Abstract
Previous structural studies suggested that ribosomal translocation is accompanied by large interdomain rearrangements of elongation factor G (EF-G). Here, we follow the movement of domain IV of EF-G relative to domain II of EF-G using ensemble and single-molecule Förster resonance energy transfer (smFRET). Our results indicate that ribosome-free EF-G predominantly adopts a compact conformation that can also, albeit infrequently, transition into a more extended conformation in which domain IV moves away from domain II. By contrast, ribosome-bound EF-G predominantly adopts an extended conformation regardless of whether it is interacting with pre- or posttranslocation ribosomes. Our data suggest that ribosome-bound EF-G may also occasionally sample at least one more compact conformation. GTP hydrolysis catalyzed by EF-G does not affect the relative stability of the observed conformations in ribosome-free and ribosome-bound EF-G. Our data support a model suggesting that, upon binding to a pretranslocation ribosome, EF-G moves from a compact to a more extended conformation. This transition is not coupled to, but likely precedes both GTP hydrolysis and mRNA/tRNA translocation.
Keywords: ribosome, tRNA translocation, smFRET, conformational dynamics, viomycin
Introduction
After the addition of a new amino acid to a growing polypeptide chain, peptidyl and deacylated tRNAs together with associated mRNA codons are translocated from A and P to P and E sites of the ribosome, respectively. The translocation of tRNA and mRNA is catalyzed by a universally conserved ribosome-dependent GTPase (EF-G in prokaryotes and EF-2 in eukaryotes) 1. Despite years of studies, the molecular mechanism of translocation is not fully understood, likely due to the complexity of structural rearrangements in ribosomes, tRNAs, and EF-G involved in the process. Translocation comprises stepwise movement of tRNA: first, the acceptor ends of the tRNAs move relative to the large (50S) ribosomal subunit, and then their anticodon ends move on the small (30S) ribosomal subunit, coupled with mRNA movement 2. tRNAs likely adopt additional intermediate conformations and positions between these two main steps of translocation3; 4; 5; 6; 7. tRNA and mRNA translocation is accompanied by a number of conformational changes in the ribosome: rotational movements between subunits, swiveling motion of the head domain of the small subunit, and inward-outward movement of the L1 stalk of the large subunit8; 9; 10. These conformational changes are all coordinated with each other 11.
It is likely that EF-G also undergoes a number of structural rearrangements during translocation. EF-G is a five-domain protein containing ~700 amino acids 12; 13 (Fig. 1a). Domain I comprises G’ and G subdomains; the latter hydrolyzes GTP and is structurally similar to G domains in other G proteins. The elongated domain IV of EF-G was shown to be critical for the catalysis of translocation 14; 15. In the structure of the posttranslocation ribosome, in which the A site of the ribosome is not occupied by tRNA, domain IV is docked into the 30S A site 9; 16. By contrast, in the structure of the pretranslocation ribosome containing tRNA in the A site, the tip of domain IV of EF-G is located ~20Å away from the A site 5. Hence, domain IV of EF-G likely promotes translocation of the A-site tRNA by the movement toward the 30S A site that mostly results from the rotation of EF-G as a whole around the GTPase active site of domain I and the conserved sarcin-ricin loop of 23S rRNA 5.
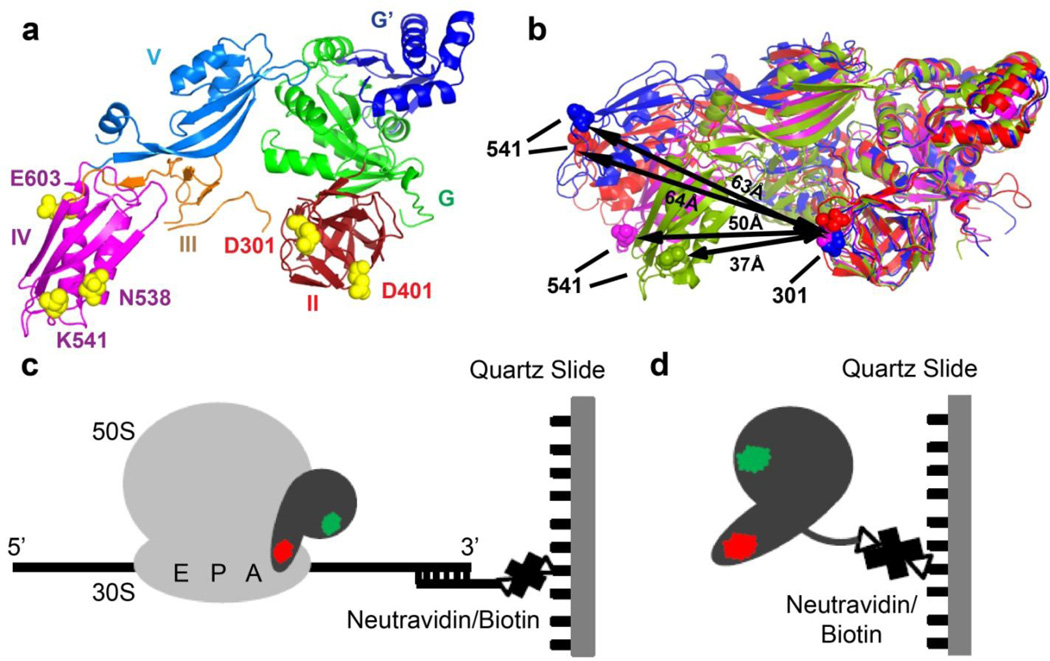
FIGURE 1. Experimental design.
(a) Residues chosen for fluorescent labeling (yellow spheres, E. coli numbering) viewed in the crystal structure of ribosome-free EF-G from T. thermophilus (PDB code 1dar12). Domains of EF-G are colored as shown. (b) Comparison of conformations in ribosome-bound and ribosome-free EF-G: EF-G (T. thermophilus) bound to the posttranslocation ribosome is shown in blue (PDB code 2wri16); EF-G (E. coli) bound to the pretranslocation ribosome in the presence of viomycin is shown in red (PDB code 3j5×5); ribosome-free EF-G•GDP (T. thermophilus) is shown in magenta (PDB code 1dar12) and ribosome-free EF-G (S. aureus) is shown in green (PDB code 2×ex17). Domains I and II of the EF-G structures were superimposed. Distance between the α-carbon atoms of residues 541 and 301 (E.coli numbering, shown as spheres) used for fluorescent labeling in respective structures is indicated by black arrows. (c) During smFRET imaging, EF-G doubly-labeled with donor (green) and acceptor (red) dyes was bound to ribosomes, which were immobilized by hybridization of the mRNA 3’ tail to a biotin-derivatized DNA oligonucleotide tethered via neutravidin to a microscope slide. (d) In smFRET studies of ribosome-free EF-G, EF-G doubly-labeled with donor (green) and acceptor (red) dyes was immobilized via C-terminal biotinylated-tag.
In addition to the movement relative to the ribosome, ribosome-bound EF-G likely undergoes interdomain rearrangement during the translocation cycle. Between pre- and posttranslocation conformations of EF-G, two superdomains formed by domains III, IV and V and I and II, respectively, move relative to each other resulting in a shift of the tip of domain IV by 10–15 Å 5 (Fig. 1b). More dramatic interdomain rearrangement is observed when the crystal and cryo-EM structures of ribosome-bound EF-G are compared to the crystal structures of ribosome-free EF-G (Fig. 1b). In contrast to the extended, elongated conformations that EF-G adopts in pre- and posttranslocation ribosomes, ribosome-free EF-G assumes more compact conformations where superdomain III-IV-V is positioned closer to superdomain I-II. These rearrangements result in a movement of the tip of domain IV by ~30 Å 9; 16; 17. In addition, different structures of ribosome-free EF-G show variability in the relative orientation of superdomains I-II and III-IV-V. When compared to the structure of ribosome-free EF-G from Thermus thermophilus 12, ribosome-free EF-G from Staphylococcus aureus 17 (Fig. 1b) displays a movement of superdomain III-IV-V by 25 Å in a direction orthogonal to the movement observed between ribosome-bound and ribosome-free EF-G from T. thermophilus.
