Abstract
Myosin binding protein-C is a thick filament protein of vertebrate striated muscle. The cardiac isoform (cMyBP-C) is essential for normal cardiac function, and mutations in cMyBP-C cause cardiac muscle disease. The rod-shaped molecule is composed primarily of 11 immunoglobulin- or fibronectin-like domains, and is located at 9 sites, 43 nm apart, in each half of the A-band. To understand how cMyBP-C functions, it is important to know its structural organization in the sarcomere, as this will affect its ability to interact with other sarcomeric proteins. Several models have been proposed, in which cMyBP-C wraps around, extends radially from, or runs axially along the thick filament. Our goal was to define cMyBP-C orientation by determining the relative axial positions of different cMyBP-C domains. Immuno-electron microscopy was performed using mouse cardiac myofibrils labeled with antibodies specific to the N- and C-terminal domains and to the middle of cMyBP-C. Antibodies to all regions of the molecule, except the C-terminus, labeled at the same nine axial positions in each half A-band, consistent with a circumferential and/or radial rather than an axial orientation of the bulk of the molecule. The C-terminal antibody stripes were slightly displaced axially, demonstrating an axial orientation of the C-terminal 3 domains, with the C-terminus closer to the M-line. These results, combined with previous studies, suggest that the C-terminal domains of cMyBP-C run along the thick filament surface, while the N-terminus extends towards neighboring thin filaments. This organization provides a structural framework for understanding cMyBP-C’s modulation of cardiac muscle contraction.
Keywords: cMyBP-C, cardiac muscle contraction, cardiac muscle disease, cardiac muscle structure, cardiac muscle regulation
1. Introduction
Myosin binding protein C (MyBP-C) is an accessory protein of the thick filaments of vertebrate skeletal and cardiac muscle 2; 3; 4; 5. It is an elongated, flexible molecule with a molecular weight of ~ 140 kDa, a length of ~ 40 nm, and a width of ~ 3 nm. It consists principally of a linear string of 10 kDa immunoglobulin-like (Ig) and fibronectin-like (Fn) domains (Fig. 1) 3; 6; 7. Three isoforms (fast skeletal, slow skeletal and cardiac) are encoded by different genes 3; 6; 8. The skeletal isoforms have a total of ten Ig and Fn domains, numbered C1-C10 from the N-terminus, while the cardiac isoform (cMyBP-C) has an extra N-terminal domain (C0) 3; 8. A proline- and alanine-rich (PA) domain occurs at the N-terminus in the skeletal isoform and between C0 and C1 in the cardiac isoform, and there is a conserved MyBP-C-specific domain (the M-domain) between C1 and C2 in all isoforms. Four serines in the M-domain of cMyBP-C can be phosphorylated by a variety of protein kinases 4; 8; 9. In vivo, phosphorylation occurs in r o to β-adrenergic stimulation, enhancing cardiac contractility 4; 8; 9; 10; 11; 12. Mutations in cMyBP-C are one of the most common causes of inherited hypertrophic cardiomyopathy (HCM) 3; 13, and alterations in cMyBP-C phosphorylation levels have been implicated in heart failure and HCM 14.
Figure 1. Domain organization of cardiac MyBP-C.
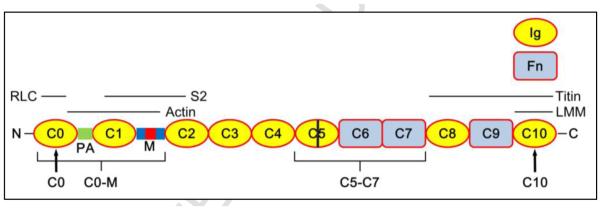
Ig and Fn domains numbered from N-terminus, at left. PA, ProAla-rich domain (green); M, M-domain (blue), containing phosphorylation sites (red); bar in C5, cardiac-specific insert. Black labels beneath molecule are positions of epitopes of the 4 antibodies used in this study; C0 and C10 antibodies are specific for peptides near the N- and C-termini; C0-M and C5-C7 antibodies preferentially recognize these regions of the molecule. Labels above cMyBP-C indicate regions of binding to the RLC, S2, LMM, actin and titin.
cMyBP-C is located in a series of nine regularly spaced, narrow, transverse stripes, 43 nm apart, in the middle third (the C-zone) of each half of the A-band. These are the outer 9 of a total of eleven non-myosin stripes in the vertebrate striated muscle half A-band (Fig. 2;15; 16; 17; 18; 19). Three or four domains at the C-terminus (~C7-C10) anchor MyBP-C to the thick filament backbone at each site by binding to the light meromyosin (LMM) tails of myosin and/or to titin 20; 21; 22. At the other end of the molecule, the N-terminal region, including the M-domain, can also bind to myosin in vitro, via the regulatory light chain (RLC) 23, and via the subfragment 2 (S2) region of the tail, near its junction with the heads 24; 25; 26; 27; 28, an interaction that is abolished by phosphorylation of the M-domain 4; 29; 30. Surprisingly, this same region also binds to actin filaments in vitro, and this binding is likewise reduced by phosphorylation 5; 24; 31; 32; 33; 34; 35; 36; 37. The physiological significance of binding to actin and S2 is not fully understood, although it could potentially affect thick-thin filament interaction and thus play a key role in cMyBP-C’ modulation of filament sliding and muscle contraction 38; 39; 40.
