Abstract
Molecular mechanisms governing the maintenance and proliferation of dorsal root ganglia (DRG) progenitors are largely unknown. Here we reveal that the Hippo pathway regulates the expansion of DRG progenitors and glia during mammalian DRG development. The key effectors of this pathway, transcriptional coactivators Yap and Taz, are expressed in DRG progenitors and glia during DRG development but are at least partially inhibited from activating transcription. Aberrant YAP activation leads to overexpansion of DRG progenitor and glial populations. We further show that the Neurofibromatosis 2 (Nf2) tumor suppressor inhibits Yap during DRG development. Loss of Nf2 leads to similar phenotypes as does YAP hyperactivation, and deleting Yap suppresses these phenotypes. Our study demonstrates that Nf2-Yap signaling plays important roles in controlling the expansion of DRG progenitors and glia during DRG development.
Keywords: the peripheral nervous system, neural crest, satellite glia, sensory neurons
INTRODUCTION
The dorsal root ganglia (DRG) are clusters of sensory neurons and glia found at the dorsal root of the spinal nerves. They transmit sensory information from the body to the central nervous system (CNS) (Zigmond et al., 1999). Neurons and glia in the DRG are derived from neural crest (NC) cells, whose precursors reside within the dorsal neural tube (Le Douarin and Smith, 1988). Upon delaminating from the neural tube around embryonic day (E) 8.5 in mice, some NC cells migrate ventrally between somites and the neural tube and coalesce to generate the DRG (Frank and Sanes, 1991; Serbedzija et al., 1992). During migration and after condensation into a ganglion, multipotent NC cells (here we refer to the migrating ones as migratory NC cells and those within the DRG as DRG progenitors) become committed to neuronal or glial fates and give rise to sensory neurons and satellite glia in DRG (Frank and Sanes, 1991; Ma et al., 1999; Rifkin et al., 2000; Serbedzija et al., 1992). In mice, starting from around E9.5, a subset of multipotent NC cells give rise to sensory neurons in DRG in two temporally distinct but overlapping waves, which are regulated by basic helix-loop-helix transcription factors Neurogenin (Ngn) 2 and Ngn1 (Ma et al., 1999; Rifkin et al., 2000). The first wave of neurogenesis, dependent on Ngn2 activity, is completed by E11.5 in mice and mostly gives rise to large-diameter neurotrophic tyrosine kinase receptor (Trk) B-positive (TrkB+) mechanoceptive and TrkC+ proprioceptive neurons. Ngn1-dependent second wave of neurogenesis occurs between E10.5 and E13.5 and mainly produces small-diameter TrkA+ nociceptive neurons (Marmigère and Ernfors, 2007). All sensory neurons express markers Isl1/2 and Tuj1. Satellite glia are first detected around E11 by the early glial marker brain lipid binding protein (Blbp) (Kurtz et al., 1994). Although factors regulating DRG neurogenesis and gliogenesis have been studied extensively (Marmigère and Ernfors, 2007; Woodhoo and Sommer, 2008), those governing the maintenance and proliferation of multipotent DRG progenitors are largely unknown.
The Hippo pathway is an evolutionarily conserved signaling pathway that regulates organ growth and tumorigenesis (Yu and Guan, 2013; Zhao et al., 2011). Central to this pathway are kinases Mst1/2 and Lats1/2, which phosphorylate transcriptional coactivators Yap and Taz (here referred to as Yap/Taz), resulting in their cytoplasmic retention. Inactivation of Mst1/2 or Lats1/2 kinases or overexpression of Yap leads to accumulation of unphosphorylated Yap, which translocates into the nucleus and activates genes that promote proliferation and survival. Nf2/Merlin is an upstream activator of the Hippo pathway (Hamaratoglu et al., 2006; Milton et al., 2010; Zhang et al., 2010; Zhao et al., 2007). Encoded by the tumor suppressor gene Neurofibromatosis 2, Nf2 is a close relative of the Ezrin/Radixin/Moesin (ERM) protein family, which links the cytoskeleton to membrane proteins (Bretscher et al., 2002). In addition to its functions in maintaining cell-cell contact and cell polarity, Nf2 mediates contact-dependent inhibition of cell proliferation in vitro and controls tissue homeostasis and tumorigenesis in vivo (Li et al., 2012).
The Hippo-Yap/Taz pathway controls self-renewal and expansion of mouse and human embryonic stem cells (Lian et al., 2010; Varelas et al., 2008) and tissue-specific stem/progenitor cells (Camargo et al., 2007; Lee et al., 2010; Zhang et al., 2010). We previously showed that Yap regulates neural progenitor cell number during vertebrate CNS development (Cao et al., 2008) and recently found that Nf2 inhibits Yap/Taz to limit the expansion of the neural progenitor pool during mammalian brain development (Lavado et al., 2013). In Drosophila, the Yap ortholog Yki promotes the expansion of optic lobe neuroepithelial and glial cells (Reddy and Irvine, 2011; Reddy et al., 2010). Taken together, these studies establish Yap as an important regulator of the sizes of CNS neural progenitor and glial populations.
Here we investigated the function of Yap and Nf2 during mouse DRG development. We found that Yap/Taz are expressed in migratory NC cells, DRG progenitors, and the glial lineage but not in the neuronal lineage. Elevation of YAP expression in DRG progenitors and glial cells expands these cell populations. Furthermore, we found that Nf2 inhibits Yap during DRG development. In the absence of Nf2, the numbers of DRG progenitors and glial cells are increased and that of neurons is reduced, mimicking the phenotypes of YAP gain-of-function mutants. Deletion of Yap in the Nf2 conditional knockout (cKO) background suppresses these phenotypes. We further show that Nf2-Yap signaling regulates progenitor expansion during the development of another NC derivative, the sympathetic ganglia (SG), in a similar fashion. Our findings provide novel insights into the function of Nf2-Yap signaling during NC development.
MATERIALS AND METHODS
Animals
All animal experiments were performed in accordance with the guidelines set by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital (SJCRH). The TetO-YAP line (Camargo et al., 2007) was provided by Thijn R. Brummelkamp and Fernando D. Camargo (Children’s Hospital, Boston, MA). Sox10-rtTA line (Ludwig et al., 2004) was a gift from Michael Wegner (Universität Erlangen-Nürnberg, Erlangen, Germany). Wnt1-Cre (Stock No: 007807) and Axin2-rtTA (Stock No: 016997) lines were obtained from the Jackson Laboratory. The YapFlox/Flox line (Xin et al., 2011) was provided by Eric N. Olson (University of Texas Southwestern Medical Center, Dallas, TX). The Nf2Flox/Flox line has been described previously (Giovannini et al., 2000). An Nf2Δ allele was generated by breeding Nf2Flox/Flox mice with the Ella-Cre line (Stock No: 003724, Jackson Laboratory). As both Nf2 and Wnt1-Cre are on Chromosome 11, a recombined allele, Nf2Δ;Wnt1-Cre, was generated by extensive breeding. For BrdU pulse-labeling, timed pregnant mice were given intraperitoneal injections of BrdU (10 mg/ml in 0.9% saline) at a dose of 100 mg/kg. Doxycycline was delivered in drinking water at 500 mg/L at the time points indicated.
Histology and immunostaining
Embryos younger than E15.5 were fixed with 4% paraformaldehyde in PBS overnight at 4°C. Embryos at E15.5 or older were intracardially perfused with 4% paraformaldehyde and post-fixed overnight at 4°C. Samples were embedded in OCT and sectioned at 16-μm thickness. Immunostainings were performed according to standard protocols. Primary antibodies were as follows: Blbp (Millipore, ABN14; rabbit, 1:500), BrdU (Abcam, ab6326; rat, 1:100), Isl1/2 (Developmental Studies Hybridoma Bank, Clone # 39.4D5; mouse, 1:500), Ki67 (Vector Laboratories, VP-RM04; rabbit, 1:250), Nf2 (Sigma, HPA003097; rabbit, 1:1000 and amplification with Invitrogen TSA kit), p75 (Promega, G3231; rabbit, 1:2000), S100β (Sigma, S2532; mouse, 1:100 and amplification with Invitrogen TSA kit), Sox10 (Santa Cruz, sc-17342; goat, 1:250), TrkA (provided by Louis Reichardt, University of California, San Francisco, CA; rabbit, 1:1000), Tuj1 (Covance, MMS-435P; mouse, 1:1000), Yap/Taz (Cell Signaling, 8418; rabbit, 1:200 and amplification with Invitrogen TSA kit), and pYap (Cell Signaling, 4911; rabbit, 1:200). Secondary antibodies were from Invitrogen and Jackson ImmunoResearch. Fluorescent images were acquired with a Zeiss LSM 510 or 780 confocal microscope.