Taken together, comparative analysis of ribosome-bound and ribosome-free EF-G structures and molecular modeling simulations of EF-G dynamics 18; 19 suggest that EF-G undergoes large interdomain rearrangements during ribosomal translocation. In addition, EF-G is a G-protein whose G-domain shares a number of common structural features with the motor domain of motor proteins such as myosin and kinesin20. For example, similar to molecular motors, G-proteins contain γ-phosphate sensing switch loops I and II, whose conformational changes are relayed though the rest of the protein structure20. Hence, it is tempting to hypothesize that EF-G promotes tRNA translocation via the movement of superdomain III-IV-V relative to superdomain I-II in a manner similar to the power stroke of the lever arm of molecular motors15; 21. Indeed, restricting the conformational dynamics of EF-G by introducing an intramolecular disulfide crosslink between domains I and V was shown to abolish EF-G translocation activity, suggesting the importance of interdomain rearrangements for translocation22. However, the exact role of interdomain movement in EF-G during translocation remains elusive. It is not clear which conformations observed in structural studies are actually sampled by ribosome-free and ribosome-bound EF-G in solution. For instance, some previously described conformations could be induced by crystal packing and are not stable in solution. Moreover, the relative stability of EF-G conformations has not been explored. In particular, it is not clear whether ribosome-bound EF-G adopts, at least transiently, the compact conformation observed in ribosome-free EF-G structures. Most importantly, it remains unclear if the interdomain structural rearrangements of EF-G are induced by GTP binding to free EF-G, EF-G•GTP binding to the ribosome or GTP hydrolysis, and/or whether rearrangements are coupled to tRNA/mRNA translocation. Here, we address these questions by following the movement of domain IV relative to domain II of ribosome-free and ribosome-bound EF-G using ensemble and single-molecule molecule Förster resonance energy transfer (smFRET). Our results suggest that EF-G moves between compact and extended conformations. Surprisingly, inhibition of translocation by the antibiotic viomycin or replacement of GTP with non-hydrolysable analogues do not affect the stability of the extended and compact conformations of ribosome-bound EF-G, suggesting that large-scale interdomain rearrangements of EF-G are not directly coupled to GTP hydrolysis or tRNA/mRNA translocation, but are induced by the interaction of EF-G with the ribosome.
Results
Construction of fluorescently labeled EF-G variants
The interdomain dynamics of EF-G were followed using FRET between fluorophores attached to domains II and IV of EF-G (Fig. 1a). Cysteine residues were introduced in domain II (positions 301 or 401) and domain IV (positions 538, 541 or 603) of a cysteine-free mutant of EF-G from Escherichia coli 23. Positions chosen for cysteine substitutions were not conserved among EF-G homologues. Furthermore, replacement of the three naturally occurring cysteines and introduction of cysteines at positions 301 and 541 were previously shown not to perturb EF-G function 23. In addition, the α-carbon atoms of residues chosen for labeling were ~50 Å apart from each other in the structure of ribosome-free EF-G from T. thermophilus (Fig. 1a). This inter-dye distance nears R0 (the distance between the donor and acceptor fluorophore pair at which the efficiency of energy transfer is 0.5, equal to 56 Å for the Cy3/Cy5 pair 24), allowing changes in distance to be measured with optimal sensitivity. Our pre-steady-state kinetics measurements of translocation rates demonstrated that fluorophore conjugation to each of the five positions used for labeling did not affect the translocation activity of EF-G (Supplemental Table S1). Thus, three double-cysteine mutants were created: D301C/K541C, D301C/E603C and D401C/N538C. Purified double-cysteine variants of EF-G were labeled with equimolar amounts of maleimide derivatives of the donor (Cy3) and acceptor (Cy5) fluorescent dyes. This labeling strategy produced doubly-labeled EF-G variants with mixed donor-acceptor orientations between the two positions. No significant changes in translocation activity were observed for EF-G_301-603(Cy3/Cy5) and EF-G_301-541(Cy3/Cy5) under multiple-turnover conditions (i.e. where the concentration of ribosome was higher than that of EF-G) using a puromycin assay (Supplemental Fig. S1 and Supplemental Table S2). The translocation activity of EF-G_401-538(Cy3/Cy5) decreased ~2-fold. Hence, fluorescently-labeled EF-G constructs retained high translocation activity.
Interdomain movements in EF-G were investigated using ensemble and single-molecule FRET. In order to probe conformations of ribosome-bound EF-G using smFRET, ribosomal complexes were immobilized on a microscope slide/cover slip via a biotin-derivatized DNA oligonucleotide annealed to the mRNA and imaged using total internal reflection (TIR) microscopy (Fig. 1c). When interdomain dynamics of ribosome-free EF-G were studied in smFRET experiments, EF-G was directly tethered to the miscroscope slides (Fig. 1d). For this purpose, a 15 amino-acid polypeptide derived from the biotin carboxyl carrier protein (BCCP) subunit of acetyl-coA carboxylase 25 was added to the EF-G sequence between the C-terminus of D301C/E603C mutant and the C-terminal 6-His purification tag. This EF-G construct was biotinylated in vitro by biotin ligase. Our pre-steady-state kinetics measurements of translocation rates showed that addition of the biotinylated tag to EF-G decreased EF-G translocation activity ~2-fold (Supplemental Table S1). Under multiple-turnover conditions, K1/2 of fluorescently-labeled and biotinylated construct EF-G_301-603(Cy3/Cy5)_biotin decreased ~4-fold when compared to unlabeled wild-type EF-G (Supplemental Fig. S1 and Table S2) indicating that EF-G_301-603(Cy3/Cy5)_biotin retained high ribosome-binding and translocation activities.
Observation of interdomain rearrangements of EF-G using ensemble FRET
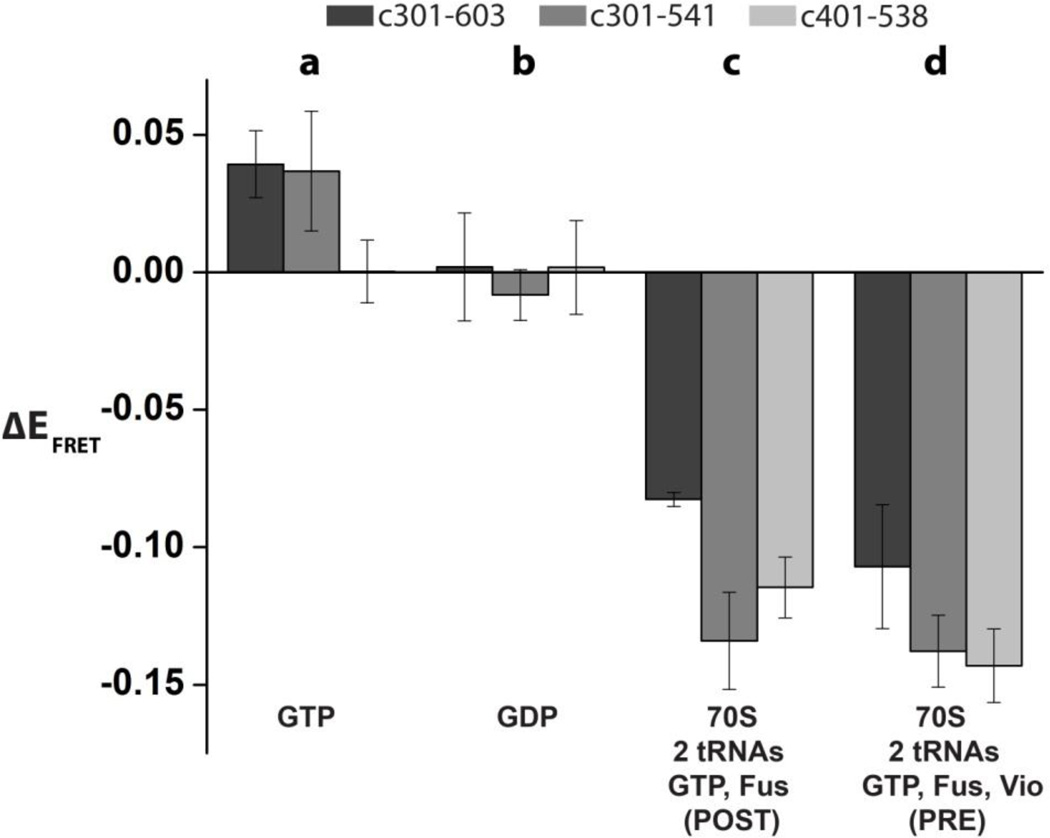
We first probed the interdomain rearrangements of EF-G using three fluorescently-labeled EF-G constructs (EF-G_301-603(Cy3/Cy5), EF-G_301-541(Cy3/Cy5) and EF-G_401-538(Cy3/Cy5)) in ensemble FRET measurements. GTP binding resulted in a small but nevertheless reproducible increase in FRET in EF-G_301-603(Cy3/Cy5) and EF-G_301-541(Cy3/Cy5) relative to the FRET values observed in ligand-free EF-G (Fig. 2a). However, the addition of GTP had no effect on the FRET in EF-G_401-538(Cy3/Cy5). Hence, in agreement with earlier X-ray light scattering measurements 26, GTP binding does not result in major interdomain rearrangements in EF-G. Binding of GDP to EF-G did not change the efficiency of energy transfer relative to nucleotide-free EF-G in all three constructs (Fig. 2b).
FIGURE 2. Changes in ensemble FRET between domains IV and II of EF-G induced by nucleotide and ribosome binding.
Increases or decreases in FRET efficiency relative to ribosome-free and nucleotide-free EF-G (ΔE=Ecomplex-Efree) are shown upon GTP binding (a); GDP binding (b), binding to the 70S ribosome containing deacylated tRNAMet in the P site and N-acetyl-Met-Phe-tRNAPhe in the A in the presence of GTP and fusidic acid (c); binding to the 70S ribosome containing deacylated tRNAMet in the P site and N-acetyl-Met-Phe-tRNAPhe in the A site, in the presence of GTP, fusidic acid and viomycin (d). Ensemble FRET measurements were taken for EF-G_301-603(Cy3/Cy5), EF-G_301-541(Cy3/Cy5), and EF-G_401-538(Cy3/Cy5). Error bars show standard deviations for changes in FRET efficiency were calculated from triplicate measurements.
