Abstract
MicroRNAs (miRNAs) act as an oncogene or a tumor suppressor by negatively regulating target genes. Genetic variants in miRNA genes confer susceptibility to cancer and risk of death in cancer patients. The aim of this study was to investigate whether miRNA polymorphisms were associated with survival in breast cancer patients. Five miRNA polymorphisms (miR-26a1 rs7372209, miR-125a rs12976445, miR-218 rs11134527, miR-423 rs6505162, and miR-608 rs4919510) were genotyped in 196 breast cancer patients. We found that miR-125a rs12976445 was significantly associated with survival in codominant, recessive, and dominant models. However, only association under the codominant model remained significant after adjustment for lymph node metastasis, TNM stage, estrogen receptor, and progesterone receptor. Furthermore, this effect remained in stratification analysis. In conclusion, our results provide evidence that miR-125a rs12976445 may serve as a prognostic biomarker for breast cancer. Further large-scale studies are required to confirm these findings.
Keywords: MicroRNA, miR-125a, polymorphism, breast cancer, prognosis
Introduction
Breast cancer remains the most frequently diagnosed cancer and the leading cause of cancer-related death worldwide in women [1], despite the improvement in adjuvant treatment and screening. Although there are varieties of treatment options for breast cancer, the choice of therapy in the metastatic setting remains limited. Some biomarkers, such as estrogen receptor (ER), progesterone receptor (PR), have been widely used to help guide treatment of breast cancer. However, many patients suffer a cancer relapse and eventually die of metastatic disease. Therefore, it is important to identify reliable prognostic biomarkers that can distinguish patients with a favorable or poor prognosis.
MicroRNAs (miRNAs) are a class of endogenous small non-coding RNA found in animals, plants, and other diverse eukaryotes as well as a number of DNA viruses. miRNAs negatively regulate gene expression at the post-transcriptional level by binding to the 3’-UTR of their target genes. To date, there are 2588 human mature microRNAs in miRBase 21 database [2]. Since a single miRNA can target hundreds of genes, miRNAs participate in the regulation of almost every cellular process. Aberrant expression of miRNA is involved in numerous human disease, including cancer [3-5]. Many studied have demonstrated that dysregulation of miRNAs not only affects carcinogenesis, but also promotes cancer progression and metastasis [3,6-8]. miRNAs have great potential to serve as biomarkers for cancer detection, diagnosis and prognosis [6,7,9-11].
Previous studied have revealed that single nucleotide polymorphisms (SNPs) in miRNA genes affect mature miRNA expression and disrupt the ability of miRNAs to target genes [12-15]. miRNA SNPs are not only responsible for susceptibility to cancer, but also influence cancer prognosis [9,16-18]. In the present study, we examined the relationship between five miRNA SNPs (miR-26a1 rs7372209, miR-125a rs12976445, miR-218 rs11134527, miR-423 rs6505162, and miR-608 rs4919510) and survival in breast cancer patients in a Chinese Han population.
Materials and methods
Patients
We retrospectively collected data of breast cancer patients after surgical resection of the primary tumor between 2001 and 2003. A total of 196 breast cancer patients were included in the final analysis after excluding patients with a history of cancer, other than breast cancer, or without survival status or clinical information. All specimens were archived in the Biobank of National Engineering Center for Biochip at Shanghai. The written informed consent was obtained from all patients before surgery, and the study protocol was approved by the Ethics Committees of Taizhou People’s Hospital and National Engineering Center for Biochip at Shanghai.
Genotyping
Genomic DNA was extracted from five sections (5 µm each) of each FFPE tissue specimen using QIAamp DNA FFPE Tissue Kit (Qiagen, German) according to the manufacturer’s protocol. The concentration and quality of genomic DNA was measured by NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, DE, USA) and then stored at -80°C until use. All SNPs were genotyped using polymerase chain reaction-ligation detection reaction (PCR-LDR) method as described previously [19].