Western blot analysis
DRG were dissected from E13.5 embryos. Total DRG lysates were prepared in 20 mM HEPES (pH 7.4), 150 mM NaCl, 2% SDS, and 5% glycerol supplemented with the Halt protease and phosphatase inhibitor cocktail (Thermo Scientific). The following primary antibodies were used: Nf2 (Sigma, HPA003097; rabbit, 1:1,000) and GAPDH (Sigma, G8795, mouse; 1:10,000).
In situ hybridization
In situ hybridization was performed as described (Schaeren-Wiemers and Gerfin-Moser, 1993). Ngn1 probe was provided by Andy Groves (Baylor College of Medicine Houston, TX) (Raft et al., 2007). Ngn2 and TrkB probes were provided by Qiufu Ma (Harvard Medical School, Boston, MA) (Ma et al., 1999). A probe specific for the TrkC tyrosine kinase domain was designed based on Fagan et al., 1996. Total RNA was extracted from E13.5 wild-type mouse brain using TRIzol reagent (Invitrogen) and reverse transcribed by using the SuperScript III cDNA kit (Invitrogen). A 738 bp-long cDNA fragment encoding a portion of the kinase domain was amplified using forward primer CATCAAGAGGAGAGATATCGTGTTGAAGAG and reverse primer GGTCTCTTCTAGACACGGCC, where underlined sequences show the restriction enzyme sites integrated into the fragment by PCR (EcoRV and XbaI, respectively). The fragment was cloned into pBluescript plasmid at EcoRV and XbaI sites. Antisense probe was generated by linearizing the vector with EcoRV and transcribing with T3 RNA polymerase. Images were acquired with a Zeiss Axio Imager M2 microscope equipped with a Zeiss AxioCam MRc camera.
Quantification and statistical analysis
Cell counting was performed using the ImageJ software. Three embryos for each genotype were quantified. Quantifications were performed on sections at similar rostrocaudal levels. Absolute numbers shown in bar graphs are mean number of cells per section. All values shown are mean ± s.e.m. P-values were calculated with two-tailed, unpaired t-test.
RESULTS
Yap/Taz expression pattern in the developing DRG
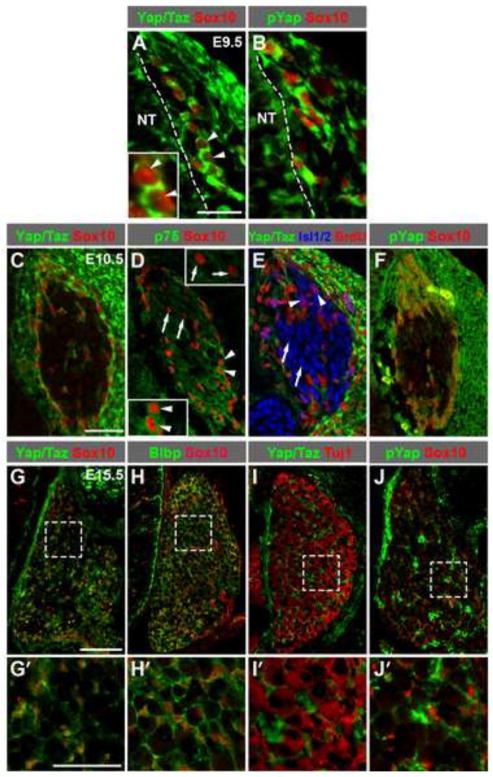
To begin investigating the potential function of the Hippo pathway during DRG development, we first examined the expression pattern of Yap/Taz in mouse embryos using an antibody that recognizes both Yap and Taz. At E9.5, Yap/Taz were detected in Sox10+ migrating NC cells (Kuhlbrodt et al., 1998; Paratore et al., 2001) (Fig. 1A). At E10.5, when migrating NC cells have coalesced into the forming DRG, Yap/Taz were expressed in all Sox10+ cells (Fig. 1C), a population consisting of p75+ DRG progenitors (Fig. 1D, arrowheads) and p75− glial precursors which retain Sox10 expression but have not yet started expression of the glial marker Blbp (Hu et al., 2011; Sonnenberg-Riethmacher et al., 2001) (Fig. 1D, arrows). Cells of the neuronal lineage, marked by Isl1/2, did not express Yap/Taz (Fig. 1E), whether they were proliferating BrdU+ neuronal precursors (Fig. 1E, arrowheads, 2-hour BrdU pulse) or BrdU− postmitotic neurons (Fig. 1E, arrows). At E15.5, Yap/Taz expression persisted in Sox10+ cells (Fig. 1G,G′), almost all of which coexpressed the glial marker Blbp at this stage (Fig. 1H,H′). Neurons, marked by Tuj1, did not express Yap/Taz (Fig. 1I,I′). In summary, during mouse DRG development, Yap/Taz are expressed in migratory NC cells, DRG progenitors, glial precursors, and satellite glia, but not in neuronal precursors or post-mitotic neurons.
Fig. 1. Yap/Taz are expressed in migratory NC cells, DRG progenitors, glial precursors, and satellite glia during DRG development.
(A) At E9.5, Yap/Taz are expressed in Sox10+ migratory NC cells. Two cells marked by arrowheads are magnified in the inset. (C–E) At E10.5, Yap/Taz are expressed in Sox10+ cells (C) consisting of p75+ DRG progenitors (D, arrowheads, lower left corner inset) and p75− glial precursors (D, arrows, upper right corner inset). Neuronal precursors (E, arrowheads, Isl1/2+BrdU+, 2-hour BrdU pulse) or postmitotic neurons (E, arrows, Isl1/2+BrdU−) do not express Yap/Taz. (G–I′) At E15.5, Yap/Taz are expressed in Sox10+ cells (G,G′), almost all of which coexpress the glial marker Blbp (H,H′). Tuj1+ neurons do not express Yap/Taz (I,I′). (B,F,J,J′) S112-phosphorylated form of Yap (pYap) is detected in Sox10+ cells at each stage. Dashed lines in A,B mark the neural tube (NT) border. Panels labeled with primes are the magnified view of the boxed areas in corresponding panels. Scale bars: 25 μm in A,G′; 50 μm in C; 100 μm in G.
Throughout the developmental stages examined, Yap/Taz were present in the cytoplasm (Fig. 1A,C,G,G′), suggesting that their transcriptional activity is at least partially inhibited. Indeed, the S112-phosphorylated form of Yap (S127 in human YAP), which is the product of the Hippo kinase cascade and is unable to activate transcription, was readily detectible in Sox10+ cells at each stage (Fig. 1B,F,J,J′).
