Abstract
Isolation of high-quality RNA from ribonuclease-rich tissue such as mouse pancreas presents a challenge. As a primary function of the pancreas is to aid in digestion, mouse pancreas may contain as much a 75 mg of ribonuclease. We report modifications of standard phenol/guanidine thiocyanate lysis reagent protocols to isolate RNA from mouse pancreas. Guanidine thiocyanate is a strong protein denaturant and will effectively disrupt the activity of ribonuclease under most conditions. However, critical modifications to standard protocols are necessary to successfully isolate RNA from ribonuclease-rich tissues. Key steps include a high lysis reagent to tissue ratio, removal of undigested tissue prior to phase separation and inclusion of a ribonuclease inhibitor to the RNA solution. Using these and other modifications, we routinely isolate RNA with RNA Integrity Number (RIN) greater than 7. The isolated RNA is of suitable quality for routine gene expression analysis. Adaptation of this protocol to isolate RNA from ribonuclease rich tissues besides the pancreas should be readily achievable.
Keywords: Molecular Biology, Issue 90, pancreas, mouse, RNA integrity, ribonuclease, RNA isolation, gene expression
Introduction
Isolation of RNA with high integrity is required for routine molecular biology experiments such as northern blotting 9, qRT-PCR 1 or gene expression profiling 5. Most contemporary methods of RNA isolation are based upon modifications of the guanidine thiocyanate protocols 2, 3. Guanidine thiocyanate is a strong protein denaturant and will effectively disrupt the activity of ribonuclease under most conditions. The popular method of Chomczynski and Sacchi 3 combined phenol to the guanidine thiocyanate lysis solution, reducing the isolation time to about 4 hr. Many commercially-available RNA extraction reagents are based upon the Chomczynski and Sacchi method 3.
Ribonuclease rich tissues such as human or mouse pancreas presents an additional challenge for isolating RNA. Mouse pancreas may contain up to 75 mg of ribonuclease 6 some of which will be released during disruption of the pancreatic tissue resulting in tissue autolysis. Modifications of the guanidine thiocyanate protocols have been successfully used to isolate RNA from pancreas with high integrity 2, 4, 7, 10. Infusion of RNA stabilization reagent into mouse pancreas facilitated isolation of RNA with high integrity 4, 7, 10, however infusion of solutions into the pancreas requires great skill, may require specialized instruments such as a dissecting microscope and disrupting tissue architecture during the procedure may result in cell lysis. Perfusing tissue with RNA stabilization reagent might interfere with other applications including protein isolation and histological staining. Furthermore, this technique is not suitable for isolating RNA from human pancreas. Other protocols require the preparation of specific solutions by the investigator 2.
We report a protocol to isolate RNA with high integrity from mouse pancreas. The protocol uses elements of previously published methods and is largely based upon the standard phenol/guanidine thiocyanate-based lysis reagent protocols. It does not require the preparation of specialized solutions nor does it require infusion of reagents into the pancreas. Critical steps for successful RNA isolation include a high phenol/guanidine-based lysis reagent to tissue ratio, removal of undigested tissue prior to phase separation and inclusion of a ribonuclease inhibitor to the resulting RNA solution. Using these and other modifications, we typically isolate RNA with RIN greater than 7 to be used for routine gene expression analysis.
Protocol
1. Preparation
The following practices adhere to the policies set by the Institution’s Animal Care and Use Committee (IACUC).
It is recommended to isolate no more than 6 mouse pancreases in any given day. Have all surgical instruments clean and autoclaved, one set per animal.
Label all microcentrifuge tubes. Four 2 ml tubes per animal.
Place 8 ml of lysis reagent in 50 ml centrifuge tubes on ice. Note: While the purification kit comes with a guanidine thiocyanate phenol reagent that is suitable for pancreas isolation, lysis reagent is used because of the large volumes that are necessary per isolation.
Dispense about 10 ml of the following solutions into 15 ml centrifuge tubes for cleaning the homogenizer’s blades. Ethanol, 70% (2 tubes), HPLC grade water (1 tube) and HPLC grade water containing approximately 200 µl of RNase Away or equivalent ribonuclease inactivator.
2. Surgery, Pancreas Dissection and Homogenization
Place 1-2 ml of isoflurane into a jar containing cotton gauze or paper towels. Mice are placed on a platform that prevents them from reaching the source of the isoflurane. Add fresh isoflurane to the jar prior to anesthetizing each mouse.
Gently place the mouse into the jar containing the isoflurane. Cap the jar and monitor the effects of the anesthesia by gently rotating the jar back and forth. The animal is anesthetized once it is in unable to stand in its’ own.
