Abstract
Objectives
We hypothesized that bright spots in intravascular optical coherence tomography (IVOCT) images may originate by co-localization of plaque materials of differing indices of refraction (IR). To quantitatively identify bright spots, we developed an algorithm that accounts for factors including tissue depth, distance from light source, and signal-to-noise ratio. We used this algorithm to perform a bright spot analysis of IVOCT images, and compared these results with histologic examination of matching tissue sections.
Background
Although bright spots are thought to represent macrophages in IVOCT images, studies of alternative etiologies have not been reported.
Methods
Fresh human coronary arteries (n=14 from 10 hearts) were imaged with IVOCT in a mock catheterization laboratory then processed for histologic analysis. The quantitative bright spot algorithm was applied to all images.
Results
Results are reported for 1599 IVOCT images co-registered with histology. Macrophages alone were responsible for only 23% of the bright-spot positive regions, though they were present in 57% of bright-spot positive regions. Additional etiologies for bright spots included: cellular fibrous tissue (8%), interfaces between calcium and fibrous tissue (10%), calcium and lipid (5%), and fibrous cap and lipid pool (3%). Additionally, we showed that large pools of macrophages in CD68 histology sections correspond to dark regions in comparative IVOCT images; this is due to the fact that a pool of lipid-rich macrophages will have the same index of refraction as a pool of lipid and thus will not cause bright spots.
Conclusions
Bright spots in IVOCT images are correlated to a variety of plaque components that cause sharp changes in the index of refraction. Algorithms that incorporate these correlations may be developed to improve the identification of some types of vulnerable plaque and allow standardization of IVOCT image interpretation.
Keywords: Intravascular optical coherence tomography, quantitative analysis bright spots, macrophages
IVOCT is the highest resolution technique available to image vulnerable plaque in coronary arteries (1). In 2003, Tearney and colleagues published a quantitative study that demonstrated bright spots in IVOCT images could represent macrophages (2). They showed that regions with an increased normalized standard deviation (NSD) correlated with areas in human aortic plaque that stained positively for macrophages in immunohistochemical studies. These findings were used to interpret subsequent clinical and animal IVOCT studies (3–7). However, the finding that macrophages cause increased NSD has not been validated histologically by other research groups (8).
Concerns have been raised about the specificity of bright spots in identifying macrophages, despite the use of shadowing behind the bright spots as a secondary confirmation of macrophage identification (1). For example, other plaque components appear to cause bright spots, including fibrin accumulations (9), neoatherosclerosis in previously placed stents (9), elastic lamina, cholesterol crystals, and microcalcifications (1,10). Furthermore, the NSD method was designed to be accurate only in fibrous caps and does not apply to the detection of macrophages in deeper arterial structures (2).
Tearney proposed that regions of high NSD represent areas in which the optical index of refraction (IR) has a higher heterogeneity; macrophages appear as bright spots because of the difference in IR between them and the surrounding fibrous tissue (2). Using this hypothesis, we propose that bright spots can arise from constituents other than macrophages in which a sharp change in IR occurs at interfaces between plaque components. This sharp change in IR occurs in multiple scenarios: lipid [IR 1.33] mixed with calcified cores [IR 1.65], lipid mixed with fibrous tissue [IR 1.47], and cellular fibrous tissue rich in proteoglycans [IR 1.33]—each of which may generate bright spots in IVOCT images based on this hypothesis.
We developed an algorithm that can be applied to the entire depth of the artery to enable quantitative identification of bright spots in IVOCT images of human atherosclerotic plaque. In the present study, we used this algorithm to analyze bright spots in images of human atherosclerotic plaque and compared these results to corresponding histologic sections. In addition, we further examined the hypothesis that abrupt changes in plaque components of differing IR result in the generation of bright spots in IVOCT images.
METHODS
Specimens
10 human hearts (from 3 women and 7 men) at autopsy within 24 hours of death were examined. The average age at death was 65 ± 11 years. The cause of death was cardiac in 6 cases. We imaged 14 coronary arteries (n=10, left anterior descending artery [LAD]; n=4 right coronary artery [RCA]). The IRB at the University of Texas approved this study.
Imaging procedure
The human heart catheterization laboratory was recreated with a custom IVOCT system (Volcano Corporation, San Diego, CA) to access the raw signal data. The IVOCT system has a 1310 nm swept source laser (HSL-1000, Santec, Hackensack, NJ) and a bandwidth of 80 nm scanning at a repetition rate of 34 kHz. The measured free-space axial resolution was 20 μm with a 2.8 mm scan depth. The IVOCT signal was sampled with a linear k-space clock to allow real-time OCT image acquisition and display. A fluoroscopy system (GE Medical Systems) and a chamber designed to maintain the tissue at 37°C were used. Left and right coronary 6F guide catheters were sewn into the coronary ostia, 0.014 inch guide-wire access to the coronary arteries was gained under fluoroscopic guidance, and a stent was deployed 80 mm from the guide catheter tip as a fiduciary marker. IVOCT pullbacks were acquired from the stent to the guide catheter (80 mm total pullback length). The LAD and RCA were imaged. Following imaging, the RCA and LAD were perfusion-fixed with formalin at 100 mm Hg. The left circumflex artery was not imaged due to its tortuosity in the ex vivo heart.
Histology
The LADs and RCAs were perfusion-fixed with 10% neutral-buffered formalin, excised from each heart, individually radiographed on a Faxitron MX-20 (Faxitron Bioptics LLC, Tucson AZ), and decalcified overnight with Cal-Rite (Richard Allen Scientific) if necessary. The arterial segments were sliced into 2–3 mm–thick rings and further processed on a Tissue-Tek Vacuum Infiltration Processor (Sakura Finetek USA, Torrance, CA) for standard paraffin-embedded sections. An average of 25 rings was generated from each artery. Serial tissue sections (5 μm thick) were cut at 150-μm intervals and stained with hematoxylin and eosin (H&E), modified Movat’s pentachrome, and Von Kossa. Anti-CD68 (Dako North America, Inc, Carpinteria, CA) and anti-α-smooth muscle cell-actin (Sigma-Aldrich, St. Louis, MO) antibodies were used in immunohistochemical studies to identify macrophages and smooth muscle cells, respectively.
