SUMMARY
Plant stem cells in the shoot apical meristem (SAM) and root apical meristem (RAM) provide for postembryonic development of above-ground tissues and roots, respectively, while secondary vascular stem cells sustain vascular development1–4. WUSCHEL (WUS), a homeodomain transcription factor expressed in the rib meristem of the SAM, is a key regulatory factor controlling stem cell populations in the Arabidopsis SAM5–6 and is thought to establish the shoot stem cell niche via a feedback circuit with the CLAVATA3 (CLV3) peptide signaling pathway7. WUSCHEL-RELATED HOMEOBOX5 (WOX5), specifically expressed in root quiescent center (QC), defines QC identity and functions interchangeably with WUS in control of shoot and root stem cell niches8. WOX4, expressed in Arabidopsis procambial cells, defines the vascular stem cell niche9–11. WUS/WOX family proteins are evolutionarily and functionally conserved throughout the plant kingdom12 and emerge as key actors in the specification and maintenance of stem cells within all meristems13. However, the nature of the genetic regime in stem cell niches that centers on WOX gene function has been elusive, and molecular links underlying conserved WUS/WOX function in stem cell niches remain unknown. Here we demonstrate that the Arabidopsis HAIRY MERISTEM (HAM)family transcription regulators act as conserved interacting co-factors with WUS/WOX proteins. HAM and WUS share common targets in vivo and their physical interaction is important in driving downstream transcriptional programs and in promoting shoot stem cell proliferation. Differences in the overlapping expression patterns of WOX and HAM family members underlie the formation of diverse stem cell niche locations, and the HAM family is essential for all of these stem cell niches. These findings establish a new framework for the control of stem cell production during plant development.
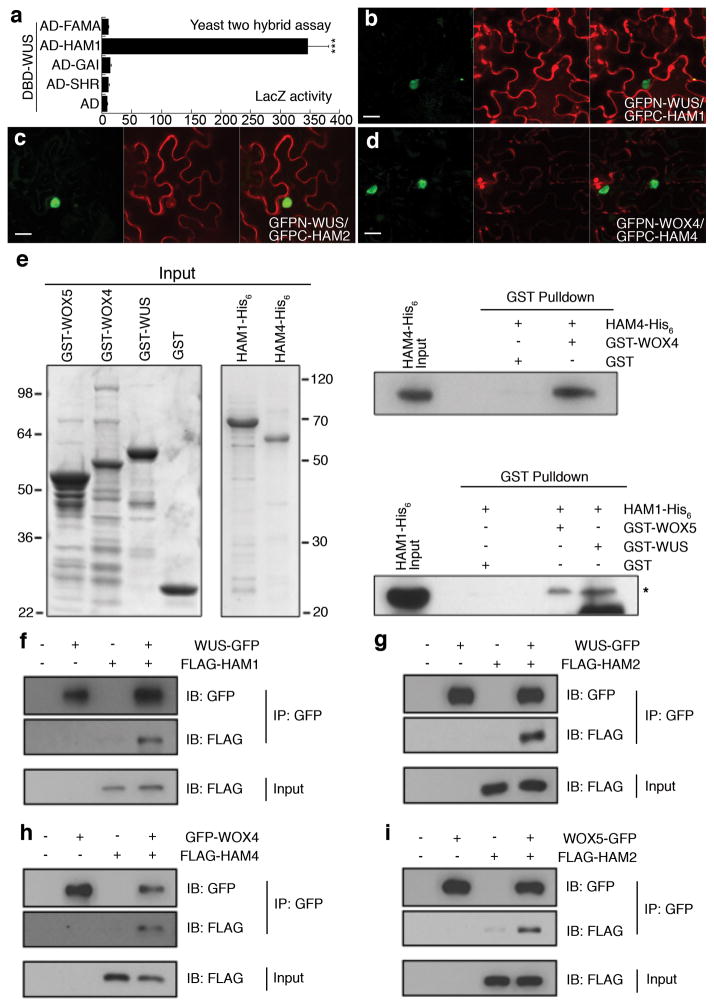
To identify the molecular mechanism underlying WUS functions in stem cells, we screened for WUS-interacting transcription co-factors using yeast-two-hybrid assays with a transcription factor library14, and found HAIRY MERISTEM1 (HAM1) strongly and specifically interacting with WUS(Fig. 1a). HAM genes, encoding GRAS domain transcription regulators, contribute to shoot stem cell function in petunia and Arabidopsis15–17. Four HAM genes (HAM1-HAM4) have been identified in Arabidopsis16, and further yeast assays revealed that WUS also interacted with three other HAM family members (Extended Data Fig. 1a). WUS-HAM associations were confirmed by bimolecular fluorescence complementation (BiFC) assays in tobacco (Nicotiana benthamiana), where WUS and HAM were fused to the N- and C-terminal halves of green fluorescent protein (GFP), respectively. Strong GFP fluorescence in nuclei was observed when GFPn-WUS was co-transformed with GFPc-HAM (Fig. 1b–c, Extended Data Fig. 1b–e). WOX4 and WOX5 also interacted with HAM protein sin BiFC assays (Fig. 1d, Extended Data Fig. 1f–q). These WOX-HAM interactions were further confirmed through in vitro pull-down assays, where glutathione S-transferase (GST)-WOX4 but not GST bound HAM4-His6, and GST-WUS but not GST bound HAM1-His6 (Fig. 1e). Interactions in planta were then tested using co-immunoprecipitation assays in tobacco, where WUS-GFP bound FLAG-HAM1 (Fig. 1f) and FLAG-HAM2 (Fig. 1g), GFP-WOX4 bound FLAG-HAM4 (Fig. 1h), and WOX5-GFP boundFLAG-HAM2 (Fig. 1i). In short, with multiple approaches, our work revealed physical interactions between HAM and WUS/WOX family members.
Figure 1. WUS/WOX and HAM family proteins physically interact.
(a) LacZ activity of yeast-two-hybrid assays. Error bar = mean ± sem (n=3biological replicates). ***, P<0.001(two-tailed t-test). (b–d) Bimolecular fluorescence complementation in tobacco. Panels (left to right): GFP; propidium iodide (PI) staining; merged channels. Scale bar = 20μm.(e) SDS-PAGE of input recombinant proteins stained by Coomassie Blue (left), and pull down of His6-tagged HAM proteins through GST-tagged WUS/WOX proteins detected by immunoblotting with anti-His antibody (right). Asterisk: HAM1-His6band. (f–j) Co-immunoprecipitation of WUS-GFP and FLAG-HAM1 (f), WUS-GFP and FLAG-HAM2 (g), GFP-WOX4 and FLAG-HAM4(h), WOX5-GFP and FLAG-HAM2(i) (see Methods).
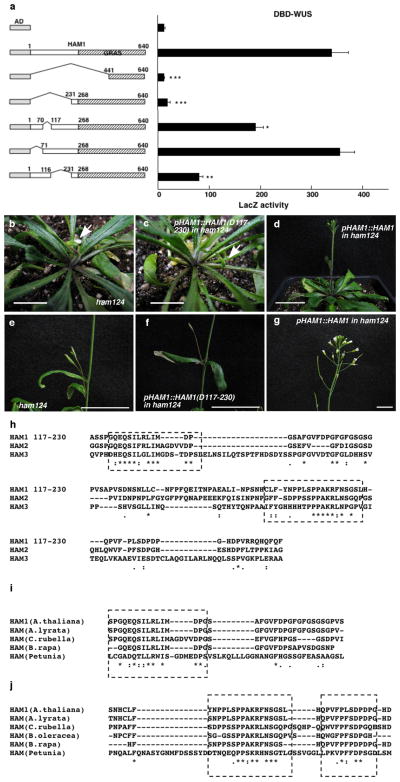
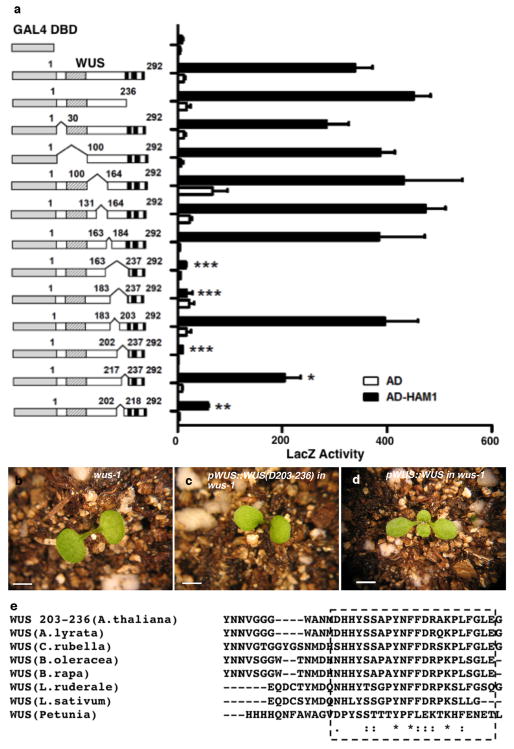
We next constructed various deleted derivatives of HAM1 and WUS for yeast-two-hybrid assays to identify essential regions for their interactions. Deleting amino acids from 117 to 230(D117-230) in HAM1abolished the interaction (Extended Data Fig. 2a). This N-terminal fragment is important for HAM1 function in stem cell maintenance, asHAM1 (D117-230)did not complement the ham1;2;4early termination phenotype, while full-length HAM1 driven by the same HAM1 promoter did(Extended Data Fig. 2b–g), and it is conserved in HAM proteins from Arabidopsis and across different plant species (Extended Data Fig. 2h–j). Deletion analyses of WUS identified a C-terminal region required for interaction with HAM1 (Extended Data Fig. 3a), which is also required for WUS function (Extended Data Fig. 3b–d) and is conserved in different plant species (Extended Data Fig. 3e).
