SUMMARY
Proteasomes and lysosomes constitute the major cellular systems that catabolize proteins to recycle free amino acids for energy and new protein synthesis. Tripeptidyl peptidase II (TPPII) is a large cytosolic proteolytic complex that functions in tandem with the proteasome-ubiquitin protein degradation pathway. We found that autosomal recessive TPP2 mutations cause recurrent infections, autoimmunity, and neurodevelopmental delay in humans. We show that a major function of TPPII in mammalian cells is to maintain amino acid levels, and that TPPII-deficient cells compensate by increasing lysosome number and proteolytic activity. However, the overabundant lysosomes derange cellular metabolism by consuming the key glycolytic enzyme hexokinase-2 through chaperone-mediated autophagy. This reduces glycolysis and impairs the production of effector cytokines including IFN-γ and IL-1β. Thus, TPPII controls the balance between intracellular amino acid availability, lysosome number, and glycolysis, which is vital for adaptive and innate immunity and neurodevelopmental health.
INTRODUCTION
Protein degradation occurs continuously within cells. This removes misfolded or damaged proteins and generates free amino acids for protein synthesis or energy production via glutaminolysis (Schutz, 2011). Mammalian cells utilize two principal pathways: proteasomes, which are protein complexes that recognize and degrade ubiquitinated proteins within the cytosol, and lysosomes, which are membrane-bound organelles containing acid hydrolases that are fed substrate by endosomal and autophagic vesicles (Ciechanover, 2005). Evidence suggests that these pathways can cross-compensate to maintain balanced proteolysis and amino acid homeostasis (Korolchuk et al., 2010). In both pathways, proteins are first degraded into long oligopeptides from which N-terminal tripeptides are then trimmed by tripeptidyl peptidases (TPP). These tripeptides are further cleaved by dipeptidyl peptidases and aminopeptidases to generate free amino acids (Tomkinson, 1999).
There are two types of TPP in eukaryotic cells, TPPI and TPPII. TPPI is a lysosomal acid protease, whereas TPPII is a cytosolic protease that forms a giant multi-subunit complex acting downstream of proteasomes (Schonegge et al., 2012; Tomkinson, 1999). By trimming long oligopeptides, TPPII was thought to be principally important in producing antigenic peptides that bind to major histocompatibility complex (MHC) class I molecules for presentation to CD8 T cells (Reits et al., 2004). However, the development and function of CD8 T cells was largely unaffected by genetic deletion of Tpp2 in mice, even during experimental viral infections (Kawahara et al., 2009). By contrast, other TppII-deficient mouse strains exhibited either embryonic lethality (McKay et al., 2007) or an immunosenescent phenotype characterized by declining thymic output and progressive loss of CD4 and CD8 T cells (Huai et al., 2008). Thus, the physiological role for TPPII in proteolysis, amino acid homeostasis, and metabolism in mammals remains obscure. Furthermore, although humans with loss-of-function mutations in TPP1 develop a lysosomal storage disease called classical late-infantile neuronal ceroid lipofuscinosis (Tomkinson, 1999), whether TPP2 mutations cause human disease is unknown.
In the immune system, innate and adaptive cells quickly and coordinately respond to invading pathogens and inflammatory signals. The biosynthetic and bioenergetic demands of the responding leukocytes are extreme because of the sudden requirements for cell growth, trafficking, proliferation, and effector functions. To support this burst of anabolic activity, cellular metabolism radically reorients towards aerobic glycolysis (MacIver et al., 2013; Pearce and Pearce, 2013). Although less efficient in generating ATP, glycolysis generates intermediate metabolites that support biosynthetic pathways for effector functions including cytokine production (Chang et al., 2013; Shi et al., 2011). It is thus not surprising that metabolic reprogramming is an integral part of leukocyte activation, and that a complex regulatory network links nutrient availability with a concerted immune response. Unraveling this complexity is important because of the potential to target metabolic pathways for modulating pathological immune responses. To this end, we have studied patients with a metabolic immunodeficiency caused by TPP2 mutations.
RESULTS
Human disease Caused by Loss of TPPII Activity
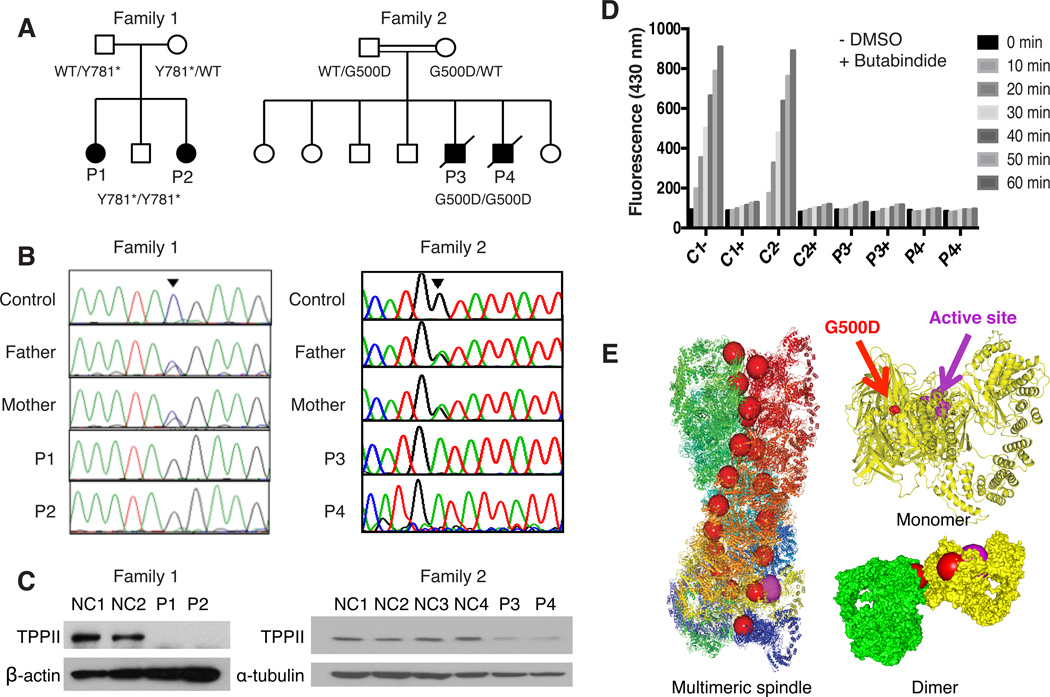
We identified four patients from two families, affected by combined immunodeficiency, severe autoimmunity, and developmental delay (Figure 1A, Table 1, and Data S1), with biallelic loss-of-function mutations in TPP2. Except for P2, who was diagnosed by screening in early infancy, patients presented in early childhood with recurrent bacterial and viral infections of the respiratory tract and middle ear. All three tested patients showed markedly decreased circulating T, B, and natural killer lymphocytes (Figure S1A), including severely reduced naïve T cells, and hypergammaglobulinemia (Figure S1B and data not shown). Patients had severe, intractable autoimmunity, manifesting as antibody-mediated destruction of red blood cells, platelets, and neutrophils (in all patients, Figure S1C), central nervous system lupus erythematosus with stroke (in P1), and hepatitis with autoantibodies (in P4). All patients had mild to moderate developmental delay in acquiring motor, language, and social skills. We assumed an autosomal recessive disorder was present, as siblings P1 and P2 originated from a geographically isolated, small indigenous population, and sibship P3 and P4 from a consanguineous union. Diagnostic testing excluded known primary immunodeficiency disorders, so we performed whole exome sequencing (WES) of genomic DNA from P1 and her healthy parent, and WES with homozygosity mapping in P3 (Figure S2A). Each patient bore a novel homozygous single nucleotide variant in TPP2, for which their parents were heterozygous carriers. Sanger sequencing of TPP2 confirmed that P1 and P2 were homozygous for the nonsense mutation c.2343C>G, p.Tyr781*, while P3 and P4 were homozygous for the missense mutation c.1499G>A, p.Gly500Asp (Figure 1B).
Figure 1. Autosomal Recessive Loss-of-function Mutations in Human TPPII deficiency.
(A) Patients’ pedigrees. (B) Sanger sequencing showing the mutations. (C) Immunoblots for TPPII in T cells from P1 and P2 (left) or fibroblasts from P3 and P4 (right). (D) TPPII enzymatic activity in fibroblast lysates from two healthy controls, P3 and P4, incubated for the indicated minutes without (−) or with (+) added TPPII inhibitor BUTA. (E) Structural representations of TPPII highlighting G500D (red spheres) and the active site (purple spheres). Shown are ribbon representations of multimeric spindle and monomer, and surface representation of dimer of yellow and green monomers (Schonegge et al., 2012). Experiments were repeated at least twice for (C) and 3 times for (D). See also Figure S2.
Table 1.