IVOCT and histology co-registration
Each histologic ring was matched to a respective IVOCT frame. Co-registration was performed between IVOCT images and histological sections based on the following: (1) 2 fiducial landmarks—a stent deployed at the distal end of the pullback and the sewn-in guide catheter at the proximal edge—that were visible in IVOCT images, fluoroscopy, and radiography before histopathological processing, and (2) the physical position of IVOCT images in the pullbacks measured against the estimated distance in microns from the fiducial landmarks in the tissue sections. Additionally, anatomical landmarks (e.g., arterial branches or calcification patterns when present) and luminal geometric features further aided co-registration. Two researchers independently co-registered the IVOCT images and histology, discrepancies were discussed to find agreement between both co-registrations.
In IVOCT images, bright bands <65 μm thick that covered diffusely shadowed regions were identified as TCFAs. Histologic TCFAs were identified by fibrous caps <65 μm thick that covered lipid or necrotic cores.
Histologic composition of bright spot-containing areas
Regions within the arterial wall that elicit bright spots after application of the algorithm were first categorized by whether macrophages were present or not. Next each of these macrophage-positive or macrophage-negative bright spot sources were classified into the following 4 broad categories: (1) hypocellular or acellular collagen-rich fibrous tissue (mesh-like collagen-rich areas mixed with lipid, or the fibrous cap of fibrocalcific plaques); (2) cellular fibrous tissue (as found in intimal thickening or early lesions with high smooth muscle and proteoglycan content); (3) cholesterol clefts within necrotic cores; and (4) areas of layering or interface (as observed in remodeled plaque ruptures; at the interfaces between calcium and surrounding tissue; between lipid and calcium in fibrocalcific plaques; at the interface of necrotic or lipid cores and the overlying fibrous cap; at neovascularization sites and the media; or at the elastic lamina intimal/medial or medial/adventitial interface).
Bright spot quantitative detection
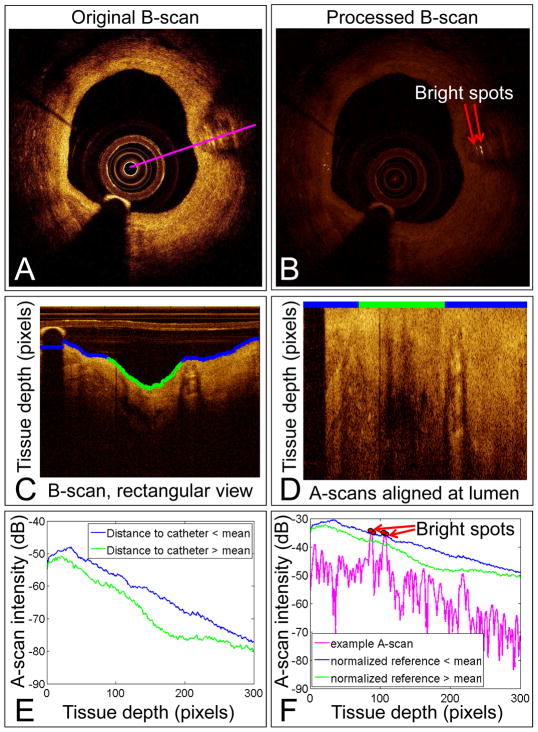
The detection method is outlined in Figure 1. First, we measured the distance between the lumen edge and the catheter for each A-scan per frame. Next, the mean of those distances was calculated for each frame. To account for variations in signal intensity that occur as the catheter moves closer or further away from the lumen, we calculated 2 reference A-scans by averaging all A-scans that were less than or greater than the mean distance to the catheter. Then, to account for varying SNR, the reference A-scans were normalized (divided by the difference between the maximum and minimum values of each frame). We compared each A-scan to the averaged and normalized reference A-scan that corresponded to whether its catheter to lumen edge distance was less than or greater than the mean; this provided a threshold to identify bright spots based on tissue depth, distance from catheter, and SNR of the IVOCT system and catheter.
Figure 1. Bright spot quantification method.
An original B-scan (A). Algorithm-processed B-scan showing the identified bright spots (B). B-scan from (A) converted to a rectangular view (C). The blue and green line marks the tissue lumen identified by the algorithm. The blue tissue edge identifies A-scans that are closer than the mean distance to the catheter; the green tissue edge identifies A-scans further than the mean distance to the catheter. Original B-scan with A-scans aligned at the blue and green line from (C) (D). Averaged A-scans from the blue and green regions in (C) (E). The intensity required to be considered a bright spot decreases with tissue depth and increases with closeness to the catheter. An example A-scan shown in magenta (A) compared to the reference value (F). Pixels with intensity greater than the reference value are marked in red and labeled as bright spots in (B). dB=decibels.
Statistics
Four statistical analyses were performed: 1) inter and intra-observer variability between two expert IVOCT readers who evaluated the unprocessed IVOCT frames for identification of bright spots, 2) joint probability of agreement between one expert IVOCT reader and the bright spot algorithm, 3) sensitivity and specificity of one expert IVOCT reader for identifying macrophages compared to the gold standard of histology, and 4) sensitivity and specificity of the bright spot algorithm for identifying macrophages compared to the gold standard of histology. Sensitivity and specificity calculations were performed at two depths, superficial (< 100 μm) and deep (>100 μm).
RESULTS
We imaged 14 coronary arteries (n=10, LAD; n=4, RCA) from 10 human hearts, generating 300 IVOCT images per vessel. After application of the algorithm, we observed a total of 2206 IVOCT frames with bright spots. 1111 of the IVOCT frames with bright spots were co-registered with histology, and 1700 regions within these 1111 frames caused distinct sources of bright spots. 488 IVOCT frames without bright spots were co-registered to histology for negative control. See the Supplement for the morphologic classification of the arteries. When looking at only the raw, unprocessed IVOCT frames for identification of bright spots between 2 expert IVOCT readers, intra-observer variability was 88% and inter-observer variability was 76%.