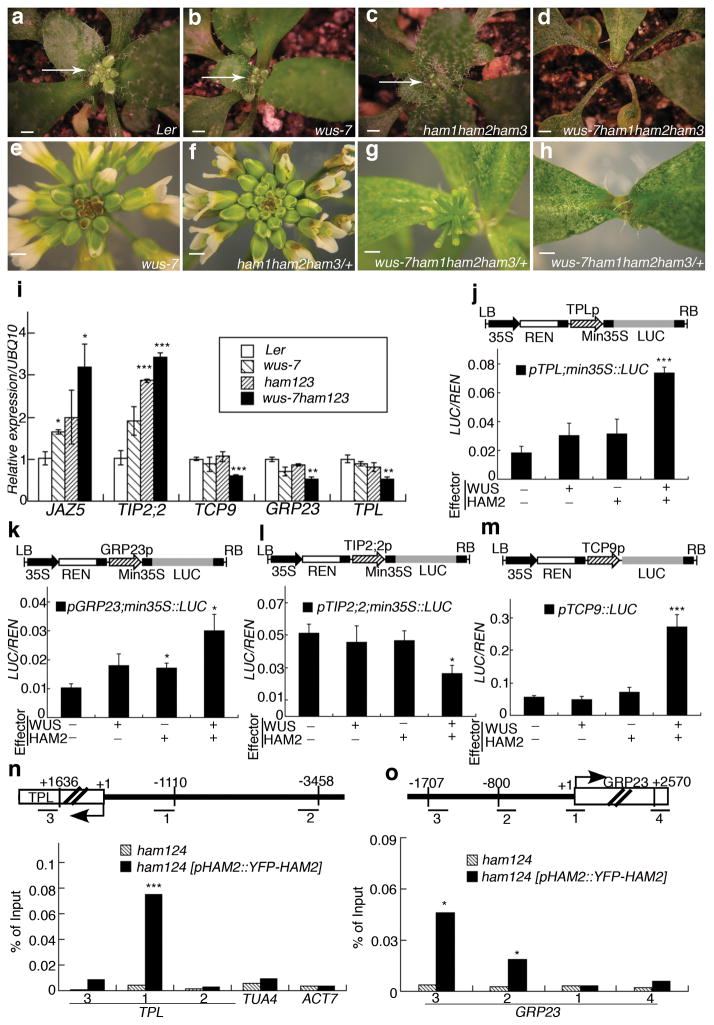
To dissect roles of the HAM-WUS interaction in controlling shoot stem cell niches, genetic interactions were analyzed between ham1;2;3 (lacking function of three of four HAM genes) and the weak wus allele wus-7(missense mutant) which forms a functional shoot apex18, similar to wild type in terms of vegetative and inflorescence meristems (Fig. 2a, b, e). Different fromwus-7single mutants (Fig. 2b) or ham1;2;3 triple mutants (Fig. 2c), wus-7;ham1;2;3 quadruple mutants display early termination of vegetative meristem development(Fig. 2d), thus resembling wus complete loss of function (null) mutants5. This effect also occurred in wus-7/wus-7;ham1/ham1;ham2/ham2;ham3/+ plants, where 41 out of 45 plants showed strong termination of inflorescence and floral meristems, with only leaves (Fig. 2h) or barren pedicels (flowers without carpels) (Fig. 2g) left at the top of the main shoot, a phenotype typical of wus-1 null mutants5, but never observed in wus-7 (Fig. 2e) or ham1/ham1, ham2/ham2, ham3/+ (Fig. 2f) plants. Secondary inflorescence meristems initiated from axillary meristems inwus-7/wus-7;ham1/ham1;ham2/ham2;ham3/+ also terminated prematurely (Extended Data Fig. 4a–b). Additionally, 3 out of 4wus-7/wus-7;ham1/ham1;ham2/ham2;ham4/+ plants displayed inflorescence meristem termination and lacked carpels (Extended Data Fig. 4c). A dose-dependent enhancement of stem cell termination was evident in wus-7;ham1/+;ham2/+;ham3/+, and wus-7; ham1/+;ham2/ham2;ham3/ham3 backgrounds (Extended Data Fig. 4d–f), demonstrating a functional interdependence between WUS and HAM family members in vivo. Down regulation of HAM1, HAM2 and HAM3 in a ham4 shoot meristem, through activation of microRNA171reported to target HAM1, HAM2 and HAM3 genes19, led to terminated vegetative development (Extended Data Fig. 4g–h)similar to the wus-1phenotype, suggesting that WUS alone is not sufficient to maintain SAMs in the absence of HAM activity. Lastly, the wus-1;ham1;2;3 quadruple homozygote resembles a wus-1single mutant in several aspects including the vegetative meristem (Extended Data Fig. 4i–l), suggesting that WUS and HAM genes could act together at the SAM. All these genetic data are consistent with the hypothesis that WUS and HAM function as partners in shoot meristem maintenance.
Figure 2. WUS and HAM family genes cooperatively control the shoot stem cell niche and co-regulate a common gene set.
Shoot apices (a–d) (arrows) and inflorescence structures(e–h)of plants in indicated genotypes. Bars = 2 mm in (a–h).(i) RT-PCR quantification of WUS and HAM target gene expression in indicated genotypes. Error bar = mean ± sem(n=3 biological replicates). (j–m) LUC/REN activity in tobacco cells co-transformed with different reporter constructs (structured above each graph) and indicated effectors(see Methods). Min35S: 60 base pair 35S minimum element, REN: Renilla luciferase, LUC: firefly luciferase, LB/RB: T-DNA left or right border. Error bar = mean ± sem(n=3biological replicates). (n–o) Chromatin Immunoprecipitation of HAM2 protein with TPL or GRP23 chromatin regions, with ampliconlocations (Bars with numbers) diagrammed above each graph. The ChIP experiments were repeated three times using independent biological replicates with similar results, and one representative data set is shown.*, P<0.05; **, P<0.01; ***, P<0.001(two-tailed t-test) in (i–o).
In addition to genetic interactions, the molecular function of the WUS-HAM interaction was further investigated. First, quantitative RT-PCR results (Fig. 3i) demonstrated that HAM proteins regulate expression of a set of genes including JAZ5, TIP2;2, TCP9, GRP23 and TPL, which were reported to be directly regulated by WUS20. These WUS downstream targets were misregulated in wus-7 or ham1;2;3 triple mutant in similar manners, and wus-7 and ham1;2;3 synergistically regulated their expression (Fig. 2i), consistent with functional physical (Fig. 1) and genetic (Fig. 2a–h) interactions between WUS and HAM. Then, dual luciferase assays were conducted in planta to confirm the direct effects of WUS-HAM on target gene expression. Compared to empty vector controls, the target genes examined were moderately (Fig. 2j–k) or barely (Fig. 2l–m) regulated by WUS or HAM alone, but were dramatically affected when WUS and HAM were combined(Fig. 2j–l), indicating a role for WUS-HAM interaction in regulating their transcription activities. Lastly, Chromatin Immunoprecipitation (ChIP) experiments demonstrated in vivo association of YFP-HAM2 proteins with TPL (Fig. 2n) and GRP23 promoters (Fig. 2o), genomic regions similar to that reported to associate with WUS protein in vivo20, supporting the notion that HAM family members are functional WUS cofactors in controlling the shoot stem cell niche via regulation of common target genes.
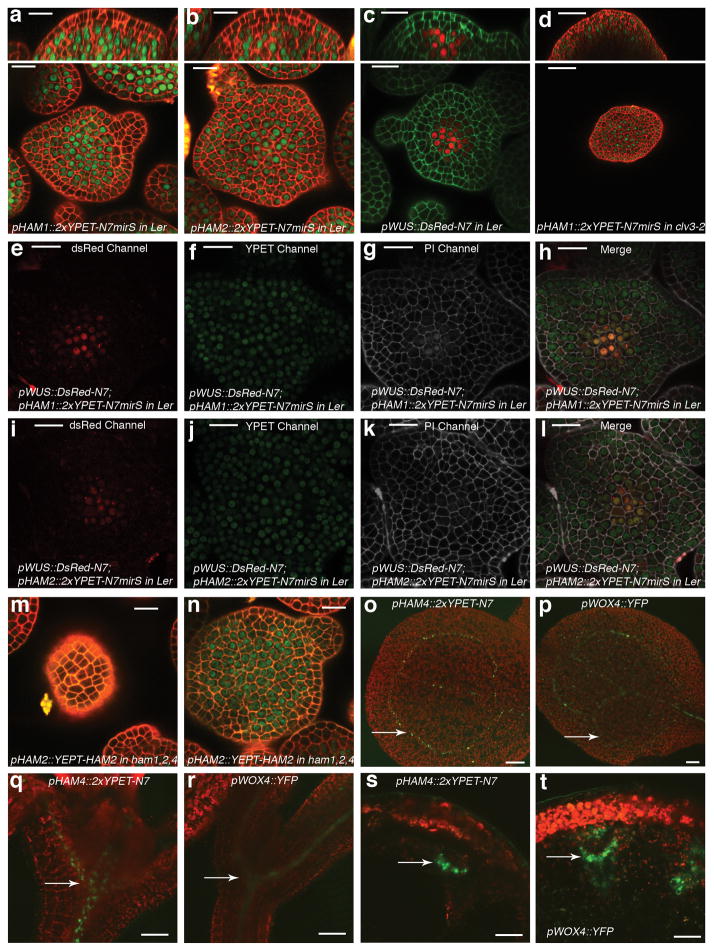
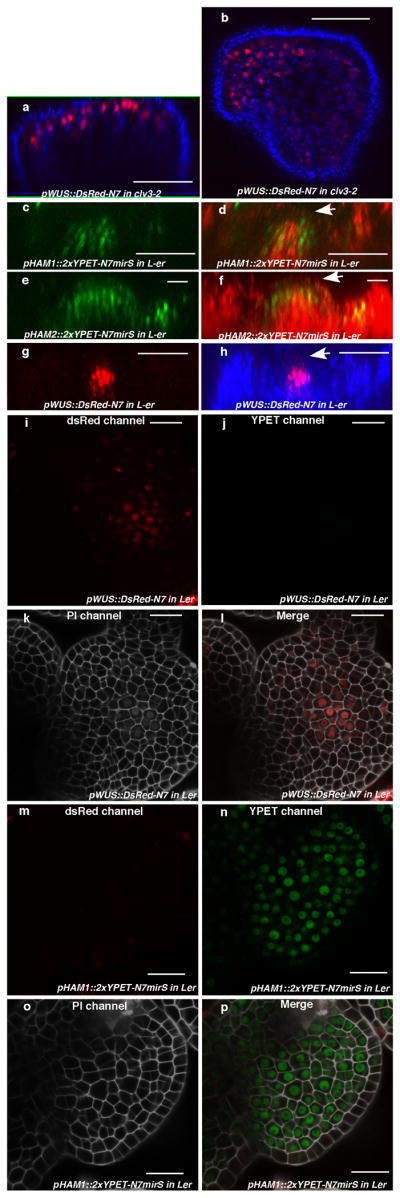
Figure 3. HAM and WUS/WOX expression domains overlap.