Clinical Features of TPPII-deficient patients
| Patient | P1 | P2 | P3 | P4 |
|---|---|---|---|---|
| Demographics | 10 year old First Nations female | 18 month old First Nations female | 11 year old British Pakistani male | 3 year old British Pakistani male |
| Immunodeficiency | ||||
| -Respiratory tract | Recurrent lower respiratory tract infections (including cytomegalovirus, adenovirus) with bronchiectasis, recurrent otitis media | Lower respiratory tract infections (including adenovirus, Aspergillus fumigatus), recurrent otitis media | Recurrent lower respiratory tract infections with bronchiectasis, recurrent otitis media complicated by mastoiditis | Recurrent lower respiratory tract infections, recurrent otitis media |
| -Other infections | Recurrent orolabial herpes simplex virus type 1 | None | Pneumococcal sepsis, hemorrhagic varicella, acute hepatitis A, persistent cytomegalovirus | Acute hepatitis A, cytomegalovirus |
| Autoimmunity | ||||
| -Cytopenias | Autoimmune hemolytic anemia, immune thrombocytopenic purpura, neutropenia | Neutropenia | Autoimmune hemolytic anemia, immune thrombocytopenic purpura | Autoimmune hemolytic anemia, immune thrombocytopenic purpura, neutropenia |
| -Other autoimmunity | Central nervous sytem lupus erythematosus (ANA+) with stroke due to suspected vasculitis | None | None | Autoimmune hepatitis (ANA+ SMA+) leading to end-stage liver disease |
| Developmental delay | Yes | Yes, assessment ongoing | Yes | Yes |
| Outcome | Alive | Alive, recently underwent hematopoietic stem cell transplantation | Died of adenovirus encephalitis after hematopoietic stem cell transplantation | Died of complications after orthotopic liver transplantation |
ANA, anti-nuclear autoantibodies; SMA, anti-smooth muscle autoantibodies. See also Figure S1 and Data S1.
TPPII protein is normally expressed at high levels in lymphocytes (Figure S2B) but was absent from T cells from both P1 and P2, consistent with nonsense-mediated mRNA decay (Figure 1C). In contrast, TPPII protein was expressed by dermal fibroblasts and peripheral blood mononuclear cells (PBMC) from P3 and P4, although at levels 60–80% less than controls (Figure 1C and Figure S2B). The residual protein lacked exopeptidase activity sensitive to the TPPII-specific inhibitor butabindide (BUTA, Rose et al., 1996) (Figure 1D). To model the effect of the p.G500D missense mutation we considered the recently solved quaternary structure of human TPPII (Schonegge et al., 2012). TPPII exists as a giant (~6 MDa) spindle-shaped cytoplasmic protein complex made up of two twisted strands composed of stacked dimers that align to form a series of internal catalytic chambers (Figure 1E). Glycine at residue 500, which is conserved across all species (Figure S2C), lies outside the active site at the hydrophobic interface between strands of stacked dimers (Figure 1E). We predicted that substitution by a charged amino acid would disrupt this “molecular clamp,” thereby impairing multimerization and enzymatic function. In keeping with this hypothesis, native gel electrophoresis revealed loss of high molecular weight complexes of TPPII protein and associated enzymatic activity in lysates from P3 and P4 (Figure S2D).
TPPII Has a Major Role in Amino Acid Homeostasis
During normal cellular homeostasis, cytosolic free amino acids are derived from recycling of intracellular proteins through the proteasomal or lysosomal proteolytic pathways, and from extracellular transport (Barnes et al., 1992). Because TPPII is a cytosolic protease that acts downstream of the proteasome, we hypothesized that TPPII-deficiency would impair amino acid recycling. Indeed, total levels of intracellular free amino acids were markedly decreased within 4 hours of TPPII inhibition by BUTA treatment of mouse embryonic fibroblasts (MEF), at values ~20% to ~70% of controls (Figure 2A). This decrease affected all measurable individual amino acids (Figure 2B and data not shown), suggesting no selective effect on amino acids or transporters that recognize structurally related classes of amino acids (Taylor, 2014). However, by 6 to 12 hours of BUTA treatment, intracellular free amino acids had returned to normal or higher levels (Figure 2A), similar to chronically TppII-deficient knockout (KO) MEF cells (Figures S3A and S3B). Thus, a major function of TPPII is to maintain amino acid levels in the cell, and during chronic TPPII inhibition a compensatory mechanism maintains amino acid homeostasis.
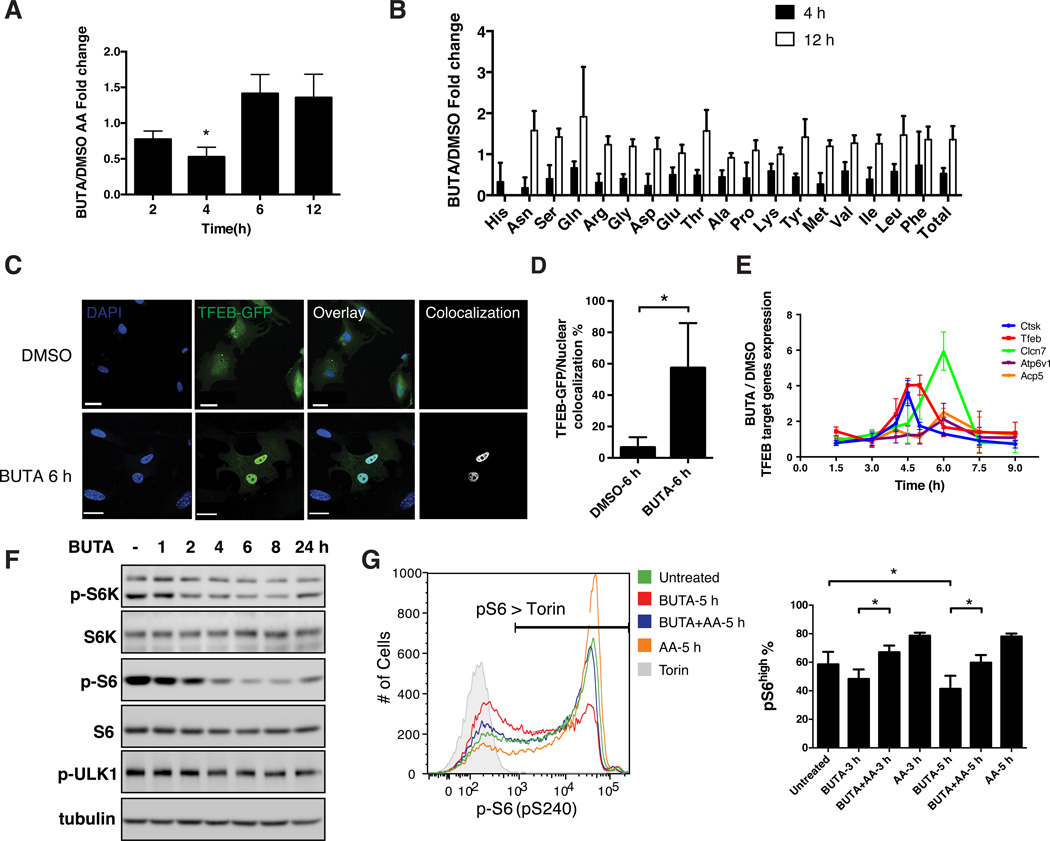
Figure 2. TPPII Regulates Intracellular Amino Acid Homeostasis.
(A) Each bar shows the ratio of the quantity of total free amino acids (AA) in wildtype MEF cells treated with BUTA (200 µM) for the indicated hours (h), to the quantity of total AA in the same cells treated with the dimethyl sulfoxide (DMSO) vehicle, as determined by ultra-performance liquid chromatography (UPLC). (B) Normalized concentrations of intracellular free AA in MEF after 4 h or 12 h of treatment with BUTA or DMSO as in (A). (C) Confocal microscopy showing colocalization (white) of GFP-tagged TFEB (green) and DAPI (blue) in transfected MEF treated with BUTA or DMSO for 6 h. Scale bars, 30 µm (BUTA), 40 µm (DMSO). (D) Quantitation of (C). (E) Normalized expression of TFEB-regulated gene transcripts, measured by quantitative RT-PCR, in MEF treated with BUTA or DMSO for the indicated times. (F) Immunoblot showing kinetics of S6K, S6, and ULK1 phosphorylation in BUTA-treated primary human fibroblasts. (G) Flow cytometric detection of reduced pS6 in Jurkat T cells treated with BUTA, and rescue by supraphysiologic AA (mTOR inhibitor torin, negative control); percentages of pS6 positive cells quantified (right). Data in (A, B, D, E, G) are represented as mean ± SD from 3 independent experiments. *P < 0.05, as calculated by one sample t-test (A), unpaired two-tailed Student’s t-test (D) or repeated measures ANOVA followed by Tukey’s test (G). Images in (C, F, G) are representative of 3 independent experiments. See also Figure S3.