Macrophage detection by the bright spot algorithm (Table 1)
Table 1.
Histologic composition of algorithm identified bright spots when macrophages are present.
| Bright Spot Source | N (%)* |
|---|---|
| Macrophage-rich areas with fibrous tissue | 391 (23.0) |
| Cholesterol clefts in necrotic cores | 16 (0.9) |
| Areas of layering or interface | |
| Interface of old and new fibrous tissue | 25 (1.5) |
| Intimal/medial or adventitial/medial interface | 39 (2.3) |
| Calcium/fibrous interface | 130 (7.6) |
| Calcium/lipid interface | 98 (5.8) |
| Fibrous cap/lipid pool interface | 219 (12.9) |
| Neovascularization/media interface | 48 (2.8) |
Data are given as percentage of total bright spot regions in all IVOCT images (N=1700).
Using our quantitative algorithm and histologic examination, we characterized the role of macrophages in the origin of bright spots in IVOCT images (Figure 2). Macrophages alone were responsible for bright spots in 391 regions (23%). A combination of macrophages and other etiologies were the source of bright spots in an additional 574 regions (34%). In total, macrophages were present in 57% of bright-spot positive regions. See the Supplement for a description of how NSD images were generated and for a discussion of the NSD method compared to our bright spot method for detection of macrophages.
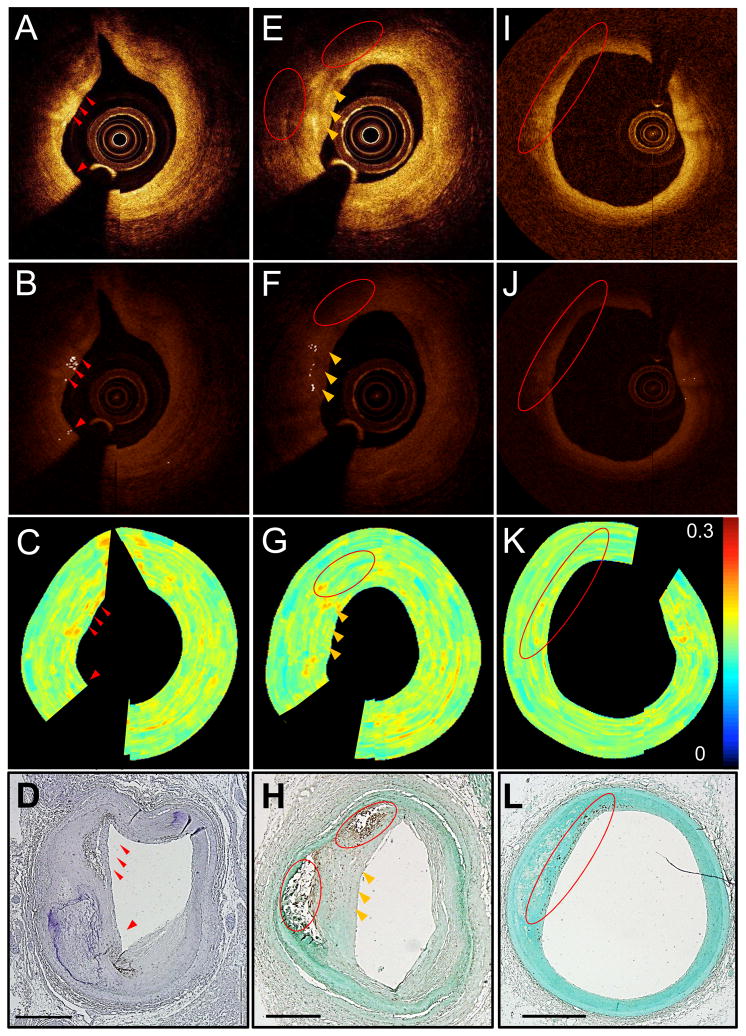
Figure 2. Macrophages depicted by our quantitative bright spot detection method.
Unprocessed IVOCT image with visible bright spots (red arrows) (A). Algorithm-processed IVOCT image with bright spots identified (B). Normalized standard deviation (NSD) image (C). CD68 stain showing that bright spots were caused by macrophages (D). Unprocessed IVOCT image with bright spots (yellow arrows) and shadows (red circles) caused by macrophages (E). Algorithm-processed IVOCT image with bright spots identified (F). NSD image (G). CD68 stain showing that bright spots came from a region of macrophage positivity and that the red circled macrophage pool was depicted as a shadow in (E) and (F) (H). Unprocessed IVOCT image from a region with macrophages far from the catheter (I). Algorithm-processed IVOCT image showing that bright spots were not found in the CD68+ region (J). NSD image (K). CD68 stains showing macrophage positivity in the red circle (L); this region was too far from the catheter for the signal to identify bright spots. All scale bars are 1 mm.