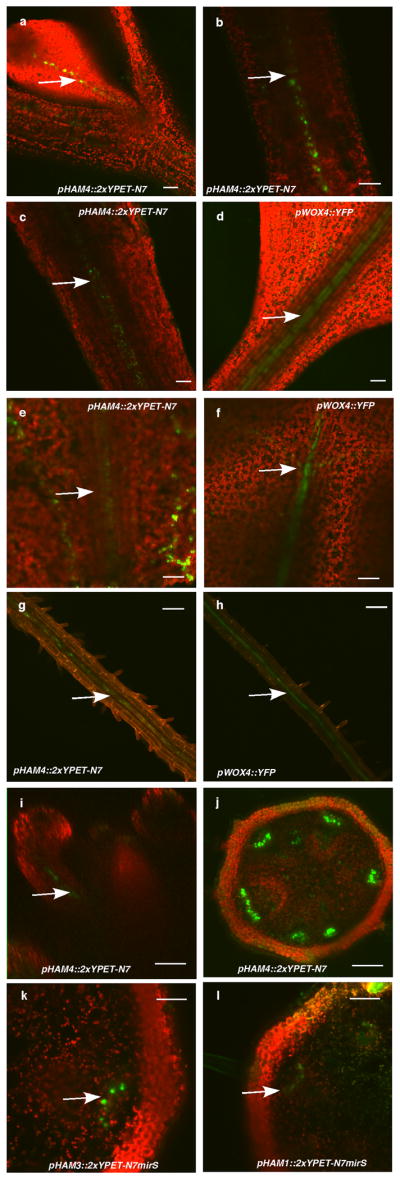
Expression of (a) pHAM1::2xYPET-N7microRNAsensitive marker (pHAM1::2xYPET-N7mirS) (green), (b) pHAM2::2xYPET-N7mirS (green), (c) pWUS::DsRed-N7 (red) in L-er inflorescence meristem (IM), and (d) pHAM1::2xYPET-N7mirS (green) marker in a clv3-2 IM. Orthogonal (upper panels) and transverse section (lower panels) views of the same plant are shown. (e–l) Overlapping expression patterns of pWUS::DsRed-N7 with pHAM1::2xYPET-N7mirS or pHAM2::2xYPET-N7mirS in the same shoot meristems (see Methods). Panels (from left to right): dsRed (red); YPET (green); PI (gray); merged channels. (m–n) Expression of pHAM2::YPET-HAM2 translation almarker (green) in L1 (m) and L3 (n) of the same ham1,2,4 SAM.(o–t) Overlapping expression patterns of pHAM4::2xYPET-N7and pWOX4::YFP (green, arrows) in the provascular and procambium cells in cotyledons (o–p), seedlings (q–r), and stem transverse sections (s–t). PIcounter stain: red (a–b, d, m–n), green (c), gray (g–h, k–l). Chlorophyll autofluorescence: red (o–t). Bars = 50 μm in (d, s–t); 200 μm in (o); 100 μm in (p–r); 20 μm in (a–c, e–n).
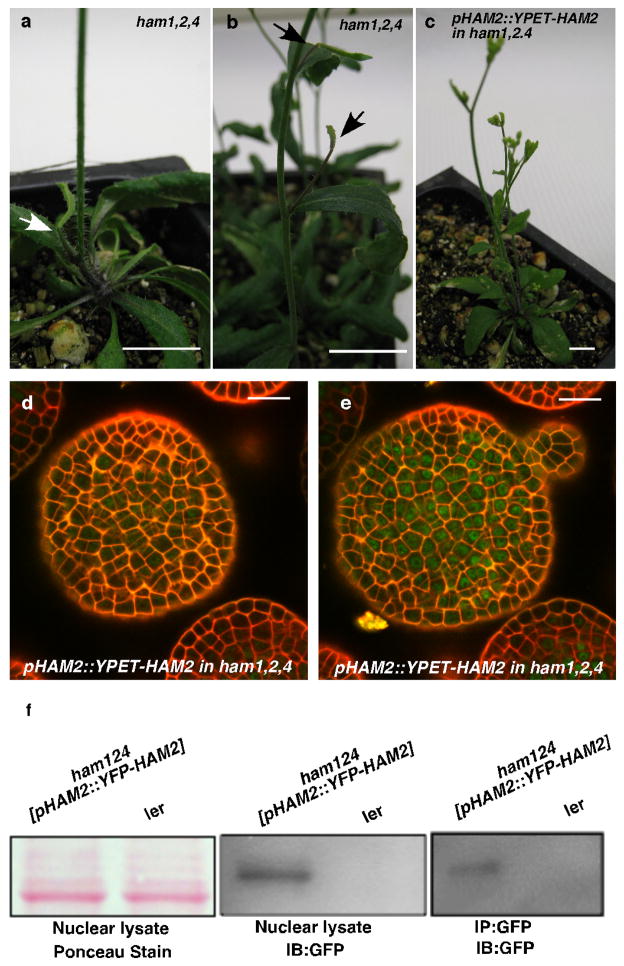
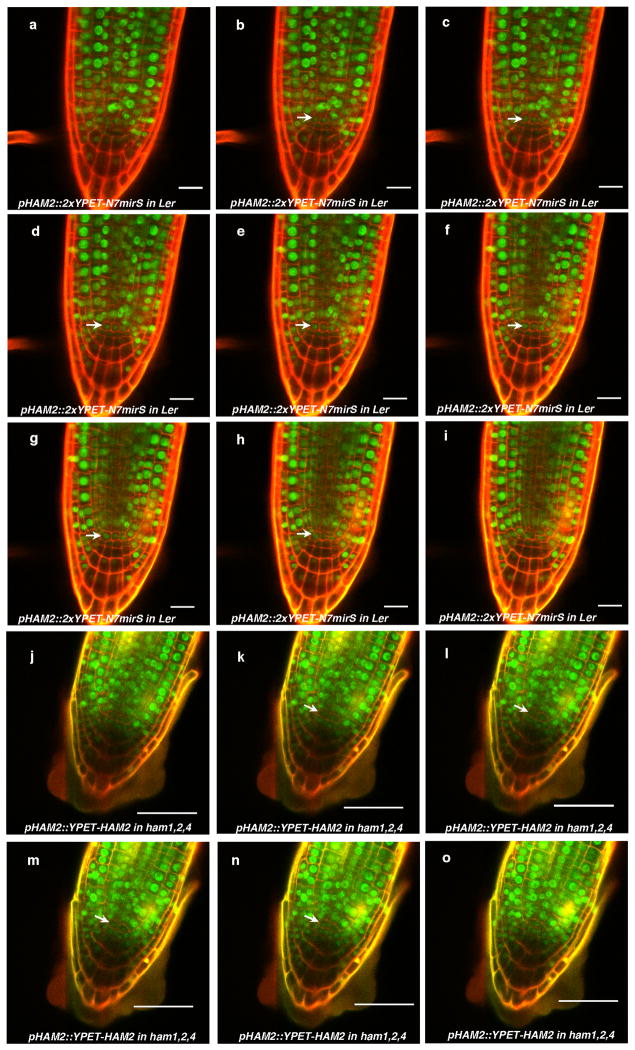
Consistently with physical and genetic interactions between HAM and WOX members, visualization of HAM and WUS/WOX fluorescent transcriptional reporters revealed that WOX and HAM family expression overlapped in planta. In vegetative (Extended Data Fig. 5c–h)and inflorescence meristems (Fig. 3a–c), HAM1 and HAM2 expression overlapped with that of WUS in the rib meristem. HAM1is expressed in rib meristem and peripheral zone but not in the L1 or L2 layers of the central zone (Fig. 3a, Supplementary video. 1), while HAM2 expression peaks within the center of the rib meristem (Fig. 3b). Similarly to WUS (Extended Data Fig. 5a, b), HAM1 is negatively controlled by the CLV signaling, asHAM1 is expressed throughout clv3-2 meristems (Fig. 3d). We imaged the WUSandHAM1 or HAM2 reporters in the same SAMs (Fig. 3e–l). Although expressed broadly, signals from HAM1 (Fig. 3f) or HAM2 (Fig. 3j) overlap with WUS signals (Fig. 3e, i) in the same rib zone cells (Fig. 3h, l, Extended Data Fig. 5i–p). Since WUS protein was reported to move in the SAM from its site of transcription in the rib domain21, the WUS and HAM1/HAM2 interaction domain in SAMs could be broader than their transcriptional domain overlap. We also examined the HAM2 translational reporter, pHAM2::YPET-HAM2, in the ham1,2,4 SAM (Fig. 3m–n, Extended Data Fig. 6), which completely complements the ham1,2,4 triple mutant (Extended Data Fig. 6 a–c), and it showed a pattern similar to the HAM2 transcriptional reporter: strong signal in the center starting from L3 and low or no signal in the L1 (Fig. 3m–n, Extended Data Fig. 6d–e). Taken together, the co-localization of WUS and HAM1/HAM2 in SAMs is consistent with functional WUS-HAM1/HAM2 interactions (Fig. 1, 2).
HAM4 and WOX4 co-express in the provascular or procambial cell types of various tissues (Fig. 3o–t, Extended Data Fig. 7). In stem transverse sections, HAM4 expresses specifically in the procambium, overlapping with WOX4expression, as well as with theHAM3 and HAM1 expression domains (Fig. 3s–t, Extended Data Fig. 7j–l). The tightly co-regulated spatial and temporalHAM4 and WOX4 expression patterns are consistent with a WOX4-HAM4 interaction module (Fig. 1h). Both HAM2 transcriptional and translational reporters (Extended Data Fig. 8) are expressed in root meristem cells including the QC, overlapping with the QC-specific WOX5 expression domain8, consistent with the previous reports of cell-type specific transcriptome analyses22,23, and indicating the possibility of WOX5-HAM2 interactions in roots. Our finding that both WUS and WOX5 interact with HAM2 may be partially accounted for the fact that WUS and WOX5 are interchangeable in controlling SAMs and RAMs8. Taken together, distinct and overlapping expression patterns of HAM and WOX members indicate that specific HAM-WOX pairs function within different stem cell niches throughout the plant.
To address the importance of the entire HAM family in the control of stem cell niches, we generated a ham1;2;3;4 quadruply homozygous mutant. Compared to wild type, ham1;2;3;4 plants displayed growth arrest at the early seedling stage, containing short roots and terminated shoots with two small leaf-like structures 26 days after germination (DAG)(Fig. 4a–c, Extended Data Fig. 9a–d); the shoot apices exhibited valley-like shapes at 26 DAG, lacking functional meristems(Fig. 4d); the hypocotyl transverse sections showed clear vascular defects, and the vascular bundles had reduced numbers of xylem vessels, fibers(dark blue-stained) and phloem cells (red-stained), consistent with a reduction in the stem cell activity necessary for generating these cell types (Fig. 4e–f). Moreover, midveins in ham1;2;3;4 leaf-like tissues did not differentiate but instead accumulated a dark-staining cell mass (arrow indicated), resembling ground tissue cells (Extended Data Fig. 9e–f). This is similar to but much stronger than the reported WOX4 RNAi phenotype10. Root meristematic activity is also severely compromised in ham multiple mutants. The QC and columella stem cells (CSC) in ham1;2;3;4 mutants displayed enlarged and irregular shapes (Extended Data Fig. 9g–h), and with incomplete penetrance, the CSCs in ham1;2;3 mutant differentiate(Extended Data Fig. 9i–l), resembling reported defects in wox5 mutants8. However, the root phenotype of ham1;2;3 or ham1;2;3;4 is much more severe than that of wox5 mutant, suggesting that HAM regulates root meristem development through not only direct interaction with WOX5 but also WOX5-independent pathways. In summary, in diverse meristems, ham1;2;3;4 mutants display defects that share similarities with mutants lacking WOX activities, supporting the idea that HAM proteins are co-factors for WUS/WOX family-mediated stem cell niche maintenance. Given the evolutionary conservation of plant meristem cell niches and the WOX/HAM gene families12, 16, and the fact that WOX-HAM interactions exist in higher plants besides Arabidopsis(Extended Data Fig. 10), this work establishes new basis for studying stem cell niches in Arabidopsis, and provides a paradigm for meristem cell control regimes likely to be universal in higher plants.