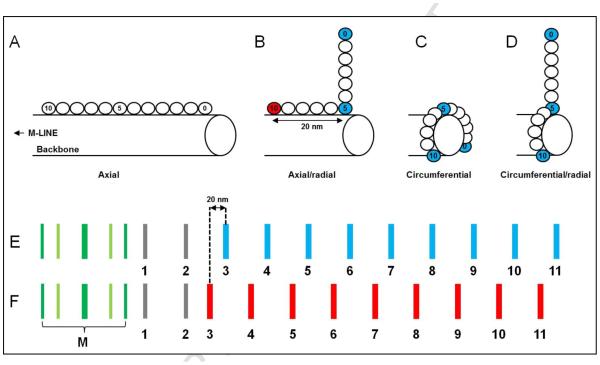
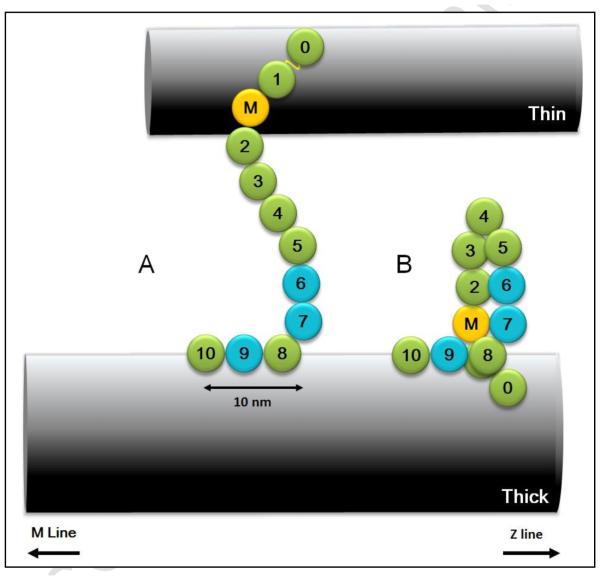
Figure 2. Examples of cMyBP-C arrangement and antibody-labeling patterns, demonstrating the principle of the labeling experiment.
(A) Fully axial arrangement of cMyBP-C. (B) C-terminal axial, N- terminal radial; C10 closer to M-line. (C) Fully circumferential, C0-C10 all involved in collar. (D) C-terminal circumferential, N-terminal radial. (E, F) Examples showing how positions of antibody stripes within the 43 nm repeat can depend on whether cMyBP-C is axially or transversely (either radially or circumferentially) oriented. Red and blue show the nine stripes (numbered 3-11 of the 11 total nonmyosin stripes in each half A-band 17) that are strongly labeled by anti-cMyBP-C (Fig. 4C-F). Stripes 1 and 2 are unlabeled or weakly labeled. With a transverse orientation (e.g. (C) and (D)) all antibodies would label at the same axial position (e.g. the blue stripe array in (E) due to labeling of blue epitopes in C, D). However, if part or all of the molecule is axially organized (e.g. (A), (B)), different domains would label at different axial positions, the relative positions of the stripe arrays depending on the length of the axial component and its orientation (towards or away from the M-line). For example, in (B) the blue domains (C0 and C5) could (as shown here) be situated ~20 nm further from the M-line than the red domain (C10), resulting in the relative labeling patterns shown in (E) and (F). In this case, the stripes labeled by antibodies to C0 and C5 (blue stripes) would lie ~20 nm further from the M-line (as in E) than the corresponding stripes labeled by anti-C10 (as in F). The same concepts can apply to other possible domain distributions. The actual distance of each of the stripes 3-11 from the center of the M-line was measured for each antibody, producing the results in Table S1. From these results, the average position of all nine stripes was calculated (Table 1), and can be thought of as indicating the position of stripe 7 (see 2.3; Materials and Methods).
To understand the structural basis of MyBP-C function at the molecular level, knowledge of its organization in the sarcomere is essential. Several models have been proposed based on MyBP-C’ in vitro interactions with other sarcomeric proteins, and on electron microscopy, X-ray diffraction, and other techniques. These have led to differing suggestions as to how MyBP-C might function 41; 42; 43; 44. A key aspect of any model is the orientation of the rod-like and flexible molecule with respect to the thick filament, as different orientations will constrain MyBP-C’ potential binding partners and modes of functioning. For example, based on measurements of MyBP-C periodicity, it has been suggested, at one extreme, that the ~ 40 nm-long molecule runs axially along the thick filament, spanning most or all of the distance between the 43 nm- spaced stripes (Fig. 2A). With this scenario, it was suggested that MyBP-C might co-polymerize with myosin in a way that led to precise determination of thick filament length 44; 45; 46. Models where the MyBP-C N-terminus interacts with S2, potentially modulating head movement, also show an approximately axial orientation 43. At the other extreme, yeast two-hybrid studies of cMyBP-C have suggested intermolecular interactions between domains C5 and C8 and between C7 and C10 41, which led to the proposal that the C-terminal half of the molecule is perpendicular to the thick filament, wrapping around the filament backbone to form a narrow collar (Fig. 2C, D) 41. This could function, for example, to clamp the LMM regions of myosin in position, stabilizing the filament, or to influence myosin head motions. Similar interactions have been detected in yeast two-hybrid studies of fast skeletal MyBP-C, but not slow skeletal MyBP-C 47. Other experiments, including in vitro binding and motility assays, show that N-terminal domains of MyBP-C can interact with F-actin filaments 34; 35; 36; 37; 39; 48, suggesting a sarcomeric organization in which MyBP-C, anchored to the thick filament by its C-terminal domains, may extend its central and N-terminal domains radially and bind to a neighboring thin filament (Fig. 2B, D). Some models combine aspects of more than one of these orientations.