Statistical analyses
Genotype frequencies of all SNPs were test for Hardy-Weinberg equilibrium using a goodness-of-fit Chi square test with 1 df. Codominant, recessive and dominant models were used for each SNP. For the codominant model, the common homozygote was used as reference group. The primary endpoint was overall survival (OS), which was calculated from the day of surgery to the date of the last follow-up or date of death due to any cause. OS estimates were calculated by using the Kaplan-Meier method, and the differences in OS between genotypes were compared by using log-rank tests. Hazard ratio (HR) and their 95% confidence intervals (CIs) of the effect for each SNP on the OS of breast cancer were calculated by using Multivariate Cox-proportional hazard model and were adjusted for lymph node metastasis (LNM), TNM stage, estrogen receptor status, and progesterone receptor status. All analyses were performed using SPSS software package (SPSS Inc., Chicago, IL, USA). All analyses were two-sided, and a P value < 0.05 was considered as statistically significant.
Results
Patient characteristics
The clinical and pathologic characteristics of 196 breast cancer patients included in the analyses were listed in Table 1. The median age at diagnosis was 51 years (range, 29-83 years). Among these patients, 115 (58.7%) were ER-positive, 94 (48.0%) were PR-positive. The duration of follow-up ranged from 2.0 to 160.0 months with a median follow-up of 114.0 months. During the follow-up period, 55 (28.1%) patients were died. LNM, TNM stage, ER status, and PR status were significantly associated with survival time of patients (log-rank P < 0.05).
Table 1.
Clinical characteristics of breast cancer patients
| Characteristic | Number | % |
|---|---|---|
| Age (year) | 53.3 ± 12.7 | |
| Family history of cancer | ||
| Yes | 59 | 30.1 |
| No | 124 | 63.3 |
| Known | 13 | 6.6 |
| Menopausal | ||
| Premenopausal | 104 | 53.1 |
| Postmenopausal | 92 | 46.9 |
| LNM | ||
| Negative | 79 | 40.3 |
| Positive | 113 | 57.7 |
| Unknown | 4 | 2.0 |
| TNM | ||
| I | 22 | 11.2 |
| II | 112 | 57.1 |
| III | 56 | 28.6 |
| Unknown | 6 | 3.1 |
| ER status | ||
| Positive | 115 | 58.7 |
| Negative | 61 | 31.1 |
| Unknown | 20 | 10.2 |
| PR status | ||
| Positive | 94 | 48.0 |
| Negative | 83 | 42.3 |
| Unknown | 19 | 9.7 |
Association between miRNA SNPs and OS
Genotype frequencies of miR-26a1 rs7372209 deviated from Hardy Weinberg equilibrium (P = 0.001), and thus miR-26a1 rs7372209 was excluded from further analysis. The associations of miRNA SNPs with OS of breast cancer patients were presented in Table 2. In the univariate analysis of various clinical features for OS, LNM (HR = 2.200, 95% CI 1.215-3.984, P = 0.009), TNM stage III (HR = 2.609, 95% CI 1.521-4.475, P < 0.001), positive ER (HR = 0.379, 95% CI 0.216-0.666, P = 0.001), positive PR (HR = 0.446, 95% CI 0.249-0.798, P = 0.007), and miR-125a rs12976445 T allele (TT vs CC, HR = 4.024, 95% CI 1.544-10.483, P = 0.004; TT+CT vs CC, HR = 1.709, 95% CI 1.006-2.901, P = 0.047; TT vs CC+CT, HR = 3.357, 95% CI 1.338-8.422, P = 0.010) were associated with worse survival (Figure 1). In the multivariate analysis, only TNM stage (HR = 1.661, 95% CI 1.132-2.436, P = 0.009) and miR-125a rs12976445 TT genotype (HR = 3.964, 95% CI 1.070-14.686, P = 0.039) were independent unfavorable prognostic factors for breast cancer patients.