Figure 4. HAM family members are essential for various plant stem cell activities.
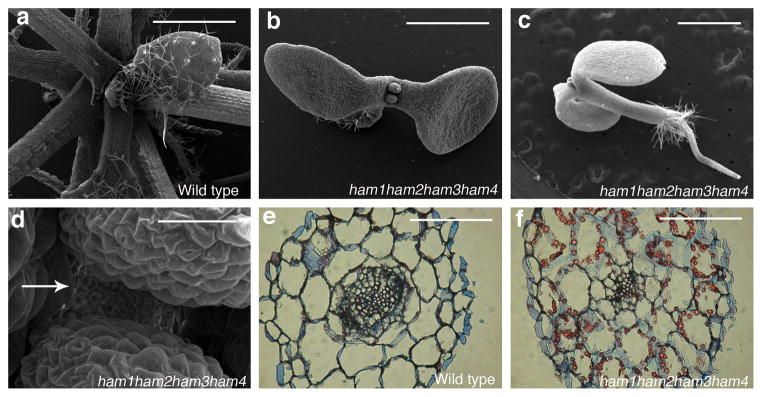
Scanning electron microscopic imaging of WT (a) and ham1, ham2, ham3, ham4 (b–d) seedlings (26 DAG). Arrow: ham1;2;3;4lacking a functional SAM.(e–f) Transverse sections of WT and ham1;2;3;4hypocotyls (7 DAG). Bars = 1 mm in (a–c, e–f), 50 μm in (d).
Methods
Plant materials and growth conditions
Arabidopsis thaliana plants were grown in a sunshine soil/vermiculite/perlite mixture under continuous light at 20°C. The mutant lines ham1;2;3(triply homozygous for mutant alleles of ham1-1, ham2-1, and ham3-1), ham1;2;4(triply homozygous for mutant alleles of ham1-1, ham2-1, and ham4-1), wus-7, wus-1, clv3-2 were previously described5, 16, 18, 24. wus-7;ham1;2;3, wus-7;ham1;2;4, wus-1;ham1;2;3, and ham1;2;3;4 mutants were generated through genetic crosses, and identified based on PCR genotyping in the F2 segregating population. Different mutant combinations in an er background were chosen for genetic and morphological analyses. All of the phenotypes were confirmed from multiple independent segregation lines to control for differences in ecotype background. The PCR genotyping was performed as previously described16, 18. Reporter lines for pWUS::DsRed-N7 and pWOX4::YFP were previously reported11, 25.
Yeast two-hybrid assay
Yeast transformation and beta-galactosidase assays were performed following the manufacturer’s instructions (Clontech). Full-length cDNAs for WUS, HAM1, HAM2, HAM3, and HAM4 were cloned into pENTR/D/TOPOor pCR8 (Invitrogen), and then WUS cDNA was Gateway cloned to pDEST32, and HAM1, HAM2, HAM3, and HAM4 cDNAs were Gateway cloned into pDEST22 using standard LR reactions (Invitrogen). All of the deletion derivatives for WUS or HAM1 were generated through overlapping PCR with the primers listed in methods, cloned into pENTR/D-TOPO or pCR8, and cloned into pDEST32 or pDEST22 through LR recombination (Invitrogen). All clones were sequenced to confirm they were in-frame and the corresponding deletions before being transformed into yeast. The bait and prey vectors were transformed into yeast strain MaV203, and three single transformed colonies per genotype were used as triplicate for the LacZ liquid assay in 96 Deep well plates (Thermo) and OD reading was recorded in 96-well plate reader (Tecan). The LacZ activity was calculated as (OD420×1000)/(OD600 × cell volume in μl × assay time in minutes) following the yeast two hybrid handbook (Clontech), including a standard error from three biological replicates.
BiFC
For BiFC experiments, full-length Arabidopsis WUS, WOX4, WOX5, HAM1, HAM2, HAM3, HAM4, BARD1 and FAMA cDNA Gateway clones were recombined into vectors containing each half of GFP (N or C terminus) to generate the fusion proteins (GFPn-WUS, GFPn-WOX4, GFPn-WOX5, GFPn-BARD1, GFPn-FAMA, GFPc-HAM1, GFPc-HAM2, GFPc-HAM3, GFPc-HAM4, GFPc-BARD1, GFPc-FAMA) as previously described26. Plasmid pairs for testing the specific interactions (such as GFPn-WUS and GFPc-HAM1) were co-transformed together with the P19 silencing suppressor27 into Nicotiana benthamiana leaves through Agrobacterium infiltration. The infiltrated tobacco leaves were stained with propidium iodide (PI) and imaged using a Zeiss LSM 510 Meta confocal microscope two days after infiltration. Green GFP signals in nuclei (which demonstrate the physical interaction) and red PI staining signals (which indicate tobacco cell structure) were captured at the same time from different detection channels. A 488nm laser line was used to stimulate GFP and PI. A 505–530 band pass filter was used to collect GFP signal and a 585–615 band pass filter was used to collect PI signal. BARD1, a nuclear-localized protein, was included as a negative control. FAMA, a bHLH transcription factor, which has been demonstrated to interact with bHLH transcription factors28, was used as an additional negative control. The positive signals for each pair were confirmed with four independent biological replicates, and representative image sareshown in the figures. The same method was also used for tomato (Solanum lycopersicum) proteins, including GFPn-tomato WUS, GFPn- tomato WOX4, and GFPc-tomato HAM.
Co-immunoprecipitation and Western Blot analysis
WUS or WOX5cDNA in pCR8 was recombined to pMDC8329 to generate a WUS-GFP or WOX5-GFP fusion clone. FLAG-HAM1, FLAG-HAM2 and FLAG-HAM4 were PCR amplified with primers 5′-CACCATGgactacaaggacgacgatgacaagggcggtggaagtCCCTTATCCTTTGAAAGGTTTCAAGG -3′, 5′-CTAACATTTCCAAGCAGAGACAGTAACAAGTTC-3′, and with primers 5′-CACCATGgactacaaggacgacgatgacaagggcggtggaagtCCCCTGCCCTTTGAGCAATTT-3′, 5′-TTAACATTTCCAAGCTGAGACAGTA-3′, and with primers 5′-CACCATGgactacaaggacgacgatgacaagggcggtggaagtAAAATCCCTGCATCATCTCCTC -3′, 5′-CTAAAACCGCCAAGCTGATGTGGCAACAAG-3′, respectively. GFP DNA was amplified and sub-cloned in front of WOX4 cDNA in frame to generate a GFP-WOX4 fragment. FLAG-HAM1, FLAG-HAM2, FLAG-HAM4, and GFP-WOX4 were then recombined into pMDC3229, respectively. For co-immunoprecipitation of WUS-GFP with FLAG-HAM1, WUS-GFP with FLAG-HAM2, GFP-WOX4 with FLAG-HAM4, or WOX5-GFP with FLAG-HAM2 in Nicotiana benthamiana, the constructs were introduced into Nicotiana benthamiana leaves through Agrobacterium infiltration. The leaves were harvested two days after infiltration and frozen in liquid nitrogen. For the immunoprecipitation of YFP-HAM2 in Arabidopsis, the shoot apices from the transgenic plants pHAM2::YFP-HAM2 in ham1,2,4 were harvested. The nuclei from Arabidopsis or tobacco were isolated, and then lysed with RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 3 mM DTT, 2 mM NaF and 1 mM NaVO3, or 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS for Co-IP GFP-WOX4 with FLAG-HAM4) containing protease inhibitors cocktail (Roche) and 200 μM PMSF by incubation on ice for 30 min followed by brief sonication. Clear lysates were mixed with diluting buffer containing PMSF and protease inhibitors cocktail (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 3 mM DTT, 2 mM NaFand 1 mM NaVO3, or 50 mM Tris-HCl, pH 8.0, 60 mM NaCl for Co-IP GFP-WOX4 with FLAG-HAM4) (1:5, v:v), immunoprecipitated with GFP-Trap® agarosebeads (ChromoTek), and the beads were washed three times with the diluting buffer in spin columns (BioRad). The recovered proteins were eluted from the beads by boiling in 2× SDS sample buffer, separated by SDS-PAGE and transferred to nitrocellulose membrane (Millipore). Proteins were detected using anti-GFP antibody (Roche, Cat #11814460001), anti-FLAG antibody (Sigma, Cat # F1804), and HRP conjugated anti-mouse antibody (Promega, Cat # W4021). The co-immunoprecipitation experiments were repeated twice with similar results.
Protein expression constructs and protein purification
WOX4 cDNA was amplified with primers 5′-CATAGAATTCATGAAGGTTCATGAGTTTTCGAA -3′ and 5′-AGTTGCGGCCGCTCATCTCCCTTCAGGATGGAGAGGA -3′(restriction enzyme sites are underlined), and cloned in frame in pGEX-4T-1 with Eco RI and Not I sites. WOX5 cDNA was amplified with primers 5′-ATTTCCCGGGTATGTCTTTCTCCGTGAAAGGTCG -3′ and 5′-AGTTGCGGCCGCTTAAAGAAAGCTTAATCGAAGATCT -3′(restriction enzyme sites are underlined), and cloned in frame in pGEX-4T-1 with XmaI and Not I sites. WUS cDNA was amplified with primers 5′-CATAGAATTCATGGAGCCGCCACAGCATCAG -3′ and 5′-AGTTGCGGCCGCCTAGTTCAGACGTAGCTCAAGA -3′(restriction enzyme sites are underlined), and cloned in frame in pGEX-4T-1 with Eco RI and Not I sites. HAM1-His6 tag was generated from PCR with primers 5′-CATAGAATTCATGCCCTTATCCTTTGAAAGGTTTCAAGG -3′ and 5′-AGTTGCGGCCGCCTAGTGATGATGATGATGATGACATTTCCAAGCAGAGACAGTAACAAGTTCTT -3′(restriction enzyme sites are underlined, and HIS6 coding sequence is italic), and cloned in frame with thrombin cutting site in pGEX-4T-1 with Eco RI and Not I. HAM4-His6 tag was generated from PCR with primers 5′-CATAGAATTCATGAAAATCCCTGCATCATCTCCTC -3′ and 5′-AGTTGCGGCCGCCTAGTGATGATGATGATGATGAAACCGCCAAGCTGATGTGGCAACAAG -3′(restriction enzyme sites are underlined, and HIS6 coding sequence is italic), and cloned in frame with thrombin cutting site in pGEX-4T-1 with Eco RI and Not I. All proteins were expressed in Rosetta™ E. Coli (Novagen®) by inducing with 0.4 mM IPTG at 16 °C for 2–4 hours. GST-WOX4, GST-WOX5 and GST were purified on glutathione resin. HAM1 was purified on glutathione resin followed by digestion with thrombin and chromatography on S200 resins as described previously30–31. HAM4 was purified on glutathione resin followed by digestion with thrombin and removal of the GST associated with glutathione resin as described previously31.