For confirmation of anesthesia, remove the animal from the jar and apply a firm toe pinch. If the mouse does not react to the toe pinch, proceed to step 2.5.
If the mouse reacts to the toe pinch, then repeat steps 2.2 to 2.3.
Place the anesthetized animal on the bench top in the prone position. Dislocate the mouse’s cervix by tightly placing the left hand on the mouse’s cervix. Using the right hand grasp the mouse’s tail and quickly pull upwards at approximately a 45° angle to dislocate the cervix.
Secure the carcass to a surface (a level piece of styrofoam covered with paper towels) by piercing each of the animal’s four paws into the flat surface using 25 gauge hypodermic needles.
Using sterile surgical instruments, cut the midsection of the mouse open and quickly remove the pancreas from the mouse. Use the forceps to hold the pancreas and cut it from the spleen and small intestine using spring scissors. Be careful not to stretch the pancreas during the removal from the mouse. Disruption of the tissue could release ribonuclease. Note: The time it takes to remove the pancreas from the mouse is critical. A skilled person should be able to dissect the pancreas in about 1 min or less.
Place the entire pancreas into a 50 ml tube containing 8 ml of ice cold lysis reagent. Cap the tube and briefly shake to immerse the tissue into the reagent and proceed to the next step without delay. The ratio of lysis reagent to tissue is critical. See Results section for a comparison of the RNA integrity isolated using different volumes of lysis reagent.
Homogenize for 5 sec. It is important to press the homogenizer blades to the bottom of the centrifuge tube to allow the tissue to be pulled into the blades for efficient homogenization.
3. Removal of Undigested Tissue
Note: Following homogenization, small bits of tissue will remain in the lysis reagent. It is more important to quickly lyse the tissue leaving some tissue undigested rather than over homogenizing and risk degradation of RNA.
Transfer 2 ml of lysis reagent containing the lysed pancreas to a 2 ml microcentrifuge tube. Collect two microcentrifuge tubes of homogenate per isolation. Discard the remaining homogenate or store at -80 °C for future studies.
Centrifuge at 4 °C, 12,000 × g for 5 min to pellet the undigested tissue. This is a critical step as carryover of undigested tissue into subsequent solutions of RNA will likely contaminate the sample with ribonuclease (step 5.13).
Transfer 1 ml of the supernatant into a 2 ml microcentrifuge tube. Proceed with the isolation immediately and store the replicate tube at -80 °C for future use. Place all tubes containing homogenate on wet ice while proceeding to the next step.
Dispose of the remaining lysis reagent into an appropriate chemical waste receptacle.
4. Cleaning of the Homogenizer Blades
Before proceeding to the next isolation, clean the homogenizer blades by placing them in 10 ml of 70% ethanol. Activate the homogenizer for about for 20 sec to remove any chunks of tissue that may be caught in the blades.
Use a 200 µl pipet tip to remove any pieces that remain in the homogenizer blades.
Repeat step 4.1 in water, general Rnase inactivator and 70% ethanol. Dry the homogenizer shaft with a clean Kim wipe.
Repeat step 2.1 for the pancreas isolation of the remaining animals, keeping RNA isolated from each animal separate rather than pooling the RNA.
5. RNA Isolation
The following section is identical to the purification kit protocol. The advantage of the purification kit is that it will isolate total RNA including small RNAs.
If using frozen homogenate solution, thaw on wet ice and let stand at RT for 5 min.
Add 200 µl of chloroform, cap the tube and shake vigorously by hand for 15 sec. Let the tube stand at RT for 2-3 min.
Centrifuge for 15 min at 12,000 x g, 4 °C.
Transfer the upper aqueous layer into a microcentrifuge tube. Add 1.5 volumes of isopropanol and mix thoroughly by pipetting up and down several times.
Transfer up to 700 μl of the sample into a spin column that is placed in a 2 ml collection tube. Close the lid and centrifuge at 8,000 x g for 15 sec at RT.
Discard the flow-through.
Repeat steps 5.5-5.6, using the remainder of the sample.
Add 700 μl Buffer RWT to the spin column. Centrifuge for 15 sec at 8,000 x g. Discard the flow-through.
Pipet 500 μl Buffer RPE into the spin column. Centrifuge for 15 sec at 8,000 x g to wash the column. Discard the flow-through.
Add 500 μl Buffer RPE to the spin column. Centrifuge for 2 min at 8,000 x g to dry the column.
Transfer the spin column to a 1.5 ml microcentrifuge tube. Pipet 80 μl ribonuclease-free water directly onto the spin column membrane. Centrifuge for 1 min at 8,000 x g to elute the RNA.