Table 2.
Univariate and multivariate Cox regression analysis of overall survival in breast cancer patients
| Characteristics | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age (years), ≤ 50 vs > 50 | 0.612 (0.355-1.054) | 0.077 | ||
| Family history of cancer, Yes vs No | 0.650 (0.399-1.247) | 0.195 | ||
| Menopausal, Premenopausal vs Postmenopausal | 1.404 (0.819-2.409) | 0.217 | ||
| LNM, Positive vs Negative | 2.200 (1.215-3.984) | 0.009 | 1.127 (0.494-2.568) | 0.776 |
| TNM, III vs I+II | 2.609 (1.521-4.475) | < 0.001 | 1.661 (1.132-2.436) | 0.009 |
| ER, Positive vs Negative | 0.379 (0.216-0.666) | 0.001 | 0.519 (0.238-1.129) | 0.098 |
| PR, Positive vs Negative | 0.446 (0.249-0.798) | 0.007 | 0.685 (0.308-1.525) | 0.355 |
| rs11134527 | ||||
| AA | 1 | |||
| AG | 1.456 (0.658-3.220) | 0.354 | ||
| GG | 1.187 (0.660-2.135) | 0.567 | ||
| Dominant model | 1.321 (0.646-2.701) | 0.445 | ||
| Recessive model | 1.245 (0.714-2.170) | 0.441 | ||
| rs12976445 | ||||
| CC | 1 | 1 | ||
| CT | 1.526 (0.876-2.658) | 0.135 | 1.618 (0.884-2.960) | 0.119 |
| TT | 4.024 (1.544-10.483) | 0.004 | 3.964 (1.070-14.686) | 0.039 |
| Dominant model | 1.709 (1.006-2.901) | 0.047 | 1.702 (0.953-3.041) | 0.072 |
| Recessive model | 3.357 (1.338-8.422) | 0.010 | 2.134 (0.631-7.224) | 0.223 |
| rs6505162 | ||||
| CC | 1 | |||
| AC | 0.636 (0.154-2.636) | 0.533 | ||
| AA | 0.660 (0.358-1.215) | 0.182 | ||
| Dominant model | 0.657 (0.367-1.175) | 0.157 | ||
| Recessive model | 0.723 (0.176-2.968) | 0.653 | ||
| rs4919510 | ||||
| CC | 1 | |||
| GG | 1.424 (0.652-3.112) | 0.375 | ||
| CG | 1.318 (0.721-2.408) | 0.370 | ||
| Dominant model | 1.344 (0.759-2.382) | 0.311 | ||
| Recessive model | 1.210 (0.610-2.402) | 0.585 | ||
Figure 1.
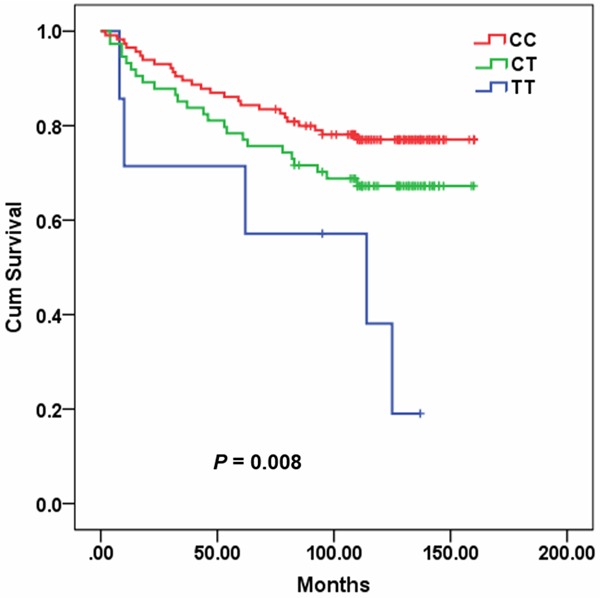
Plot of overall survival curve in breast cancer patients according to rs12976445 genotype.