In vitro pull-down assay
GST-WOX4, GST-WOX5, GST-WUS or GST were immobilized on glutathione resin and incubated with HAM1-His6 or HAM4-His6 for 30 min at 4 °C. The glutathione resin was then washed three times and processed for SDS-PAGE analysis and western blot analysis using antibody to His-tag (Qiagen, Cat. # 34660). The pull-down experiments were repeated twice with similar results.
Transactivation assay in tobacco
A 60 base pair minimal 35S fragment (the −60 minimal promoter) was amplified and cloned with Bam HI /Nco I sites into the pGREEN800II LUC32 to generate a pGREEN800II-60LUC. TPL promoter was PCR amplified from Col-0 genomic DNA with primers 5′-AACAGGTACCGAACGCTTCGTTTCATTAGTTTATC -3′ and 5′-AATAGGATCCGTTTTCTCTCACTTCCTTAAAAGACT -3′ (restriction enzyme sites are underlined)and cloned with Kpn I /Bam HI sites into the pGREEN800II-60LUC. TIP2;2 promoter was amplified with primers 5′-AACAGGTACCCGAGTGAAGCAGATTGGGAGAGAA -3′ and 5′-AATACTGCAGTTTGATCCGACAAAATAACTCTGTT -3′ and cloned with KpnI /PstI sites into the pGREEN800II-60LUC. GRP23 promoter was amplified with primers 5′-AACAGGTACCCAGGTGTGATTGTCAATAGACTACG -3′ and 5′-AACAGATATCGGTGGAGGGAAAATGATTTAGGGTT -3′ and cloned with KpnI /Eco RV sites into the pGREEN800II-60LUC. TCP9 promoter was amplified with primers 5′-AACAGGTACCGTATGCTGATGGTAGGCAAAAGTT-3′ and 5′-AATACTGCAGTAAAATATAGCTGAGAGAAAACG-3′ and cloned with KpnI /PstI sites into the pGREEN800II LUC. The different reporter constructs (dual-luciferase reporter with different gene promoters) and indicated effectors (empty effector vector or WUS or HAM2, or WUS together with HAM2) were introduced into Nicotiana benthamiana leaves through Agrobacterium infiltration. The activities from firefly luciferase (LUC) and Renilla luciferase (REN) were quantified two days after infiltration with a Dual Luciferase Assay kit (Promega), and luminescence was recorded using a 96-well dual injection luminometer (Tecan). The LUC activity was normalized to the REN activity (LUC/REN). The means and standard errors of LUC/REN were calculated from three independent biological replicates.
Plasmid constructions for the transgenic plants
It has been reported that HAM1, HAM2 and HAM3 are targeted and repressed by the microRNA171 family19. To generate new microRNA-sensitive fluorescence reporters for HAM1, HAM2 and HAM3, an approach similar to that in a previous report33 was used. Briefly, a 2xYPET-N7mirS fragment was generated through PCR amplification, which contains a 2x version of YPET with a N7 nuclear localization sequence (2xYPET-N7) followed by 26 base pairs of microRNA target sequence (GCAAGGGATATTGGCGCGGCTCAATC) from the HAM family. These 26 base pairs are recognized and targeted by the microRNA171 family19, 34.
For the construction of the pHAM1::2xYPET-N7mirS reporter, a 4 kb AscI fragment containing the HAM1 promoter was amplified from Col-0 genomic DNA with primers 5′-TACAGGCGCGCCTTTCCCTCACTTTTTCTTACATTGAA-3′ and 5′-TACAGGCGCGCCACGCCTCCTCAACAACACAGAGTAA-3′(restriction enzyme sites are underlined), and cloned 5′ of the 2xYPET-N7mirS fragment. The fused DNA fragment was introduced into the pMOA34 binary vector35. For the construction of pHAM2::2xYPET-N7mirS, 3122 bp HAM2 promoter was amplified with 5′-TACAGTTTAAACAGCAGGACATATCTAAACCAGAAGTT-3′ and 5′-TACAGTTTAAACGACCAATCTTACAGAGTCAGAAAGAG-3′ (restriction enzyme sites are underlined) and cloned in front of 2xYPET-N7mirS; and 1149 bp HAM2 3′-untranslated sequence was PCR amplified with 5′-TACAGGCGCGCCGACGAAAAAGGAGGATATTTTCACGGT-3′ and 5′-TACAGGCGCGCCACTATGTTTCCATGTACTGTGGGATA-3′(restriction enzyme sites are underlined) and cloned 3′ of the 2xYPET-N7mirS construct, then the fused DNA fragment was cloned into pMOA34. For the construction of pHAM3::2xYPET-N7mirS, 3816 bp HAM3 promoter was amplified with 5′-TACAGTTTAAACTTTATAAGACTTGCTATGGTCGTGAG-3′ and 5′-TACAGTTTAAACTGCAGACGATAAAAAATAGTGTATT-3′ (restriction enzyme sites are underlined) and cloned before 2xYPET-N7mirS; and 1755 bp HAM3 3′-untranslated sequence was PCR amplified with 5′-TACAGGCGCGCCTTTCCACCGGAGTTTCAATTATTAAA -3′ and 5′-TACAGGCGCGCCTTAGTTGAAGGACAAATAACACCAAA -3′ (restriction enzyme sites are underlined) and cloned 3′ of the 2xYPET-N7mirS fragment, then the fused DNA fragment was introduced into pMOA34. The double reporter lines including pWUS::DsRed-N7; pHAM1::2xYPET-N7mirS line and pWUS::dsRed-N7; pHAM2::2xYPET-N7mirS line were generated through genetic crosses.
For the construction of pHAM4::2xYPET-N7 reporter, 6413 bp HAM4 promoter was amplified with primers 5′-TACAGGCGCGCCAAATATAAAATAGAATCAAACAAAGTTGGTAAC-3′ and 5′-CAAAGGCGCGCCGTGTTGTGTGTTAAGAAGAAAGAAAGGTGGAGCCTTT-3′(restriction enzyme sites are underlined), and cloned 5′ of a 2xYPET-N7 fragment, then the fused DNA fragment was cloned into pMOA34.
For the complementation of wus-1, a full-length WUS or WUS derivative without base pairs encoding amino acids from 203 to 236 was cloned into pMOA36 binary vector, together with a 4.4 kb of WUS upstream regulatory sequence and 1.5 kb WUS 3′-untranslated sequence. The construct was introduced into wus-1/+ plants using the floral dip method. For the complementation of ham1,2,4, HAM1 or HAM1 derivative without 117–230 was cloned into pMOA34 binary vector, with a 3949 bp of HAM1 upstream regulatory sequence and 1387 bp HAM1 3′-untranslated sequence. The construct was introduced into ham1,2,4 plants using the floral dip method.
To generate a microRNA171 expression construct in shoot meristems, the microRNA171 DNA was amplified with 5′-CACCTGAGCGCACTATCGGACATCAAA-3′ and 5′-TAAACGCGTGATATTGGCAC-3′ and cloned into pMOA36 together with a 4.4 kb of WUS upstream regulatory sequence and 1.5 kb WUS 3′-untranslated sequence. The construct was introduced into the ham4 mutant through the floral dip method. Five independent transgenic plants (pWUS::mirRNA171 in ham4) showing terminated vegetative meristems were identified.
Confocal imaging of fluorescence reporters in living plants
All of the fluorescent reporters were imaged by using a Zeiss LSM 510 Meta confocal microscope, except for the fluorescent reporters in inflorescence meristems and HAM2 fluorescent reporters in the roots, which were imaged by using a Zeiss LSM 780 Meta confocal microscope. Zeiss LSM software was used for reconstructing the Z-stacks for a projection view. Laser and filter settings were used as described previously36–38. To image HAM4 and WOX4 reporters, the cotyledons, first leaf, hypocotyls and roots from 7-d-old seedlings and stems from 1-cm bolting plants were used. To image dsRED, YPET and PI simultaneously in SAMs, the multi tracking mode in the ZEISS LSM 780 was used. dsRed was excited using a 561 nm laser line in conjunction with 571–589 nm collection; the YPET was excited using a 514 nm laser line in conjunction with a 519–549 nm collection; and PI was excited using a 514 nm laser with 631–673 nm collection. There is no spectral bleed-through of DsRed into the YPET collection channel, nor YPET into the DsRed collection channel under these settings, and for better display, all images from dsRed channel are equally enhanced with the same scale and all images from the PI channel are uniformly enhanced to the similar intensity using Image J software.
Histology
The wild type and ham1;2;3;4 seedlings were fixed in 4% paraformaldehyde, dehydrated and embedded in Paraplast X-tra (Fisher). The samples in wax were sectioned at 8 microns, de-waxed and dehydrated, and the slides were stained with Alcian blue together with Safranin O (red) as previously described39, to detect non-lignified cell walls and lignified cell walls, respectively.
Real-time RT-PCR analysis
Total RNA was isolated from 10-day old plants with roots, hypocotyls and leaves dissected off, using the RN easy Kit (Qiagen). SuperScript III reverse transcriptase (Invitrogen) was used to synthesize the first-strand cDNA with oligo (dT) primer and 1 μg of total RNA at 50°C for 1 h. Quantitative PCR was then performed with the Sensi Mix SYBR Hi-ROX Kit (Bioline) on Roche Real-Time PCR machine following the manufacturer’s instruction. The thermal cycling program was 95°C for 10 min, followed by 45 cycles of 95°C for 10 s, 56°C for 30 s, 72°C for 40s, and a one-cycle dissociation stage at 95°C for 15 s, 60°C for 1 min, and 97°C for 15 s. The primers used in quantitative RT-PCR were JAZ5, 5′-GAAAGACAGAGCTGTGGCTAGG -3′ and 5′-TTGGCCTTCTTCAATCTTCATAATA -3′; TIP2;2, 5′-ACCAATGGCGAGAGCGTACCG -3′ and 5′-ATGAAACCGATAGCAATTGGAG -3′; TCP9, 5′-ACCTCCTTTACAAGTTGTTCCAAG -3′ and 5′-TGAAGCTCTTGTTTCTCGTATATCTC -3′; GRP23, 5′-AGACAGCTAGCCATCAGCAGTCAC -3′ and 5′-AGTTCCTCAACTCCACTACCTTTTT -3′; TPL, 5′-AGCTAGTCTCAGCAATTCAAA -3′ and 5′-AGGCTGATCAGATGCAGAGG -3′; and UBQ10, 5′-AACAATTGGAGGATGGTCGT -3′ and 5′-TTCCAGGGAAGATGAGACG-3′. Fold change was calculated as 2ΔΔCt and standard error was calculated from three biological replicates, and each biological replicate was examined in triplicate.