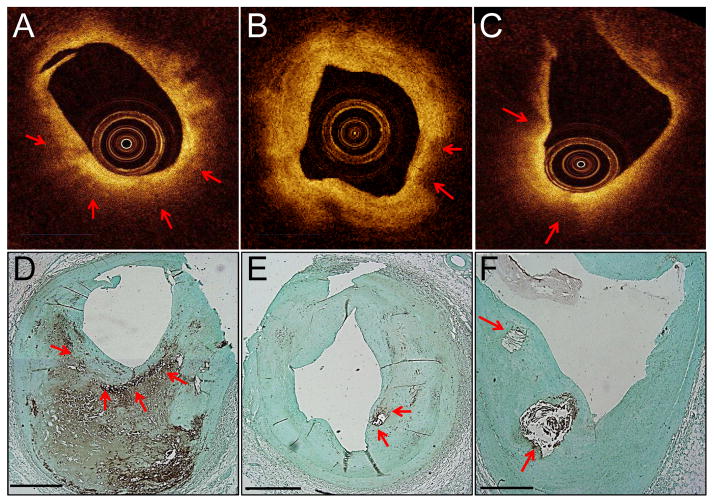
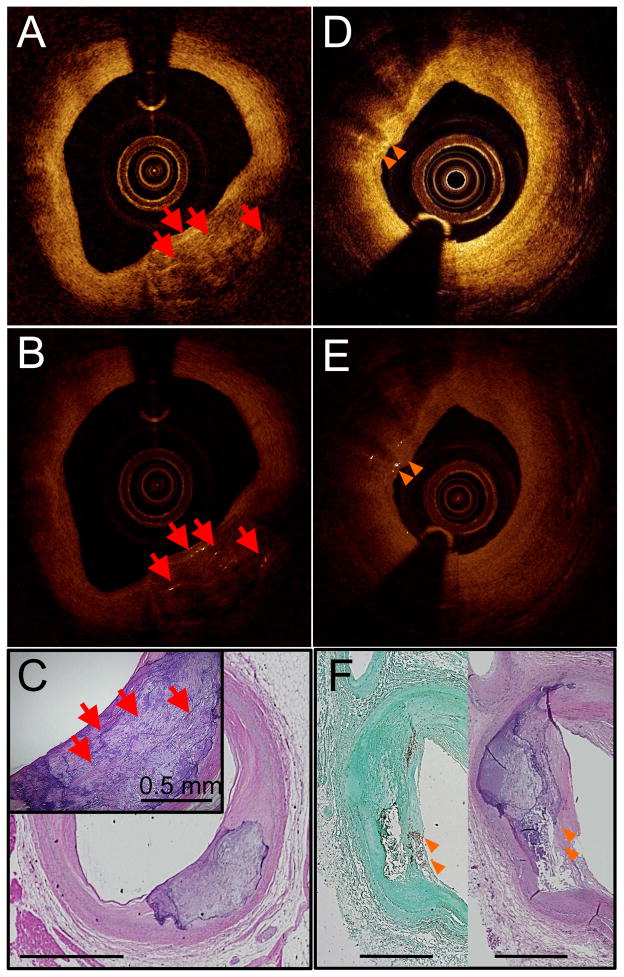
Large, dense CD68+ areas on histology corresponded to dark regions (Figs. 2 and 3) in the associated IVOCT image (125 of the CD68+ regions). Additionally, we identified 61 regions in which macrophages were located too deep in the tissue or too far from an eccentric catheter position to be visualized by IVOCT (Fig. 2I). There were also 89 regions of CD68 positivity in histologic sections that did not cause bright spots and thus were not identified by the bright spot algorithm. Lastly, in 186 regions, macrophages were depicted as bright spots that caused superficial shadowing. Of these 186, 115 (62%) were found in regions where calcium was co-localized with the macrophages (Fig. 4D) and 4 were in regions where cholesterol crystals were co-localized with the macrophages.
Figure 3. Macrophages that appear as dark regions.
Macrophages and macrophage shadows in IVOCT images. Unprocessed IVOCT images (A–C). CD68 stain verifying the presence of macrophages (D–F). Red arrows mark macrophage-rich regions that appear as dark IVOCT regions, not bright spots. All scale bars are 1 mm.
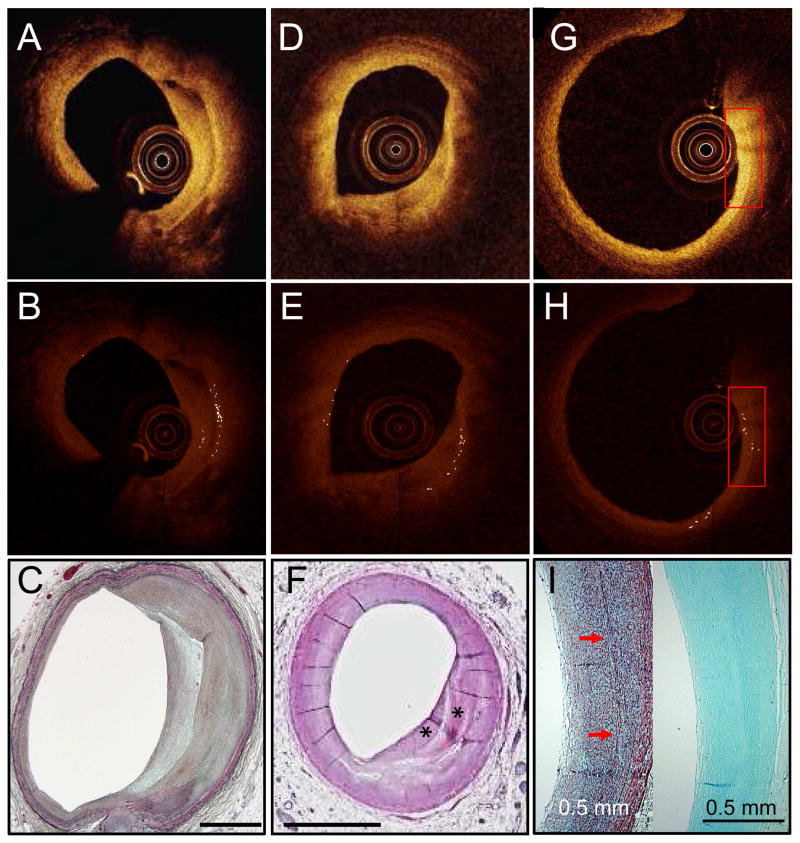
Figure 4. Bright spots found near calcium.
Unprocessed IVOCT image with bright spots caused by calcium mixed with islands of lipid (red arrows) (A). Algorithm-processed IVOCT image with bright spots identified (B). H&E stain (C). Unprocessed IVOCT image with bright spots that cause shadows like macrophages (orange arrows) (D). Algorithm-processed IVOCT image with bright spots identified (E). CD68 (left) and H&E (right) stains showing that macrophages co-mingled with calcium are the cause of the bright spots with shadowing (F). All scale bars are 1 mm unless otherwise noted.