Stratified analysis
To further evaluate the effect of miR-125a rs12976445 on OS of breast cancer patient, stratification analyses were performed according to age, family history of cancer, menopausal, LNM, TNM stage, PR, and ER. In the different subgroups of patients, there was significant relationship between miR-125a rs12976445 and OS of breast cancer patients under codominant, recessive, or dominant models (Table 3).
Table 3.
Stratification analysis of rs12976445 genotype associated with survival of breast cancer patients
| Characteristics | Codominant model | Dominant model | Recessive model | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | |
| Age (years) | ||||||
| ≤ 50 | 4.212 (0.932-19.048) | 0.062 | 1.350 (0.573-3.179) | 0.492 | 3.975 (0.924-17.108) | 0.064 |
| > 50 | 3.854 (1.113-13.346) | 0.033 | 2.043 (1.037-4.022) | 0.039 | 2.919 (0.890-9.571) | 0.077 |
| Family history of cancer | ||||||
| Yes | 2.283 (0.281-18.565) | 0.440 | 1.036 (0.328-3.268) | 0.952 | 2.364 (0.305-18.321) | 0.410 |
| No | 4.925 (1.133-21.406) | 0.033 | 1.995 (1.044-3.810) | 0.036 | 3.674 (0.882-15.303) | 0.074 |
| Menopausal | ||||||
| Premenopausal | 3.566 (0.818-15.555) | 0.091 | 1.389 (0.702-2.749) | 0.346 | 3.204 (0.766-13.411) | 0.111 |
| Postmenopausal | 4.773 (1.309-17.404) | 0.018 | 2.170 (0.937-5.023) | 0.071 | 3.915 (1.157-13.251) | 0.028 |
| LNM | ||||||
| Positive | 3.502 (1.189-10.318) | 0.023 | 1.718 (0.923-3.197) | 0.088 | 2.936 (1.044-8.259) | 0.041 |
| Negative | 4.602 (0.560-37.789) | 0.155 | 1.782 (0.646-4.915) | 0.264 | 3.675 (0.478-28.255) | 0.211 |
| TNM | ||||||
| I+II | 5.941 (1.342-26.303) | 0.019 | 1.447 (0.690-3.035) | 0.328 | 5.588 (1.316-23.727) | 0.020 |
| III | 2.343 (0.652-8.415) | 0.192 | 2.505 (1.133-5.539) | 0.023 | 1.676 (0.501-5.613) | 0.402 |
| ER | ||||||
| Positive | 3.223 (0.706-14.719) | 0.131 | 2.095 (0.919-4.779) | 0.079 | 2.413 (0.566-10.294) | 0.234 |
| Negative | 2.868 (0.367-22.430) | 0.315 | 1.045 (0.483-2.260) | 0.911 | 3.003 (0.399-22.578) | 0.285 |
| PR | ||||||
| Positive | 4.449 (0.897-22.065) | 0.068 | 3.197 (1.199-8.525) | 0.020 | 2.583 (0.594-11.237) | 0.206 |
| Negative | 3.558 (0.462-27.379) | 0.223 | 1.014 (0.497-2.068) | 0.970 | 3.580 (0.480-26.684) | 0.213 |
Discussion
In the present study, we investigated the association between miRNA SNPs and OS in breast cancer patients. Our findings revealed that miR-125a rs12976445 TT genotype was significantly related to increased risk of mortality in breast cancer patients compared with those carrying the CC genotypes. Furthermore, multivariate Cox multivariate analysis demonstrates that miR-125a rs12976445 was an independent risk factor for poor survival in breast cancer.