Chromatin immunoprecipitation(ChIP)
For the construction of pHAM2::YFP-HAM2 (pHAM2::YPET-HAM2), the YFP variant YPET was amplified and cloned in front of HAM2 cDNA in frame to generate YFP-HAM2 fragment. Then the 3122 bp HAM2 promoter was amplified with 5′-TACAGTTTAAACAGCAGGACATATCTAAACCAGAAGTT-3′ and 5′-TACAGTTTAAACGACCAATCTTACAGAGTCAGAAAGAG-3′ (restriction enzyme sites are underlined) and cloned in front of YFP-HAM2 and 1149 bp HAM2 3′-untranslated sequence was amplified with 5′-TACAGGCGCGCCGACGAAAAAGGAGGATATTTTCACGGT-3′ and 5′-TACAGGCGCGCCACTATGTTTCCATGTACTGTGGGATA-3′ and cloned 3′ of the YFP-HAM2. Then the whole fused DNA fragment (pHAM2-YFP-HAM2-HAM2 3′UTR) was cloned into the binary vector pMOA34. The construct was introduced into ham1,2,4 plants using the floral dip method, and the complemented ham1,2,4 [pHAM2::YFP-HAM2] line was selected for the western blot, GFP IP (shown in Extended Data Fig. 6f) and ChIp experiments. ChIP followed by a quantitative real-time PCR approach was used to investigate the in vivo association of HAM2 with the TPL and GRP23 promoters as described previously40 with some modifications. In general, 2 g of ham1,2,4 (negative control) or ham1,2,4 [pHAM2::YFP-HAM2] plants was harvested and fixed with 1% formaldehyde under vacuum. Nuclei were isolated and lysed, and chromatin was sheared to an average size of 500 bp by sonication 7 times for 20 seconds each with a Branson Sonifier. Samples were kept on ice during sonication and were cooled for 1 min between sonication pulses. The sonicated chromatin served as input. Immunoprecipitations (IPs) were performed with GFP-Trap® Agarose beads (Chromotek) at 4°C following the manufacturer’s procedure. The precipitated DNA was isolated and purified, and served as a template for PCR. Quantitative PCR was performed as described in real-time RT-PCR analysis. The relative enrichment for each immunoprecipitated amplicon (from TPL or GRP23 promoter) from GFP-TRAP is presented as ChIP/Input ratio, and TUA4 and ACT7 (ACTIN7) amplicons are also included to serve as negative controls. The ChIp experiments have been conducted three times using independent biological replicates with similar results, and one representative data set with two technical replicates is presented. The primer pairs used in ChIp-PCR are as follows: TPL Amplicon1, 5′-GCAATTGGCTCTTCAATGTC -3′ and 5′-GGACGGAGATCTAACGGCTA -3′; TPL Amplicon2, 5′-CCATATGACCGGGATATGAGA -3′ and 5′-GGGATATGTCGCTTTCCATT -3′; TPL Amplicon3, 5′-TTGAGTCAGGGCTCATCTCC -3′ and 5′-CTTTCGCGAGAACCAACTTC-3′; GRP23 Amplicon1, 5′-ACCATCGTCATTGGTTTCGT -3′ and 5′-GGAGGTGACTGAGAGACATGG -3′; GRP23 Amplicon2, 5′-CAACAAATTCCTGTTTTCACGTT -3′ and 5′-CGAAAATGTTCGAACTGCAT -3′; GRP23 Amplicon3, 5′-CGCCATCGCCTAAAAGTAAA -3′ and 5′-TTTGTTGGCTAGGCATAGGG -3′; GRP23 Amplicon4, 5′-AGACAGCTAGCCATCAGCAGTCAC -3′ and 5′-AGTTCCTCAACTCCACTACCTTTTT -3′; TUA4 Amplicon, 5′-CTTTGGTCTTTAGCAGGTTC -3′ and 5′-CCCATCTGTATATAACGACAC -3′; ACTIN7 Amplicon, 5′-TGCTTGTTATGTGATTCGATCC -3′ and 5′-GATCGACAGAAGCGAGAAGAAT -3′.
Staining
mPS-PI staining and root imaging of the staining was performed as previously described 41.
SEM
For scanning electron microscopy, tissue was placed in 1.2% glutaraldehyde in 0.025M phosphate buffer (sodium phosphate, pH 6.8), vacuum was applied for 10 min, and tissue was fixed overnight at 4°C. Tissue was then rinsed twice with 0.025M phosphate buffer for 1 h, postfixed with 0.5% osmium tetroxide in 0.025Mphosphate buffer for 24 h at room temperature, and moved through an increasing ethanol series (20% increments), each increment lasting a minimum of 1 h and ending with two exchanges of 100% ethanol. Ethanol was removed by critical point drying with a critical point drier (SAMDRI), and tissue was mounted to stubs with double-sided adhesive tape and sputter coated with gold-palladium alloy using a Hummer Sputtering System (Anatech). Samples were examined with a Hitachi 4700 scanning electron microscope.
Primers used for cDNA clones and deletion constructions
HAM1c/5CACC (5′-CACCATGCCCTTATCCTTTGAAAGGTTTCAAGG-3′); HAM1c/3 (5′-ACATTTCCAAGCAGAGACAGTAACAAG-3′); HAM1c5/231 (5′-CCGTTTTATCACAACAACCAG-3′); HAM1c5/441 (5′-GAAAATCTCAAAACATTCG-3′); HAM1D71-116/5 (5′-AGTCCTCTCGCTTCTTATTCTGCTTCTTCTCCTGGTCAAGAGC-3′); HAM1D71-116/3 (5′-GCTCTTGACCAGGAGAAGAAGCAGAATAAGAAGCGAGAGGACT-3′); HAM1c71/5CACC (5′-CACCATGTCTACCACCACCACGCTGTCTTCCTCT-3′); HAM1D117-230/5 (5′-GATGATCTTGACGGTGTTCTCTCTCCGTTTTATCACAACAACCAGCAA-3′); HAM1D117-230/3 (5′-TTGCTGGTTGTTGTGATAAAACGGAGAGAGAACACCGTCAAGATCATC-3′); HAM4c/5 (5′-ATGAAAATCCCTGCATCATCTCCTC-3′); HAM4c/3 (5′-AAACCGCCAAGCTGATGTGGCAACA-3′); WUSc5/1 (5′-ATGGAGCCGCCACAGCATCAG-3′); WUSc5/30 (5′-TACACGTGTCGCCAGACCAG-3′); WUSc5/100 (5′-AGATTCAACGGAACAAACATGAC-3′); WUSc5/171 (5′-GCAAGCTCAGGTACTGAATGT-3′); WUSc/3stop (5′-CTAGTTCAGACGTAGCTCAAGA-3′); WUSc3/236 (5′-ACCTTCTAGACCAAACAGAGG-3′); WUSc3/292 (5′-GTTCAGACGTAGCTCAAGAGAAGC-3′); WUSD164-183/5 (5′-TAACAAGCCATATCCCAGCTTCAATGGCTACATGAGTAGCCATG-3′); WUSD164-183/3 (5′-CATGGCTACTCATGTAGCCATTGAAGCTGGGATATGGCTTGTTA-3′); WUSD101-163/5 (5′-GGCTCGTGAGCGTCAGAAGAAGAGAAATAACGGGAATTTAAATCATGCAA-3′); WUSD101-163/3 (5′-TTGCATGATTTAAATTCCCGTTATTTCTCTTCTTCTGACGCTCACGAGCC-3′); WUSD132-163/5 (5′-TATCATCCTCTACTTCACCATCATAATAACGGGAATTTAAATCATGCAA-3′); WUSD132-163/3 (5′-TTGCATGATTTAAATTCCCGTTATTATGATGGTGAAGTAGAGGATGATA-3′); WUSD184-236/5 (5′-AATGTGGTGTTGTTAATGCTTCTCATCAAGAAGAAGAAGAATGTGG-3′); WUSD184-236/3 (5′-CCACATTCTTCTTCTTCTTGATGAGAAGCATTAACAACACCACATT-3′); WUSD164-236/5 (5′-TAACAAGCCATATCCCAGCTTCCATCAAGAAGAAGAAGAATGTG-3′); WUSD164-236/3 (5′-CACATTCTTCTTCTTCTTGATGGAAGCTGGGATATGGCTTGTTA-3′); WUSD184-202/5 (5′-AATGTGGTGTTGTTAATGCTTCTTACAACAACGTAGGTGGAGGAT-3′); WUSD184-202/3 (5′-ATCCTCCACCTACGTTGTTGTAAGAAGCATTAACAACACCACATT-3′); WUSD203-236/5 (5′-TGGAACAAGACTGTTCTATGAATCATCAAGAAGAAGAAGAATGTGG-3′); WUSD203-236/3 (5′-CCACATTCTTCTTCTTCTTGATGATTCATAGAACAGTCTTGTTCCA-3′); WUSD218-236/5 (5′-GGGCAAACATGGATCATCATTACCATCAAGAAGAAGAAGAATGTGG-3′); WUSD218-236/3 (5′-CCACATTCTTCTTCTTCTTGATGGTAATGATGATCCATGTTTGCCC-3′) WUSD203-217/5 (5′-TGGAACAAGACTGTTCTATGAATTCATCTGCACCTTACAACTTCTT-3′); WUSD203-217/3 (5′-AAGAAGTTGTAAGGTGCAGATGAATTCATAGAACAGTCTTGTTCCA-3′); WOX4c/5CACC (5′-CACCATGAAGGTTCATGAGTTTTCGAA-3′); WOX4c/3stop (5′-TCATCTCCCTTCAGGATGGAGAGGA-3′); WOX5c/5CACC (5′-CACCATGTCTTTCTCCGTGAAAGGTC-3′); WOX5c/3 (5′-AAGAAAGCTTAATCGAAGATCT-3′); TomatoHAM/5CACC (5′-CACCATGATTGTAATACCTCAAAGTAATAA-3′); TomatoHAM/3stop (5′-TTAAAAGAAAATCTCTTCTGGCTTCAGA-3′); ToamtoWUS/5CACC (5′-CACCATGGAACATCAACACAACATAGAAGA-3′); TomatoWUS/3stop (5′-TTAGGGGAAAGAGTTGAGAGTAAGT-3′); TomatoWOX4/5CACC (5′-CACCATGTACATGGGATCATCATCAGGAAG-3′); TomatoWOX4/3stop (5′-TCATCTCATGCCTTCTGGATGCAATG-3′)
Extended Data
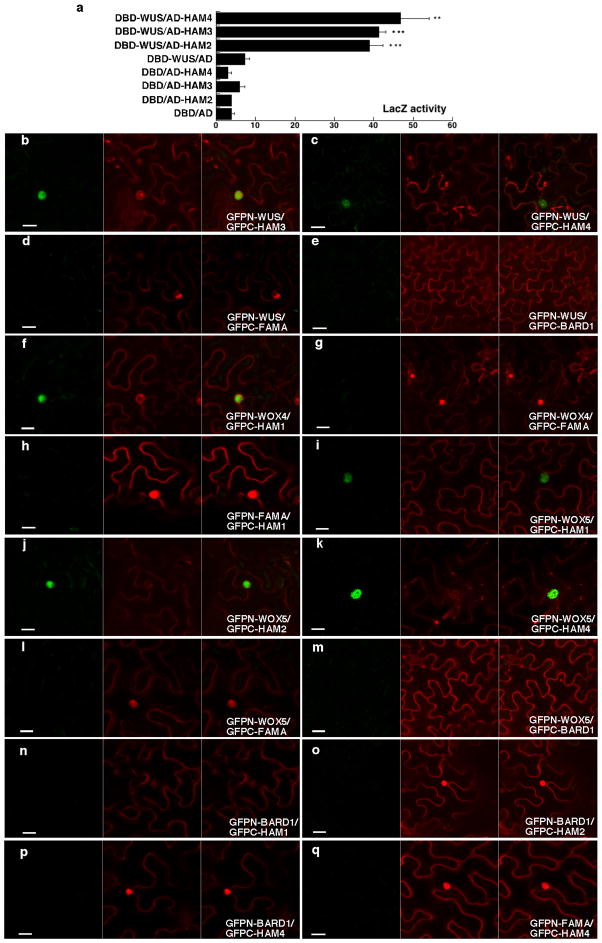
Extended Data Figure 1.