YAP overexpression in NC precursors, migratory NC cells, and DRG progenitors leads to overexpansion of DRG progenitors
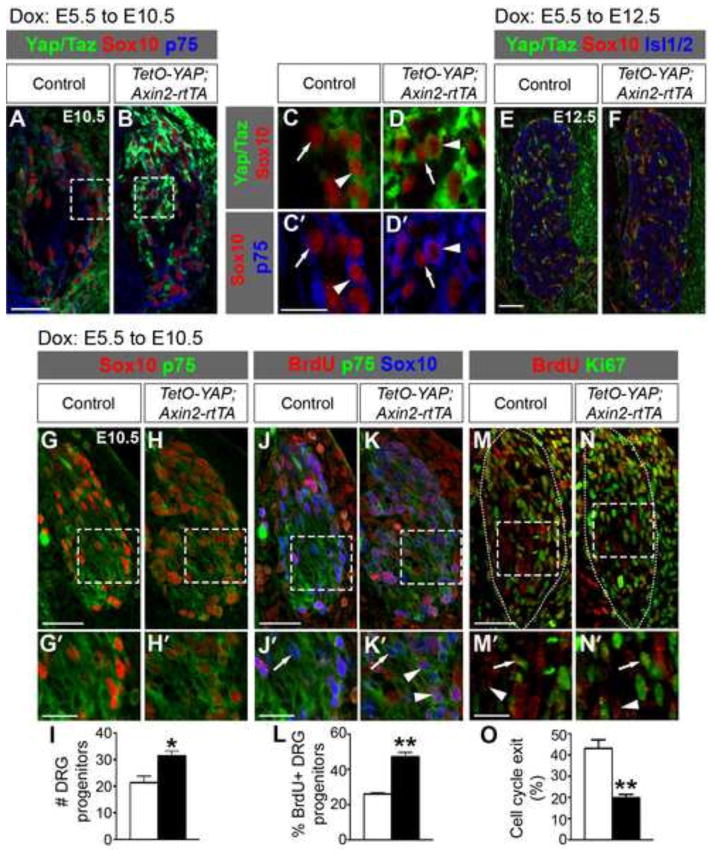
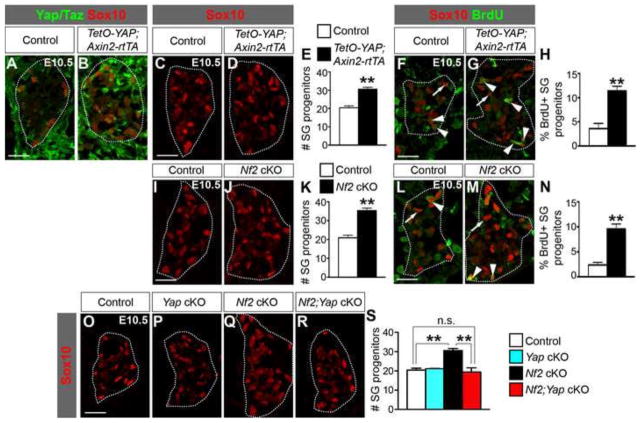
To understand the functional significance of keeping Yap/Taz suppressed, we decided to examine the consequences of their aberrant activation on DRG development. To this end, we utilized a mouse line that carries a doxycycline (Dox)-dependent allele of human YAP1 (TetO-YAP1S127A, here referred to as TetO-YAP) (Camargo et al., 2007). Mating these mice with those expressing the tetracycline-dependent transactivator rtTA under the control of the Axin2 promoter (Axin2-rtTA) (Yu et al., 2007) and providing dams with Dox enable overexpression of the S127A mutant version of YAP, which presumably has enhanced nuclear localization compared to the wild-type protein due to the lack of phosphorylation and therefore inhibition by the Hippo kinase cascade. The Axin2 promoter is active in NC precursors within the dorsal neural tube starting at E8.5 and in migratory NC cells (Yu et al., 2007). To examine the effect of YAP overexpression at early stages of DRG development, we gave Dox-containing water to pregnant females starting from E5.5. At E10.5, YAP was overexpressed in the DRG of TetO-YAP;Axin2-rtTA double transgenic embryos and the overexpression was restricted to Sox10+p75+ DRG progenitors (Fig2. A–D′, arrowheads) and Sox10+p75− glial precursors (Fig2. A–D′, arrows). At E12.5, YAP was no longer overexpressed in TetO-YAP;Axin2-rtTA DRG (Fig. 2E,F) despite the presence of Dox, which is likely due to the downregulation of the Axin2 promoter. Thus, TetO-YAP;Axin2-rtTA embryos experienced elevated YAP activity in NC precursors, migratory NC cells, DRG progenitors, and glial precursors before E12.5.
Fig. 2. YAP overexpression in NC precursors, migratory NC cells, and DRG progenitors expands the DRG progenitor pool.
Dox-containing water was given to pregnant females starting from E5.5 and embryos were collected at E10.5 and E12.5. (A–D′) At E10.5, YAP is overexpressed in Sox10+ cells in TetO-YAP;Axin2-rtTA DRG. C,C′ and D,D′ are magnified views of the boxed regions in A and B, respectively. Arrowheads: Sox10+p75+ DRG progenitors; arrows: Sox10+p75− glial precursors. (E,F) At E12.5, YAP is no longer overexpressed in TetO-YAP;Axin2-rtTA DRG. (G–I) The number of Sox10+p75+ DRG progenitors is increased in TetO-YAP;Axin2-rtTA DRG compared to controls. Panels labeled with primes are the magnified view of the boxed areas in corresponding panels. (J–L) Quantification of BrdU+ cells among Sox10+p75+ DRG progenitors after a 2-hour BrdU pulse shows increased proliferation of DRG progenitors in TetO-YAP;Axin2-rtTA embryos. Arrowheads: BrdU+ DRG progenitors; arrows: BrdU− DRG progenitors. (M–O) Quantification of Ki67− cells among BrdU+ cells after a 24-hour BrdU pulse shows decreased cell-cycle exit in TetO-YAP;Axin2-rtTA DRG. Arrowheads: BrdU+Ki67− cells; arrows: BrdU+Ki67+ cells. *P<0.05, **P<0.01. Scale bars: 50 μm in A,E,G,J,M; 25 μm in C′,G′,J′,M′.
At E10.5, the DRG of TetO-YAP;Axin2-rtTA embryos contained significantly more Sox10+p75+ DRG progenitors (Fig. 2G–I) than did the DRG of control embryos. To determine the cellular basis underlying these phenotypes, we analyzed the proliferation of DRG progenitors. We labeled S-phase cells with BrdU 2 hours before harvesting the embryos and quantified the percentage of BrdU+ DRG progenitors (BrdU+Sox10+p75+, Fig. 2J–K′, arrowheads) within the progenitor population (Sox10+p75+). The fraction of proliferating (BrdU+) DRG progenitors was markedly higher in TetO-YAP;Axin2-rtTA embryos than in controls (Fig. 2L). To measure cell-cycle exit, we labeled S-phase cells with BrdU 24 hours prior to harvesting the embryos and quantified the fraction of BrdU+ cells that no longer expressed the proliferating-cell marker Ki67 (BrdU+Ki67−/BrdU+ cells) (Fig. 2M–N′), which indexes the fraction of cells that have exited the cell-cycle within the past 24-hour period. Cell-cycle exit was significantly reduced in TetO-YAP;Axin2-rtTA DRG (Fig. 2O). Taken together, these results demonstrate that YAP augments the proliferation capacity of DRG progenitors during the early stages of DRG development.
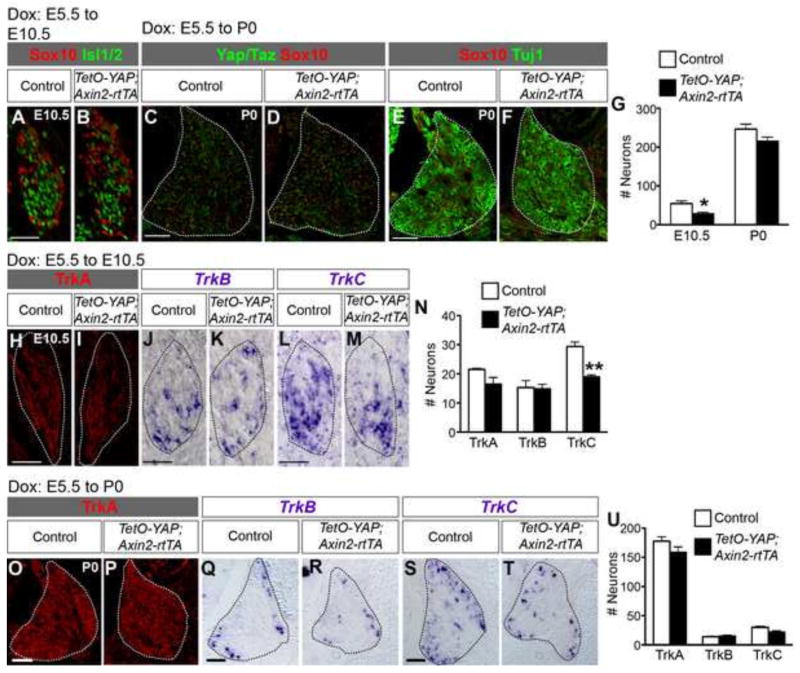
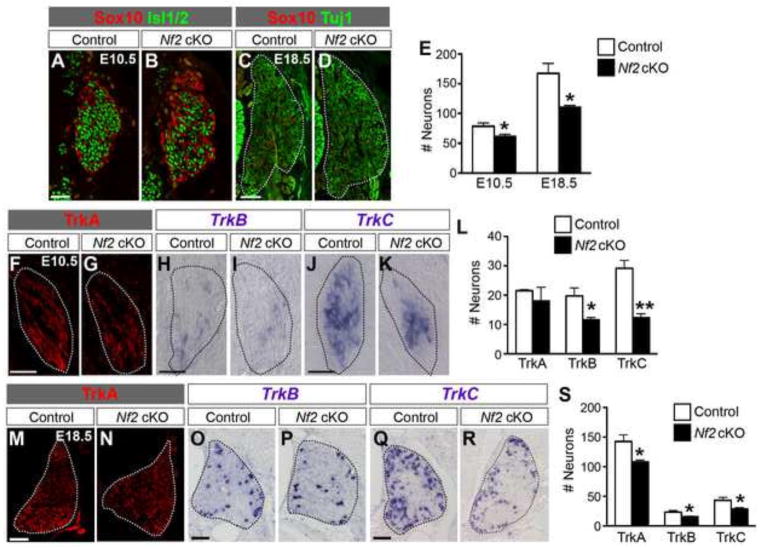
To determine whether YAP hyperactivation at early stages of DRG development affects neurogenesis, we examined the neuronal population at two stages of DRG development. At E10.5, TetO-YAP;Axin2-rtTA DRG contained fewer Isl1/2+ neurons (Fig. 3A,B,G) than did the DRG of control embryos. The number of TrkC+ neurons was significantly reduced whereas the numbers of TrkA+ or TrkB+ neurons were unchanged in TetO-YAP;Axin2-rtTA DRG (Fig. 3H–N). At postnatal day 0 (P0), when YAP was no longer overexpressed in TetO-YAP;Axin2-rtTA DRG (Fig. 3C,D), both the total number of neurons (Fig. 3E–G) and the numbers of each subtype of neurons (Fig. 3O–U) were restored to control levels. These results suggest that YAP hyperactivation at early stages of DRG development delays the production of TrkC+ neurons.