miR-125a, located at chromosome 19q13.41, plays important roles both in organ development and in the adult tissues [5,20,21]. Growing evidence has demonstrated that miR-125a is implicated in the pathogenesis of human cancer, including breast, gastric and lung cancers [4,22-24]. miR-125a functions as an oncogene or a tumor suppressor depending on the cellular context. For example, miR-125a was found downregulated in some types of cancer, including breast cancer [4], gastric cancer [22,25], leukemia [5,26], lung cancer [23,24], and medulloblastoma [27], where its overexpression inhibited cancer cell proliferation and induced apoptosis. Furthermore, the effect of miR-125a on growth, migration and apoptosis of multiple myeloma cells is p53-dependant [21]. On the other hand, Kim et al. found that miR-125a was upregulated in diffuse large B-cell lymphoma, leading to enhanced NF-κB activity by targeting TNFAIP3 and thus promoting the development of a malignant and anti-apoptotic phenotype in B cells [28]. The expression level of miR-125a is not only associated with prognosis of cancer patients [22,23], but also affects the sensitivity of cancer cell lines to anticancer drugs [26,29].
Previous study revealed that germline mutation of miR-125a is associated with breast cancer tumorigenesis [30]. There are two SNPs (rs41275794 and rs12976445) in the pre-miR-125a gene in Chinese Han population [14], which are in complete linkage disequilibrium with each other. Rs12976445 contributes to the susceptibility to autoimmune thyroid diseases [31] and recurrent pregnancy loss [14]. Moreover, rs12976445 is associated with poor prognosis in patients with esophageal squamous cell carcinoma [9]. In this study, we found that patients carrying rs12976445 TT genotype had poor prognosis. Recent studies demonstrated that the T allele of rs12976445 impairs mature miRNA processing and expression of mature miR-125a, leading to increased expression of target genes, such as ERBB2 and LIFR [14,15]. Amplification/overexpression of ERBB2 is related to a more aggressive behavior and a poor prognosis in breast cancer [32,33], which is predictive biomarkers of trastuzumab. In addition, higher level of miR-125a implied better effect of gefitinib on nasopharyngeal carcinoma cells [29]. Accordingly, miR-125a rs12976445 may influence the effectiveness of trastuzumab.
In conclusion, our findings suggest that miR-125a rs12976445 are possible prognostic biomarkers for breast cancer patients. Further larger-scale and well-designed clinical studies are warranted to validate these findings.
Acknowledgements
This work was supported by the Fund for International Scientific Cooperation of Shanghai Committee of Science and Technology, China (grant No. 13440701500).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munker R, Calin GA. MicroRNA profiling in cancer. Clin Sci (Lond) 2011;121:141–158. doi: 10.1042/CS20110005. [DOI] [PubMed] [Google Scholar]
- 4.Min W, Wang B, Li J, Han J, Zhao Y, Su W, Dai Z, Wang X, Ma Q. The expression and significance of five types of miRNAs in breast cancer. Med Sci Monit Basic Res. 2014;20:97–104. doi: 10.12659/MSMBR.891246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaham L, Binder V, Gefen N, Borkhardt A, Izraeli S. MiR-125 in normal and malignant hematopoiesis. Leukemia. 2012;26:2011–2018. doi: 10.1038/leu.2012.90. [DOI] [PubMed] [Google Scholar]
- 6.Wu C, Cao Y, He Z, He J, Hu C, Duan H, Jiang J. Serum levels of miR-19b and miR-146a as prognostic biomarkers for non-small cell lung cancer. Tohoku J Exp Med. 2014;232:85–95. doi: 10.1620/tjem.232.85. [DOI] [PubMed] [Google Scholar]