The sensitivity and specificity of the bright spot algorithm compared to an expert IVOCT reader for identification of macrophages with histology as a gold standard are summarized in Table 2. The IVOCT reader used unprocessed IVOCT images for the analysis. The algorithm was more sensitive to bright spots (80%), implying that the presence of macrophages was more often correctly identified by the algorithm than by an expert reader; but less specific (49%), as anticipated since the algorithm detects sources of bright spots due to etiologies other than macrophages. This is also supported by the joint probability of agreement that was calculated between the algorithm and the expert reader (53%), which reflects the fact that the algorithm identified regions of macrophages that the expert IVOCT reader would have missed.
Table 2.
Sensitivity (SE) and specificity (SP) of the bright spot algorithm compared to an expert IVOCT reader for identification of macrophages defined by histology.
| Algorithm | Expert Reader | |||
|---|---|---|---|---|
| Macrophage Locations | SE (%) | SP (%) | SE (%) | SP (%) |
| All | 80 | 49 | 46 | 76 |
| Superficial (<100 μm) | 74 | 49 | 42 | 76 |
| Deep (>100 μm) | 87 | 49 | 52 | 76 |
Algorithm identified bright spots not co-localized with macrophages (Table 3)
Table 3.
Histologic composition of algorithm identified bright spots when macrophages are not present.
| Bright Spot Source | N (%)* |
|---|---|
| Fibrous tissue | |
| Hypocellular or acellular | 141 (8.3) |
| Cellular | 138 (8.1) |
| Cholesterol clefts in necrotic cores | 2 (0.1) |
| Areas of layering or interface | |
| Interface of old and new fibrous tissue | 61 (3.6) |
| Intimal/medial or adventitial/medial interface | 90 (5.3) |
| Calcium/fibrous interface | 162 (9.5) |
| Calcium/lipid interface | 83 (4.9) |
| Fibrous cap/lipid pool interface | 45 (2.6) |
| Neovascularization/media interface | 12 (0.7) |
Data are given as percentage of total bright spot regions in all IVOCT images (N=1700).
Bright spots were also associated with fibrous tissue (cellular: 8%; Fig. 5G, acellular: 8%); areas of plaque layering between old and new fibrous tissue in remodeled plaques (4%; Fig. 5A); calcified lipid cores with non-calcified remaining lipid (5%; Fig. 4A); and the fibrous cap and lipid pool interface (3%; Fig. 5D).
Figure 5. Bright spots found in fibrous tissue.
Unprocessed IVOCT image from a region with fibrous tissue layering that generated bright spots (A). Algorithm-processed IVOCT image with bright spots identified (B). Movat’s stain showing layering suggestive of a healed rupture (C). Unprocessed IVOCT image from a region with an interface between fibrous tissue and a long, narrow lipid core (D). Algorithm-processed IVOCT image with bright spots identified (E). H&E stain showing the fibrous components (asterisks) (F). The lipid core appears as a clear band. Unprocessed IVOCT image from a region of cellular fibrous intimal thickening that is rich in smooth muscle cells and proteoglycans (G). Algorithm-processed IVOCT image with bright spots identified (H). Movat’s stain (left) of the region within the inset showing elastin layers (arrows) (I); CD68 stain (right) showing that this region is CD68 negative. All scale bars are 1 mm unless otherwise noted.
Algorithm identified bright spots co-localized with TCFA (Table 4)
Table 4.
Histologic composition of algorithm identified bright spots in TCFA when macrophages are present.
| Bright Spot Source | N (%)* |
|---|---|
| Macrophage-rich areas with fibrous tissue | 51 (31) |
| Cholesterol clefts in necrotic cores | 4 (2) |
| Areas of layering or interface | |
| Interface of old and new fibrous tissue | 6 (4) |
| Intimal/medial or adventitial/medial interface | 9 (5) |
| Calcium/fibrous interface | 13 (8) |
| Calcium/lipid interface | 19 (12) |
| Fibrous cap/lipid pool interface | 54 (33) |
| Neovascularization/media interface | 9 (5) |
Data are given as percentage of total regions in IVOCT images with bright spots that are co-localized with CD68 positivity (N=165).
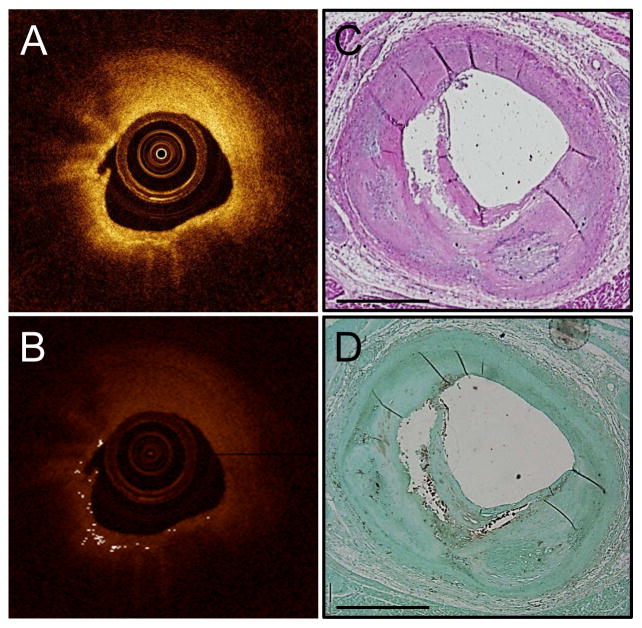
Bright spots occurred in N=175 regions of IVOCT frames morphologically classified as TCFA. Of these, 165 regions were also co-localized with CD68 positivity. Figure 6 demonstrates bright spots originating in a TCFA; it can be observed that the bright spots in this case are caused by both macrophages in the fibrous cap and the fibrous cap interface with the lipid core. Not described in a Table, there were only 10 regions of bright spots in TCFA IVOCT frames that were not co-localized with macrophages; of these regions, the source of bright spots was the fibrous cap and lipid or necrotic core interface (N=4), the fibrous cap and calcium border (N=3), and acellular fibrous tissue (N=3).