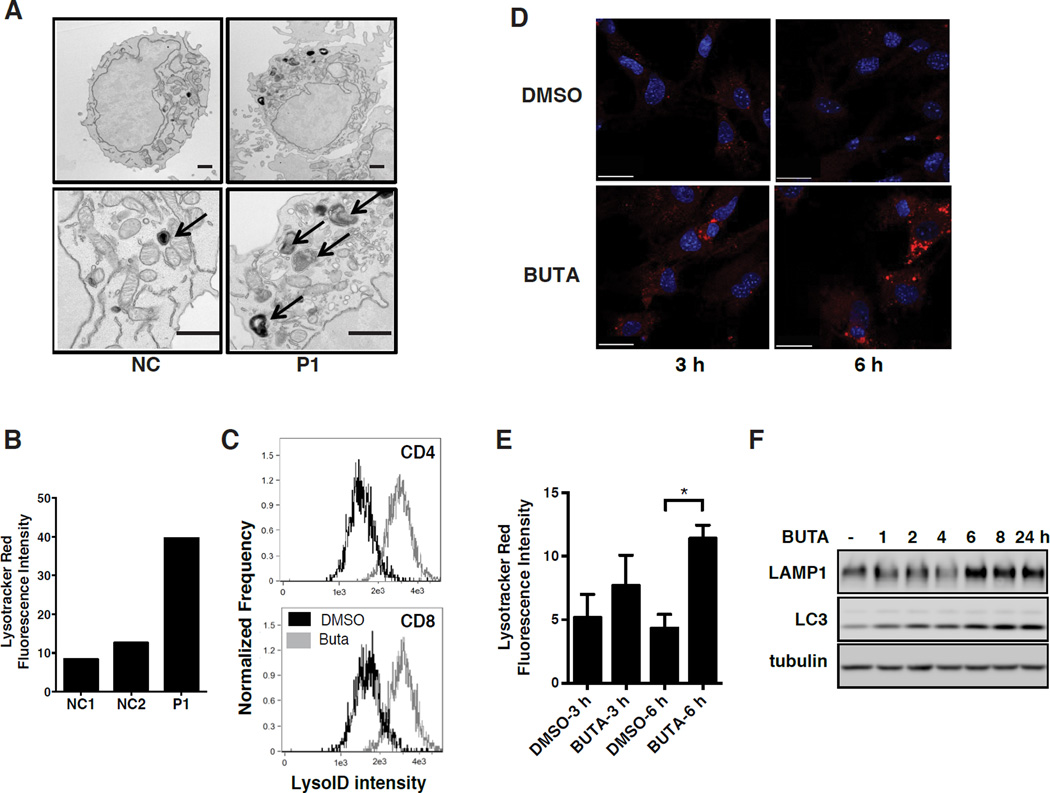
Figure 3. Loss of TPPII Leads to Compensatory Lysosome Biogenesis.
(A) Transmission electron micrographs of T cells from P1 and a healthy normal control. Arrows, electron-dense lysosomal structures. Scale bar, 500 nm. (B) Quantification of Lysotracker Red fluorescence intensity from confocal microscopy of T cells as in (A). (C) Imagestream analysis of lysosome content by LysoID red staining of normal human T cells, activated in the presence of BUTA or DMSO. (D) Confocal microscopy showing Lysotracker Red staining of lysosomes and DAPI staining of nuclei (blue) after treatment of MEF with DMSO or BUTA for the indicated h. Scale bar, 30 µm. (E) Quantification of (D). (F) Simultaneous immunoblotting of LAMP1 and LC3 following BUTA treatment of human fibroblasts for the indicated h. All experiments were repeated 3 times except for (A) which was repeated twice, and show representative images. Data in (E) are represented as mean ± SD from 3 independent experiments. *P < 0.05, as calculated by unpaired two-tailed Student’s t-test. See also Figure S4.
Amino acid deprivation causes the transcription factor TFEB to translocate from the cytosol to the nucleus, where it functions as a master regulator of genes involved in lysosomal biogenesis and autophagy to preserve intracellular amino acid homeostasis (Roczniak-Ferguson et al., 2012; Settembre et al., 2011). We therefore monitored TFEB subcellular localization and target gene transcription upon BUTA treatment. Because commercially available antibodies were unreliable for detecting endogenous TFEB, transfected GFP-tagged TFEB was used. Coincident with the acute decrease in intracellular free amino acids after TPPII inhibition (Figure 2A), we found TFEB translocated into the nuclei of MEF (Figures 2C and 2D) or the human neuroblastoma cell line SH-SY5Y (Figure S3C). Furthermore, in untransfected MEF, BUTA upregulated endogenous TFEB-regulated transcripts, including Tfeb itself, within 4 to 6 hours of treatment, before returning to baseline (Figure 2E). We did not detect elevated TFEB-regulated transcripts in cells constitutively deficient in TPPII (Figure S3D). Thus, maximal amino acid reduction precedes maximal TFEB induction, which in turn precedes restored amino acid homeostasis.
TFEB nuclear localization is normally inhibited by mTOR when growth and nutritional signals are adequate (Roczniak-Ferguson et al., 2012). We found that basal mTOR activity, as measured by phosphorylation of either S6K or S6, was markedly diminished in fibroblasts after 2 to 8 hours of BUTA treatment (Figures 2F and S3E) when TFEB activity was highest. Acute TPPII inhibition also decreased mTOR activity in the Jurkat T cell line and PHA-stimulated T cell blasts, which could be overcome by supraphysiological extracellular amino acids (Figures 2G and S3F). Remarkably, mTOR activity recovered when BUTA treatment was extended beyond 18 hours (Figures 2F, S3E, and S3G), and mTOR activity was normal in a THP1 macrophage cell line stably transduced with TPP2 shRNA (Figure S3H). Such chronically TPPII-inhibited cells nevertheless showed exaggerated sensitivity to partial amino acid starvation (Figure S3G). Together, our results indicate that acute TPPII inhibition caused amino acid depletion, leading to mTOR deactivation, TFEB nuclear translocation, and TFEB-dependent gene induction, thereby adapting the cells to chronic TPPII inhibition.
Compensatory Lysosomal Biogenesis in the Absence of TPPII Activity
Lysosomes are a key regulator of amino acid homeostasis (Bar-Peled and Sabatini, 2014), so we hypothesized that the new metabolic state established in the absence of TPPII activity was achieved through a TFEB-dependent increase in lysosomal activity. We observed increased abundance of lysosomes in T cells from P1 (Figures 3A and 3B), above levels expected from T cell receptor (TCR) stimulation alone (Valdor et al., 2014; and data not shown). Similar effects were observed after BUTA treatment of normal T cells (Figure 3C), human fibroblasts (Figures 3D and 3E), MEF, and various transformed cell lines (293T, A549, SH-SY5Y, HeLa) (Figure S4A), or when TPPII was knocked down in THP1 cells stably expressing TPP2 shRNA (Figure S4B). The lysosomal markers LAMP1 and cathepsin B were increased in fibroblasts derived from P3 and P4, as well as in control fibroblasts treated with BUTA (Figures 3F, S4C, and data not shown). Measures of lysosomal function, such as acid phosphatase and β-N-acetyl-glucosaminidase activity, were also commensurately elevated in liver tissues from Tpp2 KO mice (Figures S4D and S4E). Confirming that this lysosomal expansion was caused by inhibition of cytoplasmic proteolysis, we could mimic the effect by treating MEF with the proteasome inhibitor MG132, which blocks the pathway upstream of TPPII (Figure S4F). Thus, cells of multiple lineages experience impaired cytosolic amino acid recycling when TPPII activity is deficient, and then recover through an mTOR- and TFEB-regulated compensatory lysosomal biogenesis that restores amino acid homeostasis.
Lysosomal Overactivity Induced by TPPII Deficiency Impairs Glycolysis
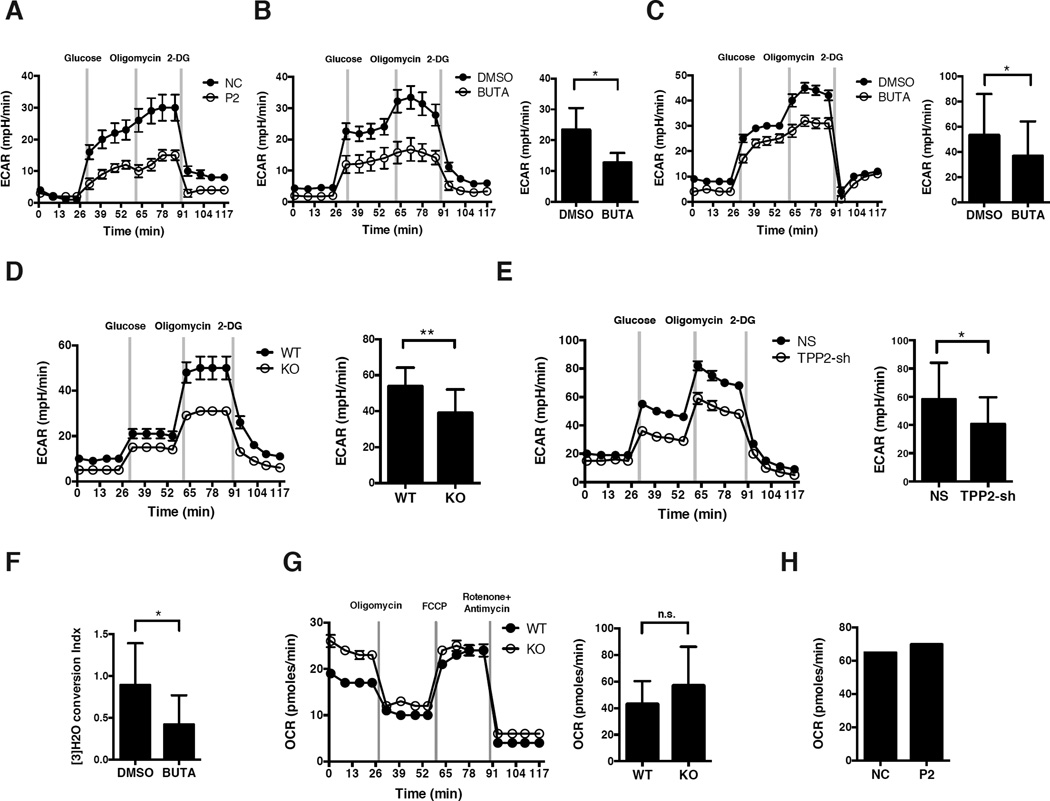
Our patients’ neurodevelopmental delay, altered amino acid homeostasis, their expanded lysosomal compartment, and in the eldest patient (P3) an unusual pattern of intracranial calcification involving basal ganglia and subcortical U fibers (Figure S5A), all implied a metabolic disorder. Because metabolic reprogramming is crucial for activated proliferating lymphocytes, we evaluated glycolysis by measuring extracellular acidification rate (ECAR) after providing glucose to TPPII-deficient or -replete cells. We also measured the effect of adding oligomycin (to block oxidative phosphorylation and shunt to glycolysis), as well as 2-deoxy-D-glucose (2-DG, to inhibit glycolysis). TPPII deficiency reduced basal and/or maximal glycolysis in activated T cells from P2 (Figure 4A), as well as BUTA-treated activated CD4 or CD8 T cells from controls (Figures 4B and 4C). We observed similar effects in CD4 T cells isolated from Tpp2 KO mice (Figure 4D), THP1 cells stably transduced with TPP2 shRNA (Figure 4E), or in SH-SY5Y cells after BUTA treatment (Figure S5B). By contrast, no defect was evident in unstimulated naïve T cells that have yet to switch from mixed fuel oxidative phosphorylation to aerobic glycolysis (Figure S5C). In BUTA-treated activated CD4 T cells, we confirmed the glycolytic defect by showing impaired conversion of radiolabeled glucose to water (Figure 4F). We next evaluated oxidative phosphorylation in activated T cells by measuring oxygen consumption rate (OCR) at baseline, followed by oligomycin (to calculate ATP production), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP, to determine maximal respiration), and rotenone plus antimycin (to calculate basal respiration) and found oxidative phosphorylation was unaffected by TPPII deficiency, implying that glutamine utilization was intact (Figures 4G and 4H).