Interaction between WUS/WOX and HAM family transcriptional regulators. (a) LacZ activity in yeast-two-hybrid assays testing interactions between WUS and HAM2, HAM3 or HAM4. Error bar = mean ± sem (n=3biological replicates). **, P<0.01; ***, P<0.001(two-tailed t-test, compared to DBD-WUS/AD). (b–o) Bimolecular fluorescence complementation (BiFC) analyses in tobacco transient assays with HAM and WOX family genes. Tobacco was co-transformed with GFPn-WUS and GFPc-HAM3 (b), or GFPn-WUS and GFPc-HAM4 (c), or GFPn-WUS and GFPc-FAMA (d), or GFPn-WUS and GFPc-BARD1 (e), or GFPn-WOX4 and GFPc-HAM1 (f), or GFPn-WOX4 and GFPc-FAMA (g), or GFPn-FAMA and GFPc-HAM1 (h), or GFPn-WOX5 and GFPc-HAM1 (i), or GFPn-WOX5 and GFPc-HAM2 (j), or GFPn-WOX5 and GFPc-HAM4 (k), or GFPn-WOX5 and GFPc-FAMA (l), or GFPn-WOX5 and GFPc-BARD1 (m), or GFPn-BARD1 and GFPc-HAM1 (n), or GFPn-BARD1 and GFPc-HAM2 (o), or GFPn-BARD1 and GFPc-HAM4 (p), or GFPn-FAMA and GFPc-HAM4 (q). BARD1 and FAMA proteins are both included as negative controls. Left panel: GFP channel; middle panel: propidium iodide (PI) staining channel; right panel: merged channels. Scale bar = 20 μm.
Extended Data Figure 2.
An N-terminal region of HAM1 is important for WUS-HAM1 interactionand is essential for HAM1 function in stem cell maintenance. (a) Yeast two-hybrid assay of interactions between WUS and various deleted derivatives of HAM1. Deleting amino acids 117 to 230(D117-230) from HAM1 compromised the WUS-HAM1 interaction. Left panel: box diagrams of the HAM1 derivatives. Shaded boxes indicate the GRAS domains. Numbers indicate amino acid residues. Error bar = mean ± sem (n=3biological replicates). *, P<0.05; **, P<0.01; ***, P<0.001(two-tailed t-test, compared to AD-HAM1 full-length). (b–g) The complementation of the ham1;2;4 triple mutant requires amino acids 117-230. The early termination phenotype of ham1;2;4(b, e) was not complemented by HAM1 (D117-230) driven by a HAM1 promoter and 3′UTR (c, f), but was fully complemented by wild type HAM1(d, g). Arrows in (b, c) indicate the early-terminated inflorescences. (h–j) Amino acid sequence alignment of the HAM1 N-terminal domains (117–230) using Clustal Omega. (h) Sequence alignment of the N-terminal domains among three Arabidopsis HAM members. (i) Sequence alignment of partial N-terminal domains in HAM from A.thaliana, A.lyrata, C.rubella, B.rapa, and Petunia. (j) Sequence alignment of partial HAM1 N-terminal domains in HAM from A.thaliana, A.lyrata, C.rubella, B.oleracea, B.rapa, and Petunia. Asterisks: same amino acids; dots: similar amino acids. The conserved regions are boxed. Bars = 10 mm in (b, c), and (g), 40 mm in (d), 20 mm in (e, f).
Extended Data Figure 3.
A C-terminal region of WUS is important for WUS-HAM1 interaction and is essential for WUS function in stem cell maintenance. (a) Yeast-two-hybrid assay of interactions between HAM1 and various deleted WUS derivatives. Deleting amino acids 203 to 236 (D203-236) from WUS greatly compromised the WUS-HAM1 interaction. Left panel: box diagrams of the deleted WUS derivatives; shaded boxes: the homeodomain; three black boxes: the acidic domains, the WUS box and the EAR motif, respectively. Numbers indicate amino acid residues. Error bar = mean ± sem (n=3biological replicates).*, P<0.05; **, P<0.01; ***, P<0.001(two-tailed t-test, compared to DBD-WUS full-length). (b–d) WUS function requires the same region that is important for WUS-HAM1 interaction. The terminated shoot meristem phenotype of wus-1 (b) was not complemented by WUS (D203-236) driven by WUS promoter and 3′UTR (c), and was fully complemented by the wild type WUS (d). (e) Amino acid sequence alignment of C-terminal regionsof WUS from Arabidopsis thaliana, A.lyrata, Capsella rubella, Brassica oleracea, B.rapa, Lepidium ruderale, L.sativum, and Petunia, using Clustal Omega. Asterisks: same amino acids; dots: similar amino acids. The conserved regions are boxed. Bars = 2 mm in (b–d).
Extended Data Figure 4.
Genetic interaction between WUS and HAM family members. (a, b) The secondary inflorescence meristems initiated from axillary meristems in wus-7; ham1;2 homozygotes with ham3/+ terminate prematurely. (c)wus-7; ham1;2 homozygotes with ham4/+ display early termination of the main inflorescence meristem and lack of carpels in flowers (indicated by arrow). (d–f) WUS and HAM family members interact genetically in a dose-dependent manner. wus-7 (d) formed functional shoot apices and normal stature, but wus-7; ham1/+; ham2/+; ham3/+ (e) enhanced the wus-7 phenotype, and wus-7; ham1/+; ham2; ham3 (f) showed stronger enhancement, with reduced flower numbers and plant stature, and elongated vegetative stage, resembling a wus strong allele. Plants are 36 days after germination. (g–h) Down-regulation of HAM1, HAM2 and HAM3 in ham4 shoot meristems leads to an early termination phenotype. Compared to wild type (Col) (g), pWUS::mir171 in ham4 (h) showed terminated vegetative meristems. (i–l) WUS is required for functions of HAM1, HAM2 and HAM3. At 11 days after germination, compared to Ler wild type (i) and ham1;2;3 (k) which formed functional vegetative meristem and leaf primordia, wus-1; ham1;2;3 (l) displays terminated vegetative meristems similar to wus-1 (j). Bars = 2 mm.
Extended Data Figure 5.
Expression of HAM1, HAM2 and WUS in the SAMs. (a–b) WUS expression in clv3-2. Orthogonal (a) andtop (b) views of pWUS::DsRed-N7 expression (red)and chlorophyll autofluorescence (blue) in the same clv3-2 inflorescence meristem. (c–h) Comparison between expression patterns of HAM1, HAM2 and WUS in vegetative meristems.(c) Orthogonal view of pHAM1::2xYPET-N7mirS expression (green) in Ler vegetative meristem. (d) Orthogonal view of pHAM1::2xYPET-N7mirS expression (green) together with chlorophyll autofluorescence (red) in the same vegetative meristem shown in (c), indicating that HAM1 is expressed in the rib meristem. (e) Orthogonal view of pHAM2::2xYPET-N7mirS expression (green) in Ler vegetative meristem. (f) Orthogonal view of pHAM2::2xYPET-N7mirS expression (green) together with chlorophyll autofluorescence (red) in the same vegetative meristem shown in (e), indicating that HAM2 is highly expressed in the rib meristem. (g) Orthogonal view of pWUS::DsRed-N7 expression (red) in Ler vegetative meristem. (h) Orthogonal view of pWUS::DsRed-N7 expression (red) together with chlorophyll autofluorescence (blue) in the same vegetative meristem shown in (g), indicating that WUS is expressed in the rib meristem. Arrows indicate the positions of L1 cell layer. (i–p) Control images confirming the specificity of confocal spectral settings for Fig. 3(e–l). The SAMs from pWUS::DsRed-N7 line (i–l) or pHAM1::2xYPET-N7mirS line (m–p) were imaged from the same three separated channels used in Fig. 3 (e–l). There is no spectral bleed-through of YPET signal into the ds Red channel (m), nor ds Red signal into the YPET channel (j). (i, m)ds Red channel (red); (j, n) YPET channel (green); (k, o) PI staining channel (gray); (l, p) merged all three channels. Bars = 50μm in (a–d, g–h), 20 μm in (e–f, i–p).
Extended Data Figure 6.
pHAM2::YFP-HAM2 (pHAM2::YPET-HAM2)complemented the ham1,2,4 mutant and was expressed in the center of SAMs. The early termination phenotype ofham1;2;4(a, b) was completely complemented by YPET-HAM2 driven by the HAM2 promoter and 3′UTR (c), indicating the promoter used for HAM2 transcriptional and translational reporters are functional and the fusion protein (YPET-HAM2) is also functional in vivo. Arrows in (a, b) indicate early-terminated apices. Bars = 10 mm (a–c). (d–e) Different Z sections from the same SAM in ham1,2,4 [pHAM2::YPET-HAM2] plant for Fig. 3 (m–n) show expression of pHAM2::YPET-HAM2 translationalmarker (green) in L2 (d) and L3 (e), together with PI as counter stain (Red). Bars = 20 μm (d–e). (f) Immunoblot with anti-GFP antibody validates the presence of YFP-HAM2 (YPET-HAM2) in both nuclear lysate and nuclear proteins immunoprecipitated with GFP-Trap from ham1,2,4 [pHAM2::YFP-HAM2] line used in ChIP experiment (Fig. 2, n and o).
Extended Data Figure 7.
Expression patterns of HAMgenesin comparison to WOX4. (a) pHAM4::2xYPET-N7 (green, arrow indicated) is expressed in procambium cells of the first leaf. (b) pHAM4::2xYPET-N7 (green, arrow indicated) is expressed in vasculature in the 7-day-old hypocotyl. (c–h) Comparison of pHAM4::2xYPET-N7 (green, arrow indicated) and pWOX4::YFP (green, arrow indicated) expression patterns in vasculature cells in the 7-d-old leaf petiole (c–d),20-d-old leaf petiole (e–f) and 7-d-old root (g–h). (i) Orthogonal view of pHAM4::2xYPET-N7 (green, arrow indicated) expression in flower vasculature. (j) Procambium-specific expression of pHAM4::2xYPET-N7 in stems from 1cm bolting plants. (k–l) Procambium-specific expression of pHAM3::2xYPET-N7mirS (k) and pHAM1::2xYPET-N7mirS (l) in transverse sections of stems from 1 cm bolting plants. Red in (a–f, i–l) represents chlorophyll autofluorescence, and represents PI staining in (g–h). Bars = 50μm in (a, h–i, k–l); 100μm in (b–g, j).