Figure 6. Bright spots in TCFA.
Unprocessed IVOCT image that was classified as TCFA by histology (A). Algorithm-processed IVOCT image with bright spots identified (B). H&E stain showing a TCFA (C). CD68 stain showing macrophages at the fibrous cap and lipid pool interface (D).
DISCUSSION
We have confirmed that IVOCT bright spots can be caused by macrophages—however, we have also identified new alternative etiologies. Our findings suggest that IVOCT bright spots can be generated in areas characterized by sharp changes in IR. Our results support the principle that spatial gradients in IR are responsible for enhanced light scattering that results in bright regions in IVOCT images. Thus, most bright spots in IVOCT images are not caused by macrophages, but originate from a mixture of atherosclerotic components that have maximal differences in optical IR. Of the sources of bright spots identified, all were found in regions with known sharp gradients of IR. In the case of layering between old and new fibrous tissue, the co-localization of different types of collagen fibers or the degree of maturation is responsible for the shift in IR (12). In particular, after a rupture has healed, collagen type III is replaced by type I and results in a band of high backscattering signal between the layers of tissue—this is likely due to the greater optical density of collagen type I in comparison to type III (1).
Some macrophages appear dark
Our finding that large pools of macrophages appear dark supports the observation that homogeneous material in plaque, even groups of macrophages as shown in this study, can have a homogeneous IR and, thus, would not be expected to cause bright spots. Moreover, this finding implies that a large pool of macrophages that have engulfed lipid (foam cells) will appear dark on IVOCT images, similar to lipid pools, and may not be easily identified by IVOCT (Figs. 2 and 3). Thus, the juxtaposition of foam cells with a low IR next to the fibrous cap with a higher IR may have been the origin of bright spots identified in previous studies (2).
An alternative hypothesis for explaining why not all macrophages appear as bright spots in IVOCT images involves differences in macrophage subsets. M1 macrophages, considered the “classic” phenotype, are thought to be pro-inflammatory and engulf lipid to form foam cells, whereas M2 macrophages, considered anti-inflammatory, contain smaller vesicles of engulfed lipid (13) and a higher density of mitochondria (14). Thus, M1 macrophage foam cells may appear as shadows or dark regions because of the large amount of intracellular lipid, but M2 macrophages may appear bright because of a higher density of light-scattering mitochondria (15,16). Further study of how M1 and M2 macrophages appear in IVOCT images is needed. Additionally, combining IVOCT with other imaging techniques that have a higher specificity for lipid, fibrous tissue and macrophages, such as two-photon luminescence (17) or fluorescence lifetime imaging (18), may provide enhanced contrast for distinguishing between the subtypes of macrophages.
Macrophages alone are responsible for few regions of brightness
While 57% of all regions with algorithm-defined bright spots were CD68+, 34% of all regions were CD68+ that were also co-localized with other tissue components which caused bright spots in the absence of any macrophages (see Table 3). It is uncertain whether bright spots in the presence of both macrophages and those other components were caused by the macrophages or by another etiology. Thus, only 23% of the bright spots were definitely caused by macrophages.
Mechanism of superficial shadowing caused by macrophages
In addition to appearing as bright spots in IVOCT images, macrophages can cause shadowing that may appear as a lipid pool (1,19). Although our algorithm for identifying bright spots does not directly search for shadowing behind the bright spots, we identified 186 regions of bright spots that caused shadows; 119 were located in regions characterized by the co-localization of microcalcification or cholesterol crystals and macrophages (Figs. 4D–F). We believe shadowing was actually caused by microcalcification and/or cholesterol crystals. Considering Mie scattering, which describes the way light scatters from symmetric objects, smaller features with higher IRs will cause increased shadowing. Thus, small cholesterol crystals and microcalcifications, both of which have high IRs compared with the other plaque components, can cause shadows. In addition to ingesting lipid, macrophages can engulf microcalcifications (20) and cholesterol crystals (21). Macrophages alone do not have optical properties that would cause a shadow, unless they have engulfed a micro-calcification or plaque component that has an IR substantially higher than that of lipid. The high IR of cholesterol clefts is also consistent with the bright spots observed within necrotic cores. Thus, we propose that the type and distribution of engulfed material affects the shadowing by macrophages.
Algorithm identified bright spots in TCFAs
Most of the bright spot regions found in TCFA IVOCT frames (165 of 175 regions) were co-localized with macrophages. Interestingly, the majority were caused by either macrophage-rich areas with fibrous tissue or the fibrous cap/lipid pool interface, which is also where macrophages are often found. Thus it can be difficult to distinguish when bright spots found in TCFAs are caused by macrophages or the fibrous cap/lipid pool interface.
Statistics regarding identification of macrophages
The algorithm was more sensitive at detecting the true presence of macrophages than an expert reader, demonstrating its value. This result is supported by the joint probability of agreement between the expert IVOCT reader and the algorithm—only 53%, which quantifies the finding that the algorithm identified regions of macrophages that the expert IVOCT reader missed. The reduced specificity of the algorithm was anticipated because it also detects other causes of bright spots than macrophages, due to differences in IR of co-mingled plaque components, the hypothesis of the paper. The increased specificity (76%) of the IVOCT reader shows that an expert reader can distinguish between bright spots caused by macrophages and bright spots caused by other sources. One way to increase the accuracy for identification of macrophages is to allow an expert reader to sort through the bright spot processed images, and disregard images that have bright spots obviously not caused by vulnerable plaque morphologies. Future advances in multi-modal or more advanced image processing methods could discard regions of brightness from non-vulnerable plaque types automatically. For example, the accuracy of identifying vulnerable plaque could be improved by combining IVOCT with a novel technique that provides biochemical specificity such as time-resolved fluorescence (22) or Raman spectroscopy (23).