Fig 3. YAP overexpression in NC precursors, migratory NC cells, and DRG progenitors leads to a transient reduction in neuron number.
(A–G) The number of neurons is reduced in TetO-YAP;Axin2-rtTA DRG at E10.5 but not changed at P0. Neurons are labeled by Isl1/2 at E10.5 and Tuj1 at P0. At P0, YAP is no longer overexpressed in TetO-YAP;Axin2-rtTA DRG (C,D). (H–N) At E10.5, the number of TrkC+ neurons is reduced but the numbers of TrkA+ and TrkB+ neurons are unchanged in TetO-YAP;Axin2-rtTA DRG. (O–U) At P0, the numbers of each subtype of neurons are unchanged in TetO-YAP;Axin2-rtTA DRG. *P<0.05, **P<0.01. Scale bars: 50 μm in A,H,J,L; 100 μm in C,E,O,Q,S.
YAP overexpression in DRG progenitors and glial cells expands these populations
Because the Axin2 promoter was downregulated in DRG by E12.5 (Fig. 2E,F), we were unable to analyze the effect of aberrant YAP activation on DRG development at later stages. We therefore crossed TetO-YAP mice with those expressing rtTA under the control of the Sox10 promoter (Sox10-rtTA) (Ludwig et al., 2004), which is first turned-on in migratory NC cells around E8.5 and stays active in DRG progenitors and the glial lineage during DRG development. Dox administration starting from E7.5 led to increased YAP levels in Sox10+ DRG progenitors and glia in the DRG of TetO-YAP;Sox10-rtTA embryos at E12.5 (Fig. 4A,B). These DRG contained significantly more Sox10+p75+ DRG progenitors (Fig. 4C–E, arrowheads) and Sox10+p75− glia (Fig. 4C–D′,F, arrows) than did control DRG. The number of Isl1/2+ neurons, however, was unperturbed at either E12.5 (Fig. 4G–I) or E11.5 (Fig. S1A–E). Furthermore, the numbers of each subtype of neurons were similar between control and TetO-YAP;Sox10-rtTA DRG (Fig. S1F–L). Thus, YAP hyperactivation in Sox10+ NC cells expands the DRG progenitor and glial populations but does not affect neurogenesis.
Fig. 4. YAP overexpression in DRG progenitors and glial cells expands these populations.
(A–I) Dox-containing water was given to pregnant females starting from E7.5 and embryos were collected at E12.5. (A,B) YAP is overexpressed in Sox10+ cells in E12.5 TetO-YAP;Sox10-rtTA DRG. (C–F) At E12.5, TetO-YAP;Sox10-rtTA DRG have more Sox10+p75+ DRG progenitors (arrowheads in C′,D′) and Sox10+p75− glia (arrows in C′,D′) compared to controls. Panels labeled with primes are the magnified view of the boxed areas in corresponding panels. (G–I) The number of Isl1/2+ neurons is not changed in TetO-YAP;Sox10-rtTA DRG at E12.5. (J–N) Dox-containing water was given to pregnant females starting from E12.5 and embryos were collected at E18.5. (J–K′) YAP is overexpressed in Sox10+ cells in E18.5 TetO-YAP;Sox10-rtTA DRG. (L–M′) Almost all Sox10+ cells express the glial marker S100β. (N) The number of glial cells, quantified by counting Sox10+ cells, is increased in TetO-YAP;Sox10-rtTA DRG compared to controls. *P<0.05, **P<0.01, ***P<0.001. Scale bars: 50 μm in A,C,G,J′,L′; 25 μm in C′; 100 μm in J,L.
To examine the effect of YAP overexpression in the glial lineage specifically, we started Dox delivery at E12.5, when most Sox10+ cells in control embryos have differentiated into the glial lineage (Fig. 4E,F; ~14 DRG progenitors vs. ~100 glia in control embryos) (Hu et al., 2011). At E18.5, YAP was overexpressed in Sox10+ cells in TetO-YAP;Sox10-rtTA DRG (Fig. 4J–K′) and almost all Sox10+ cells expressed the glial marker S100β (Fig. 4L–M′). We quantified the number of glia by counting Sox10+ cells. TetO-YAP;Sox10-rtTA DRG contained significantly more glial cells at E18.5 than did controls (Fig. 4N). These results demonstrate that aberrant YAP activation in glial cells causes overexpansion of this population.
Nf2 is expressed in the developing DRG
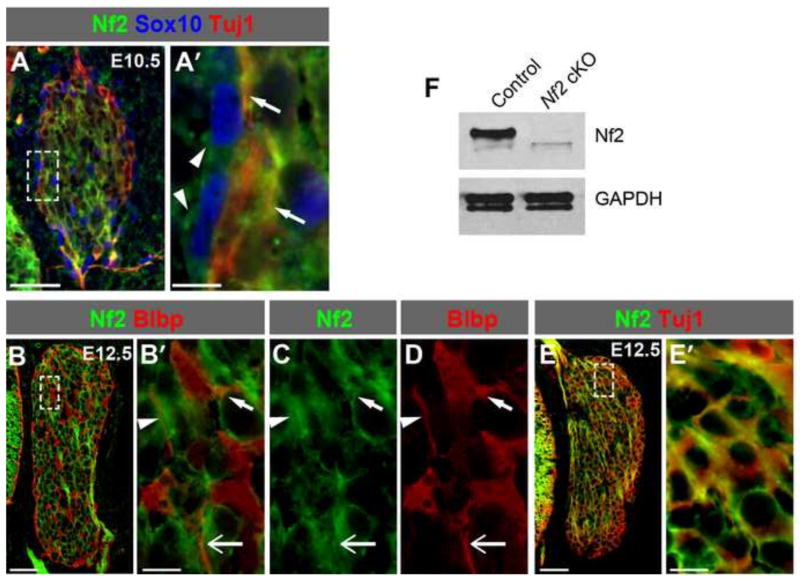
So far, our data demonstrate that Yap/Taz are expressed in DRG progenitors and glial cells during normal DRG development and that aberrant YAP activation in these cells causes overexpansion of these cell populations. We next sought to identify upstream regulators of Yap/Taz. We hypothesized that Nf2 is an inhibitor of Yap during DRG development. The Nf2 transcript is widely expressed during mouse embryogenesis (Gutmann et al., 1995; Huynh et al., 1996), and high levels of Nf2 promoter activity are seen in migrating NC cells (Akhmametyeva et al., 2006). We performed immunostaining to monitor the expression and localization of Nf2 proteins in the developing DRG. At E10.5, Nf2 proteins were detected in the soma of Sox10+ DRG progenitors and glial precursors (Fig. 5A,A′, arrowheads) and in the soma and processes of Tuj1+ sensory neurons (Fig. 5A,A′, arrows). At E12.5, Nf2 was present in Blbp+ satellite glia (Fig. 5B–D; arrows and arrowheads mark three examples of glial processes) and Tuj1+ sensory neurons (Fig. 5E,E′). Thus, Nf2 is expressed in all cell types in the developing DRG and is coexpressed with Yap/Taz in DRG progenitors and the glial lineage (see Fig. 1).