Figure 4. Loss of TPPII Activity Impairs Glycolysis.
(A to E) Glycolytic flux, measured as ECAR of cells at baseline, and after adding glucose, oligomycin, and 2-DG, and/or quantitation by total area under the curve (AUC) of histograms. (A) Activated T cells from P2 or healthy control. (B) Human naïve CD4 T cells, activated for 3 days in the presence of BUTA (200 µM) or DMSO. (C) Human naïve CD8 T cells stimulated for 4 days as in (B). (D) Naïve CD4 T cells from Tpp2 KO or WT mice, activated for 3 days. (E) THP1 cells, stably transduced with TPP2 shRNA or NS shRNA. (F) Glycolysis as measured by [5-3H]-glucose conversion to [5-3H]-water in cells stimulated as in (B). (G, H) Oxidative phosphorylation as measured by OCR at baseline and after adding oligomycin, FCCP, and rotenone plus antimycin, and/or quantitation by AUC. (G) Naïve CD4 T cells from Tpp2 KO or WT mice, stimulated as in (D). (H) T cells from P2 or control as in (A). For representative histograms, data shows mean ± SD of 6 replicate wells. Data showing AUC represents mean ± SD from at least 3 independent experiments, except for (H) which was from one experiment. *P < 0.05, **P < 0.01, n.s. non-significant, as calculated by unpaired Student’s t-test. See also Figures S5 and S6.
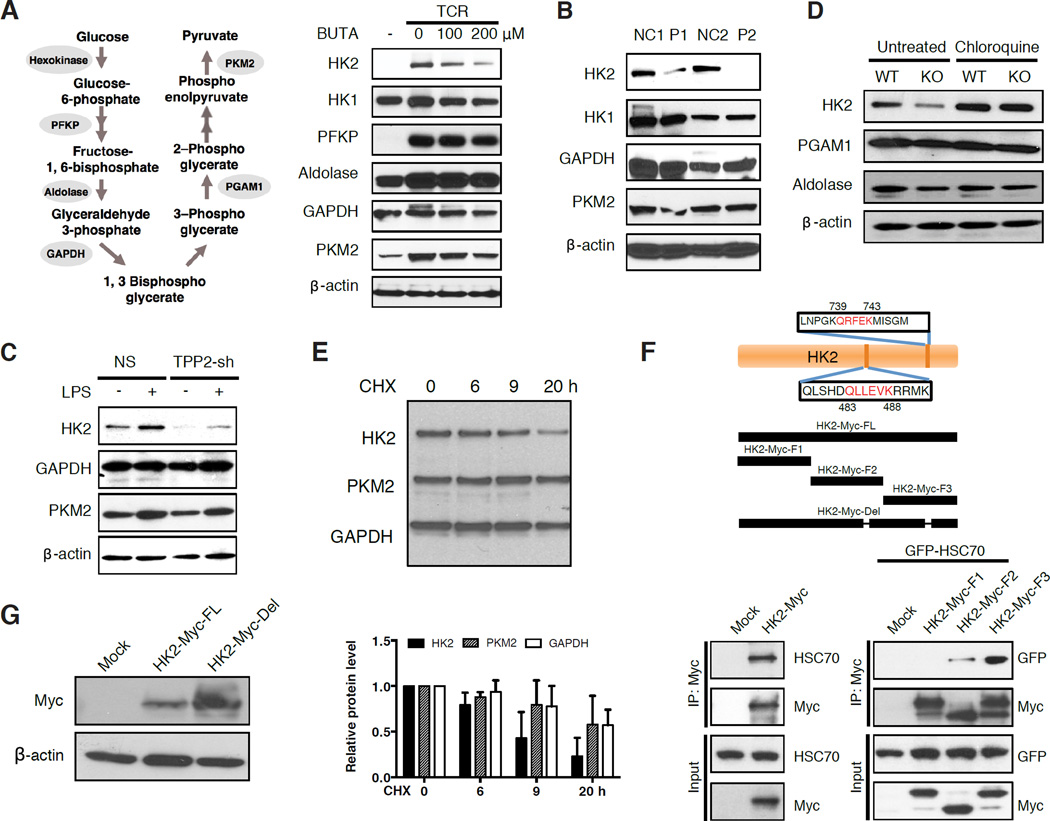
In mammalian cells, glycolysis proceeds through a series of enzymatic reactions that convert glucose to pyruvate and ATP (Figure 5A). We hypothesized that TPPII deficiency reduced glycolytic flux by decreasing glycolytic enzyme activity at one or more of these steps (Figure 5A). Consistent with previous observations that T cell activation upregulates transcripts for glycolytic enzymes (Stentz and Kitabchi, 2004; Wang et al., 2011), immunoblotting revealed that CD3/CD28 stimulation of human naïve CD4 T cells from healthy donors induced the expression of hexokinase-2 (HK2), phosphofructokinase (PFKP), pyruvate kinase (PKM2), and more modestly that of HK1, aldolase, phosphoglycerate mutase-1 (PGAM1), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Figure 5A); addition of IL-2 did not further alter levels (data not shown). Remarkably, treatment with BUTA greatly attenuated the induction of HK2 and PKM2 proteins, while minimally affecting HK1, PFKP, aldolase, GAPDH, and PGAM1 (Figure 5A and data not shown). By contrast, activated T cells from both P1 and P2 showed markedly decreased HK2, but no consistent decreases in HK1 or PKM2 (Figure 5B). Furthermore, activated T cells from Tpp2 KO mice showed decreased HK2 and aldolase, but not PGAM1 or PKM2 (Figures 5C and S5D). Similarly, TPP2 shRNA-transduced THP1 showed decreased HK2 at baseline and after lipopolysaccharide (LPS) stimulation (Figure 5D). Thus, only HK2 was consistently decreased. Because hexokinase performs the rate-limiting first step in glycolysis, this lack of HK2 could explain the profound reduction in glycolytic flux when TPPII was defective, although other glycolytic enzymes may be involved.
Figure 5. Targeting of HK2 for Lysosomal Degradation Compromises Glycolysis.
(A) Schematic of glycolytic enzymes (left) that were immunoblotted (right). Lysates were from human naïve CD4 T cells, activated for 3 days or not with the indicated concentrations of BUTA. (B) Similar to (A), except that lysates were from activated T cells from P1 and P2 or two healthy normal controls, cultured for 14 days in IL-2. (C) Similar to (B), except that lysates were from purified naïve CD4 T cells from KO or WT mice, after activation for 4 days, and without or with chloroquine for 6 hours. (D) Similar to (A), except that lysates were from THP1 cells stably transduced with TPP2 or non-specific shRNA, treated with PMA for 3 hours, and then without or with LPS for 6 hours. (E) Stability of HK2, PKM and GAPDH in cycloheximide (CHX)-treated A549 cells as assessed by densitometric scanning of immunoblots. (F) Schematic diagram showing the predicted KFERQ-like lysosomal targeting peptide motifs in the HK2 protein sequence, HK2 fragments F1, F2, and F3, and full-length HK2 mutant lacking both lysosomal targeting motifs (HK2-Myc-del). Below, co-immunoprecipitation of full-length HK2 but not GFP empty vector (mock) with endogenous HSC70 in SH-SY5Y cells (left), or HK2 fragments F1 to F3 with overexpressed GFP-HSC70 in 293T cells (right). (G) Immunoblot showing HK2 levels following transfection into 293T cells of HK2-Myc-del. Immunoblots are representative of experiments repeated at least 3 times. Data in (E) are represented as mean ± SD from 3 independent experiments. See also Figure S5.
