TS, Lee JC, Zambidis ET, Reijo-Pera R, Cooke JP. Human induced pluripotent stem cell-derived endothelial cells exhibit functional heterogeneity. Am J Transl Res. 2013;5:21–35. [PMC free article] [PubMed] [Google Scholar]
- 12.Rufaihah AJ, Huang NF, Jame S, Lee JC, Nguyen HN, Byers B, De A, Okogbaa J, Rollins M, Reijo-Pera R, Gambhir SS, Cooke JP. Endothelial cells derived from human ipscs increase capillary density and improve perfusion in a mouse model of peripheral arterial disease. Arterioscler Thromb Vasc Biol. 2011;31:e72–79. doi: 10.1161/ATVBAHA.111.230938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 14.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM, Thomson JA. Chemically defined conditions for human ipsc derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, Carpenter MK. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 16.Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA, Chen G. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat Protoc. 2012;7:2029–2040. doi: 10.1038/nprot.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge X, Wang IN, Toma I, Sebastiano V, Liu J, Butte MJ, Reijo Pera RA, Yang PC. Human amniotic mesenchymal stem cell-derived induced pluripotent stem cells may generate a universal source of cardiac cells. Stem Cells Dev. 2012;21:2798–2808. doi: 10.1089/scd.2011.0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navarrete EG, Liang P, Lan F, Sanchez-Freire V, Simmons C, Gong T, Sharma A, Burridge PW, Patlolla B, Lee AS, Wu H, Beygui RE, Wu SM, Robbins RC, Bers DM, Wu JC. Screening drug-induced arrhythmia events using human induced pluripotent stem cell-derived cardiomyocytes and low-impedance microelectrode arrays. Circulation. 2013;128(Suppl 1):S3–13. doi: 10.1161/CIRCULATIONAHA.112.000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Sun N, Bruce MA, Wu JC, Butte MJ. Atomic force mechanobiology of pluripotent stem cell-derived cardiomyocytes. PLoS One. 2012;7:e37559. doi: 10.1371/journal.pone.0037559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iyer RK, Chiu LL, Radisic M. Microfabricated poly (ethylene glycol) templates enable rapid screening of triculture conditions for cardiac tissue engineering. J Biomed Mater Res A. 2009;89:616–631. doi: 10.1002/jbm.a.32014. [DOI] [PubMed] [Google Scholar]
- 21.Caspi O, Lesman A, Basevitch Y, Gepstein A, Arbel G, Habib IH, Gepstein L, Levenberg S. Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circ Res. 2007;100:263–272. doi: 10.1161/01.RES.0000257776.05673.ff. [DOI] [PubMed] [Google Scholar]
- 22.Sekine H, Shimizu T, Hobo K, Sekiya S, Yang J, Yamato M, Kurosawa H, Kobayashi E, Okano T. Endothelial cell coculture within tissue-engineered cardiomyocyte sheets enhances neovascularization and improves cardiac function of ischemic hearts. Circulation. 2008;118(Suppl 14):S145–152. doi: 10.1161/CIRCULATIONAHA.107.757286. [DOI] [PubMed] [Google Scholar]
- 23.Kawaguchi N, Hatta K, Nakanishi T. 3d-culture system for heart regeneration and cardiac medicine. Biomed Res Int. 2013;2013:895967. doi: 10.1155/2013/895967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pontes Soares C, Midlej V, de Oliveira ME, Benchimol M, Costa ML, Mermelstein C. 2d and 3d-organized cardiac cells shows differences in cellular morphology, adhesion junctions, presence of myofibrils and protein expression. PLoS One. 2012;7:e38147. doi: 10.1371/journal.pone.0038147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akins RE Jr, Rockwood D, Robinson KG, Sandusky D, Rabolt J, Pizarro C. Three-dimensional culture alters primary cardiac cell phenotype. Tissue Eng Part A. 2010;16:629–641. doi: 10.1089/ten.tea.2009.0458. [DOI] [PMC free article] [PubMed] [Google Scholar]