We next determined if EF-G binding to the pre- and posttranslocation ribosome resulted in movement between the domains of EF-G. We incubated fluorescently-labeled EF-G with pretranslocation ribosomes containing dipeptidyl N-acetyl-Met-Phe-tRNAPhe in the A site and deacylated tRNAMet in the P site in the presence of GTP and fusidic acid (Fus), an antibiotic which does not interfere with one round of translocation or GTP hydrolysis, but instead inhibits EF-G release after GTP hydrolysis27. These experiments were performed in the absence (Fig. 2c) or presence (Fig. 2d) of viomycin, an antibiotic which is known to inhibit EF-G-induced translocation and enhance EF-G binding affinity to the ribosome 5; 28; 29. In the absence of viomycin, EF-G induces translocation and remains bound to posttranslocation ribosomes, while in the presence of viomycin EF-G becomes trapped in pretranslocation ribosomes5; 28; 30. EF-G binding to the pre- (Fig. 2d) and posttranslocation ribosomes (Fig. 2c) resulted in a decrease in FRET of varying degrees in all doubly-labeled constructs when compared to ribosome-free EF-G. These results confirm that, consistent with X-ray crystal and cryo-EM structures9; 16; 31; 32; 33,34, ribosome-bound EF-G indeed adopts a more extended conformation than ribosome-free EF-G. Because of signal averaging in ensemble FRET experiments, it is not clear whether the variations in FRET efficiency observed between different EF-G-ribosomal complexes correspond to discrete conformations of EF-G or to differences in the extent of EF-G binding. The interpretation of these experiments is also complicated by the stabilizing effect of viomycin on EF-G binding28; 30. Thus, ensemble FRET experiments do not reveal whether EF-G undergoes large interdomain movement during translocation. Nevertheless, when taken together, ensemble FRET experiments indicate that, in solution, ribosome-free EF-G adopts a significantly more compact conformation than ribosome-bound EF-G.
Ribosome-free EF-G predominantly adopts a compact conformation
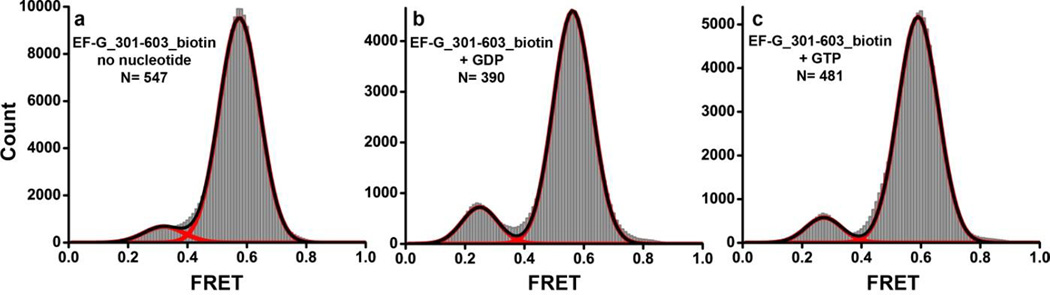
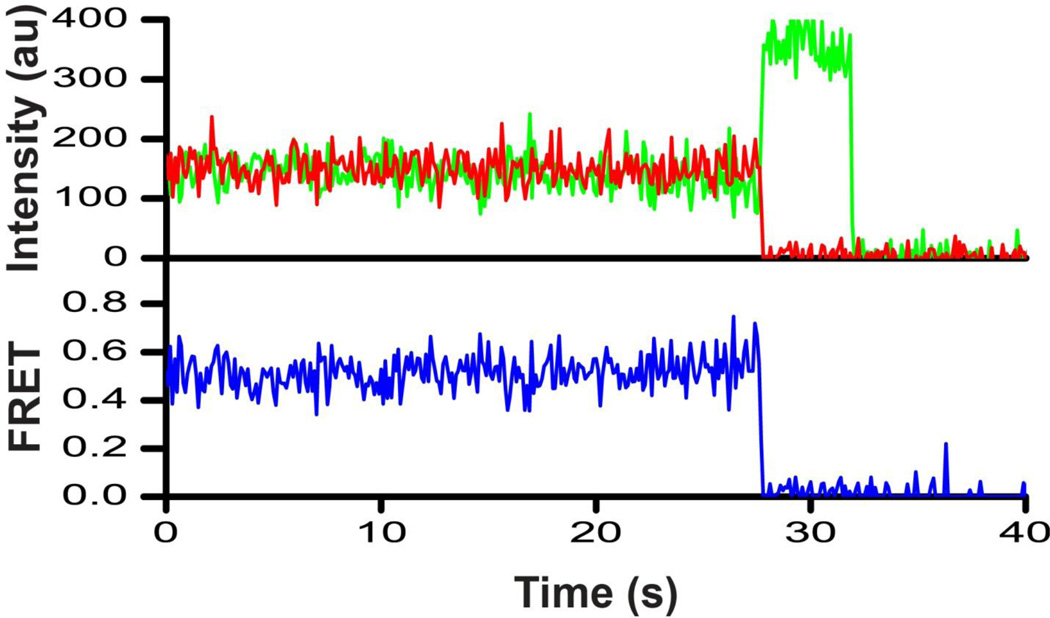
Given that ensemble experiments demonstrated the capability of designed FRET pairs to detect interdomain rearrangement in EF-G, we next examined the conformation of EF-G using smFRET and TIR microscopy. We first probed the conformational dynamic spectrum of ribosome-free EF-G. EF-G_301-603(Cy3/Cy5)_biotin was tethered to the slide via a biotinylated tag engineered at the C-terminus of EF-G (Fig. 1d). FRET distribution histograms compiled from several hundred smFRET traces showed a single peak centered at ~0.6 (Fig. 3a). Although the majority (97%) of smFRET traces displayed no fluctuations (Fig. 4), ~3% of traces showed transient sampling of a lower (~0.3) FRET state (Supplemental Fig. S2a). We next investigated the effect of nucleotide binding on EF-G dynamics. GDP binding to EF-G resulted in a slight stabilization of the 0.3 FRET state as the fraction of EF-G molecules showing the 0.3 FRET state increased from 6% in apo EF-G to 13% in EF-G•GDP (Fig. 3b, Supplemental Fig. S2b). A similar distribution of FRET values (91% of population in 0.6 FRET state and 9% in 0.3 FRET state) was observed when EF-G was incubated with GTP (Fig. 3c, Supplemental Fig. S2c), suggesting that EF-G•GTP and EF-G•GDP do not significantly differ in either sampled conformations or conformational dynamics. The inter-fluorophore distances derived from the observed 0.6 FRET state (Supplemental Table S3) were in good agreement with the distances between the α-carbon atoms of the respective residues in the crystal structure of apo- and GDP-bound EF-G from T. thermophilus 12–13. It should be noted that inferring distances from FRET values is prone to significant error because of uncertainties in the determination of fluorophore orientation factor κ2 and Förster radius R0: the parameters used in distance calculations 35. Nevertheless, the 0.6 FRET efficiency observed with ribosome-free EF-G directly immobilized onto the microscope slide is consistent with the compact EF-G conformation observed in the crystal structures of ribosome-free EF-G 12; 13. Our smFRET data also indicate that ribosome-free EF-G can sample, albeit infrequently, a more extended conformation corresponding to a 0.3 FRET state, in which domain IV is moved away from domain II (Supplemental Fig. S2). However, FRET distribution histograms indicate that this extended conformation is much less stable than the compact conformation corresponding to the 0.6 FRET state, and is not uniquely induced nor stabilized by GTP binding.
FIGURE 3. Histograms showing distribution of FRET values in single ribosome-free EF-G molecules in the presence or absence of nucleotides.
EF-G_301-603(Cy3/Cy5)_biotin was imaged in the absence of nucleotides (a); presence of GDP (b) and GTP (c). N is the number of single-molecule traces compiled for each histogram. Red lines show Gaussian fits. The black line represents the sum of Gaussians.
FIGURE 4. Representative smFRET trace for EF-G_301-603(Cy3/Cy5)_biotin imaged in the absence of nucleotides.
The trace shows fluorescence intensities observed for Cy3 (green) and Cy5 (red) and the calculated apparent FRET efficiency (blue).