To explore the mechanism of HK2 depletion, we first determined that HK2 mRNA was not decreased in TPPII-defective cells (Figure S5E). Next, we found that HK2 protein could colocalize with lysosomes (Figure S5F). Because TPPII-deficiency caused lysosomal expansion (Figures 3A to 3E, S4A, and S4B) and lysosomal expansion induced by forced TFEB overexpression decreased HK2 protein levels (Figure S5G), we postulated that HK2 was targeted for lysosomal degradation. Indeed, we observed that treating Tpp2 KO T cells with lysosomal inhibitors restored HK2 levels (Figures 5C and S5H). Moreover, when protein synthesis was blocked by cycloheximide, HK2 underwent more rapid protein turnover than other glycolytic enzymes (Figure 5E). The glycolytic enzymes aldolase, GAPDH, PGAM1, and PKM2 undergo chaperone-mediated autophagy (CMA) whereby the chaperone HSC70 mediates entry through the lysosomal LAMP2A receptor of substrates containing a KFERQ-like lysosomal degradation targeting peptide motif (Dice, 1990; Kaushik et al., 2011). PKM2 and GAPDH each have one experimentally validated motif, whereas HK2 has two such motifs (Figure 5F). To test their functionality, we overexpressed Myc-tagged HK2 fragments containing either the central QLLEVK motif (residues 483 to 488) or the C-terminal QRFEK motif (residues 760 to 764), or an N-terminal fragment lacking both motifs. Co-immunoprecipitations showed that full-length HK2 interacted with HSC70, as did fragments containing either of two lysosomal targeting motifs (F2, F3); however, co-immunoprecipitation of the N-terminal fragment lacking both motifs (F1) was decreased (Figure 5F). Moreover, given that overexpression of the HK2 fragment F2 showed partial degradation, its interaction with HSC70 was probably underestimated. Deletion of both lysosomal targeting motifs resulted in a non-degradable HK2 protein that was expressed to higher levels (Figure 5G). We also considered the possibility that macroautophagy contributed to the loss of HK2 in TPPII-deficient cells, but observed only slightly increased expression of LC3-II, a marker of autophagosomes, albeit levels increased with starvation (Figures S5I and S3G). Furthermore, normal cells, under the stress of amino acid starvation, showed only a minor decrease in HK2 by comparison with TPPII inhibition, suggesting that the response to internal amino acid starvation differs from external amino acid starvation (Figure S5H). Thus, HK2’s targeted entry into and degradation within lysosomes, which was markedly increased by TPPII deficiency, provides a molecular explanation for decreased glycolysis.
Reduced Glycolysis Impairs Adaptive and Innate Immunity in TPPII Deficiency
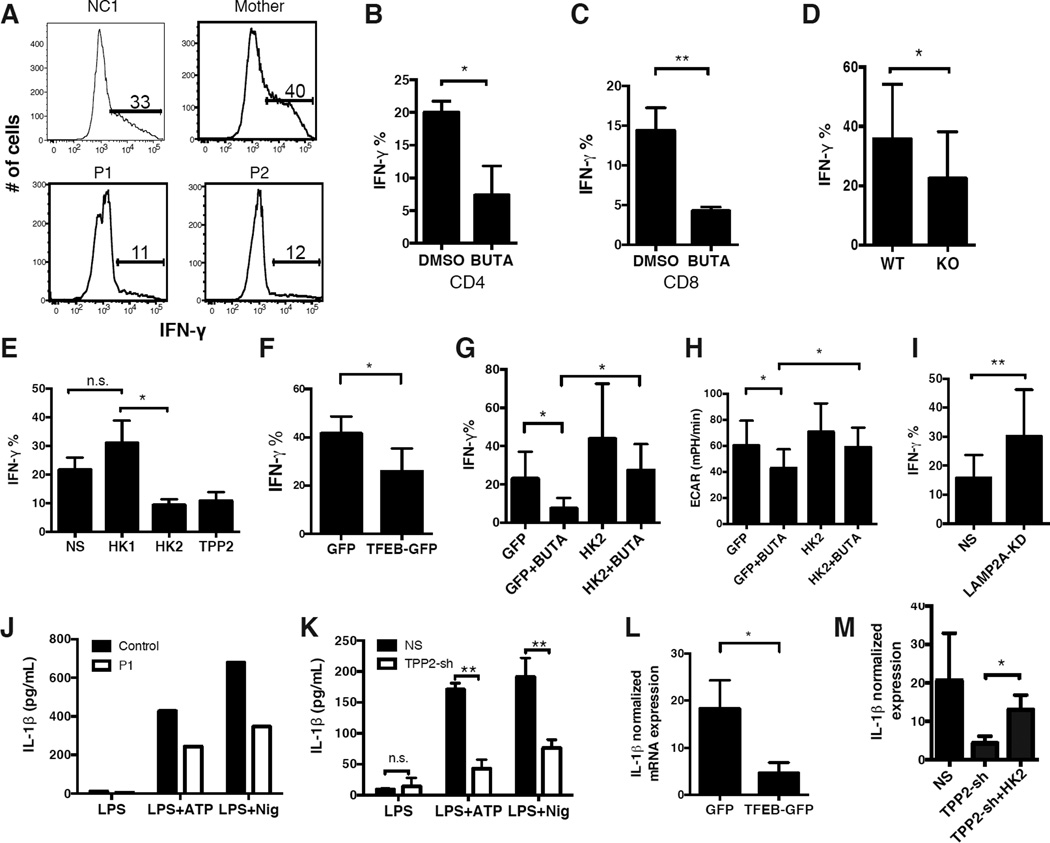
Oxidative phosphorylation is required for early activation of naïve T cells, and T cell proliferation can be fueled by either oxidative phosphorylation or aerobic glycolysis (Chang et al., 2013). Accordingly, the preservation of oxidative phosphorylation in TPPII-deficient T cells (Figures 4G and 4H) was consistent with their normal upregulation of activation markers and near normal proliferation after TCR stimulation (Figures S6A to S6G). However, a rapid transition to aerobic glycolysis is required to meet the increased biosynthetic and bioenergetic needs of effector functions in activated T cells, especially the acquisition of IFN-γ and IL-2 production in CD4 T cells (Chang et al., 2013). We therefore predicted that the glycolytic defect in TPPII-deficient T cells (Figures 4A to 4D, and 4F) would reduce their ability to produce effector cytokines. As expected, we found that naïve CD4 cells from P1 and P2 expressed less IFN-γ after activation (Figure 6A). Inhibiting TPPII by BUTA in activated CD4 or CD8 T cells from healthy normal control subjects reduced IFN-γ post-transcriptionally (Figures 6B, 6C, and S7A) and also reduced IL-2 (Figures S7B and S7C), with no effects on TNF-α production (Figure S7D). Moreover, mouse Tpp2 KO naïve CD4 T cells expressed less IFN-γ even when differentiated under TH1 polarizing conditions with exogenously added IL-2 (Figure 6D). These effector functions were specifically decreased because other cytokines such as IL-4 and IL-17 were unaffected by TPPII deficiency (Figures S7E and S7F). Consistent with our metabolic model, TPPII inhibition of IFN-γ production was recapitulated by siRNA silencing of HK2 but not HK1 (Figure 6E), or by overexpressing TFEB which decreases HK2 protein (Figures 6F and S5G). Chloroquine did not restore glycolysis or cytokine production, probably because undegraded HK2 remained sequestered and non-functional within lysosomes (Figure S5F and data not shown). Importantly, overexpression of WT HK2 in TPPII-inhibited T cells restored both glycolysis and IFN-γ expression (Figures 6G, 6H, and S7G). Rescue also occurred when a non-degradable mutant HK2 (lacking both lysosomal targeting motifs) was overexpressed, despite its partially impaired enzymatic activity (Figures S7H and S7I). Finally, when the receptor for CMA substrates was reduced using LAMP2A siRNA, HK2 levels and IFN-γ production were increased in TPPII-replete cells (Figures 6I and 5G), suggesting that CMA also tonically regulates HK2. Thus reduced glycolysis in TPPII-deficiency results from increased lysosomal degradation of HK2 via CMA, and leads to impaired T cell function.
Figure 6. Abnormal Metabolism Impairs Adaptive and Innate Immune Functions in TPPII Deficiency.