Add 1 µl Rnase inhibitor to the RNA solution. Store at -80 °C or proceed to 5.13.
To validate the integrity of the RNA, first calculate the RNA concentration and 260/280 ratio using a spectrophotometer.
Dilute a portion of the RNA solution to approximately 500 ng/µl with molecular biology grade water.
Determine the RNA Integrity (i.e., RIN) using capillary electrophoresis analysis. Approximately 5 µl of RNA solution is sufficient for the analysis.
Aliquot the RNA solution (see Figure 2) and store at -80 °C.
Representative Results
The total RNA yield from 1 ml of lysis homogenate is 20-40 µg. OD 260/280 ratios are typically around 2.0 and the RIN are consistently greater than 7.0. If the RIN is ≤6, the isolation will need to be repeated. Occasionally, a RIN that is higher than 8.0 is achieved.
The two most commonly used methods of euthanizing mice prior to removal of the pancreas are CO2 asphyxiation or inhalation of isoflurane. Both techniques are followed by cervical dislocation. Since it is possible that the chemical agent and/or procedure time might influence RNA integrity, the RIN of mouse pancreatic RNA isolated using both techniques was compared. Mice were anesthetized using CO2 asphyxiation or isoflurane inhalation; both techniques were followed by cervical dislocation. The time of anesthetizing the animals prior to cervical dislocation was approximately 1 to 2 min longer for CO2 asphyxiation compared to isoflurane. RNA isolated following inhalation of isoflurane produced a RIN of 7.5±0.31 compared to a RIN of 2.2±0.3 (p<10-4, mean ± SD, triplicate isolations) for CO2 asphyxiation. The method of euthanasia has a dramatic impact on RNA integrity and the technique of isoflurane inhalation is preferred to CO2 asphyxiation.
The integrity of RNA isolated from mouse pancreases that were homogenized in 2, 4 and 8 ml of lysis reagent were compared. We hypothesized that increasing the volume of lysis reagent will lead to a greater degree of ribonuclease inactivation. Other than the volume of lysis reagent, all conditions were identical to those listed in the protocol. There was a direct correlation between the volume of lysis reagent used in the homogenization and RNA integrity (Figure 1). The RIN of RNA isolated using 2, 4 and 8 ml of lysis reagent was 4.2, 6.5 and 7.4, respectively (mean, triplicate isolations). Therefore 8 ml of lysis reagent should be used in this protocol.
For some experiments, it may be necessary to collect both RNA and protein from the same pancreas. For example, RNA may be used for qRT-PCR and protein may be visualized using immunohistochemistry. If this is the case, it is recommended to cut the pancreas in half from head to tail. This is important since the pancreas differs anatomically from head to tail. To demonstrate if any differences in the RNA integrity exists between RNA isolated from either half of the pancreas, the following experiment was done. RNA was isolated from the first half of the pancreas. The RIN determined from this RNA was compared to that of RNA isolated from the second half of the pancreas after it sat at RT for about 30 sec during the workup of the initial half. The RIN for the initial half of pancreas was 7.4±0.20 compared to a RIN of 6.9±0.55 for the subsequent half. While cutting the pancreas in this manner does not appear to release ribonuclease and digestion of RNA, if the pancreas is to be sectioned, it is recommended to use the first half for RNA analysis and the second half for protein.
Lysis reagent disrupts protein structure and therefore reduces ribonuclease activity. Due to the large amount of ribonuclease that is present in the pancreas, it is possible that some active ribonuclease remains in the lysis homogenate and this ribonuclease will equilibrate to the aqueous phase. Any ribonuclease that remains in the last RNA solution will impede integrity. To overcome this, we added a ribonuclease inhibitor to the aqueous solution of RNA. The RNA integrity was compared from an RNA solution that contained ribonuclease inhibitor to one that did not. Upon one freeze thaw, the RIN for the solution of RNA with inhibitor was 7.3±0.15 compared to 2.9±0.65 (Figure 2, p<0.001, mean ± SD, triplicate isolations) for the solution that did not contain inhibitor. Another issue is the possibility that repeated freeze thawing will denature the ribonuclease inhibitor rendering it ineffective. To examine this possibility, a solution of RNA containing ribonuclease inhibitor was repeatedly frozen and thawed. The RNA solutions that were frozen and thawed up to 5 times still produced a RIN of 7 or higher while those solutions that were frozen and thawed 6 times or higher produced a RIN below 7 (Figure 2). It is recommended to aliquot the RNA solution prior to freezing and limit the number of freeze thaw cycles.