EF-G adopts the extended conformation in single pre- and posttranslocation ribosomes
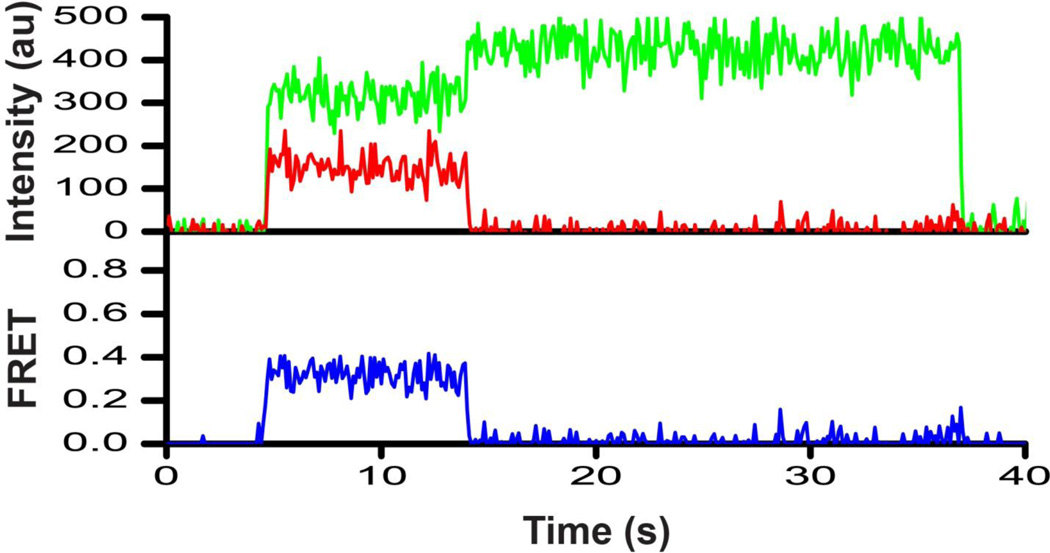
We next asked whether the compact and extended conformations corresponding to the 0.6 and 0.3 FRET states, respectively, observed in ribosome-free EF-G are also sampled by EF-G bound to the ribosome. Only single-molecule traces showing Cy5 photobleaching events leading to reciprocal increase of Cy3 fluorescence were used to compile FRET distribution histograms. Thus, it was critical that EF-G disassociation from the ribosome was slower than the acceptor photobleaching. We used Fus to extend EF-G residence time on the ribosome because in the absence of Fus (with GTP alone), EF-G binds and rapidly dissociates from the ribosome within the time resolution of single-molecule measurements (dwell time 100–350 ms)36. A pretranslocation complex containing dipeptidyl N-acetyl-Met-Phe-tRNAPhe in the A site and deacylated tRNAMet in the P site was immobilized on a microscope slide/cover slip via a biotin-derivatized DNA oligonucleotide annealed to the mRNA (Fig. 1c). Then, doubly-labeled EF-G was flowed into the slide in the presence of GTP and Fus. Under these experimental conditions EF-G hydrolyses GTP; dipeptidyl- and deacylated tRNAs rapidly (5–20 s−1) translocate from the A and P to the P and E sites, respectively, and EF-G•GDP•Fus binds the posttranslocation ribosome37; 38; 39. Single-molecule traces showed EF-G binding events recognizable by the appearance of fluorescence signals (Fig. 5).
FIGURE 5. Representative smFRET trace for posttranslocation EF-G-ribosome complex.
EF-G_301-603(Cy3/Cy5) was incubated with pretranslocation ribosomes containing tRNAMet in the P site and N-acetyl-Met-Phe-tRNAPhe in the A site in the presence of GTP and fusidic acid. The trace shows fluorescence intensities observed for Cy3 (green) and Cy5 (red) and the calculated apparent FRET efficiency (blue).
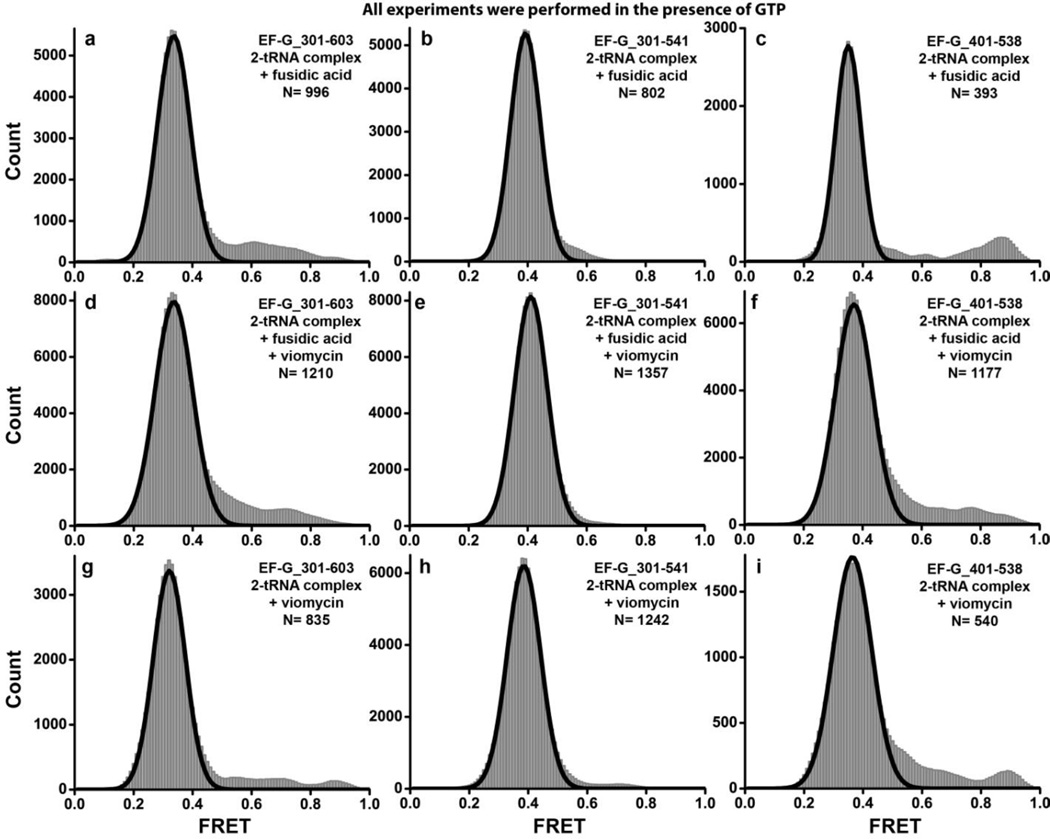
Time traces of individual EF-G_301-603(Cy3/Cy5) molecules bound to the posttranslocation ribosome and FRET distribution histograms built from several hundred molecules showed a single predominant low (0.3) FRET state (Fig. 6a). This low FRET state is significantly different from the stable compact conformation (0.6 FRET) observed for ribosome-free EF-G_301-603(Cy3/Cy5)_biotin directly bound to the microscope slide via biotinylated tag, and similar to the low FRET state transiently sampled by immobilized ribosome-free EF-G. Likewise, EF-G_301-603(Cy3/Cy5)_biotin showed the same (0.3) FRET state when it was added to immobilized ribosomes pre-incubated with an excess of free biotin to saturate the biotin binding sites of neutravidin and prevent direct immobilization of biotin-tagged EF-G to the slide (Supplemental Fig. S3). This experiment demonstrates that our untagged and biotin-tagged EF-G constructs adopt the same ribosome-bound conformation and exhibit similar structural dynamics. Furthermore, smFRET data indicate that when EF-G is bound to the posttranslocation ribosome (corresponding to the 0.3 FRET state) superdomain III-IV-V moves away from superdomain I-II when compared to the predominant conformation of ribosome-free EF-G (corresponding to the 0.6 FRET state). This conclusion is consistent with the results of ensemble FRET experiments (Fig. 2) and further corroborated by experiments with the other two doubly-labeled constructs we tested, EF-G_301-541(Cy3/Cy5) and EF-G_401-538(Cy3/Cy5), which showed a 0.4 FRET state when bound to the posttranslocation ribosome (Fig. 6b, c). The inter-fluorophore distances derived from the observed apparent FRET values for the low (0.3–0.4) FRET state observed for all constructs tested (Supplemental Table S3) were similar to the distances between the α-carbon atoms of the respective residues in the crystal structure of EF-G bound to a posttranslocation ribosome in the non-rotated, classical state conformation16. Thus, it is likely that the 0.3–0.4 FRET state corresponds to the extended EF-G conformation observed in the crystal structure of the posttranslocation ribosome.
FIGURE 6. Histograms showing distribution of FRET values in pre- and posttranslocation EF-G-ribosome complexes.
EF-G_301-603(Cy3/Cy5) (a, d, g), EF-G_301-541(Cy3/Cy5) (b, e, h), and EF-G_401-538(Cy3/Cy5) (c, f, i) were incubated with pretranslocation ribosomes containing tRNAMet in the P site and N-acetyl-Met-Phe-tRNAPhe in the A site. Experiments were performed in the presence of GTP and fusidic acid (a–f); absence (a–c) or presence of viomycin (d–i); absence of fusidic acid (g–i). N is the number of single-molecule traces compiled for each histogram. Black lines show Gaussian fits.