Fig. 5. Nf2 is expressed in the developing DRG.
(A,A′) At E10.5, Nf2 is detected in the soma of Sox10+ DRG progenitors and glia (arrowheads) and in the soma and processes of Tuj1+ neurons (arrows). Panels labeled with primes are the magnified view of the boxed areas in corresponding panels. (B–D) At E12.5, Nf2 is detected in Blbp+ satellite glia. Arrows and arrowheads point to three examples of glial processes. (E,E′) At E12.5, Nf2 is expressed in Tuj1+ neurons. (F) Western blot analysis shows that Nf2 is no longer detected in E13.5 Nf2 cKO DRG. Scale bars: 50 μm in A,B,E; 10 μm in A′,B′,E′.
Loss of Nf2 leads to similar defects in DRG development as does YAP hyperactivation
To test our hypothesis that Nf2 is an inhibitor of Yap during DRG development, we conditionally deleted Nf2 in the NC lineage using Wnt1-Cre, which is expressed in NC precursors in the dorsal neural tube starting from E8.5 (Danielian et al., 1998). We confirmed that Nf2 proteins were no longer detected in Nf2 cKO (Nf2Flox/Δ;Wnt1-Cre) DRG at E13.5, the earliest stage we could collect sufficient amount of tissues for western blot analysis (Fig. 5F).
Because YAP overexpression affected DRG progenitors, we first examined the effect of Nf2 deletion on this population. At E10.5, the number of Sox10+p75+ DRG progenitors was significantly increased in Nf2 cKO embryos (Fig. 6A–C). The percentage of proliferating DRG progenitors within the progenitor population, measured by BrdU incorporation after a 2-hour pulse (BrdU+Sox10+p75+/Sox10+p75+ cells), was also significantly increased (Fig. 6D–F), whereas cell-cycle exit during the 24-hour period between E9.5 and E10.5 was reduced (BrdU+Ki67−/BrdU+ cells) (Fig. 6G–I). At E12.5, most Sox10+ cells in both control and Nf2 cKO DRG no longer expressed p75 (Fig. 6J–K′, arrows), and the numbers of Sox10+p75+ DRG progenitors in control and Nf2 cKO DRG were similar (Fig. 6L). These phenotypes closely resemble those of TetO-YAP;Axin2-rtTA embryos (see Fig. 2G–O), thus supporting our hypothesis that Nf2 is a Yap inhibitor during DRG development.
Fig. 6. Loss of Nf2 leads to similar defects in DRG development as does YAP hyperactivation.
(A–C) At E10.5, Nf2 cKO DRG have more Sox10+p75+ DRG progenitors than controls. Panels labeled with primes are the magnified view of the boxed areas in corresponding panels. (D–F) Quantification of BrdU+ cells among Sox10+p75+ DRG progenitors after a 2-hour BrdU pulse shows increased proliferation in Nf2 cKO. Arrowheads: BrdU+ DRG progenitors; arrows: BrdU− DRG progenitors. (G–I) Quantification of Ki67− cells among BrdU+ cells after a 24-hour BrdU pulse shows decreased cell-cycle exit in Nf2 cKO. Arrowheads: BrdU+Ki67− cells; arrows: BrdU+Ki67+ cells. (J–L′) At E12.5, the number of Sox10+p75+ DRG progenitors and the percentage of DRG progenitors among all DRG DAPI+ cells (DAPI staining not shown) in Nf2 cKO are similar to that in controls. Arrows: Sox10+p75− glial cells. (M–Q) The number of glial cells is increased in Nf2 cKO DRG at E12.5 and E18.5. Glia are Sox10+Blbp+ at E12.5 and Sox10+S100β+ at E18.5. *P<0.05, **P<0.01. Scale bars: 50 μm in A,D,G,J,M; 25 μm in A′,D′,G′,J′; 100 μm in O.
We next analyzed gliogenesis and neurogenesis in Nf2 cKO DRG. Glia are Sox10+Blbp+ at E12.5 and Sox10+S100β+ at E18.5 (Fig. 6M–P). We quantified glia by counting Sox10+ cells at these stages. This approach gives a close estimate of the quantity of glial cells because, from E12.5 and onward, Sox10+p75+ DRG progenitors constitute less than 5% of the cells in DRG and their numbers were similar in control and Nf2 cKO DRG (Fig. 6J–L′). At E12.5 and E18.5, the number of glia was significantly increased in Nf2 cKO DRG (Fig. 6Q). The number of neurons, marked by Isl1/2 at E10.5 and Tuj1 at E18.5, was reduced in Nf2 cKO DRG (Fig 7A–E). At E10.5, the numbers of TrkB+ and TrkC+ neurons were significantly reduced but the number of TrkA+ neurons was unchanged (Fig. 7F–L). At E18.5, the numbers of all three subtypes of neurons were reduced in Nf2 cKO DRG (Fig 7M–S). The expansion in the glial population at E12.5 and E18.5 and reduction in neuron numbers at E10.5 in Nf2 cKO DRG resembles the effects of YAP overexpression (see Fig. 2–4), reinforcing our hypothesis that Nf2 is a Yap inhibitor in the developing DRG.
Fig. 7. Deletion of Nf2 leads to a reduction in neurons.
(A–E) The number of neurons, marked by Isl1/2 at E10.5 and by Tuj1 at E18.5, is reduced in Nf2 cKO DRG. (F–L) The numbers of TrkB+ and TrkC+ neurons are reduced but the number of TrkA+ neurons is not changed in Nf2 cKO DRG at E10.5. (M–S) The numbers of each subtype of neurons are reduced in Nf2 cKO DRG at E18.5. *P<0.05, **P<0.01. Scale bars: 50 μm in A,F,H,J; 100 μm in C,M,O,Q.
Yap deletion rescues Nf2 cKO phenotypes in DRG
To further confirm that Nf2 functions by inhibiting Yap, we generated Nf2 and Yap double cKO animals (Nf2Flox/Δ;Wnt1-Cre;YapFlox/Flox, herein called Nf2;Yap cKO). Yap cKO (YapFlox/Flox;Wnt1-Cre) and Nf2;Yap cKO embryos all died around E10.5 with massive forebrain bleeding (data not shown), probably due to defects in cerebral vessel formation. We therefore focused our analyses at E10.5. Yap deletion alone in the NC lineage did not affect DRG progenitor (Fig. 8A,B,E) or neuronal populations (Fig. 8F,G,J). However, deletion of Yap in the Nf2 cKO background suppressed the expansion of DRG progenitors (Fig. 8A,C–E) and rescued the reduction in the neuronal population (Fig. 8F,H–J). These results strongly support that Nf2 is an inhibitor of Yap during DRG development.
Fig. 8. Yap deletion rescues the early DRG defects of Nf2 cKO embryos.
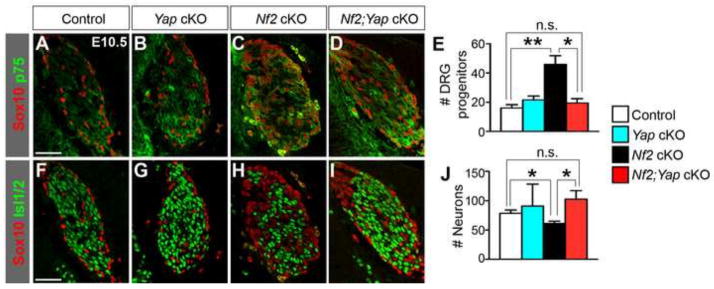
Yap deletion alone does not affect the numbers of Sox10+p75+ DRG progenitors (A,B,E) and Isl1/2+ neurons (F,G,J). However, it suppresses the expansion of DRG progenitors (A–E) and reduction in neurons (F–J) in Nf2 cKO DRG. *P<0.05, **P<0.01, n.s., not significant. Scale bars: 50 μm.