These different models cannot all be correct. The narrowness of the MyBP-C stripes (~7 nm) in longitudinal sections of the A-band 17; 19; 49 suggests that the bulk of the elongated molecule runs transversely (i.e. circumferentially or radially; Fig. 2B-D), rather than axially, consistent with X-ray diffraction data from intact muscle 42; 46; 50, and with electron tomographic data of sectioned sarcomeres 51; 52. When skeletal and cardiac myofibrils are labeled with polyclonal antibodies to MyBP-C, the labeled stripes are also relatively narrow 15; 16; 19, further suggesting a transverse rather than an axial arrangement for the majority of the molecule. However, it is not certain that the whole of MyBP-C is visualized in unlabeled sarcomeres, nor that the polyclonal antibodies used in labeling experiments recognized epitopes along the entire molecule rather than a local, strongly antigenic region. In addition, while X-ray and electron tomographic studies provide information on the approximate distribution of mass in the sarcomere, they do not reveal the identity of the mass, nor the locations of specific regions of a molecule. Thus previous studies do not unambiguously reveal MyBP-C organization. Furthermore, because most of these studies used skeletal muscle, there is additional uncertainty concerning the organization of the cardiac isoform.
Here we use domain-specific antibodies combined with immuno-EM to directly localize the axial positions of known domains of cMyBP-C in cardiac myofibrils, thus helping to distinguish between the different models. A similar approach was used to reveal the locations of specific domains of M-protein, myomesin and titin in skeletal muscle 53. Our antibodies specifically recognized the C-terminal domain (designated C10 antibodies), the middle of the molecule (C5-C7), the N-terminal region (C0-M), and the N-terminal domain (C0) of cMyBP-C (Fig. 1) (details of antibody preparation and specificity are provided in Materials and Methods and Supplemental Discussion (1)). The relative positions of these antibodies provide information on the axial locations of the two ends and the approximate middle of cMyBP-C at each MyBP-C stripe in the sarcomere, thus defining the orientation of the elongated molecule. The concept is illustrated in Fig. 2. If labeling with all antibodies occurs at the same axial position for each stripe, this will imply a transverse (radial or circumferential) organization for cMyBP-C (for example, the blue domains and stripes in Fig. 2C-E), while if they are in different positions, this will signify that some or all of the molecule is axially arranged (e.g., the blue vs. red domains and stripes in Fig. 2B,E,F). Our results show that all epitopes, except the C-terminus, are within 5 nm of each other axially. This suggests a model in which the C-terminal 3 domains run axially, while the bulk of the molecule is transverse to the filament axis.
2. Results
2.1. Immunofluorescence microscopy
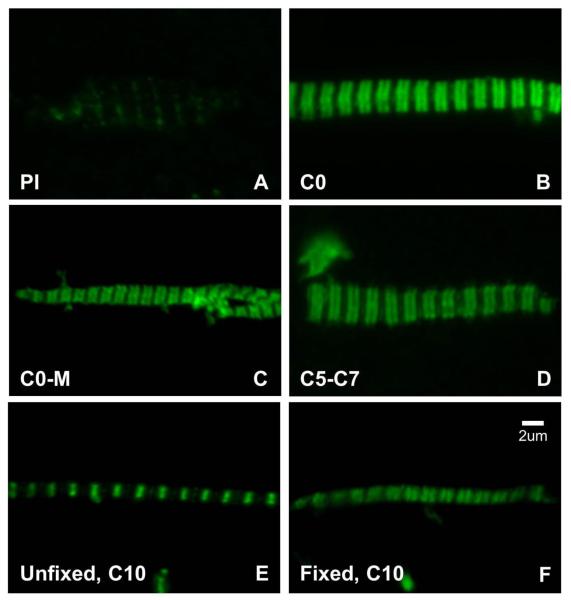
Before carrying out ultrastructural observation of labeled myofibrils, we first analyzed antibody localization, binding capability and specificity by immunofluorescence microscopy (Fig. 3). Pre-immune (control) serum weakly labeled myofibrils at the Z-line (Fig. 3A), but nowhere else. In myofibrils that had not been fixed before labeling, antibodies to C0, C0-M and C5-C7 all specifically labeled the A-band (Fig. 3B-D), in two bands whose center-to-center spacing was ~0.67 ± 0.03 μm (mean and standard deviation for C0, n=52). Surprisingly, however, antibodies to the C10 domain labeled the Z-line rather than the A-band (Fig. 3E), a result also obtained using a commercial antibody to the C-terminus (Santa Cruz MYBPC3 sc-50114). Given that the C-terminal region is responsible for anchoring MyBP-C to the thick filament by binding to LMM and titin 20; 21; 22, and that the other antibodies labeled the A-band, a simple interpretation is that the C10 domain is masked by binding to these other components, and that unbound antibodies bind non-specifically to the Z-line (like the pre-immune serum). We tested this idea by carrying out antibody labeling of myofibrils that had first been fixed with glutaraldehyde, which, we reasoned, could potentially alter epitope availability or reactivity 54. C10 labeling was now found in the A-band and was absent from the Z-line (Fig. 3F), consistent with this interpretation. Labeling of fixed myofibrils with the three other antibodies gave the same staining pattern to that obtained without fixation. An alternative explanation for the absence of C10 labeling with unfixed myofibrils is that binding of antibody to C10 causes dissociation of cMyBP-C from the filament by competing with LMM and titin for C10 binding. However, incubation of unfixed myofibrils with C10 antibody, followed by antibody to C0-M or C5-C7, showed C0-M and C5-C7 A-band labeling by immunofluorescence and by immuno-EM (data not shown), implying that cMyBP-C is present, even though not detected by anti-C10.