- 7.Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H, Yu H. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res. 2014;6:391–401. [PMC free article] [PubMed] [Google Scholar]
- 8.Adams BD, Kasinski AL, Slack FJ. Aberrant Regulation and Function of MicroRNAs in Cancer. Curr Biol. 2014;24:R762–R776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C, Li M, Hu C, Duan H. Prognostic role of microRNA polymorphisms in patients with advanced esophageal squamous cell carcinoma receiving platinum-based chemotherapy. Cancer Chemother Pharmacol. 2014;73:335–341. doi: 10.1007/s00280-013-2364-x. [DOI] [PubMed] [Google Scholar]
- 10.Yu H, Gao G, Jiang L, Guo L, Lin M, Jiao X, Jia W, Huang J. Decreased expression of miR-218 is associated with poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2013;6:2904–2911. [PMC free article] [PubMed] [Google Scholar]
- 11.Yu H, Duan B, Jiang L, Lin M, Sheng H, Huang J, Gao H. Serum miR-200c and clinical outcome of patients with advanced esophageal squamous cancer receiving platinum-based chemotherapy. Am J Transl Res. 2013;6:71–77. [PMC free article] [PubMed] [Google Scholar]
- 12.Lin J, Huang Y, Zhang X, Chen J, Sheng H. Association of miR-146a rs2910164 with childhood IgA nephropathy. Pediatr Nephrol. 2014;29:1979–86. doi: 10.1007/s00467-014-2818-3. [DOI] [PubMed] [Google Scholar]
- 13.Gao X, Yang L, Ma Y, Yang J, Zhang G, Huang G, Huang Q, Chen L, Fu F, Chen Y, Su D, Dong Y, Ma X, Lu C, Peng X. No association of functional variant in pri-miR-218 and risk of congenital heart disease in a Chinese population. Gene. 2013;523:173–177. doi: 10.1016/j.gene.2013.03.119. [DOI] [PubMed] [Google Scholar]
- 14.Hu Y, Liu CM, Qi L, He TZ, Shi-Guo L, Hao CJ, Cui Y, Zhang N, Xia HF, Ma X. Two common SNPs in pri-miR-125a alter the mature miRNA expression and associate with recurrent pregnancy loss in a Han-Chinese population. RNA Biol. 2011;8:861–872. doi: 10.4161/rna.8.5.16034. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann TP, Korski K, Ibbs M, Zawierucha P, Grodecka-Gazdecka S, Jagodzinski PP. rs12976445 variant in the pri-miR-125a correlates with a lower level of hsa-miR-125a and ERBB2 overexpression in breast cancer patients. Oncol Lett. 2013;5:569–573. doi: 10.3892/ol.2012.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang B, Liu C, Diao L, Wang C, Guo Z. A polymorphism at the microRNA binding site in the 3’ untranslated region of C14orf101 is associated with non-Hodgkin lymphoma overall survival. Cancer Genet. 2014;207:141–146. doi: 10.1016/j.cancergen.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Du M, Lu D, Wang Q, Chu H, Tong N, Pan X, Qin C, Yin C, Wang M, Zhang Z. Genetic variations in microRNAs and the risk and survival of renal cell cancer. Carcinogenesis. 2014;35:1629–1635. doi: 10.1093/carcin/bgu082. [DOI] [PubMed] [Google Scholar]
- 18.Zhang N, Huo Q, Wang X, Chen X, Long L, Jiang L, Ma T, Yang Q. A genetic variant in pre-miR-27a is associated with a reduced breast cancer risk in younger Chinese population. Gene. 2013;529:125–130. doi: 10.1016/j.gene.2013.07.041. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Chen Q, He C, Mao W, Zhang L, Xu X, Zhu J, Chen B. Polymorphisms on 8q24 are associated with lung cancer risk and survival in Han Chinese. PLoS One. 2012;7:e41930. doi: 10.1371/journal.pone.0041930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo S, Lu J, Schlanger R, Zhang H, Wang JY, Fox MC, Purton LE, Fleming HH, Cobb B, Merkenschlager M, Golub TR, Scadden DT. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci U S A. 2010;107:14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leotta M, Biamonte L, Raimondi L, Ronchetti D, Martino MT, Botta C, Leone E, Pitari MR, Neri A, Giordano A, Tagliaferri P, Tassone P, Amodio N. A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells in vitro. J Cell Physiol. 2014;229:2106–16. doi: 10.1002/jcp.24669. [DOI] [PubMed] [Google Scholar]