Nf2-Yap signaling controls the expansion of sympathetic ganglia (SG) progenitors
So far we have shown that Nf2-Yap signaling controls the expansion of DRG progenitors. Since Yap/Taz are expressed in migrating NC cells at E9.5 (Fig. 1A), we asked whether progenitors in other NC derivatives are also regulated by Nf2-Yap signaling. To this end, we examined the development of SG, which are a part of the autonomic nervous system that regulates the homeostasis of the internal environment of the organism (Rohrer, 2011). In control SG, Yap/Taz were expressed in Sox10+ SG progenitors (Fig. 9A) but not in Isl1/2+ sympathetic neuroblasts (data not shown). In the aforementioned TetO-Yap;Axin2-rtTA double transgenic embryos (Dox starting from E5.5), YAP was overexpressed in Sox10+ SG progenitors at E10.5 (Fig. 9A,B). At this stage, the SG of TetO-Yap;Axin2-rtTA embryos contained significantly more Sox10+ SG progenitors (Fig. 9C–E). The percentage of proliferating SG progenitors within the progenitor population, measured by BrdU incorporation after a 2-hour pulse (BrdU+Sox10+/Sox10+ cells), was also markedly increased (Fig. 9F–H). Nf2 deletion in NC cells led to similar phenotypes as YAP hyperactivation. At E10.5, the SG progenitor population was expanded in Nf2 cKO embryos (Fig. 9I–K). The fraction of proliferating SG progenitors was markedly higher in Nf2 cKO embryos than in controls (Fig. 9L–N). Finally, although Yap deletion alone did not affect the SG progenitor population (Fig. 9O,P,S), it suppressed the expansion of SG progenitors in the Nf2 cKO background (Fig. 9O,Q–S). These results demonstrate that Nf2-Yap signaling controls the expansion of SG progenitors.
Fig. 9. Nf2-Yap signaling controls the expansion of SG progenitors.
(A–H) Dox-containing water was given to pregnant females starting from E5.5 and embryos were collected at E10.5. (A–B) At E10.5, YAP is overexpressed in Sox10+ cells in TetO-YAP;Axin2-rtTA SG. (C–E) The number of Sox10+ SG progenitors is increased in TetO-YAP;Axin2-rtTA SG compared to controls. (F–H) Quantification of BrdU+ cells among Sox10+ SG progenitors after a 2-hour BrdU pulse shows increased proliferation of SG progenitors in TetO-YAP;Axin2-rtTA embryos. Arrowheads: BrdU+ SG progenitors; arrows: BrdU− SG progenitors. (I–K) At E10.5, Nf2 cKO SG have more Sox10+ SG progenitors than controls. (L–N) Quantification of BrdU+ cells among Sox10+ SG progenitors after a 2-hour BrdU pulse shows increased proliferation in Nf2 cKO SG. Arrowheads: BrdU+ SG progenitors; arrows: BrdU− SG progenitors. (O–S) Yap deletion alone does not affect the number of Sox10+ SG progenitors. However, it suppresses the expansion of SG progenitors in Nf2 cKO SG. **P<0.01, n.s., not significant. Scale bars: 50 μm.
DISCUSSION
The Hippo pathway is recently recognized as a crucial regulator of tissue growth and tumorigenesis. Here we show that, during mammalian DRG development, the key effectors of this pathway, transcriptional coactivators Yap/Taz, are expressed in DRG progenitors and glia but not in the neuronal lineage. The cytoplasmic localization of Yap/Taz proteins and the presence of the inactive, phosphorylated form of Yap indicate that their transcriptional activities are at least partially inhibited. We show that aberrant YAP activation leads to overexpansion of the DRG progenitor and glial populations. We further identified that Nf2 is an inhibitor of Yap in the developing DRG; Nf2 cKO embryos exhibit similar phenotypes as those with elevated YAP activity and these phenotypes are suppressed by Yap deletion. Our study demonstrates that Nf2-Yap/Taz signaling is an important regulator of mammalian DRG development.
Nf2-Yap signaling regulates the proliferation of DRG progenitors
Our YAP gain-of-function experiments using the TetO-YAP transgenic mouse line demonstrate that aberrant YAP activation in DRG progenitors promotes their proliferation and over-expands the progenitor pool. Loss of Nf2 in the NC lineage leads to nearly identical phenotypes. Furthermore, deleting Yap in the Nf2 cKO background suppresses overexpansion of the DRG progenitor pool. These results strongly suggest that Nf2-Yap signaling controls the size of the progenitor pool during normal DRG development.
It is unclear whether YAP overactivation promotes the maintenance of the multipotent state of DRG progenitors as it does in mouse embryonic stem cells (Lian et al., 2010). Moreover, it remains an open question whether Yap/Taz are required for normal DRG development. The early lethality of YapFlox/Flox;Wnt1-Cre embryos prevented us from thoroughly investigating Yap’s role during DRG development. Nevertheless, our preliminary analysis of E10.5 YapFlox/Flox;Wnt1-Cre DRG did not reveal any obvious defects. It is quite possible that Taz may compensate for the loss of Yap. Future studies using Yap;Taz Wnt1-Cre double cKO embryos, Yap or Yap;Taz cKO with Cre lines that act either at later stages or in more restricted lineages (e.g., Sox10-Cre (Matsuoka et al., 2005) and Plp-CreERT2 (Hari et al., 2012; Leone et al., 2003)), and neural crest stem cell cultures (Morrison et al., 2000; Paratore et al., 2001; Paratore et al., 2002; Taylor et al., 2007) prepared from gain− and loss-of-function mutant embryos will provide more insights into the function of Yap/Taz during DRG development.
One phenotypic difference between Nf2 cKO and YAP gain-of-function mutants is that, although overexpressing YAP using the Sox10-rtTA line is able to over-expand the DRG progenitor population at E12.5, Nf2 deletion fails to do so (compare Fig. 4E vs. Fig. 6L). We think this difference is likely because the level of Yap activation achieved by Nf2 deletion is lower than that obtained by YAP overexpression. We were unable to detect, by immunostaining, increased Yap/Taz nuclear localization in Nf2 cKO DRG (data not shown), which is likely because the change is too small to be quantitatively detected by immunostaining. Previously, we were able to detect increased Yap/Taz nuclear localization in Nf2 cKO brains by subcellular fractionation and quantitative western blot analysis (Lavado et al., 2013). This approach, however, is not feasible for analyzing DRG samples because the tissue amount is too limited.
The functions of Yap and Nf2 during DRG gliogenesis
By selectively elevating YAP level in the glial lineage of DRG, we show that YAP hyperactivation over-expands the glial population, probably by increasing the proliferation of glial precursors. Nf2 cKO mice exhibit a similar phenotype. Although the phenotypic similarity suggests that, as in DRG progenitors, Nf2 regulates Yap activity, we cannot yet prove this genetically due to the early lethality of YapFlox/Flox;Wnt1-Cre embryos. It remains possible that Nf2 functions independently of Yap/Taz during glia development. Indeed, previous studies have shown that Nf2 regulates CNS glial cell proliferation by inhibiting ErbB2-dependent Src signaling (Houshmandi et al., 2009) and Schwann cell proliferation and function through a number of signaling pathways (Li et al., 2012; McClatchey, 2005). These possibilities need to be addressed by more molecular and genetic studies in the future.
The functions of Yap and Nf2 during DRG neurogenesis
In addition to the effects on DRG progenitor and glial populations, YAP gain-of-function and Nf2 loss also affect the amount of DRG sensory neurons. The mutant phenotypes are quite complex. TetO-YAP;Axin2-rtTA mice show a transient reduction of early born TrkC+ neurons at E10.5 but, by P0, neuron numbers are restored to normal levels. TetO-YAP;Sox10-rtTA mice, with Dox administration starting from E7.5, do not exhibit neuron number defects at E11.5 or E12.5. Nf2 cKO mice, however, exhibit reductions of early born TrkB+ neurons and TrkC+ neurons at E10.5 and reductions of all subtypes of neurons at E18.5. Deleting Yap in the Nf2 cKO background restores the total number of neurons at E10.5. Based on these results, we postulate the following.
The genetic rescue experiment indicates that, at least before E10.5, Nf2 regulates DRG neurogenesis by inhibiting Yap. Only neurons born during the first wave of neurogenesis are affected in TetO-YAP;Axin2-rtTA and Nf2 cKO embryos, suggesting that Nf2-Yap signaling probably selectively regulates the first wave of neurogenesis but not the second wave. We have examined Ngn2 and Ngn1 expression in YAP gain-of-function and Nf2 cKO mutants but did not detect obvious differences in their expression patterns or levels (Fig. S2). It is possible that Nf2-Yap signaling regulates a neuronal differentiation step downstream of Ngn2 expression, for example the upregulation of NeuroD or Isl1/2.