Figure 3. Immunofluorescence of mouse cardiac myofibrils labeled with domain-specific antibodies against mouse cMyBP-C.
All myofibrils are fixed except (E). (A) labeled with pre-immune serum (PI); the Z-line but not the A-band is labeled. (B)-(D) labeled with C0, C0-M and C5-C7 antibodies, showing labeling of the A-band. (E, F) unfixed and fixed myofibrils labeled with C10 antibodies, showing labeling of the Z-line (E) and the A-band (F).
Mouse cardiac myofibrils lacking cMyBP-C (obtained from t/t mouse hearts 55; 56) treated with all four antibodies showed Z-line but no A-band labeling (Fig. S1A-D), confirming that A-band labeling of wild type myofibrils was specific for cMyBP-C, and that Z-line labeling with pre-immune serum and with C10 antibodies using unfixed myofibrils is due to non-specific binding of antibody to the Z-line (see Supplemental Discussion (1) for further discussion of labeling specificity).
2.2 Immuno-electron microscopy
Negatively stained, control (unlabeled) wild type myofibrils showed eleven, narrow (~ 10 nm wide), stain-excluding (white) stripes in each half of the A-band (Fig. 4A), representing non-myosin proteins 17; 19; 49; 57. The M-line and Z-line appeared as broader, unstained bands, at the center of the A-band and I-band respectively. cMyBP-C null cardiac myofibrils (negative controls, lacking cMyBP-C) showed similar M-line, A-band, I-band and Z-line appearances, but did not exhibit any of the eleven stripes in the A-band (Figs. 4B, 5).
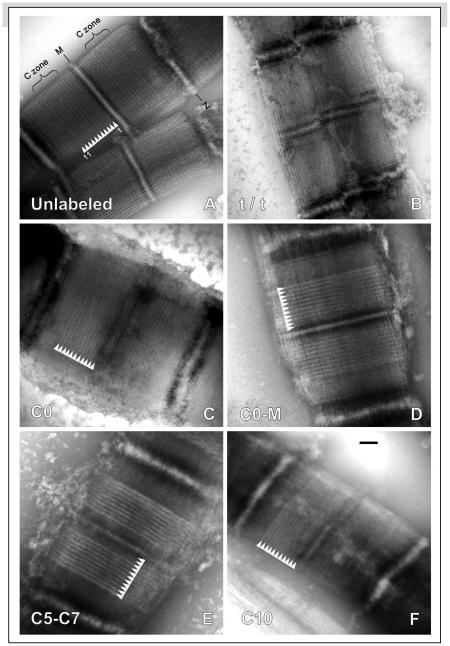
Figure 4. Electron micrographs of cMyBP-C localization in fixed myofibrils labeled with specific antibodies.
(A) Control myofibril without antibody shows 11 narrow stripes in each half A-band (arrowheads, cf. Fig. 2E). (B) Unlabeled tit myofibril (cMyBP-C absent) shows no stripes in the A-band. (C-F) Myofibrils labeled with antibodies to CO, CO-M, C5-C7, and C10 show clear intensification of MyBP-C stripes (stripes 3-9). For CO-Mand C5-C7, only primary antibodies were used, while for epitope-specific antibodies (CO and C10), myofibrils were also labeled with secondary antibody. Stripes 3-11 strongly labeled with antibody; stripe 1 unlabeled; stripe 2 often weakly labeled (see Supplemental Discussion (2)). Note: in all cases where labeling occurred both with and without fixation, the labeling pattern was similar; 10 we therefore show only the results obtained with fixation. Scale bar = 200 nm.
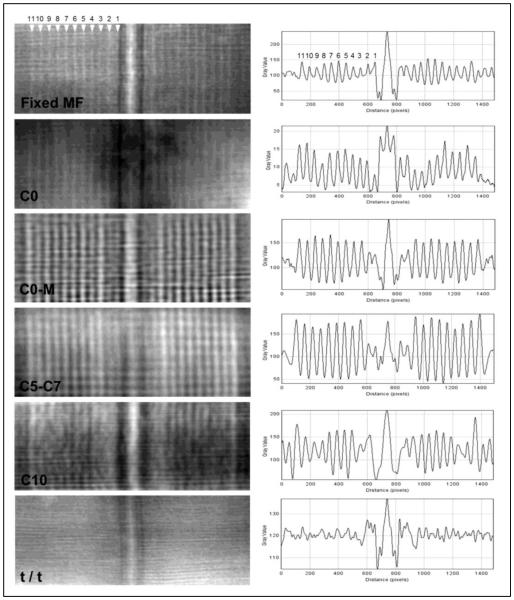
Fig. 5. Examples of filtered images of individual negatively stained myofibrils together with their profile plots (used to make measurements of stripe position).
Each filtered image shows a clear M-line and 9-11 stripes in each half of the A-band. In the profile plots, the antibody-labeled myofibrils show 9 strong peaks (stripes 3-11) in the C-zone, and in some cases a peak on stripe 2. The control myofibril (Fixed MF) shows 11 weak peaks, while the cMyBP-C-deficient (t/t) myofibril shows no peaks in the C-zone. For measuring stripe position, the periodicity of the peaks seen in these profiles was used as an internal standard, assumed to be 43 nm.