Since a similar distribution of FRET values is observed for the three doubly-labeled constructs, perturbations of the fluorescent properties of the dyes due to local environmental effects is unlikely to contribute to our FRET measurements. To further validate this conclusion, we checked for changes in quantum yield of the acceptor dye using ensemble fluorescence measurements since the apparent FRET efficiency depends on the acceptor quantum yield, but not on the donor quantum yield 40. We found no significant changes in fluorescence intensity of the acceptor dye (Cy5) attached to positions 301, 401, 538, 541 and 603 occurred upon EF-G binding to the pretranslocation or posttranslocation ribosome (Supplemental Fig. S4). Thus, the observed changes in the apparent FRET efficiency are not due to alterations of the quantum yield of the acceptor dye.
Although EF-G bound to the posttranslocation ribosome predominantly showed the 0.3–0.4 FRET state, ~2–3 % of traces displayed excursions into higher (0.5–0.9) FRET states (Supplemental Fig. S5). Although traces exhibiting such transitions were too rare to be quantitatively analyzed, their presence suggested that ribosome-bound EF-G can also sample more compact conformations, in which domain IV moves toward domain II of EF-G. We next aimed to determine whether these compact conformations correspond to the pretranslocation intermediate of the EF-G-ribosome complex. To probe the pretranslocation conformation of EF-G in smFRET experiments, we pre-incubated the pretranslocation ribosome containing dipeptidyl tRNA in the A site and deacylated tRNA in the P site with the antibiotic viomycin before the addition of fluorescently labeled EF-G. This was necessary because real-time observation of conformational transitions coupled to translocation in smFRET experiments is hampered by the estimated rate of translocation of 5–20 s−1; 15; 39; 41; 42. Thus, dwell times of EF-G in the pretranslocation state are expected to be 50–200 ms, which are near or below the time resolution of our smFRET measurements (100 ms). Viomycin strongly inhibits translocation but does not interfere with EF-G binding to the ribosome or GTP hydrolysis by EF-G 28; 29. Recently, a pretranslocation EF-G-ribosome complex stabilized by viomycin was visualized by cryo-EM 5. Biochemical experiments showed that in the presence of viomycin, at least half of the ribosomes remain in the pretranslocation state after incubation with EF-G and GTP for dozens of minutes30.
When doubly-labeled EF-G was added to pretranslocation ribosomes pre-incubated with viomycin the low (0.3–0.4) FRET state was again predominantly observed for EF-G_301-603(Cy3/Cy5), EF-G_301-541(Cy3/Cy5) and EF-G_401-538(Cy3/Cy5 (Fig. 6d–f). Likewise, when smFRET experiments were performed in the presence of viomycin alone (without fusidic acid), the same low (0.3–0.4) FRET state was observed for all three fluorescent EF-G constructs (Fig. 6g–i). Although comparison of the posttranslocation conformation of EF-G 16 with the cryo-EM structure of EF-G trapped in a pretranslocation ribosome in the presence of viomycin 5 reveals a shift of superdomain III-IV-V relative to domains II and I, this rearrangement is relatively small and does not result in a significant change in distance between the residues of domain IV and II used in this work for fluorescent labeling (Supplemental Table S3). Hence, consistent with existing structural data, our smFRET results suggest that EF-G mainly adopts extended conformations in both pre- and posttranslocation ribosomes. Nevertheless, ~6% of smFRET traces for EF-G bound to the pretranslocation ribosome in the presence of viomycin also showed transitions between 0.3–0.4 and higher (0.5–0.7) FRET states, suggesting a transient sampling of more compact conformations of EF-G. However, in the presence of the translocation inhibitor viomycin, no stabilization of compact conformations of EF-G was observed when compared to the distribution of FRET values for EF-G bound to the posttranslocation ribosome in the absence of viomycin. Thus, the rearrangement of EF-G from the compact to the extended conformation likely precedes and is not directly coupled to mRNA/tRNA translocation.
Movement of EF-G between the compact and extended conformation is not directly coupled to GTP hydrolysis
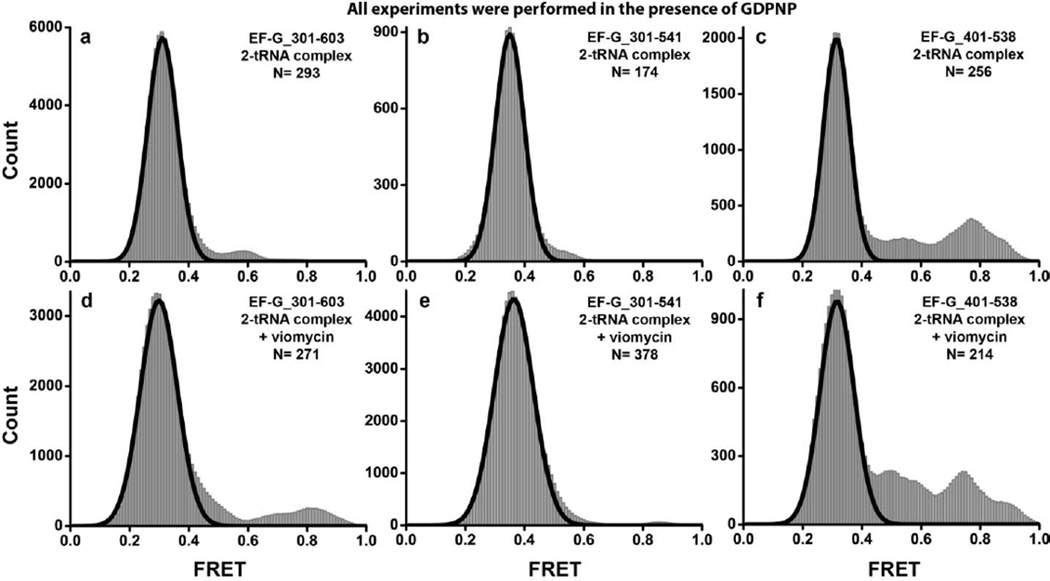
GTP hydrolysis and subsequent inorganic phosphate (Pi) release were proposed to play an important role in EF-G interdomain rearrangements 15; 22. The smFRET experiments described above were performed in the presence of GTP, viomycin and fusidic acid. Neither viomycin nor fusidic acid inhibit GTP hydrolysis15; 43. Moreover, viomycin does not affect Pi release41, while fusidic acid produces a modest (~four-fold) decrease in single-turnover Pi release38; 43. Since the rate of Pi release in the presence of fusidic acid and viomycin is 5 s−1 and 20 s−1, respectively38; 41, GTP hydrolysis and subsequent Pi release are expected to be within the time scale of smFRET data acquisition. Thus, possible interdomain rearrangements of EF-G coupled to GTP hydrolysis and Pi release could have been missed in our experiments due to time-resolution limits. In order to stabilize EF-G in its pre-hydrolysis conformation, we replaced GTP and Fus with a non-hydrolysable analogue of GTP, GDPNP. Published biochemical data suggest that EF-G•GDPNP induces translocation and remains bound to the ribosome in the posttranslocation conformation37. If the transition of EF-G from the compact conformation observed in ribosome-free EF-G to the extended conformation adopted by ribosome-bound EF-G is directly coupled to GTP hydrolysis, then the replacement of GTP with GDPNP should result in the stabilization of a compact EF-G state.
When EF-G_301-603(Cy3/Cy5)•GDPNP was added to pretranslocation ribosomes in the absence of translocation inhibitors, a predominant low 0.32 FRET state was observed in FRET distribution histograms (Fig. 7a). Likewise, binding of the two other doubly-labeled constructs (EF-G_301-541(Cy3/Cy5) and EF-G_401-538 (Cy3/Cy5)) to the ribosome in the presence of GDPNP led to the appearance of a single low (0.3–0.35) FRET state (Fig. 7b, c). Similar to experiments performed in the presence of GTP and Fus, ~2 % of traces for each of the three EF-G constructs showed excursions into higher (0.5–0.7) FRET states. When the pretranslocation complex was pre-incubated with viomycin before the addition of EF-G•GDPNP, a predominant ~0.3 FRET state (Fig. 7d–f) and rare sampling (in ~5% of traces) of higher FRET states were again observed in all three fluorescently-labeled EF-G constructs. Thus, replacing GTP and Fus with GDPNP did not change EF-G structural dynamics or distribution of FRET states in both pre- and posttranslocation ribosomes. We also took an alternative approach by making a GTPase-deficient variant of EF-G to test the role of GTP hydrolysis in the interdomain movement of EF-G. A substitution of a conserved histidine (H92 in E.coli EF-G) of the switch loop II with alanine, which was shown to inhibit GTPase activity of EF-G21; 30; 44, was introduced into the EF-G_301-541(Cy3/Cy5) construct. When EF-G(H92A)_301-541(Cy3/Cy5)•GTP was added to pretranslocation ribosomes either in the presence or absence of viomycin, no stabilization of the higher FRET states was observed, and a predominant low 0.3 FRET state was detected (Supplemental Fig. S6). Therefore, our smFRET data do not provide evidence that either GTP hydrolysis or the following phosphate release are directly coupled to interdomain rearrangements of EF-G.
FIGURE 7. Histograms showing distribution of FRET values in different EF-G-ribosome complexes assembled in the presence of GDPNP.