Algorithm identified bright spots at the lipid and calcium interface
We found that regions with lipid intermingled in fibrocalcific plaque can also generate bright spots. We believe the underlying reason for this finding is that fibrocalcific plaques show rich signal heterogeneity within the calcified cores. Lipid cores initially develop microcalcifications, which have been associated with vulnerable plaques (24). These microcalcifications coalesce into larger calcifications; the intermingling of lipid with a low IR and microcalcifications with a high IR is responsible for the complexity and heterogeneity of some calcification sites. Once these cores become homogeneous calcified plates (25), the bright reflections may resolve. However, homogeneous calcified plates in the absence of residual pools of lipid are not frequently found in human atherosclerosis and are not commonly seen during OCT imaging.
Advantages of our bright spot algorithm
Because current methods for interpreting IVOCT images are qualitative, interpretation varies widely. This is especially true when identifying thin-capped fibroatheroma (TCFA) because distinguishing lipid from calcium can be difficult (26–28), and macrophages can cause shadowing that falsely appears as a lipid core (19). Developing algorithms to quantify plaque composition is critical if IVOCT is to be used for accurately identifying plaque composition. Furthermore, several optical properties must be considered when identifying IVOCT bright spots. Light attenuates through tissue at an exponential rate dependent upon both depth and tissue composition, and the intensity of light reflections varies with distance from the catheter. Finally, the signal-to-noise ratio (SNR) will vary between IVOCT images and pullbacks due to differences in the power of laser sources, manufacturing variability between catheters, and other clinical variables such as residual blood in the field. Thus, tissue depth, distance from the catheter, and the SNR are factors that should be considered when identifying IVOCT bright spots and all are taken into account with the bright spot algorithm presented here.
Limitations
While our study used algorithm-defined bright spots within IVOCT images to focus our histology examination, the opposite was not performed. Second, as with all imaging studies, histologic co-registration is complex, and the possibility of error exists. Additionally, the histologic sections were 5-μm thick, whereas the IVOCT images were separated by 270 μm of tissue. However, serial sectioning throughout the regions of interest greatly improved the accuracy of our co-registration. Lastly, distortions due to histologic processing are always possible sources of error in co-registration and IVOCT image interpretation.
Conclusions
We have developed a novel quantitative technique to identify bright spots in IVOCT images. Our findings indicate that not all bright spots are caused by macrophages; rather they can be generated by a combination of plaque components that create sharp changes in the IR. Moreover, we found that macrophage foam cells can be seen as dark regions on IVOCT images. Our study underscores the importance of developing more discerning algorithms. Software that incorporates our quantitative technique may improve the identification of some types of vulnerable plaque and may enable the standardization of IVOCT image interpretation.
Supplementary Material
Acknowledgments
Funding sources: The Veterans Health Administration Merit Grant I01 BX000397. The Clayton Foundation, Houston, TX. Janey and Dolph Briscoe Division of Cardiology, San Antonio, TX. NIH T32 HL007446. American Heart Association 13POST17080074. The Biomedical Engineering Advancement fund in at the University of Texas at Austin.
Abbreviations list
- IVOCT
Intravascular optical coherence tomography
- IR
Index of refraction
- NSD
Normalized standard deviation
Footnotes
Disclosures: The IVOCT system used in these studies was provided by Volcano Corporation, San Diego, CA.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tearney GJ, Regar E, Akasaka T, et al. Consensus standards for acquisition, measurement, and reporting of intravascular optical coherence tomography studies: a report from the International Working Group for Intravascular Optical Coherence Tomography Standardization and Validation. J Am Coll Cardiol. 2012;59:1058–72. doi: 10.1016/j.jacc.2011.09.079. [DOI] [PubMed] [Google Scholar]
- 2.Tearney GJ, Yabushita H, Houser SL, et al. Quantification of macrophage content in atherosclerotic plaques by optical coherence tomography. Circulation. 2003;107:113–9. doi: 10.1161/01.cir.0000044384.41037.43. [DOI] [PubMed] [Google Scholar]
- 3.MacNeill BD, Jang IK, Bouma BE, et al. Focal and multi-focal plaque macrophage distributions in patients with acute and stable presentations of coronary artery disease. J Am Coll Cardiol. 2004;44:972–9. doi: 10.1016/j.jacc.2004.05.066. [DOI] [PubMed] [Google Scholar]
- 4.Tahara S, Morooka T, Wang Z, et al. Intravascular optical coherence tomography detection of atherosclerosis and inflammation in murine aorta. Arterioscler Thromb Vasc Biol. 2012;32:1150–7. doi: 10.1161/ATVBAHA.111.243626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raffel OC, Tearney GJ, Gauthier DD, Halpern EF, Bouma BE, Jang IK. Relationship between a systemic inflammatory marker, plaque inflammation, and plaque characteristics determined by intravascular optical coherence tomography. Arterioscler Thromb Vasc Biol. 2007;27:1820–7. doi: 10.1161/ATVBAHA.107.145987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raffel OC, Merchant FM, Tearney GJ, et al. In vivo association between positive coronary artery remodelling and coronary plaque characteristics assessed by intravascular optical coherence tomography. Eur Heart J. 2008;29:1721–8. doi: 10.1093/eurheartj/ehn286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chia S, Raffel OC, Takano M, Tearney GJ, Bouma BE, Jang IK. Comparison of coronary plaque characteristics between diabetic and non-diabetic subjects: An in vivo optical coherence tomography study. Diabetes Res Clin Pract. 2008;81:155–60. doi: 10.1016/j.diabres.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka A, Tearney GJ, Bouma BE. Challenges on the frontier of intracoronary imaging: atherosclerotic plaque macrophage measurement by optical coherence tomography. J Biomed Opt. 2010;15:011104. doi: 10.1117/1.3290810. [DOI] [PubMed] [Google Scholar]