Fixed or unfixed wild type mouse cardiac myofibrils were labeled separately with the four domain-specific antibodies. For both fixed and unfixed myofibrils, labeling with C0-M and C5-C7 antibodies showed clear enhancement (widening and intensification) of the outer nine stripes in each half A-band (Fig. 4D-E), similar to the labeling pattern found previously in sectioned cardiac muscle 19. In the case of the two epitope-specific antibodies, C0 and C10, no enhancement of the stripes was apparent with primary antibody alone: presumably, due to the small size of the epitope, only one antibody molecule would be able to bind to each cMyBP-C. In these cases we used a secondary antibody to enhance the signal (the same antibody used for the immunofluorescence microscopy). Following secondary antibody labeling, enhancement of the nine cMyBP-C stripes was very apparent with the N-terminal (C0) antibody (Fig. 4C) in both fixed and unfixed myofibrils. For the C10 antibody, the result depended on whether or not the myofibrils were fixed. In the case of unfixed myofibrils, there was no stripe enhancement following secondary antibody labeling. When C10 labeling was carried out after initial fixation, exposure to secondary antibody now caused enhancement of the stripes (Fig. 4F). This fixation effect is consistent with the immunofluorescence results (Fig. 3E,F) suggesting that the C10 epitope might be masked in the absence of fixation. With all four antibodies, stripe 2 was also often weakly labeled (Fig. 4 C-F) suggesting that cMyBP-C might be present on this stripe as well, but partly masked (see Supplemental Discussion (2)).
2.3 Measurement of stripe position
Visually, the axial positions of the stripes with all four antibodies appeared to be very similar. This was tested quantitatively by measuring the distance of each of stripes 3-11 from the central stripe of the M-line in labeled and unlabeled A-bands (Fig. 2E,F) using the Plot Profile function of ImageJ (Fig. 5). Table S1 shows that the average position of each of the nine stripes was approximately the same for C0, C0-M and C5-C7 antibodies, while that observed using the C10 antibodies was consistently slightly closer to the M-line.
Because the stripes are known to lie on a constant repeat 17; 42; 58, we can simplify the comparison of their relative axial positions determined using the four antibodies by averaging the locations of the nine stripes—giving, in effect, the position of the middle stripe of the nine in each half A-band (i.e. stripe 7; Fig. 2E; Table 1); averaging also reduces the experimental error in measuring the stripe positions. By comparing this position for each of the four antibodies, we can define the relative axial locations of each of the three main regions of the molecule (N-terminus, middle and C-terminus), giving insight into the overall orientation of the molecule. Two of the antibodies (C0 and C5-C7) labeled at the same axial position as each other and as the unlabeled stripes, within experimental error (3-4 nm; Table 1). The C-terminal antibody (C10) also labeled relatively close to this position, but was displaced by ~ 10 nm towards the M-line. The 1-way ANOVA test with Tukey's multiple comparison test showed that the difference between the average stripe position determined with the C10 antibody and that determined with each of the other antibodies was highly significant (p <0.01; Table 1). When the positions of each of the individual stripes (3-9) were compared using the different antibodies (Table S1), the difference between C10 labeling and labeling with each of the other antibodies was again significant for all of the stripes (p = 0.01-0.05), supporting the conclusions reached by averaging the stripe positions.
Table 1.
Average position (nm) of unlabeled and labeled stripes (mean ± S.D.)
| Antibody | Unlabeled | C0 | C0-M | C5-C7 | C10-2nd Ab |
|---|---|---|---|---|---|
|
Average
Position (nm) |
328.2 ± 2.8
( n = 33 ) |
330.3 ± 4.4
( n = 90 ) |
325.1 ± 4.3*
( n = 27 ) |
329.2 ± 3.3
( n = 63 ) |
320.2 ± 2.9**
( n = 20 ) |
The Table shows the average of the axial distances measured from the central stripe of the M-line to the peak of each cMyBP-C unlabeled or antibody-labeled stripe in fixed myofibrils (Figs. 2E,F, 5, Table S1.).
indicates significantly different from each of the three other antibody-labeled and the unlabeled stripe positions (p < 0.01, except for Unlabeled, where p < 0.05).
indicates significantly different from each of the three other antibody-labeled and the unlabeled stripe positions (p < 0.01).
The position of the C0-M stripes also differed from the others, but by a smaller amount (~ 5 nm). This difference was also significant (p <0.05) for both the averaged (Table 1) and individual (Table S1) stripe positions.
3. Discussion
3.1 Domain organization of cMyBP-C in the cardiac sarcomere
Using domain-specific antibodies, we mapped the relative axial positions of specific sites on cMyBP-C (the two ends and the middle) as they are laid out in mammalian cardiac myofibrils. The similar axial positions of C0, C0-M and C5-C7 (Tables 1, S1) suggest that the majority of the molecule (roughly C0 to C7) must run approximately transversely (radially and/or circumferentially) to the thick filament; in contrast, the small (5-10 nm) displacement of C10 from the other domains suggests that the C-terminal approximately three domains (C8-C10), each ~ 4 nm long, run roughly parallel to the filament axis (Fig. 6A). Our observations have been carried out on myofibrils in the rigor state, necessary to provide a stable cross-linked network of filaments for antibody labeling. cMyBP-C’ organization appears to be similar in the relaxed state, as the cMyBP-C stripes have similar widths and axial positions to the rigor state (see Supplemental Discussion 3). In contrast, X-ray diffraction suggests that MyBP-C becomes disordered when muscle contracts 59; 60.
Figure 6. Cartoon of possible cMyBP-C organizations in the sarcomere compatible with the antibody labeling data.