- 22.Nishida N, Mimori K, Fabbri M, Yokobori T, Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y, Mori M. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17:2725–2733. doi: 10.1158/1078-0432.CCR-10-2132. [DOI] [PubMed] [Google Scholar]
- 23.Zhu WY, Luo B, An JY, He JY, Chen DD, Xu LY, Huang YY, Liu XG, Le HB, Zhang YK. Differential Expression of miR-125a-5p and let-7e Predicts the Progression and Prognosis of Non-Small Cell Lung Cancer. Cancer Invest. 2014;32:394–401. doi: 10.3109/07357907.2014.922569. [DOI] [PubMed] [Google Scholar]
- 24.Jiang L, Chang J, Zhang Q, Sun L, Qiu X. MicroRNA hsa-miR-125a-3p activates p53 and induces apoptosis in lung cancer cells. Cancer Invest. 2013;31:538–544. doi: 10.3109/07357907.2013.820314. [DOI] [PubMed] [Google Scholar]
- 25.Guo X, Wu Y, Hartley RS. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 2009;6:575–583. doi: 10.4161/rna.6.5.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ufkin ML, Peterson S, Yang X, Driscoll H, Duarte C, Sathyanarayana P. miR-125a regulates cell cycle, proliferation, and apoptosis by targeting the ErbB pathway in acute myeloid leukemia. Leuk Res. 2014;38:402–410. doi: 10.1016/j.leukres.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferretti E, De Smaele E, Po A, Di Marcotullio L, Tosi E, Espinola MS, Di Rocco C, Riccardi R, Giangaspero F, Farcomeni A, Nofroni I, Laneve P, Gioia U, Caffarelli E, Bozzoni I, Screpanti I, Gulino A. MicroRNA profiling in human medulloblastoma. Int J Cancer. 2009;124:568–577. doi: 10.1002/ijc.23948. [DOI] [PubMed] [Google Scholar]
- 28.Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RC. MicroRNAs miR-125a and miR-125b constitutively activate the NF-kappaB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20) Proc Natl Acad Sci U S A. 2012;109:7865–7870. doi: 10.1073/pnas.1200081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Li Z, Wu L, Wang Z, Wang X, Yu Y, Zhao Q, Luo F. MiRNA-125a-5p: a regulator and predictor of gefitinib’s effect on nasopharyngeal carcinoma. Cancer Cell Int. 2014;14:24. doi: 10.1186/1475-2867-14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, Duan R, Kooy F, Sherman SL, Zhou W, Jin P. Germline mutation of microRNA-125a is associated with breast cancer. J Med Genet. 2009;46:358–360. doi: 10.1136/jmg.2008.063123. [DOI] [PubMed] [Google Scholar]
- 31.Inoue Y, Watanabe M, Inoue N, Kagawa T, Shibutani S, Otsu H, Saeki M, Takuse Y, Hidaka Y, Iwatani Y. Associations of single nucleotide polymorphisms in pri-miR-125a and the expression of mature miR-125a with the development and prognosis of autoimmune thyroid diseases. Clin Exp Immunol. 2014;178:229–35. doi: 10.1111/cei.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross JS, Fletcher JA. The HER-2/neu oncogene in breast cancer: prognostic factor, predictive factor, and target for therapy. Stem Cells. 1998;16:413–428. doi: 10.1002/stem.160413. [DOI] [PubMed] [Google Scholar]
- 33.Piccart MJ. Proposed treatment guidelines for HER2-positive metastatic breast cancer in Europe. Ann Oncol. 2001;12(Suppl 1):S89–94. doi: 10.1093/annonc/12.suppl_1.s89. [DOI] [PubMed] [Google Scholar]