(A) Flow cytometric profiles showing intracellular IFN-γ in naïve CD4 T cells from P1, P2, mother, and a healthy normal control, activated for 3 to 4 days. (B–G, I) Quantification of IFN-γ-expressing T cells studied by flow cytometry as in (A). (B) Naïve CD4 T cells from healthy normal controls were treated with BUTA or DMSO during the course of stimulation. (C) Similar to (B) except with naïve CD8 T cells. (D) Naïve CD4 T cells from KO or WT mice were stimulated similarly as in (A), while differentiated under TH1-polarizing conditions. (E) Naïve CD4 T cells from healthy normal controls, previously transfected with siRNA against HK1, HK2, or TPP2, were treated as in (A). (F) Naïve CD4 T cells from healthy normal controls, previously transfected with GFP-tagged TFEB or GFP alone, were treated as in (A). (G) Similar to (F) except that cells were transfected with GFP-tagged HK2 or empty vector, and then treated as in (B). (H) Glycolytic flux of transfected cells as in (G), expressed as AUC of measured ECAR. (I) Similar to (E) except that cells were transfected with siRNA against LAMP2A and then treated as in (A). (J) IL-1β protein secretion by stimulated monocyte-derived macrophages from P1 or a healthy normal control. (K) Similar to (J) except using THP1 cells stably expressing TPP2 shRNA or non-specific shRNA. (L–M) Real-time PCR quantitation (qRT-PCR) of (IL-1β) transcripts following LPS stimulation of primed THP1 cells, normalized to HPRT (L) or ACTB (M). (L) THP1 cells previously transfected with GFP or TFEB-GFP. (M) THP1 TPP2 or NS shRNA cells, also transfected with GFP-tagged HK2 or empty vector. All experiments were repeated three times, and show representative profiles (A and J). Data in (B–I, K–M) are represented as mean ± SD from 3 independent experiments. n.s. non-significant, *P < 0.05, **P < 0.01, n.s. non-significant, as calculated by unpaired Student’s t-test or two-way ANOVA. See also Figure S7.
Aerobic glycolysis contributes not only to adaptive immunity, but also to innate immunity (Tannahill et al., 2013). Thus, we examined macrophage inflammatory responses after priming with the TLR agonist LPS and activating with ATP or nigericin. Monocyte-derived macrophages from P1 secreted less IL-1β than control (Figure 6J), as did TPP2 shRNA-transduced THP1 cells (Figure 6K). This inhibition was likely transcriptional as shown by reduced IL1B mRNA levels upon LPS priming of TPPII-deficient cells (Figures S7J and S7K), and could be recapitulated by overexpressing TFEB (Figures 6L and S5G). By contrast, TNFA mRNA levels (TNF-α) were less affected (Figures S7J and S7K). These defects would likely contribute to impaired responses to microbes or sterile tissue damage. HK2 overexpression in the TPP2 shRNA-transduced THP1 cells restored their IL-1β expression (Figures 6M, S7L, and S7M), similar to its rescue of IFN-γ production in T cells. Together, these results define autosomal recessive TPPII deficiency as a metabolic cause of primary immune deficiency in which reduced aerobic glycolysis leads to defects in both adaptive and innate immunity.
DISCUSSION
Despite its role as a peptidase downstream of the proteasome, previous studies of TPPII have focused on its ability to process antigenic peptides for MHC class I presentation. We now show that this giant cytoplasmic proteolytic complex has a more profound influence on overall amino acid homeostasis. Our data indicate that TPPII participates in recycling cellular proteins into amino acids, and that extracellular amino acid transport inefficiently compensates for its loss unless supraphysiological levels are provided. When TPPII function is inhibited, free intracellular amino acids are transiently decreased and lysosomal activity is elevated via mTOR and TFEB as a compensatory mechanism to restore amino acid homeostasis. However, the altered amino acid equilibrium in TPPII-deficient cells remains fragile to extracellular nutrient shortage, which likely occurs intermittently in vivo despite the lysosomal compensation at baseline. Consistent with previous reports that proteasomal inhibition increases lysosome abundance (Rideout et al., 2004; Ryhanen et al., 2009), our work now reveals the essential balance between the proteasomal and lysosomal degradation pathways for normal cell performance, and that TPPII is an important linchpin in this process.
TPPII-deficient cells with intact lysosomal function maintain amino acid homeostasis at the cost of reduced metabolic fitness. We observed that lysosomal hyperproliferation causes HK2 degradation through HSC70-chaperoned lysosomal degradation. The HK2 isoenzyme is upregulated in both lymphocytes and tumor cells that utilize aerobic glycolysis (the Warburg effect) and differs from HK1 which is constitutively expressed (Robey and Hay, 2006). Because hexokinases perform a rate-limiting step in glycolysis, the targeted degradation of HK2 strongly impedes glycolytic flux in TPPII-deficient cells. Several glycolytic enzymes are degraded through CMA, which increases during amino acid starvation (Dice, 1990; Kaushik et al., 2011). CMA of HK2 was not recognized previously possibly because an experimental model in which HK2 is not induced was used (Kon et al., 2011). Glycolysis can also be regulated by PFKP and PKM2, so the latter’s decrease upon acute TPPII inhibition may further compromise glycolysis. Nevertheless, we show that when TPPII is chronically lacking, HK2 consumption alone can explain the molecular regulation of glycolysis. It is likely that the pathologies caused by TPPII deficiency in cells normally dependent on HK2 are related to the cell-specific functions that are sensitive to glycolysis.
TPPII is widely expressed in different tissues, but at relatively higher levels in the immune system and tumors such as Burkitt’s lymphoma (Balow et al., 1986; Gavioli et al., 2001). These cell types sustain high levels of aerobic glycolysis. Moreover, HK2 but not HK1 promotes growth of tumor cells through effects on aerobic glycolysis and downstream biosynthetic pathways (Gershon et al., 2013; Patra et al., 2013; Wolf et al., 2011a). In the immune system, aerobic glycolysis is required for peripheral T cell responses and for thymocyte development (Greiner et al., 1994). Activated macrophages and dendritic cells also require aerobic glycolysis to generate biosynthetic intermediates including fatty acids necessary for production of membranes and proinflammatory cytokines (Everts et al., 2014; Tannahill et al., 2013). In TPPII deficiency, adaptive and innate immune cells function poorly in elaborating IFN-γ, IL-2, and IL-1β, which explains the patients’ susceptibility to a broad range of pathogens. Furthermore, the severe immune dysregulation characteristic of this disorder, including autoimmunity, may reflect the differential sensitivity of effector functions to impaired aerobic glycolysis and secondary metabolic defects.
The role of metabolic processes in immune cell functions has been mainly elucidated through in vitro studies using pharmacological agents such as 2-DG or mice as tools in which transcriptional regulators such as HIF1α were selectively ablated. By studying a new human immunodeficiency, we now better understand the requirements and physiological consequences of metabolic reprogramming on immune functions in vivo. We find that TPPII deficiency produced less impairment of glycolytic flux than 2-DG treatment (Lampidis et al., 2006), consonant with its selective effect on HK2 that spares oxidative phosphorylation and hence T cell proliferation (Cham et al., 2008; Miller et al., 1994). Thus, our data showing unchanged IL-4 and IL-17 can be reconciled with previous reports that glycolytic blockade by 2-DG in mice interrupts not only IFN-γ, but also IL-4 and IL-17 production (Shi et al., 2011).
Finally, although hematopoietic stem cell transplantation would be expected to cure the immune abnormalities in patients with TPPII deficiency, neurometabolic correction presents a greater challenge. In the brain, TPPII degrades neuropeptides to regulate satiety (Rose et al., 1996; Wilson et al., 1993). However, HK2 is also dynamically expressed in the developing brain, mirroring the pattern of whole brain glucose consumption, which is high during early childhood before decreasing by adulthood (Gershon et al., 2013; Wolf et al., 2011b). Increased utilization of aerobic glycolysis, which predominates in brain regions associated with transcriptional neoteny, may contribute to synapse formation and growth (Goyal et al., 2014). Thus, our finding that TPPII inhibition impaired glycolysis in a neuroblastoma cell line suggests the intriguing hypothesis that the developmental delay in TPPII deficiency is mechanistically linked to defective aerobic glycolysis. Because of this unusual constellation of features we propose this disorder be called “TPP2-Related Immunodeficiency, Autoimmunity, and Neurodevelopmental delay with impaired Glycolysis and Lysosomal Expansion” (TRIANGLE) disease.
EXPERIMENTAL PROCEDURES
Study Subjects and Mice
Patients and their relatives provided written informed consent to participate in research protocols approved by the NIAID, NIH Institutional Review Board (NCT00246857) or the Newcastle and North Tyneside 1 Research Ethics Committee. Whole blood samples, primary dermal fibroblast cultures, and buffy coat cells were obtained. Mice were bred, purchased, and used under animal study protocols approved by the NIAID Animal Care Use Committee. TppII-deficient mice were from Kenneth Rock (University of Massachusetts, Worcester) (Kawahara et al., 2009). Details are provided in Extended Experimental Procedures.
Genomic Analyses
WES of genomic DNA was performed for P1 and P3, and genome-wide linkage scans for P3 and P4. Details of analyses including alignment, variant calling, and filtering are provided in Extended Experimental Procedures. Mutations were confirmed by Sanger sequencing. WES data are deposited in dbGaP.
Molecular Modeling
The HsTPPII spindle complex was used to model the G500D mutation. Details are provided in Extended Experimental Procedures.
Cell Isolation and Culture
Human PBMC, human peripheral blood T cells (including naïve CD4 or CD8 T cells), human monocyte-derived macrophages, mouse splenic naïve CD4 or CD8 T cells, MEF, human fibroblasts, THP1, 293T, A549, SH-SY5Y, Jurkat, and HeLa cell lines were used. Details are provided in Extended Experimental Procedures.