EF-G_301-603(Cy3/Cy5) (a, d), EF-G_301-541(Cy3/Cy5) (b, e), or EF-G_401-538(Cy3/Cy5) (c, f) were incubated with pretranslocation ribosomes containing tRNAMet in the P site and N-acetyl-Met-Phe-tRNAPhe in the A site in the presence of GDPNP. Experiments were performed in the absence of translocation inhibitors (ac) or the presence of viomcyin (d–f). N is the number single-molecule traces compiled for each histogram. Black lines show Gaussian fits.
Ribosome-bound EF-G samples compact conformations
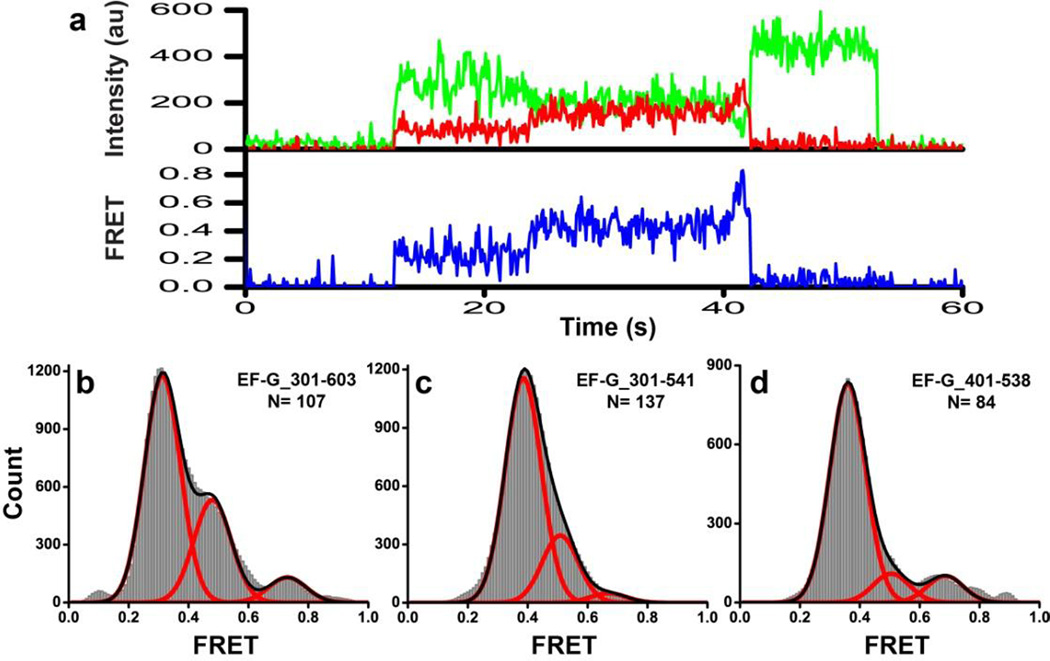
Although FRET distribution histograms for EF-G bound to various ribosomal complexes showed a single predominant low FRET state for all three fluorescently-labeled constructs, higher (>0.45) FRET values were also detected (Fig. 6 and 7). However, we cannot exclude the possibility that some of the traces displaying higher FRET values without transitions into the predominant 0.3–0.4 FRET state might represent a small fraction of non-specifically doubly-labeled EF-G. Although maleimide derivatives of fluorophores primarily react with cysteines, they are also known to modify other amino groups in proteins, albeit with very low efficiency45. Furthermore, the frequency of high FRET state sampling was too low to be quantitatively analyzed for each individual complex. In order to elucidate whether the observed high FRET values actually represented authentic conformations adopted by EF-G, we compiled FRET distribution histograms for each doubly-labeled EF-G construct (Fig. 8) by combining traces that were collected for different ribosomal complexes and showed at least one apparent transition between the predominant low (0.3–0.4) and higher FRET states (Fig. 8a, Supplemental Fig. S5). The number of such traces was still too small to enable reliable analysis of smFRET data using Hidden Markov Modeling analysis or other similar algorithms. Nevertheless, FRET distribution histograms for all three doubly-labeled constructs were best fit to three Gaussians centered at 0.3–0.4, 0.5 and 0.7 FRET, which accounted (depending on the construct) for 60–80%, 10–30% and 3–10% of time frames, respectively (Fig. 8b–d). The 0.5 FRET state is fairly abundant and likely corresponds to a compact conformation of EF-G, in which domain IV is closer to domain II when compared to the extended conformation of ribosome-bound EF-G observed in previous cryo-EM and crystal structures of the ribosome9; 16; 31; 32; 33; 34. In addition, rare excursions from the 0.3–0.4 FRET state into a higher ~0.7 FRET state (Fig. 8a, Supplemental Fig. 5b) were also observed in single-molecule FRET traces in all constructs, suggesting that EF-G might sample another unstable and more compact conformation in addition to the conformation corresponding to the 0.5 FRET state. Hence, ribosome-bound EF-G may adopt at least two, possibly three, distinct conformations corresponding to 0.3–0.4, 0.5 and 0.7 FRET states.
FIGURE 8. Ribosome-bound EF-G may sample compact conformations.
(a) A smFRET trace for EF-G_301-603(Cy3/Cy5) bound to the pretranslocation ribosome preincubated with viomycin in the presence of GTP and fusidic acid. The trace shows fluorescence intensities observed for Cy3 (green) and Cy5 (red) and the calculated apparent FRET efficiency (blue), which changes from ~0.3 to ~0.5 to ~0.7 FRET. (b–d) FRET distribution histograms compiled from single-molecules traces for ribosome-bound EF-G that show fluctuations in FRET. Histograms were built from single-molecule traces for EF-G_301-603(Cy3/Cy5) (b), EF-G_301-541(Cy3/Cy5) (c), and EF-G_401-538(Cy3/Cy5) (d) showing at least one apparent transition between the low (0.3–0.4) FRET state and a higher (0.5–0.9) FRET state. These traces were selected from traces used to compile histograms in Figures 6 and 7. N is the number of single-molecule traces compiled for each histogram. Red lines show Gaussian fits. The black line represents the sum of Gaussians.
Discussion
We followed EF-G interdomain movement using FRET between fluorophores attached to domains II and IV, which belong to the two superdomains of EF-G: I-II and III-IV-V, respectively. We aimed to address a long-standing question in the field and investigate the role of interdomain rearrangement in EF-G in translocation of tRNA and mRNA. Our smFRET experiments have allowed direct observation of rearrangement between the two superdomains of EF-G in solution that was previously inferred from comparison of various static EF-G structures. Consistent with previous crystal and cryo-EM structures, our FRET studies show that ribosome-free EF-G predominantly adopts a compact conformation corresponding to a 0.6 FRET state, while ribosome-bound EF-G mainly adopts an extended conformation corresponding to a 0.3–0.4 FRET state, in which domain IV moves away from domain II. Unexpectedly, we found no evidence of direct coupling between the transition from the compact to extended conformation of EF-G and ribosomal translocation.
Comparison of the interdomain distances derived from FRET data and crystal structures of ribosome-free EF-G suggests that the predominant conformation of ribosome-free EF-G from E.coli in solution is similar to the crystal structure of the apo-form or GDP-bound EF-G from T. thermophilus12; 13. In addition to this compact conformation, ribosome-free EF-G can also sample a more extended conformation corresponding to a 0.3 FRET state. This conformation is likely similar to the extended conformation that EF-G predominantly adopts when bound to the ribosome. Interestingly, no crystal structure has been solved to date for GTP or GTP analogue-bound wild-type ribosome-free EF-G from any organism (only a variant of EF-G containing a mutation in switch loop II of the G-domain was crystallized with GDPNP46). This fact, together with the analysis of the thermodynamics of nucleotide binding to EF-G47, suggested that EF-G adopts significantly different conformations in GDP- and GTP-bound forms. However, our data, which is consistent with early small angle X-ray scattering experiments26, show that EF-G structure and interdomain structural dynamics are very similar in GDP- and GTP-form. Nevertheless, this result does not exclude the existence of local rearrangements within domains of EF-G (such as ordering of switch loops in the G-domain) induced by exchange between GTP and GDP that might remain undetected in our experiments.
Consistent with cryo-EM and crystal structures, our smFRET data suggest that EF-G undergoes movement from the predominant compact conformation observed in ribosome-free EF-G (0.6 FRET) into a more extended conformation in the ribosome-bound state (0.3–0.4 FRET). From the outset it was not clear if this movement occurs upon binding to the ribosome, hydrolysis of GTP, inorganic phosphate release, or translocation of mRNA/tRNA. Surprisingly, stabilization of the pretranslocation state with viomycin and replacement of GTP with non-hydrolysable analogue GDPNP did not lead to a stabilization of the 0.5 or the higher (~0.7) FRET states. Hence, interdomain rearrangement within EF-G is not directly linked to GTP hydrolysis or mRNA/tRNA translocation. This contrasts with other rearrangements of EF-G and the ribosome, such as the movement of domain IV toward the A site of the small subunit resulting from the rotation of EF-G around the sarcin-ricin loop of 23S rRNA and the reverse rotation of the small subunit from the rotated into a non-rotated conformation of the ribosome, which are hampered by translocation inhibitors and, thus, directly coupled to mRNA/tRNA translocation5; 30; 39. Thus, the transition from the compact conformation, which ribosome-free EF-G adopts in solution, to the extended conformation likely occurs rapidly upon EF-G interaction with the ribosome, preceding both GTP hydrolysis and mRNA/tRNA translocation.