- 9.Ali ZA, Roleder T, Narula J, et al. Increased thin-cap neoatheroma and periprocedural myocardial infarction in drug-eluting stent restenosis: multimodality intravascular imaging of drug-eluting and bare-metal stents. Circ Cardiovascular Interv. 2013;6:507–17. doi: 10.1161/CIRCINTERVENTIONS.112.000248. [DOI] [PubMed] [Google Scholar]
- 10.Cilingiroglu M, Oh JH, Sugunan B, et al. Detection of vulnerable plaque in a murine model of atherosclerosis with optical coherence tomography. Catheter Cardiovasc Interv. 2006;67:915–23. doi: 10.1002/ccd.20717. [DOI] [PubMed] [Google Scholar]
- 11.Virmani R, Kolodgie FD, Burke AP, Farb A, Schwartz SM. Lessons from sudden coronary death - A comprehensive morphological classification scheme for atherosclerotic lesions. Arterioscler Thromb Vasc Biol. 2000;20:1262–1275. doi: 10.1161/01.atv.20.5.1262. [DOI] [PubMed] [Google Scholar]
- 12.Bornstein P, Sage H. Structurally distinct collagen types. Annu Rev Biochem. 1980;49:957–1003. doi: 10.1146/annurev.bi.49.070180.004521. [DOI] [PubMed] [Google Scholar]
- 13.Falk E, Nakano M, Bentzon JF, Finn AV, Virmani R. Update on acute coronary syndromes: the pathologists’ view. Eur Heart J. 2013;34:719–28. doi: 10.1093/eurheartj/ehs411. [DOI] [PubMed] [Google Scholar]
- 14.Tavakoli S, Zamora D, Ullevig S, Asmis R. Bioenergetic profiles diverge during macrophage polarization: implications for the interpretation of 18F-FDG PET imaging of atherosclerosis. J Nucl Med. 2013;54:1661–7. doi: 10.2967/jnumed.112.119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mourant JR, Freyer JP, Hielscher AH, Eick AA, Shen D, Johnson TM. Mechanisms of light scattering from biological cells relevant to noninvasive optical-tissue diagnostics. Appl Opt. 1998;37:3586–3593. doi: 10.1364/ao.37.003586. [DOI] [PubMed] [Google Scholar]
- 16.van der Meer FJ, Faber DJ, Baraznji Sassoon DM, Aalders MC, Pasterkamp G, van Leeuwen TG. Localized measurement of optical attenuation coefficients of atherosclerotic plaque constituents by quantitative optical coherence tomography. IEEE Trans Med Imaging. 2005;24:1369–76. doi: 10.1109/TMI.2005.854297. [DOI] [PubMed] [Google Scholar]
- 17.Wang T, Mancuso JJ, Kazmi SM, et al. Combined two-photon luminescence microscopy and OCT for macrophage detection in the hypercholesterolemic rabbit aorta using plasmonic gold nanorose. Lasers Surg Med. 2012;44:49–59. doi: 10.1002/lsm.21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phipps JE, Sun Y, Saroufeem R, Hatami N, Fishbein MC, Marcu L. Fluorescence lifetime imaging for the characterization of the biochemical composition of atherosclerotic plaques. J Biomed Opt. 2011;16:096018. doi: 10.1117/1.3626865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Soest G, Regar E, Goderie TP, et al. Pitfalls in plaque characterization by OCT: image artifacts in native coronary arteries. J Am Coll Cardiol Img. 2011;4:810–3. doi: 10.1016/j.jcmg.2011.01.022. [DOI] [PubMed] [Google Scholar]
- 20.Nadra I, Mason JC, Philippidis P, et al. Proinflammatory activation of macrophages by basic calcium phosphate crystals via protein kinase C and MAP kinase pathways: a vicious cycle of inflammation and arterial calcification? Circ Res. 2005;96:1248–56. doi: 10.1161/01.RES.0000171451.88616.c2. [DOI] [PubMed] [Google Scholar]
- 21.Rajamaki K, Lappalainen J, Oorni K, et al. Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: a novel link between cholesterol metabolism and inflammation. PLoS One. 2010;5:e11765. doi: 10.1371/journal.pone.0011765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcu L, Jo JA, Fang Q, et al. Detection of rupture-prone atherosclerotic plaques by time-resolved laser-induced fluorescence spectroscopy. Atherosclerosis. 2009;204:156–64. doi: 10.1016/j.atherosclerosis.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motz JT, Fitzmaurice M, Miller A, et al. In vivo Raman spectral pathology of human atherosclerosis and vulnerable plaque. J Biomed Opt. 2006;11:021003. doi: 10.1117/1.2190967. [DOI] [PubMed] [Google Scholar]
- 24.Maldonado N, Kelly-Arnold A, Vengrenyuk Y, et al. A mechanistic analysis of the role of microcalcifications in atherosclerotic plaque stability: potential implications for plaque rupture. Am J Physiol Heart Circ Physiol. 2012;303:H619–28. doi: 10.1152/ajpheart.00036.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bostrom K, Watson KE, Horn S, Wortham C, Herman IM, Demer LL. Bone morphogenetic protein expression in human atherosclerotic lesions. J Clin Invest. 1993;91:1800–9. doi: 10.1172/JCI116391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manfrini O, Mont E, Leone O, et al. Sources of error and interpretation of plaque morphology by optical coherence tomography. Am J Cardiol. 2006;98:156–9. doi: 10.1016/j.amjcard.2006.01.097. [DOI] [PubMed] [Google Scholar]
- 27.Rieber J, Meissner O, Babaryka G, et al. Diagnostic accuracy of optical coherence tomography and intravascular ultrasound for the detection and characterization of atherosclerotic plaque composition in ex-vivo coronary specimens: a comparison with histology. Coron Artery Dis. 2006;17:425–30. doi: 10.1097/00019501-200608000-00005. [DOI] [PubMed] [Google Scholar]
- 28.Kume T, Akasaka T, Kawamoto T, et al. Assessment of coronary arterial plaque by optical coherence tomography. Am J Cardiol. 2006;97:1172–5. doi: 10.1016/j.amjcard.2005.11.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.