To demonstrate the usefulness of this method, we performed qRT-PCR on the isolated RNA. The mean RIN from triplicate RNA isolations was 7.4±0.20 (mean±SD). Random primed cDNA was synthesized from the RNA and the expression of three housekeeping genes was measured by qPCR using standard techniques. The data shown in Figure 3 validates our ability to successfully use mouse pancreatic RNA in a standard molecular biology technique.
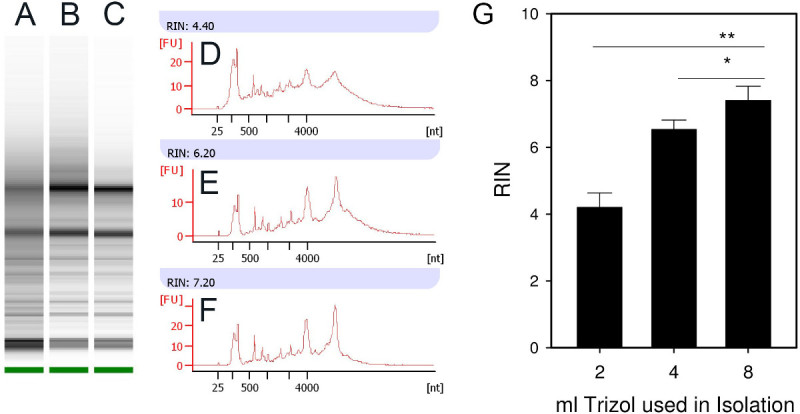 Figure 1. High lysis reagent to tissue ratio improves RNA integrity. The integrity of RNA isolated from mouse pancreases that was homogenized in 2 (A, D), 4 (B, E) or 8 (C, F) ml of lysis reagent was compared. Other than the volume of lysis reagent, all conditions were identical to those in described in the protocol herein. A direct correlation exists between the volume of lysis reagent used in the homogenization and RNA integrity. (G) Mean±SD, triplicate isolations. p<0.05, **p<0.001. Please click here to view a larger version of this figure.
Figure 1. High lysis reagent to tissue ratio improves RNA integrity. The integrity of RNA isolated from mouse pancreases that was homogenized in 2 (A, D), 4 (B, E) or 8 (C, F) ml of lysis reagent was compared. Other than the volume of lysis reagent, all conditions were identical to those in described in the protocol herein. A direct correlation exists between the volume of lysis reagent used in the homogenization and RNA integrity. (G) Mean±SD, triplicate isolations. p<0.05, **p<0.001. Please click here to view a larger version of this figure.
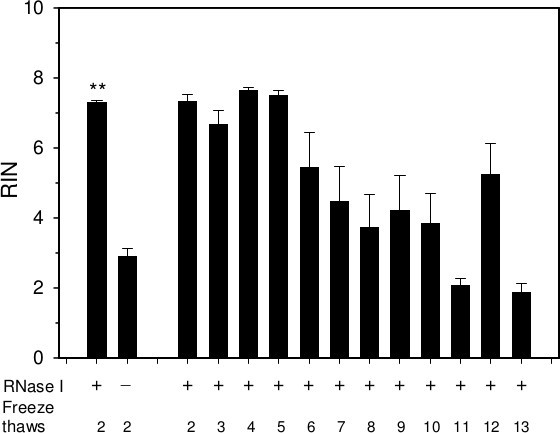 Figure 2. RNA eventually degrades upon repeated freeze thawing. RNA isolated from mouse pancreas was repeatedly frozen and thawed in the presence or absence of ribonuclease inhibitor (RNase I). Mean±SEM, **p<0.001.
Figure 2. RNA eventually degrades upon repeated freeze thawing. RNA isolated from mouse pancreas was repeatedly frozen and thawed in the presence or absence of ribonuclease inhibitor (RNase I). Mean±SEM, **p<0.001.
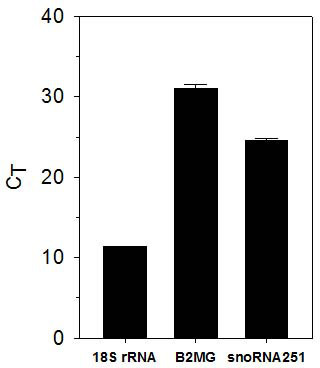 Figure 3. qRT-PCR of mouse pancreatic RNA. RNA was isolated from three mouse pancreases using the optimized protocol. The expression of 18S rRNA, β-2 microglobulin and snoRNA251 was determined using qRT-PCR (Mean±SD).