It is interesting and somewhat puzzling that our two YAP gain-of-function mouse models display different neurogenesis phenotypes: TetO-YAP;Axin2-rtTA but not TetO-YAP;Sox10-rtTA embryos exhibit a delay in neurogenesis. The key difference between these two models is the timing of rtTA expression: Axin2-rtTA expression starts in NC precursors within the neural tube (Yu et al., 2007), whereas Sox10-rtTA expression starts in migratory NC cells (Kuhlbrodt et al., 1998). Our results thus suggest that YAP hyperactivation has to occur very early during DRG development to have an impact on neurogenesis. This might explain why only TrkC+ neurons are affected in TetO-YAP;Axin2-rtTA embryos, as they are the first subtype of neurons to be born during DRG development (Ma et al., 1999). Accumulating evidence suggests that at least some NC precursors within the neural tube are already pre-specified into lineage-restricted subpopulations (George et al., 2007; Lefcort and George, 2007; Nitzan et al., 2013). It is thus possible that elevated Yap activity alters the relative proportion of these subpopulations of multipotent NC cells in the neural tube, for example by converting one subpopulation into another or by preferentially promoting the production of one subpopulation. It is also possible that elevated Yap activity promotes proliferative divisions of multipotent DRG progenitors, which then transition into glial precursors to generate glia, thus reducing neuron production, although we think this is less likely considering that TetO-YAP;Sox10-rtTA embryos do not exhibit neurogenesis defects. Lastly, the difference in the mode of action by Axin2-rtTA, which leads to transient overexpression mainly in multipotent NC cells, and Wnt1-Cre, which causes permanent gene deletion, might account for the difference between TetO-YAP;Axin2-rtTA DRG (reduction of TrkC+ neurons only) and Nf2 cKO DRG (reductions of both TrkB+ and TrkC+ neurons). In summary, more detailed in vivo lineage-tracing experiments are required to address these possibilities.
An important phenotypic difference between Nf2 cKO mutants and YAP gain-of-functions mutants is that, at the end of embryogenesis, the numbers of sensory neurons are reduced in Nf2 cKO mutants but are normal in YAP gain-of-function mutants. These results indicate that, during the later stages of DRG development, Nf2’s function in sensory neurons is independent of Yap/Taz (which are not expressed in sensory neurons). Nf2 loss might impair the proliferation of neuronal precursors or the survival of DRG neurons. The exact function of Nf2 in DRG neurons needs to be determined by future studies. Interestingly, a recent work found that, in neurons of both the CNS and the peripheral nervous system (PNS), including in DRG neurons, Nf2 promotes neurofilament phosphorylation by activating the GTPase RhoA and thereby is essential for maintaining axonal integrity (Schulz et al., 2013).
Conclusions
By using YAP gain-of-function and Nf2 loss-of-function mouse models, we demonstrate that Nf2-Yap signaling regulates the development of two NC derivatives, the DRG and the SG. The observation that significant amount of Yap/Taz proteins remain localized in the cytoplasm in Nf2 cKO DRG (see above, data not shown) suggests that other factors are involved in sequestering Yap/Taz in the cytoplasm. Consistent with this idea, although the TetO-YAP transgenic animals used here express the S127A mutant form of YAP (Camargo et al., 2007), which is thought to have increased nuclear localization than the wild-type protein because it cannot be inhibited by the Hippo kinase cascade (Yu and Guan, 2013), a large fraction of YAP-S127A proteins reside in the cytoplasm (see Fig. 2B,D and Fig. 4B,K,K′), indicating that factors besides the Hippo kinase cascade are involved in Yap/Taz suppression during DRG development. A rapidly growing list of Yap/Taz regulators are being discovered (Yu and Guan, 2013). An important task for future studies is to understand how these regulators operate in intact animals.
Supplementary Material
Dox-containing water was given to pregnant females starting from E7.5 and embryos were collected at E11.5. (A,B) YAP is overexpressed in Sox10+ cells in E11.5 TetO-YAP;Sox10-rtTA DRG. (C–E) The number of neurons, marked by Isl1/2, is not changed in TetO-YAP;Sox10-rtTA DRG. (F–L) The numbers of each subtype of neurons are not changed in TetO-YAP;Sox10-rtTA DRG at E11.5. Scale bars: 50 μm.
In situ hybridization experiments show that Ngn1 and Ngn2 expression levels or patterns are not affected in TetO-YAP;Axin2-rtTA, TetO-YAP;Sox10-rtTA, or Nf2 cKO DRG compared to controls in rostral or caudal regions at E10.5. Scale bar: 100 μm.
Highlights.
The function of the Hippo pathway during mammalian DRG development is investigated.
YAP hyperactivation expands the DRG progenitor and glial populations.
Deletion of Nf2 recapitulates Yap hyperactivation phenotypes in DRG.
Nf2 inhibits Yap during DRG development.
Nf2-Yap regulates the expansion of DRG progenitors and glia during DRG development.
Acknowledgments
We are grateful to Drs. Fernando Camargo, Thijn Brummelkamp, Michael Wegner, and Eric Olson for sharing mouse lines, Andy Groves and Quifu Ma for in situ probes, and Louis Reichardt for the TrkA antibody. We thank the Cell & Tissue Imaging Center at SJCRH for assistance with image acquisition, Cherise Guess for editorial assistance, and members of the Cao lab for assistance with animal work. This work was supported by American Lebanese Syrian Associated Charities (ALSAC). The Cell & Tissue Imaging Center is supported by SJCRH and NCI P30 CA021765-35.
Footnotes
AUTHOR CONTRIBUTIONS
X.C. conceived the project. Y.S. and X.C. designed the experiments. Y.S. performed the experiments and analyzed the results. J.P. maintained the mouse colony. M.G. generated the Nf2Flox mouse line. Y.S. and X.C. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhmametyeva EM, Mihaylova MM, Luo H, Kharzai S, Welling DB, Chang LS. Regulation of the Neurofibromatosis 2 gene promoter expression during embryonic development. Dev Dyn. 2006;235:2771–2785. doi: 10.1002/dvdy.20883. [DOI] [PubMed] [Google Scholar]
- Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 Increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Fagan AM, Zhang H, Landis S, Smeyne RJ, Silos-Santiago I, Barbacid M. TrkA, but not TrkC, receptors are essential for survival of sympathetic neurons in vivo. J Neurosci. 1996;16:6208–6218. doi: 10.1523/JNEUROSCI.16-19-06208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank E, Sanes JR. Lineage of neurons and glia in chick dorsal root ganglia: analysis in vivo with a recombinant retrovirus. Development. 1991;111:895–908. doi: 10.1242/dev.111.4.895. [DOI] [PubMed] [Google Scholar]
- George L, Chaverra M, Todd V, Lansford R, Lefcort F. Nociceptive sensory neurons derive from contralaterally migrating, fate-restricted neural crest cells. Nat Neurosci. 2007;10:1287–1293. doi: 10.1038/nn1962. [DOI] [PubMed] [Google Scholar]
- Giovannini M, Robanus-Maandag E, van der Valk M, Niwa-Kawakita M, Abramowski V, Goutebroze L, Woodruff JM, Berns A, Thomas G. Conditional biallelic Nf2 mutation in the mouse promotes manifestations of human neurofibromatosis type 2. Genes Dev. 2000;14:1617–1630. [PMC free article] [PubMed] [Google Scholar]
- Gutmann DH, Wright DE, Geist RT, Snider WD. Expression of the neurofibromatosis 2 (NF2) gene isoforms during rat embryonic development. Hum Mon Gen. 1995;4:471–478. doi: 10.1093/hmg/4.3.471. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2005;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Hari L, Miescher I, Shakhova O, Suter U, Chin L, Taketo M, Richardson WD, Kessaris N, Sommer L. Temporal control of neural crest lineage generation by Wnt/β-catenin signaling. Development. 2012;139:2107–2117. doi: 10.1242/dev.073064. [DOI] [PubMed] [Google Scholar]