Extended Data Figure 8.
Expression patterns of HAM2 transcriptional and translational reporters in root meristems. (a–i) Complete stacks of confocal sections through the root tip demonstrate that pHAM2::2xYPET-N7mirS (green) is expressed in the QC cells (arrow indicated) and cells above the QC within the root meristem. (j–o) Expression patterns of HAM2 translational reporters in ham1,2,4 root meristems. Complete stacks of confocal sections through the root tip demonstrate that pHAM2:YPET-HAM2 (green) is present in the QC cells (arrow indicated) and the cells above the QC within the root meristem in the ham1,2,4 mutant. In all figures cellular outlines were stained with PI (red). Bars = 20μm (a–i), 50μm (j–o).
Extended Data Figure 9.
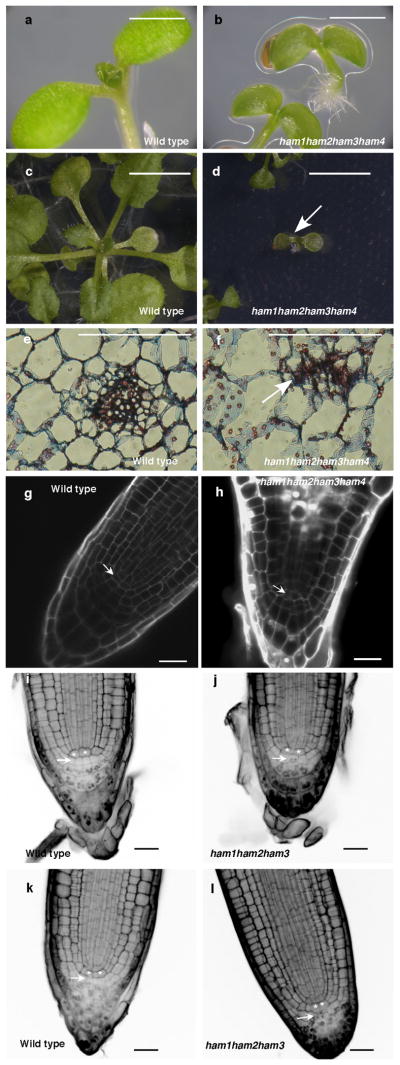
HAM family regulates various stem cell niches. (a–d) Growth arrest of ham1;2;3;4 at the seedling stage. Imaging of Ler wild type (WT) (a) and homozygous ham1;2;3;4 (b) seedlings at 7 days after germination (DAG). Imaging of WT (c) and homozygous ham1;2;3;4 (d) (arrow indicated) seedlings at 26 DAG. (e–f) Transverse section of leaves from WT (e) and ham1;2;3;4 (f) at 7 DAG. Arrow in (f) indicates undifferentiated/undetermined cell mass. (g–h) Confocal imaging of root meristem from WT (g) and ham1;2;3;4 (h) seedlings at 7 DAG. ham1;2;3;4 displayed enlarged cells with abnormal shapes at the QC (arrow indicated) and columella stem cell (CSC) positions. Cellular outlines (g–h) were visualized with PI staining (white). (i–l) mPS-PI41 stains indicate that HAM genes regulate root cell differentiation. Some CSCs (arrow indicated) undergo differentiation with starch accumulated and stained in homozygous ham1;2;3 (j, l), but none of them can be stained in L-er wild type (i, k). Asterisks mark the QC cells. Bars= 5mm (c–d), 1 mm in (a–b, e–f), 20 μm in (g–l).
Extended Data Figure 10.
Interaction between WOX and HAM homologs from tomato (Solanum lycopersicum). Bimolecular fluorescence complementation (BiFC) analyses in tobacco transient assays demonstrated that tomato WUS (Gene ID: 543793) physically interacted with a putative tomato HAM homolog (LEFL2052P11) (a) identified based on its sequence homology to HAM from Arabidopsis and Petunia (f), and tomato WOX410 (Gene ID: 100301933) physically interacted with the putative tomato HAM homolog (b). BARD1 protein is included as a negative control (c–e). Left panel: GFP channel; middle panel: PI staining channel; right panel: merged channels. Scale bar = 20 μm.(f) Amino acid sequence alignment of a putative tomato HAM, Arabidopsis HAM1 and petunia HAM using Clustal Omega. Asterisks: same amino acids; dots: similar amino acids.
Supplementary Material
Acknowledgments
The authors are grateful to Dr. R. Deshaies for his generous support with the protein purification and pull-down experiments, to Dr. D. Rees for sharing the 96-well format luminometer, to Drs. T. Laux, T. Greb, and X. Deng for sharing published reagents, to Drs. K. Sugimoto and A. Roeder for the help with the histology experiments and critical reading of the manuscript, to Dr. A. Sampathkumar for the suggestion on confocal imaging, and to A. Garda and L. Wang for technical support. Scanning electron microscopy was performed at the Applied Research Center of the College of William and Mary with technical assistance from B. Robertson. This work was funded by NIH grant R01 GM104244and by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (through Grant GBMF3406) to Elliot M. Meyerowitz, by a Caltech Gosney Postdoctoral Fellowship to Yun Zhou, by National Institutes of Health Grants GM094212, GM056006, and GM067837 to Steve A. Kay, and was aided by a grant from The Jane Coffin Childs (JCC) Memorial Fund for Medical Research to Xing Liu, a JCC fellow.
Footnotes
Author contributions
Y.Z. and E.M.M. conceived the experiments. Y.Z., X.L., E.M.E., and A.Y. performed experiments. J.L.P.-P. and S.A.K. provided the transcription factor library. Z.L.N. and P.T.T. contributed reagents. Y.Z., X.L. and A.Y. analyzed data. Y.Z. and E.M.M. wrote the manuscript and X.L., Z.L.N., A.Y. revised it. All the authors have read and approved the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Meyerowitz EM. Genetic control of cell division patterns in developing plants. Cell. 1997;88:299–308. doi: 10.1016/s0092-8674(00)81868-1. [DOI] [PubMed] [Google Scholar]
- 2.Sablowski R. The dynamic plant stem cell niches. Current opinion in plant biology. 2007;10:639–644. doi: 10.1016/j.pbi.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Miyashima S, Sebastian J, Lee JY, Helariutta Y. Stem cell function during plant vascular development. The EMBO journal. 2013;32:178–193. doi: 10.1038/emboj.2012.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinneny JR, Benfey PN. Plant stem cell niches: standing the test of time. Cell. 2008;132:553–557. doi: 10.1016/j.cell.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Laux T, Mayer KF, Berger J, Jurgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- 6.Mayer KF, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 7.Schoof H, et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 8.Sarkar AK, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- 9.Hirakawa Y, Kondo Y, Fukuda H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. The Plant cell. 2010;22:2618–2629. doi: 10.1105/tpc.110.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji J, et al. WOX4 promotes procambial development. Plant physiology. 2010;152:1346–1356. doi: 10.1104/pp.109.149641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suer S, Agusti J, Sanchez P, Schwarz M, Greb T. WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. The Plant cell. 2011;23:3247–3259. doi: 10.1105/tpc.111.087874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nardmann J, Reisewitz P, Werr W. Discrete shoot and root stem cell-promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms. Molecular biology and evolution. 2009;26:1745–1755. doi: 10.1093/molbev/msp084. [DOI] [PubMed] [Google Scholar]
- 13.van der Graaff E, Laux T, Rensing SA. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009;10:248. doi: 10.1186/gb-2009-10-12-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruneda-Paz JL, et al. A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell reports. 2014;8:622–632. doi: 10.1016/j.celrep.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuurman J, Jaggi F, Kuhlemeier C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 2002;16:2213–2218. doi: 10.1101/gad.230702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engstrom EM, et al. Arabidopsis homologs of the petunia hairy meristem gene are required for maintenance of shoot and root indeterminacy. Plant physiology. 2011;155:735–750. doi: 10.1104/pp.110.168757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulze S, Schafer BN, Parizotto EA, Voinnet O, Theres K. LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. The Plant journal. 2010;64:668–678. doi: 10.1111/j.1365-313X.2010.04359.x. [DOI] [PubMed] [Google Scholar]
- 18.Graf P, et al. MGOUN1 encodes an Arabidopsis type IB DNA topoisomerase required in stem cell regulation and to maintain developmentally regulated gene silencing. The Plant cell. 2010;22:716–728. doi: 10.1105/tpc.109.068296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 20.Busch W, et al. Transcriptional control of a plant stem cell niche. Developmental cell. 2010;18:849–861. doi: 10.1016/j.devcel.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Yadav RK, et al. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes & development. 2011;25:2025–2030. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nawy T, et al. Transcriptional profile of the Arabidopsis root quiescent center. The Plant cell. 2005;17:1908–1925. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 24.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 25.Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16529–16534. doi: 10.1073/pnas.0908122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walter M, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant journal. 2004:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 27.Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant journal. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- 28.Ohashi-Ito K, Bergmann DC. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. The Plant cell. 2006;18:2493–2505. doi: 10.1105/tpc.106.046136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierce NW, et al. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell. 2013;153:206–215. doi: 10.1016/j.cell.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harper S, Speicher DW. Current protocols in protein science. 2008;Chapter 6(Unit 6.6) doi: 10.1002/0471140864.ps0606s52. [DOI] [PubMed] [Google Scholar]
- 32.Huang X, et al. Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. The Plant cell. 2012;24:4590–4606. doi: 10.1105/tpc.112.103994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlsbecker A, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Mai YX, Zhang YC, Luo Q, Yang HQ. MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Molecular plant. 2010;3:794–806. doi: 10.1093/mp/ssq042. [DOI] [PubMed] [Google Scholar]
- 35.Barrell PJ, Conner AJ. Minimal T-DNA vectors suitable for agricultural deployment of transgenic plants. BioTechniques. 2006;41:708–710. doi: 10.2144/000112306. [DOI] [PubMed] [Google Scholar]
- 36.Heisler MG, et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Current biology. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 37.Reddy GV, Meyerowitz EM. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science. 2005;310:663–667. doi: 10.1126/science.1116261. [DOI] [PubMed] [Google Scholar]
- 38.Sugimoto K, Jiao Y, Meyerowitz EM. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Developmental cell. 2010;18:463–471. doi: 10.1016/j.devcel.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Roeder AH, Ferrandiz C, Yanofsky MF. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Current biology. 2003;13:1630–1635. doi: 10.1016/j.cub.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 40.Bowler C, et al. Chromatin techniques for plant cells. The Plant journal. 2004;39:776–789. doi: 10.1111/j.1365-313X.2004.02169.x. [DOI] [PubMed] [Google Scholar]
- 41.Truernit E, et al. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. The Plant cell. 2008;20:1494–1503. doi: 10.1105/tpc.107.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.