Three domains (C8-C10) run axially along the thick filament, while most of the molecule (C0-C7) either extends from the filament surface, binding to actin by its N-terminal region (A), or loops back, possibly binding to S2 or the RLC, close to the filament backbone (B). Assuming each domain is ~ 4 nm, the inclusive distance from C8- C10 would be ~ 12 nm, similar to the distance between the C10 antibody and the C0 and C5-C7 antibodies (Table 1). The C0-M region bound to actin is shown with the orientation suggested by 3D reconstructions of F-actin decorated with N-terminal fragments 1. This arrangement may account for the observation that C0-M antibodies appear to bind slightly closer to the M-line than C0 and C5-C7.
3.2 Relation to previous structural studies
Previous models of MyBP-C organization have been based on in vitro interactions of isolated molecules and on structural observations (EM and X-ray) of thick filaments and sarcomeres (see Introduction). However, the information available from such studies is incomplete. Interactions occurring in vitro do not necessarily occur in situ, while EM and X-ray studies do not reveal the locations of specific MyBP-C domains. Our domain-specific labeling can be used to test the different arrangements of the molecule that have been proposed based on these earlier studies.
Axial arrangement of whole MyBP-C molecule
Early X-ray diffraction models suggested that the whole of MyBP-C might run axially, functioning as a thick filament length-determining protein through co-polymerization with myosin 44; 45; 46. The transverse arrangement of the majority of cMyBP-C excludes such axial models. Length determination most likely involves the scaffolding protein, titin 61; 62.
Collar arrangement of C-terminal MyBP-C domains
Yeast two hybrid experiments showing intermolecular interaction between domains C5 and C8 and between C7 and C10 led to a model in which the C-terminal half of each cMyBP-C wraps circumferentially around the thick filament forming a collar 41. In this model, two cMyBP-C molecules overlap circumferentially at any azimuth in the collar: these would therefore have their centers ~2.5 nm apart axially (based on the ~ 2.5 nm width of an Ig domain 7, predicting < 5 nm axially between C10 and C5-C7 antibodies, not the 10 nm observed (Table 1). 3D reconstructions of isolated cardiac thick filaments provide further information on this point 63; 64. The reconstructions clearly resolve the 4-nm spaced, globular Ig and Fn domains of titin, axially arranged along the filament surface. Therefore, if the C-terminal Ig domains of MyBP-C formed a collar, they should be similarly well-resolved, and visualized in a circumferential arrangement. No such circumferential organization is observed (in fact the converse is found – see next section). The antibody labeling and 3D reconstruction results are therefore both inconsistent with a collar-like arrangement.
Axial arrangement of C-terminal MyBP-C domains
In addition to the 4-nm spaced titin domains running along the filament, the thick filament 3D reconstructions showed three further 4-nm domains, specifically at the level of the MyBP-C stripes 63; 64. These were axially arranged and interpreted as MyBP-C’ C-terminal thick filament anchoring domains, C8-C10, known to bind to light meromyosin and/or titin 20; 21; 22. The ~ 10 nm distance between C10 and the central and N-terminal domains of cMyBP-C found by antibody labeling supports this interpretation, assuming domains C7-C0 run transversely (Fig. 6A). In addition it now defines the polarity of the C-terminal region, with the C-terminus (C10) closer to the M-line. Modeling of X-ray diffraction patterns of muscle also suggested an axial arrangement of MyBP-C C-terminal domains, in this case C5-C10 42. Our antibody labeling agrees with the axial orientation, but does not support its longer extent. If C5-C7 also ran axially 42, then C5-C7 antibody stripes should be up to 20 nm from C10, not the 10 nm observed. The shorter axial extent and longer perpendicular extension implied by our labeling observations would agree equally well with the X-ray observations.
Radial arrangement of N-terminal region of MyBP-C
Extension of the bulk of MyBP-C (the middle and N-terminal half) away from the thick filament backbone is indicated by a variety of studies. Early X-ray diffraction observations suggested that the component giving rise to the “forbidden” meridional reflection on the first myosin layer line extended laterally from the thick filament backbone 46; this component was subsequently shown to be MyBP-C 58. Later X-ray modeling supported this view, suggesting that the N-terminal half of MyBP-C projected from the thick filament and bound to a neighboring thin filament 42. Recent X-ray measurements of the MyBP-C-based M1 reflection provide additional support, suggesting a perpendicular extension of MyBP-C from the thick filaments that requires the presence of actin 60. Direct, visual evidence for extension of MyBP-C from the thick filament has come from electron tomography of longitudinal sections of skeletal muscle. These studies show that the narrow MyBP-C stripes in the C-zone arise from an axially confined but perpendicularly extended density connecting thick and thin filaments 51; 52, consistent with the X-ray studies. Our antibody labeling corroborates these conclusions by demonstrating that domains C7-C0 are within 5 nm of each other axially. In the absence of evidence for a circumferential arrangement of the C7-C0 domains that would explain this axial confinement (see Collar arrangement of C-terminal MyBP-C domains), the only alternative is a radial extension (Fig. 6A).
The X-ray and tomographic investigations were carried out on skeletal muscle, while our antibody studies and the thick filament reconstructions used cardiac muscle. Taken together, the results suggest a similar organization for MyBP-C in both types of muscle. This is significant, given that skeletal MyBP-C, in addition to having less than 50% sequence identity with cMyBP-C, lacks a C0 domain and has different N-terminal interactions, phosphorylation capability, and actin and S2 binding propensity 3; 8; 65; 66.