Immunoblotting
Immunoblotting was performed according to standard methods. To assess protein degradation rates, A549 cells were transfected with myc-tagged mammalian expression constructs; 24 hours later, cycloheximide (10 µg/mL) was added and cells cultured up to 20 more hours before lysis and immunoblotting for myc. Details are provided in Extended Experimental Procedures.
TPPII Enzymatic Activity
Cell lysates were incubated with Ala-Ala-Phe-4-methylcoumaryl-7-amide (AAF-AMC) (Enzo Life Sciences), and fluorescence emission measured. Details are provided in Extended Experimental Procedures.
Native In-gel AAF-AMC Cleavage and Immunoblotting
Cell lysates were separated by NativePAGE, followed by either transfer to PVDF membranes for immunoblotting or in-gel incubation with AAF-AMC to assess TPPII enzymatic activity. Details are provided in Extended Experimental Procedures.
Cell Treatments
Human fibroblasts were treated in medium containing reduced amino acids and MEF in amino acid free media for 6 hours (USBiological Life Sciences). PBMC were stimulated with anti-human CD2/3/28 beads (Miltenyi Biotech) for T cell activation and CFSE-labeled cell proliferation assays. Cells were treated with the TPPII-specific inhibitor butabindide (Santa Cruz Biotechnology; Tocris Bioscience) at 200 or 250 µM. Purified human or mouse naïve CD4 T or CD8 cells were stimulated with anti-CD3/CD28 antibodies; subset-polarizing recombinant cytokines and antibodies against mouse cytokines were added for T helper cell differentiation studies. Human monocyte-derived macrophages were stimulated with LPS (100 ng/mL) for 6 hours, then ATP (5 mM) or nigericin (10 mM) for 30 more minutes (all from Invivogen). THP1 cells were stimulated with phorbol 12-myristate 13-acetate (PMA, 100 ng/mL; Sigma-Aldrich) for 3 hours, then LPS (1 µg/mL) for 3 to 48 hours, and in some cases ATP (5 mM) or nigericin (10 mM) for 30 more minutes. Cells were treated for 6 hours with MG132 (5 mM, Enzo Life Science), chloroquine (100 mM), or ammonium chloride (20 mM) plus leupeptin (100 µM) (both from Sigma-Aldrich). Details are provided in Extended Experimental Procedures.
Intracellular Free Amino Acid Analyses
Samples were analyzed for intracellular free amino acids by Waters ACQUITY ultra-performance liquid chromatography (UPLC) after AccQ-Tag derivatization, or by optical absorbance after reacting with ninhydrin. Details are provided in the Extended Experimental Procedures.
Lentiviral Transductions and Transient Transfections
THP1 cells were stably transduced with lentivirus encoding shRNA against human TPP2 or control shRNA. Human CD4 naïve T cells were Amaxa nucleofected with 200 nM siRNA against LAMP2A, HK1 or HK2 (Life Technologies). Mammalian expression constructs encoding tagged forms of TFEB, HK1, HK2, PKM2, or GAPDH were transfected by nucleofection or Turbofect. Details are provided in Extended Experimental Procedures.
Transmission Electron Microscopy
Transmission electron microscopy of human cycling T cells was performed after fixation in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4. Details are provided in Extended Experimental Procedures.
Lysosomal Imaging and Quantitation
The lysosome specific dyes Lysotracker Red (Life Technology) and Lyso-ID (Enzo Life Sciences) were used to stain intracellular lysosomes for confocal microscopy. Staining for T cell surface markers, LysoID, and LC3 for autophagosomes were performed for ImageStream analyses. Details are provided in Extended Experimental Procedures.
Lysosomal Enzymatic Activity
Preparations enriched in lysosomes were collected from mouse liver tissue homogenates using the Lysosome Isolation Kit with density gradient ultracentrifugation. The fraction containing the majority of the lysosomes, as determined by immunoblotting of lysosomal proteins, was used to measure lysosomal acid phosphatase and β-N-acetyl-glucosaminidase activities (all from Sigma-Aldrich). Details are provided in Extended Experimental Procedures.
Quantitative RT-PCR
Total RNA was isolated with the RNeasy kit (QIAGEN). cDNA was synthesized from 1 µg of RNA using the iScript cDNA Synthesis Kit (Bio-Rad). All quantitative RT-PCR was performed by the SYBR green method on 7900HT machine (ABI), using 200 pg of cDNA per reaction. The expression of mRNA for genes of interest was normalized to that of β-actin or HPRT. Primer sequences are provided in Extended Experimental Procedures.
TFEB Subcellular Localization
24 hours after transfecting a C-terminal EGFP-fused TFEB plasmid (Roczniak-Ferguson et al., 2012), cells were fixed with 4% paraformaldehyde in PBS, their nuclei stained with DAPI (Enzo Life Sciences), and imaged by confocal microscopy as described above for Lysosomal imaging and quantitation. For analysis of % nuclear colocalization, single cells were analyzed by Imaris with threshold settings of 100 in both GFP and DAPI channels. Ten TFEB-GFP positive cells were analyzed per experimental group. Details are provided in Extended Experimental Procedures.
Metabolic Analyses
Glycolysis was evaluated by measuring ECAR at baseline, and after adding glucose (10 mM), oligomycin (10 µM), and 2-deoxy-D-glucose (100 mM). Oxidative phosphorylation was evaluated by measuring OCR at baseline, and after adding oligomycin (5 µM), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP, 1.5 µM), and rotenone (100 nM) plus antimycin (1 µM). Measurements were made on an XF-96 Extracellular Flux Analyzer using the XF Glycolysis or Cell Mito stress test kits (Seahorse Bioscience). Glycolytic flux was also measured by monitoring conversion of [5-3H]-glucose to [5-3H]-water. Details are provided in Extended Experimental Procedures.
Co-immunoprecipitations
24 hours after transfection of SH-SY5Y or 293T cells with GFP-HSC70 and myc-HK2 expression plasmids, supernatants from cell lysates were incubated for 8 hours at 4°C with Dynal beads (Life Technology) plus anti-myc tag antibody (Cell Signaling Technology). Beads were washed with ice-cold lysis buffer and the bound proteins eluted for SDS-PAGE and immunoblotting as described above for Immunoblotting. Details are provided in Extended Experimental Procedures.
Flow Cytometric Analyses
Standard flow cytometry methods were used to evaluate surface markers of T cell activation (CD25, CD69), intracellular cytokines (IFN-γ, IL-2, IL-4, IL-17, TNF-α), mTOR activity (phospho-S6), and cell division (CFSE dilutions). Data were acquired on a BD LSRII instrument using FACSDiva software and analyzed using FlowJo software (Tree Star). Details are provided in Extended Experimental Procedures.
ELISA Quantitation of Cytokines
IL-1β was measured in conditioned media, after treating monocyte-derived macrophages or THP1 cells as described above, using an IL-1β ELISA kit (Biolegend). Details are provided in Extended Experimental Procedures.
HK2 Rescue
24 hours after transfection with a carboxyl-tagged GFP fusion protein of HK2 or derivatives thereof, human naïve CD4 T cells were stimulated for 4 days with anti-CD3/CD28 antibodies. HK2-transfected THP1 cells were treated with PMA (100 ng/mL) for 3 hours, then LPS (1 µg/mL) for 48 more hours. Details are provided in Extended Experimental Procedures.
Statistical Analyses
The Prism 5 software package (GraphPad) was used to calculate p-values using the unpaired two-tailed Student’s t-test, one-sample t-test, one-way ANOVA, or two-way ANOVA.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the following people: for technical help, David Dorward, Louise Tee, Sonia Majri, Matt Biancalana, Alex Leney-Greene, Sam Drain, and Yaobo Xu; for clinical support, Angela Wang, Emily Jewel, Shanaz Pasha, and colleagues at the BC Children’s Hospital and the Great North Children’s Hospital; for sharing reagents, data, and equipment, Kenneth Rock, Beate Rockel, Thomas von Zglinicki, and Richard Siegel; for help with genetic investigations, Eamonn Maher and the Newcastle Exome Consortium, especially John Loughlin and Patrick Chinnery; for advice and critically reading the manuscript, Karin Engelhardt, Richard Siegel, Fengyi Wan, and Li Yu; and the patients and their families for participating in this study. This work was supported by the Intramural Research Program of the NIAID, NIH (H.C.S., M.J.L.); the UK MRC (S.H.); the Sir Jules Thorn Charitable Trust (S.H., M.S.K.); the NIHR Newcastle Biomedical Research Centre and the Newcastle upon Tyne Hospitals NHS Charity (S.H., M.S.K., V.I.K.); UK BBSRC (B.C., V.I.K.); the Canadian Institutes of Health Research (S.E.T., MOP-133691). S.E.T. is a Clinical Scholar of the Michael Smith Foundation for Health Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
W.L. generated TPP2 shRNA THP1 cells, measured intracellular free amino acids and HK activity, performed HK2 rescue, co-immunoprecipitations, and quantified cytokine production by flow cytometry and ELISA. Y.Z., and independently D.O.M., L.N.R., M.S.K., N.V.M., and H.G., discovered the patients’ TPP2 mutations and evaluated their effects on expression. D.O.M. performed native gel electrophoresis and evaluated TPPII enzymatic activity. D.O.M. discovered and W.L. characterized defective glycolysis. W.L., D.O.M., H.J., and Q.Z. evaluated T cell activation and proliferation. H.J. discovered, and W.L., S.S., A.K.S., V.I.K., B.C., and Q.Z. characterized, the lysosomal expansion. W.L., H.J., and H.M.M. performed immunoblotting of glycolytic enzymes. L.Z. performed radiotracer studies of glycolysis. W.L. and Q.Z. performed confocal microscopy. Y.Z., W.L., H.J., and H.M.M. performed qRT-PCR experiments. H.J. generated HK2 mutant expression constructs. B.C., N.R., and V.I.K. assessed mTOR activity and autophagy. J.H.L. modeled the effects of the mutation on protein structure. S.E.T., T.P., S.J.H., J.H., S.H. and A.J.C. provided clinical care and collected clinical data to characterize the patients. H.F.M. coordinated clinical study protocol and sample collection. H.C.S., M.J.L., and S.H. planned and supervised the experimental work and data analyses. Y.Z., W.L., H.C.S., and S.H. prepared the manuscript. All authors discussed and commented on the manuscript.