- Houshmandi SS, Emnett RJ, Giovannini M, Gutmann DH. The Neurofibromatosis 2 protein, Merlin, regulates glial cell growth in an ErbB2− and Src-dependent manner. Mol Cell Bio. 2009;29:1472–1486. doi: 10.1128/MCB.01392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu ZL, Shi M, Huang Y, Zheng MH, Pei Z, Chen JY, Han H, Ding YQ. The role of the transcription factor Rbpj in the development of dorsal root ganglia. Neural Dev. 2011;6:14. doi: 10.1186/1749-8104-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh DP, Tran TMD, Nechiporuk T, Pulst SM. Expression of neurofibromatosis 2 transcript and gene product during mouse fetal development. Cell Growth Differ. 1996;7:1551–1561. [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz A, Zimmer A, Schnütgen F, Brüning G, Spener F, Müller T. The expression pattern of a novel gene encoding brain-fatty acid binding protein correlates with neuronal and glial cell development. Development. 1994;120:2637–2649. doi: 10.1242/dev.120.9.2637. [DOI] [PubMed] [Google Scholar]
- Lavado A, He Y, Paré J, Neale G, Olson EN, Giovannini M, Cao X. Tumor suppressor Nf2 limits expansion of the neural progenitor pool by inhibiting Yap/Taz transcriptional coactivators. Development. 2013;140:3323–3334. doi: 10.1242/dev.096537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM, Smith J. Development of the peripheral nervous system from the neural crest. Ann Rev Cell Biol. 1988;4:375–404. doi: 10.1146/annurev.cb.04.110188.002111. [DOI] [PubMed] [Google Scholar]
- Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, Kim MC, Jeong WI, Calvisi DF, Kim JM, Lim DS. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:8248–8253. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefcort F, George L. Neural crest cell fate: to be or not to be prespecified. Cell Adh Migr. 2007;1:199–201. doi: 10.4161/cam.1.4.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone DP, Genoud S, Atanasoski S, Grausenburger R, Berger P, Metzger D, Macklin WB, Chambon P, Suter U. Tamoxifen-inducible glia-specific Cre mice for somatic mutagenesis in oligodendrocytes and Schwann cells. Mol Cell Neurosci. 2003;22:430–440. doi: 10.1016/s1044-7431(03)00029-0. [DOI] [PubMed] [Google Scholar]
- Li W, Cooper J, Karajannis MA, Giancotti FG. Merlin: a tumour suppressor with functions at the cell cortex and in the nucleus. EMBO reports. 2012;13:204–215. doi: 10.1038/embor.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LSB, Abujarour R, Ding S, Guan KL. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Schlierf B, Schardt A, Nave KA, Wegner M. Sox10-rtTA mouse line for tetracycline-inducible expression of transgenes in neural crest cells and oligodendrocytes. Genesis. 2004;40:171–175. doi: 10.1002/gene.20083. [DOI] [PubMed] [Google Scholar]
- Ma Q, Fode C, Guillemot F, Anderson DJ. Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmigère F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci. 2007;8:114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- Matsuoka T, Ahlberg PE, Kessaris N, Iannarelli P, Dennehy U, Richardson WD, McMahon AP, Koentges G. Neural crest origins of the neck and shoulder. Nature. 2005;436:347–355. doi: 10.1038/nature03837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey AI, Giovannini M. Membrane organization and tumorigenesis-the NF2 tumor suppressor, Merlin. Genes Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- Milton CC, Zhang X, Albanese NO, Harvey KF. Differential requirement of Salvador-Warts-Hippo pathway members for organ size control in Drosophila melanogaster. Development. 2010;137:735–743. doi: 10.1242/dev.042309. [DOI] [PubMed] [Google Scholar]
- Nitzan E, Krispin S, Pfaltzgraff ER, Klar A, Labosky PA, Kalcheim C. A dynamic code of dorsal neural tube genes regulates the segregation between neurogenic and melanogenic neural crest cells. Development. 2013;140:2269–2279. doi: 10.1242/dev.093294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, Anderson DJ. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 2000;101:499–510. doi: 10.1016/s0092-8674(00)80860-0. [DOI] [PubMed] [Google Scholar]
- Raft S, Koundakjian EJ, Quinones H, Jayasena CS, Goodrich LV, Johnson JE, Segil N, Groves AK. Cross-regulation of Ngn1 and Math1 coordinates the production of neurons and sensory hair cells during inner ear development. Development. 2007;134:4405–4415. doi: 10.1242/dev.009118. [DOI] [PubMed] [Google Scholar]
- Paratore C, Goerich DE, Suter U, Wegner M, Sommer L. Survival and glial fate acquisition of neural crest cells are regulated by an interplay between the transcription factor Sox10 and extrinsic combinatorial signaling. Development. 2001;128:3949–3961. doi: 10.1242/dev.128.20.3949. [DOI] [PubMed] [Google Scholar]
- Paratore C, Hagedorn L, Floris J, Hari L, Kléber M, Suter U, Sommer L. Cell-intrinsic and cell-extrinsic cues regulating lineage decisions in multipotent neural crest-derived progenitor cells. Int J Dev Biol. 2002;46:193–200. [PubMed] [Google Scholar]
- Reddy BVVG, Irvine KD. Regulation of Drosophila glial cell proliferation by Merlin-Hippo signaling. Development. 2011;138:5201–5212. doi: 10.1242/dev.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BVVG, Rauskolb C, Irvine KD. Influence of Fat-Hippo and Notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development. 2010;137:2397–2408. doi: 10.1242/dev.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin JT, Todd VJ, Anderson LW, Lefcort F. Dynamic expression of neurotrophin receptors during sensory neuron genesis and differentiation. Dev Biol. 2000;227:465–480. doi: 10.1006/dbio.2000.9841. [DOI] [PubMed] [Google Scholar]
- Rohrer H. Transcriptional control of differentiation and neurogenesis in autonomic ganglia. Eur J Neurosci. 2011;34:1563–1573. doi: 10.1111/j.1460-9568.2011.07860.x. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- Schulz A, Baader SL, Niwa-Kawakita M, Jung MJ, Bauer R, Garcia C, Zoch A, Schacke S, Hagel C, Mautner VF, Hanemann CO, Dun XP, Parkinson DB, Weis J, Schröder JM, Gutmann DH, Giovannini M, Morrison H. Merlin isoform 2 in neurofibromatosis type 2–associated polyneuropathy. Nat Neurosci. 2013;16:426–433. doi: 10.1038/nn.3348. [DOI] [PubMed] [Google Scholar]
- Serbedzija GN, Bronner-Fraser M, Fraser SE. Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development. 1992;116:297–307. doi: 10.1242/dev.116.2.297. [DOI] [PubMed] [Google Scholar]
- Sonnenberg-Riethmacher E, Miehe M, Stolt CC, Goerich DE, Wegner M, Riethmacher D. Development and degeneration of dorsal root ganglia in the absence of the HMG-domain transcription factor Sox10. Mech Dev. 2001;109:253–265. doi: 10.1016/s0925-4773(01)00547-0. [DOI] [PubMed] [Google Scholar]
- Taylor MK, Yeager K, Morrison SJ. Physiological Notch signaling promotes gliogenesis in the developing peripheral and central nervous systems. Development. 2007;134:2435–2447. doi: 10.1242/dev.005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- Woodhoo A, Sommer L. Development of the Schwann cell lineage: From the neural crest to the myelinated nerve. Glia. 2008;56:1481–1490. doi: 10.1002/glia.20723. [DOI] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, Olson EN. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HMI, Liu B, Costantini F, Hsu W. Impaired neural development caused by inducible expression of Axin in transgenic mice. Mech Dev. 2007;124:146–156. doi: 10.1016/j.mod.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond MJ, Bloom FE, Landis SC, Roberts JL, Squire LR. Fundamental Neuroscience. Academic Press; 1999. pp. 25–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dox-containing water was given to pregnant females starting from E7.5 and embryos were collected at E11.5. (A,B) YAP is overexpressed in Sox10+ cells in E11.5 TetO-YAP;Sox10-rtTA DRG. (C–E) The number of neurons, marked by Isl1/2, is not changed in TetO-YAP;Sox10-rtTA DRG. (F–L) The numbers of each subtype of neurons are not changed in TetO-YAP;Sox10-rtTA DRG at E11.5. Scale bars: 50 μm.
In situ hybridization experiments show that Ngn1 and Ngn2 expression levels or patterns are not affected in TetO-YAP;Axin2-rtTA, TetO-YAP;Sox10-rtTA, or Nf2 cKO DRG compared to controls in rostral or caudal regions at E10.5. Scale bar: 100 μm.