3.3 Functional implications of MyBP-C arrangement
We conclude that, of the differing MyBP-C models that have been proposed, the domain organization that best fits both our antibody-labeling findings and the structural data from native filaments and intact sarcomeres 42; 52; 60; 63; 64, is one in which the C-terminal ~3 domains run axially along the surface of the thick filament 63 towards the M-line, while the rest of the molecule extends laterally from the thick filament and binds to a neighboring thin filament 52 (Fig. 6A). This model is supported by additional data. The narrowness of the MyBP-C stripes, representing the projecting region of the molecule 52, suggests that the distal, flexible N-terminal end must be held in position, presumably by attachment to another myofibrillar component. Connection to actin would provide a simple explanation. This concept is reinforced by the finding that the MyBP-C-based M1 meridional X-ray reflection becomes weaker as thin filaments are pulled out of the C-zone, suggesting that regular, axially confined extensions of MyBP-C from the thick filaments require the presence of actin 60. The binding to actin in situ implied by these structural data supports the physiological relevance of in vitro data showing that N-terminal domains of cMyBP-C can bind to thin filaments, modulating both their Ca2+-sensitivity and sliding with respect to thick filaments 35; 36; 38; 39; 48.
In addition to its binding to thin filaments, there is also evidence that the cMyBP-C N-terminal region (C0, C1, M) can interact in vitro with the myosin head region (both the RLC 23 and S2 near its junction with the heads 24; 25; 26; 27; 28; 29), with potential effects on myosin function. In relaxed muscle the myosin heads and S2 lie on or close to the thick filament surface 63; 64, making it difficult to reconcile these interactions with an N-terminal region that extends away from the filament backbone. Possibly such interactions occur when myosin heads move away from the thick filament to bind to actin during contraction, although even under these circumstances molecular modeling suggests that S2 and the RLC would be no more than 5-8 nm from the thick filament backbone, and so would not be close to MyBP-C’ xt N-terminus. Models in which MyBP-C extends axially along the filament, making S2/RLC interactions possible 43, are inconsistent with our labeling data showing a transverse orientation for the bulk of the molecule. An arrangement that could partially reconcile our antibody and S2/RLC binding data is one in which MyBP-C extends from the thick filament, then loops back so that its N-terminal region interacts with S2/RLC (close to the filament backbone) at the same axial level (Fig. 6B). While this would be consistent with our domain localization data, it does not agree with the tomographic observations that show MyBP-C contacting neighboring actin filaments. Alternatively MyBP-C could interact with S2 and the RLC from a different thick filament 52, although there is so far no structural evidence that such MyBP-C contacts between thick filaments actually occur in muscle.
4. Materials and Methods
4.1. Antibodies
Four polyclonal antibodies were used in this work. Two were raised in rabbits against specific N- and C-terminal epitopes of mouse cMyBP-C [Uniprot ID O70468]: residues 2-14 (EPGKKPVSAFSKK) at the N-terminal end of the C0 domain, and residues 1200-1212 (AVRGSPKPKISWFK) at the C-terminal end of C10. These peptides were selected based on their predicted antigenic potency and the absence of any homolog within other domains of cMyBP-C or other proteins in the genome. The antibodies were purified by affinity chromatography using the respective immunogenic peptides, and characterized as previously described 67. Western blots confirmed the specificity for C0 and C10 of cardiac muscle and the absence of reactivity with skeletal MyBP-C. A third antibody was raised in rabbits against native cMyBP-C purified from rat heart 68. Epitope mapping using this serum indicated that the antibody preferentially recognized epitopes in the C0 domain with some reactivity to the M-domain and little or none with domains C3-C4, C5-C7 and C8-C10. The fourth antibody was raised in rabbits against a recombinant protein comprising mouse domains C5-C7, expressed in bacteria as described for expression of N-terminal domains of cMyBP-C 35. Soluble protein was purified under native conditions from bacterial homogenates using a NiNTA affinity column against a 7x His tag added at the N-terminus of the C5-C7 protein. Purified C5-C7 was dialyzed into PBS for production of rabbit antiserum by the Comparative Pathology Laboratory at the University of California, Davis. Western blots of DAPase-treated proteins (to remove the His tag) confirmed that the antisera and affinity purified antisera recognized expressed domains C5-C7, but not C0-C2, C0-C4, nor C8-C10.
4.2. Mouse cardiac myofibrils
Myofibrils were isolated from the hearts of 8 to 10 week old mice (129 SVE, Taconic Farms, NY). Animals were handled in accordance with the principles and procedures of the University of Massachusetts Medical School Institutional Animal Care and Use Committee (IACUC) guidelines. Mice were first anesthetized in a CO2 chamber for 6 minutes, and then euthanized by cervical dislocation. The heart was excised and cut into small slices in rigor solution (100 mM NaCl, 3 mM MgCl2, 2 mM EGTA, 1 mM NaN3, 5 mM PIPES, pH 7.0) and the slices chemically skinned in rigor solution containing 0.1% w/v saponin for 3 h at 4°C with agitation. Skinned muscles were washed with several changes of fresh rigor solution overnight at 4°C, then homogenized in rigor solution using a Polytron homogenizer (Brinkmann Instruments, Westbury, NY) at setting number 5 for 1 min (3 times for 20 sec each). The suspension was centrifuged at 800 g for 3 min to pellet the myofibrils, leaving small debris in the supernatant, and the myofibrils resuspended in rigor solution.
As a negative control, cMyBP-C null myofibrils were isolated from the hearts of 3 week old cMyBP-C(t/t) mice 56 by the same method.
Phosphorylation levels of cMyBP-C were measured using SYPRO Ruby/Pro-Q Diamond phosphoprotein staining (Life Technologies).