Our data suggest that ribosome-bound EF-G can adopt two compact conformations corresponding to the 0.5 and 0.7 FRET states (Fig. 8). Although these conformations are evidently less stable than the predominant extended conformation corresponding to the 0.3–0.4 FRET state, they might resemble authentic intermediates of translocation. Adopting the compact conformation may be required for binding of EF-G to the classical, non-rotated conformation of pretranslocation ribosomes. EF-G has been shown to transiently stabilize the rotated, hybrid-state conformation of the ribosome during translocation39; 48; 49; 50. It is not clear, however, whether EF-G binds exclusively to the spontaneously-formed rotated, hybrid-state conformation of prestranslocation ribosomes or whether it can also bind to the non-rotated conformation of pretranslocation ribosomes and then induce formation of the rotated, hybrid-state conformation as suggested by one smFRET study51. Analysis of available structures of EF-G-ribosome complexes reveals that binding of EF-G in the extended conformation to the non-rotated, classical-state pretranslocation ribosome might be hindered by the steric clash between domain IV of EF-G and peptidyl-tRNA bound in the classic A/A state5; 6; 16. This steric clash might be avoided if EF-G binds to the ribosome in the compact conformation, in which domain IV of EF-G is moved toward domain II of EF-G and away from the A site of the ribosome. In addition, a flexible connection between the superdomains of EF-G might be required to accommodate conformational rearrangements of the EF-G-ribosome complex during translocation such as intersubunit rotation or swiveling of the head domain of the small subunit. These ribosome rearrangements might be accompanied by interdomain movements of EF-G that keep EF-G in an overall-extended conformation and are more subtle than the transition from the compact to extended conformation. Further structural studies are required to fully elucidate the coordinated rearrangements of EF-G and ribosome structure during translocation.
MATERIALS AND METHODS
Materials and sample preparation
All chemicals and reagents were purchased from Sigma with the exception of neutravidin (Thermo Scientific), viomycin (USP), and puromycin (Acros Organics). The tRNAMet, and tRNAPhe were purchased from Sigma, MP Biomedicals and Chemblock. The mRNA m291, ribosomes, EF-Tu and aminoacylated tRNAs were prepared as previously described 52; 53; 54.
A cysteine-free variant of EF-G (C113D, C265A, C397S) and two single-cysteine EF-G variants (D301C and K541C) that were cloned into the pET-24 expression vector were kindly provided by Harry Noller (UCSC) 23. Three double-cysteine mutants (D301C/K541C, D301C/E603C, D401C/N538C) were created by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis System (Stratagene). The H92A substitution was introduced into the D301C/K541C variant of EF-G. Different variants of 6-histidine-tagged EF-G were expressed and purified as previously described 23. A 1:1 mixture of maleimide derivatives of the Cy3 and Cy5 (Combinix Inc) fluorescent dyes was used for labeling of the double-cysteine EF-G constructs as reported55.
A DNA sequence coding for a 15 amino-acid long polypeptide derived from the biotin carboxyl carrier protein (BCCP) subunit of acetyl-coA carboxylase (GLNDIFEAQKIEWHE) 25 was introduced into the D301C/E603C EF-G mutant using PCR. For in vitro biotinylation, 10 nmol of purified tagged EF-G was incubated with 2.5 µg of BirA enzyme (Avidity) in biotin-containing buffer provided by the manufacturer at 30 °C for 90 minutes. The biotinylated tagged EF-G construct was then labeled with Cy3/Cy5 dye in a 1:1 mixture as described above.
Biochemical assays
Multiple-turnover translocation assays, and pre-steady-state translocation kinetics measurements were performed as previously described37; 39; 42. Details of these experiments can be found in the Supplemental Materials section.
Preparation of ribosomal complexes for FRET experiments
All ribosomal complexes were assembled in polyamine buffer (50 mM Hepes-KOH, pH 7.5, 6 mM MgCl2, 150 mM NH4Cl, 6 mM β-mercaptoethanol, 2 mM spermidine, 0.1 mM spermine). Ribosomal complexes carrying a single deacylated tRNA bound to the P site were assembled by incubating 300 nM 70S ribosomes with 600 nM mRNA m291 (pre-annealed to 800 nM biotin-labeled DNA primer) and 600 nM tRNAMet at 37 °C for 20 minutes. The antibiotic viomycin (when present) was added (0.5 mM) with an additional incubation at 37 °C for 10 minutes. The pretranslocation complex was assembled by incubation of 0.5 µM 70S ribosomes with 1 µM mRNA m291 (pre-annealed to 1.3 µM biotin-labeled DNA primer) and 1 µM N-acetyl-Met-tRNAMet at 37 °C for 20 minutes followed by incubation with 1 µM EF-Tu•GTP•Phe-tRNAPhe ternary complex at 37 °C for 15 minutes. The ternary complex was pre-assembled by incubating 20 µM EF-Tu with 2 mM GTP in polyamine buffer at 37 °C for 10 minutes, followed by an additional incubation with 3 µM Phe-tRNAPhe at 37 °C for 5 minutes. The antibiotic viomycin (0.5 mM when present) was added simultaneously with the ternary complex.
Ensemble fluorescence measurements
Ensemble fluorescence measurements were taken using a FluoroMax-4 spectrofluorometer (Horiba) at 22°C. A sample volume of 30 µl was used to overfill the clear window of the 10 µl cuvette (Starna Cells). All measurements were performed in polyamine buffer containing 50 mM Hepes-KOH, pH 7.5, 6 mM MgCl2, 150 mM NH4Cl, 6 mM β-mercaptoethanol, 2 mM spermidine, 0.1 mM spermine and 0.01% Nikkol. The concentration of Cy3/Cy5-labeled EF-G was 100 nM, 70S ribosome was 200 nM, GDP or GTP was 0.5 mM, fusidic acid was 40 µM, and viomycin was 0.5 mM. Cy3 emission spectra (emission 555–800 nm) were taken by exciting fluorescence at 540 nm. Cy5 emission spectra (emission 645–800 nm) were taken by exciting fluorescence at 635 nm. The slit-widths for excitation and emission were set to 5 nm of spectral band-width. Efficiency of energy transfer in doubly-labeled EF-G construct was determined from Cy3 and Cy5 emission spectra using the ratioA method 56. Briefly, ratioA for each experiment was calculated from the ratio of the extracted integrated intensity of the acceptor (Cy5) fluorescence, which is excited both directly (by 540 nm light) and by energy transfer, divided by the integrated intensity of the acceptor excited directly by 635 nm light. FRET efficiency E was determined according to the equation56:
where γ is the fraction of EF-G molecules with both donor and acceptor label; β is fraction of EF-G molecules with acceptor and without donor label (i.e. single-acceptor and two-acceptor species*2), ε(λ) is the extinction coefficient at wavelength λ, and the subscripts D and A denote donor or acceptor. The fraction of labeling for each preparation of EF-G was determined by measuring absorbance spectra in the 300–800 nm range and assuming that labeling of each of the two cysteine residues was random.
Single-molecule FRET data acquisition and analysis
Biotin-tagged doubly-labeled EF-G was diluted in polyamine buffer (50 mM Hepes-KOH, pH 7.5, 6 mM MgCl2, 150 mM NH4Cl, 6 mM β-mercaptoethanol, 2 mM spermidine, 0.1 mM spermine) to a final concentration of 200 pM and immobilized in the sample chamber assembled from quartz microscope slides and glass cover slips coated with a mixture of m-PEG and biotin-PEG and pre-treated with neutravidin (0.2 mg/ml). GDP or GTP was 0.5 mM when present. To prevent photo-bleaching, the sample was imaged in polyamine buffer containing an oxygen-scavenging system (0.8 mg/mL glucose oxidase, 0.625% glucose, 0.02 mg/mL catalase, and 1.5 mM Trolox). Trolox was used to eliminate Cy5 blinking.
Ribosomal complexes were diluted in polyamine buffer to a final concentration of 10 nM and immobilized in the sample chamber as described above. Then Cy3/Cy5-doubly-labeled EF-G (up to 3 nM) was added to ribosome in the presence of GTP (0.5 mM) or GDPNP (3 mM). Fusidic acid was 40 µM when present. Viomycin was 0.5 mM when present. All samples were imaged in polyamine buffer containing the oxygen-scavenging system described above.