REFERENCES
- Balow RM, Tomkinson B, Ragnarsson U, Zetterqvist O. Purification, substrate specificity, and classification of tripeptidyl peptidase II. J Biol Chem. 1986;261:2409–2417. [PubMed] [Google Scholar]
- Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DM, Calvert CC, Klasing KC. Source of amino acids for tRNA acylation. Implications for measurement of protein synthesis. Biochem J. 1992;283(Pt 2):583–589. doi: 10.1042/bj2830583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cham CM, Driessens G, O'Keefe JP, Gajewski TF. Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8+ T cells. Eur J Immunol. 2008;38:2438–2450. doi: 10.1002/eji.200838289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O'Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ. 2005;12:1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- Everts B, Amiel E, Huang SC, Smith AM, Chang CH, Lam WY, Redmann V, Freitas TC, Blagih J, van der Windt GJ, et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKvarepsilon supports the anabolic demands of dendritic cell activation. Nat Immunol. 2014;15:323–332. doi: 10.1038/ni.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavioli R, Frisan T, Vertuani S, Bornkamm GW, Masucci MG. c-myc overexpression activates alternative pathways for intracellular proteolysis in lymphoma cells. Nat Cell Biol. 2001;3:283–288. doi: 10.1038/35060076. [DOI] [PubMed] [Google Scholar]
- Gershon TR, Crowther AJ, Tikunov A, Garcia I, Annis R, Yuan H, Miller CR, Macdonald J, Olson J, Deshmukh M. Hexokinase-2-mediated aerobic glycolysis is integral to cerebellar neurogenesis and pathogenesis of medulloblastoma. Cancer & metabolism. 2013;1:2. doi: 10.1186/2049-3002-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal MS, Hawrylycz M, Miller JA, Snyder AZ, Raichle ME. Aerobic glycolysis in the human brain is associated with development and neotenous gene expression. Cell metabolism. 2014;19:49–57. doi: 10.1016/j.cmet.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner EF, Guppy M, Brand K. Glucose is essential for proliferation and the glycolytic enzyme induction that provokes a transition to glycolytic energy production. J Biol Chem. 1994;269:31484–31490. [PubMed] [Google Scholar]
- Huai J, Firat E, Nil A, Million D, Gaedicke S, Kanzler B, Freudenberg M, van Endert P, Kohler G, Pahl HL, et al. Activation of cellular death programs associated with immunosenescence-like phenotype in TPPII knockout mice. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5177–5182. doi: 10.1073/pnas.0801413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Bandyopadhyay U, Sridhar S, Kiffin R, Martinez-Vicente M, Kon M, Orenstein SJ, Wong E, Cuervo AM. Chaperone-mediated autophagy at a glance. J Cell Sci. 2011;124:495–499. doi: 10.1242/jcs.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara M, York IA, Hearn A, Farfan D, Rock KL. Analysis of the role of tripeptidyl peptidase II in MHC class I antigen presentation in vivo. J Immunol. 2009;183:6069–6077. doi: 10.4049/jimmunol.0803564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon M, Kiffin R, Koga H, Chapochnick J, Macian F, Varticovski L, Cuervo AM. Chaperone-mediated autophagy is required for tumor growth. Science translational medicine. 2011;3:109ra117. doi: 10.1126/scitranslmed.3003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- Lampidis TJ, Kurtoglu M, Maher JC, Liu HP, Krishan A, Sheft V, Szymanski S, Fokt I, Rudnicki WR, Ginalski K, et al. Efficacy of 2-halogen substituted D-glucose analogs in blocking glycolysis and killing "hypoxic tumor cells". Cancer Chemoth Pharm. 2006;58:725–734. doi: 10.1007/s00280-006-0207-8. [DOI] [PubMed] [Google Scholar]
- MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay RM, McKay JP, Suh JM, Avery L, Graff JM. Tripeptidyl peptidase II promotes fat formation in a conserved fashion. EMBO reports. 2007;8:1183–1189. doi: 10.1038/sj.embor.7401086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ES, Klinger JC, Akin C, Koebel DA, Sonnenfeld G. Inhibition of murine splenic T lymphocyte proliferation by 2-deoxy-D-glucose-induced metabolic stress. J Neuroimmunol. 1994;52:165–173. doi: 10.1016/0165-5728(94)90110-4. [DOI] [PubMed] [Google Scholar]
- Patra KC, Wang Q, Bhaskar PT, Miller L, Wang Z, Wheaton W, Chandel N, Laakso M, Muller WJ, Allen EL, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013;24:213–228. doi: 10.1016/j.ccr.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reits E, Neijssen J, Herberts C, Benckhuijsen W, Janssen L, Drijfhout JW, Neefjes J. A major role for TPPII in trimming proteasomal degradation products for MHC class I antigen presentation. Immunity. 2004;20:495–506. doi: 10.1016/s1074-7613(04)00074-3. [DOI] [PubMed] [Google Scholar]
- Rideout HJ, Lang-Rollin I, Stefanis L. Involvement of macroautophagy in the dissolution of neuronal inclusions. The international journal of biochemistry & cell biology. 2004;36:2551–2562. doi: 10.1016/j.biocel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Robey RB, Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25:4683–4696. doi: 10.1038/sj.onc.1209595. [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Science signaling. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose C, Vargas F, Facchinetti P, Bourgeat P, Bambal RB, Bishop PB, Chan SM, Moore AN, Ganellin CR, Schwartz JC. Characterization and inhibition of a cholecystokinin-inactivating serine peptidase. Nature. 1996;380:403–409. doi: 10.1038/380403a0. [DOI] [PubMed] [Google Scholar]
- Ryhanen T, Hyttinen JM, Kopitz J, Rilla K, Kuusisto E, Mannermaa E, Viiri J, Holmberg CI, Immonen I, Meri S, et al. Crosstalk between Hsp70 molecular chaperone, lysosomes and proteasomes in autophagy-mediated proteolysis in human retinal pigment epithelial cells. J Cell Mol Med. 2009;13:3616–3631. doi: 10.1111/j.1582-4934.2008.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonegge AM, Villa E, Forster F, Hegerl R, Peters J, Baumeister W, Rockel B. The structure of human tripeptidyl peptidase II as determined by a hybrid approach. Structure. 2012;20:593–603. doi: 10.1016/j.str.2012.01.025. [DOI] [PubMed] [Google Scholar]
- Schutz Y. Protein turnover, ureagenesis and gluconeogenesis. International journal for vitamin and nutrition research Internationale Zeitschrift fur Vitamin- und Ernahrungsforschung Journal international de vitaminologie et de nutrition. 2011;81:101–107. doi: 10.1024/0300-9831/a000064. [DOI] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stentz FB, Kitabchi AE. Transcriptome and proteome expression in activated human CD4 and CD8 T-lymphocytes. Biochem Biophys Res Commun. 2004;324:692–696. doi: 10.1016/j.bbrc.2004.09.113. [DOI] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PM. Role of amino acid transporters in amino acid sensing. Am J Clin Nutr. 2014;99:223S–230S. doi: 10.3945/ajcn.113.070086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson B. Tripeptidyl peptidases: enzymes that count. Trends Biochem Sci. 1999;24:355–359. doi: 10.1016/s0968-0004(99)01435-8. [DOI] [PubMed] [Google Scholar]
- Valdor R, Mocholi E, Botbol Y, Guerrero-Ros I, Chandra D, Koga H, Gravekamp C, Cuervo AM, Macian F. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol. 2014;15:1046–1054. doi: 10.1038/ni.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Gibson AM, McDermott JR. Purification and characterization of tripeptidylpeptidase-II from post-mortem human brain. Neurochem Res. 1993;18:743–749. doi: 10.1007/BF00966768. [DOI] [PubMed] [Google Scholar]
- Wolf A, Agnihotri S, Micallef J, Mukherjee J, Sabha N, Cairns R, Hawkins C, Guha A. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011a;208:313–326. doi: 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A, Agnihotri S, Munoz D, Guha A. Developmental profile and regulation of the glycolytic enzyme hexokinase 2 in normal brain and glioblastoma multiforme. Neurobiology of disease. 2011b;44:84–91. doi: 10.1016/j.nbd.2011.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.