Figure 3. qRT-PCR of mouse pancreatic RNA. RNA was isolated from three mouse pancreases using the optimized protocol. The expression of 18S rRNA, β-2 microglobulin and snoRNA251 was determined using qRT-PCR (Mean±SD).
Discussion
Isolating RNA from tissues that are ribonuclease rich represents a great challenge for molecular biology experiments. Various reagents and kits are commercially available that are primarily based upon the phenol guanidine thiocyanate method of RNA extraction. Guanidine thiocyanate denatures protein and thus reduces the activity of ribonuclease. However, the sheer magnitude of ribonuclease in pancreas requires additional modifications to standard protocols of RNA isolation. A protocol is reported here that is based on the standard guanidine thiocyanate method yet contains several critical modifications as well as tips for successful isolation of RNA from ribonuclease rich tissues such as the pancreas.
Standard guidelines for isolating RNA are to be noted. These techniques are critical to successful isolation of RNA, even from specimens that are less ribonuclease-rich than pancreas. Some examples include using molecular biology grade water, ribonuclease-free consumables and disposable plastic ware rather than glass. It is recommended to change gloves in between each isolation. For someone who is inexperienced with isolating RNA, it is recommended that they gain experience with isolating RNA from ribonuclease-poor samples such as liver or from cell cultures prior to embarking on work that requires isolating RNA from pancreas. Excellent protocols exist that offer a variety of points for successfully working with RNA 8.
Several important factors for isolating RNA with high integrity from mouse pancreas are reported here. These include the volume of lysis reagent used during the homogenization and the inclusion of a ribonuclease inhibitor to the RNA solution. The method of including the ribonuclease inhibitor works well, however repeated freeze thaw cycles will eventually reduce the RIN, likely due to denaturing the ribonuclease inhibitor. It should be noted that all RNA solutions used in this study went through one freeze thaw prior to RIN determination. Another method of reducing degradation upon freeze thaw is to store the RNA solution at -20 °C in a precipitated form 2. This offers some inconveniences namely removing the ethanol and re-dissolving the pellet prior to using the RNA. Compared to the phenol/guanidine thiocyanate reagent protocols 2, advantages of the protocol reported here is the simplicity of working with lysis reagent and the purification kit that do not require preparation of specialized solutions and buffers. It should also be noted that this protocol has been successfully used to isolate small RNAs from mouse pancreas.
Additional tips include the speed of pancreas dissection and the method of anesthesia prior to euthanasia (isoflurane inhalation is preferred to CO2 asphyxiation). Several attempts at practicing the pancreas surgery are recommended before anticipating high quality RNA. Adaptation of this protocol to ribonuclease-rich tissues such as the spleen or lungs should be readily achievable.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We thank Jianhua Ling, Raymond MacDonald, Galvin Swift, Michelle Griffin, Paul Grippo and Satyanarayana Rachagani for their helpful comments, suggestions and sharing of their protocols. This work was supported by grant U01CA111294 and an Idea Development Award from the Ohio State University Intramural Research Program.
References
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Griffin M, Abu-El-Haija M, Abu-El-Haija M, Rokhlina T, Uc A. Simplified and versatile method for isolation of high-quality RNA from pancreas. Biotechniques. 2012;52:332–334. doi: 10.2144/0000113862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson Jr J, Boguski MS, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- Lenstra JA, Beintema JJ. The amino acid sequence of mouse pancreatic ribonuclease. Extremely rapid evolutionary rates of the myomorph rodent ribonucleases. European journal of biochemistry FEBS. 1979;98:399–408. doi: 10.1111/j.1432-1033.1979.tb13199.x. [DOI] [PubMed] [Google Scholar]
- Mullin AE, Soukatcheva G, Verchere CB, Chantler JK. Application of in situ ductal perfusion to facilitate isolation of high-quality RNA from mouse pancreas. Biotechniques. 2006;40:617–621. doi: 10.2144/000112146. [DOI] [PubMed] [Google Scholar]
- Rio DC, Ares M, Hannon GJ, Nilsen TW. RNA A Laboratory Manual. Cold Springs Harbor, NY: Cold Springs Harbor Laboratory Press; 2011. [Google Scholar]
- Streit S, Michalski CW, Erkan M, Kleeff J, Friess H. Northern blot analysis for detection and quantification of RNA in pancreatic cancer cells and tissues. Nat Protoc. 2009;4:37–43. doi: 10.1038/nprot.2008.216. [DOI] [PubMed] [Google Scholar]
- Zmuda EJ, Powell CA, Hai T. A method for murine islet isolation and subcapsular kidney transplantation. Journal of Visualized Experiments JoVE. 2011. [DOI] [PMC free article] [PubMed]