Single-molecule FRET measurements were essentially taken as previously described35; 49. An Olympus IX71 inverted microscope, equipped with a UPlanApo 60×/1.20w objective lens, a 532 nm laser (Spectra-Physics) for excitation of Cy3 dyes, and a 642 nm laser (Spectra-Physics) for excitation of Cy5 dyes were used. Total internal reflection (TIR) was obtained by a quartz prism (ESKMA Optics). Fluorescence emission was split into Cy3 and Cy5 fluorescence using a dual view imaging system DV2 (Photometrics) equipped with a 630 nm dichroic mirror and recorded via an Andor iXon+ EMCCD camera. Movies were recorded using the Single software (downloaded from Prof. Taekjip Ha’s laboratory website at the University of Illinois at Urbana-Champaign, physics.illinois.edu/cplc/software 35), with the exposure time set at 100 ms. Data acquisition was carried out at room temperature. We typically took up to ten 3-minute-long movies while imaging different sections of the slide for each sample. Non-specific binding was virtually absent.
Collected datasets were processed with the IDL and Matlab softwares, using scripts downloaded from a freely available source: physics.illinois.edu/cplc/software. Apparent FRET efficiencies (Eapp) were calculated from the emission intensities of donor (ICy3) and acceptor (ICy5) as follows: Eapp=ICy5/[ICy5+ICy3]. The FRET distribution histograms were built from traces that showed single-step photobleaching for both Cy5 and Cy3 dyes using a Matlab script generously provided by Prof. Peter Cornish (University of Missouri, Columbia). The y-axis shows a number of data points (100 ms frames) collected per each FRET value. Single-step photobleaching of the acceptor dye resulting in a reciprocal increase in donor fluorescence indicated the presence of an energy transfer before acceptor photobleaching. All histograms were smoothed with a five-point window and fitted to Gaussian distributions using Origin software (Origin Lab Co). Fitting residuals were random.
Supplementary Material
Highlights.
Ribosomal translocation is induced by elongation factor G (EF-G).
Several smFRET assays were developed to follow interdomain rearrangements of EF-G.
EF-G samples at least one compact and one extended conformation.
Movement between compact and extended conformations of EF-G is not coupled to, but likely precedes both GTP hydrolysis and mRNA/tRNA translocation.
A flexible connection between domains of EF-G might be required to accommodate rearrangements EF-G-ribosome complex during translocation.
ACKNOWLEDGMENTS
These studies were supported by grants from the US National Institute of Health no. GM-099719 (to D.N.E.). The authors thank Harry Noller for providing plasmids containing constructs EF-G-cys, EF-G-541C and EF-G-301C; Farzaneh Todnevis and Margaret Schoeniger for their early contributions to the project; Peter Cornish for sharing MatLab scripts and advice on TIR microscopy; and Gloria Culver for helpful discussions.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shoji S, Walker SE, Fredrick K. Ribosomal translocation: one step closer to the molecular mechanism. ACS Chem Biol. 2009;4:93–107. doi: 10.1021/cb8002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 3.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 4.Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CM. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell. 2011;44:214–224. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci U S A. 2013;110:20994–20999. doi: 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramrath DJ, Lancaster L, Sprink T, Mielke T, Loerke J, Noller HF, Spahn CM. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci U S A. 2013;110:20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 9.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 10.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 11.Frank J, Gonzalez RL., Jr Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.AEvarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, al-Karadaghi S, Svensson LA, Liljas A. Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J. 1994;13:3669–3677. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czworkowski J, Wang J, Steitz TA, Moore PB. The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J. 1994;13:3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martemyanov KA, Gudkov AT. Domain IV of elongation factor G from Thermus thermophilus is strictly required for translocation. FEBS Lett. 1999;452:155–159. doi: 10.1016/s0014-5793(99)00635-3. [DOI] [PubMed] [Google Scholar]
- 15.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 16.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–9. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Koripella RK, Sanyal S, Selmer M. Staphylococcus aureus elongation factor G--structure and analysis of a target for fusidic acid. FEBS J. 2010;277:3789–3803. doi: 10.1111/j.1742-4658.2010.07780.x. [DOI] [PubMed] [Google Scholar]
- 18.Li W, Trabuco LG, Schulten K, Frank J. Molecular dynamics of EF-G during translocation. Proteins. 2011;79:1478–1486. doi: 10.1002/prot.22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitford PC, Ahmed A, Yu Y, Hennelly SP, Tama F, Spahn CM, Onuchic JN, Sanbonmatsu KY. Excited states of ribosome translocation revealed through integrative molecular modeling. Proc Natl Acad Sci U S A. 2011;108:18943–18948. doi: 10.1073/pnas.1108363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vale RD. Switches, latches, and amplifiers: common themes of G proteins and molecular motors. J Cell Biol. 1996;135:291–302. doi: 10.1083/jcb.135.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holtkamp W, Cunha CE, Peske F, Konevega AL, Wintermeyer W, Rodnina MV. GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J. 2014;33:1073–1085. doi: 10.1002/embj.201387465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peske F, Matassova NB, Savelsbergh A, Rodnina MV, Wintermeyer W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Mol Cell. 2000;6:501–505. doi: 10.1016/s1097-2765(00)00049-6. [DOI] [PubMed] [Google Scholar]
- 23.Wilson KS, Noller HF. Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell. 1998;92:131–139. doi: 10.1016/s0092-8674(00)80905-8. [DOI] [PubMed] [Google Scholar]
- 24.Dietrich A, Buschmann V, Muller C, Sauer M. Fluorescence resonance energy transfer (FRET) and competing processes in donor-acceptor substituted DNA strands: a comparative study of ensemble and single-molecule data. J Biotechnol. 2002;82:211–231. doi: 10.1016/s1389-0352(01)00039-3. [DOI] [PubMed] [Google Scholar]
- 25.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czworkowski J, Moore PB. The conformational properties of elongation factor G and the mechanism of translocation. Biochemistry. 1997;36:10327–10334. doi: 10.1021/bi970610k. [DOI] [PubMed] [Google Scholar]
- 27.Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 28.Modolell J, Vazquez The inhibition of ribosomal translocation by viomycin. Eur J Biochem. 1977;81:491–497. doi: 10.1111/j.1432-1033.1977.tb11974.x. [DOI] [PubMed] [Google Scholar]
- 29.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 30.Salsi E, Farah E, Dann J, Ermolenko DN. Following movement of domain IV of elongation factor G during ribosomal translocation. Proc Natl Acad Sci U S A. 2014;111:15060–15065. doi: 10.1073/pnas.1410873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, Whitford PC, Onuchic JN, Yu Y, Sanbonmatsu KY, Hartmann RK, Penczek PA, Wilson DN, Spahn CM. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340:1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Petrov A, Tsai A, O’Leary SE, Puglisi JD. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat Struct Mol Biol. 2013;20:718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. Rna. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Savelsbergh A, Rodnina MV, Wintermeyer W. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA. 2009;15:772–780. doi: 10.1261/rna.1592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol. 2011;18:457–462. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung HS, Louis JM, Eaton WA. Distinguishing between protein dynamics and dye photophysics in single-molecule FRET experiments. Biophys J. 2010;98:696–706. doi: 10.1016/j.bpj.2009.12.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 42.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 43.Seo HS, Abedin S, Kamp D, Wilson DN, Nierhaus KH, Cooperman BS. EF-G-dependent GTPase on the ribosome. conformational change and fusidic acid inhibition. Biochemistry. 2006;45:2504–2514. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- 44.Cunha CE, Belardinelli R, Peske F, Holtkamp W, Wintermeyer W, Rodnina MV. Dual use of GTP hydrolysis by elongation factor G on the ribosome. Translation. 2013;1 doi: 10.4161/trla.24315. e24315-e24315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hermanson GT. Bioconjugate techniques. 2nd edition. 2008. edit. [Google Scholar]
- 46.Hansson S, Singh R, Gudkov AT, Liljas A, Logan DT. Crystal structure of a mutant elongation factor G trapped with a GTP analogue. FEBS Lett. 2005;579:4492–4497. doi: 10.1016/j.febslet.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 47.Hauryliuk V, Mitkevich VA, Eliseeva NA, Petrushanko IY, Ehrenberg M, Makarov AA. The pretranslocation ribosome is targeted by GTP-bound EF-G in partially activated form. Proc Natl Acad Sci U S A. 2008;105:15678–15683. doi: 10.1073/pnas.0807912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 51.Chen C, Stevens B, Kaur J, Cabral D, Liu H, Wang Y, Zhang H, Rosenblum G, Smilansky Z, Goldman YE, Cooperman BS. Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol Cell. 2011;42:367–377. doi: 10.1016/j.molcel.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. Observation of Intersubunit Movement of the Ribosome in Solution Using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 53.Fredrick K, Noller HF. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- 54.Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 55.Elvekrog MM, Gonzalez RL., Jr Conformational selection of translation initiation factor 3 signals proper substrate selection. Nat Struct Mol Biol. 2013;20:628–633. doi: 10.1038/nsmb.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Majumdar ZK, Hickerson R, Noller HF, Clegg RM. Measurements of internal distance changes of the 30S ribosome using FRET with multiple donor-acceptor pairs: quantitative spectroscopic methods. J Mol Biol. 2005;351:1123–1145. doi: 10.1016/j.jmb.2005.06.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.