4.3. Immunofluorescence microscopy
For both immunofluorescence and immuno-EM experiments, fixative, BSA rinses and antibody dilutions used rigor solution as the carrier. A drop of the cardiac myofibril suspension was applied to a coverglass and left for 5 min at room temperature. After attachment to the coverglass, myofibrils were fixed for 5 min with 0.1% glutaraldehyde in rigor solution (or left unfixed), then washed with 3 changes of fresh rigor solution for 15 min. After fixation, the coverglass was blocked with 1% BSA for 30 min at room temperature. After washing with 3 changes of 0.1% BSA for 15 min, followed by fresh rigor solution, the cover glass was incubated with a 1:400 dilution of antibody overnight at 4°C. The coverglass was washed with 3 changes of fresh rigor solution for 15 min, and then incubated for 1 hr at room temperature with a 1:1000 dilution of goat anti-rabbit secondary antibody tagged with Alexa 488 (Invitrogen). The coverglass was washed 3 times with rigor solution, then applied to a glass slide and examined with an Axioskop 2 Plus (Carl Zeiss, Inc. Thornwood, New York, USA) equipped with filters for fluorescein isothiocyanate (FITC). Fluorescence images were recorded using AxioVision (Carl Zeiss, Inc.) software and an AxioCam camera (Carl Zeiss, Inc.). To determine the location of antibody binding in the sarcomere, Z-lines and A-bands were identified in parallel DIC images of the same myofibrils that had been observed by immunofluorescence. Z-line identification was confirmed using antibodies to α-actinin (not shown).
4.4. Immuno-electron microscopy
A 5 μl droplet of cardiac myofibril suspension was placed on a formvar- and carbon- coated copper grid, left for 5 min, washed with several drops of rigor solution then fixed with 0.1% glutaraldehyde for 5 min (or left unfixed). The grid was blocked with 1% BSA for 30 min at room temperature, washed with 0.1% BSA for 5 min, then incubated overnight at 4°C with a 1:16 dilution of domain-specific antibody. The grid was then washed on a drop of rigor buffer for 5 min, and in the cases of the C- and N-terminal epitope antibodies, this was followed by incubation on a drop of 1:32 dilution of goat anti-rabbit Alexa 488 secondary antibody (Invitrogen) at room temperature for 1 hr.
Labeled and unlabeled myofibrils were negatively stained by first fixing with 2.5% glutaraldehyde (buffered in rigor solution, pH 7.0) for 1 min, washing with several drops of 0.1 M ammonium acetate, then staining with 2 drops of 0.5% aqueous ammonium molybdate 17. The remaining drop of stain was removed with filter paper and the grid dried quickly under a hair drier to improve spreading of the stain. Images were obtained at 120 kV on a TECNAI Spirit electron microscope (FEI, Hillsboro, OR, USA), using a Gatan Erlangshen CCD camera (Gatan, Pleasanton, CA, USA) at a pixel size of 0.86 nm in the specimen). Because of the myofibril thickness (~ 1 μm), staining was typically quite dense (Fig. 4). Nevertheless, stripe visibility was substantially improved over that seen in sectioned material 19; variations on this optimal procedure that produced lower density did so at the expense of contrast in the stripes.
4.5. Image analysis
Images were selected in which the MyBP-C stripes were clear and straight, and in the case of immunolabeling, wider, indicating the presence of antibody. The selected images were rotated to make the stripes vertical, and the distance of each stripe from the center of the M-line was measured using the ImageJ 69 Plot Profile function. In some cases, a band-pass or custom filter was used to enhance the signal 19. While this made the stripes clearer and therefore easier to measure, it did not change their peak positions or their periodicity. To compensate for possible variable specimen shrinkage and slight variations in microscope magnification, the regular periodicity of the MyBP-C stripes was used as an internal standard for each micrograph 19. The periodicity was taken to be 43 nm, based on past X-ray diffraction and EM/optical diffraction studies 17; 42; 46; 58; however, the precise value is not important as it does not affect the relative positions of the stripes, which was the key parameter in our studies. Minimizing experimental variability by using this internal calibration enabled us to measure small differences in stripe position that might otherwise have gone undetected.
4.6. Statistical analysis
For statistical comparison of the distances of the stripes from the center of the M line, one-way analysis of variance (ANOVA) with Tukey's multiple comparison test was performed using GraphPad Prism version 6.04 for Windows (GraphPad Software, La Jolla, CA).
Supplementary Material
Highlights.
The organization of cMyBP-C in the cardiac muscle sarcomere is controversial
Cardiac muscle myofibrils were labeled with domain-specific antibodies to cMyBP-C
The antibodies revealed the axial positions of different domains of cMyBP-C
Different domains were located at the same axial positions along the thick filament
We conclude that most of cMyBP-C is oriented perpendicular to the thick filament
5. Acknowledgments
We thank Drs. Christine E. Seidman and Jonathan G. Seidman for their generosity in providing the cMyBP-C(t/t) mouse model, Elaine Hoye for assistance in the development of the C5-C7 antibody, and Drs. John Woodhead, Michael Previs and David Warshaw for comments on the manuscript. We are also grateful to Drs. George Witman and Branch Craige for training on and use of their immunofluorescence microscope. This work was supported by NIH grants R01 AR034711 (RC) and P01 HL059408 (to D. Warshaw), R01 HL080367 (SPH), and R01 HL105826 and K02 HL114749 (SS) and S10RR027897 (for the purchase of the electron microscope).
Abbreviations
- MyBP-C
myosin binding protein C
- cMyBP-C
cardiac myosin binding protein C
- EM
electron microscopy
- HCM
hypertrophic cardiomyopathy
- RLC
regulatory light chain of myosin
- S2
subfragment 2 of myosin
- LMM
light meromyosin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. Disclosure statement
The authors declare that there are no conflicts of interest.
7. References
- 1.Mun JY, Gulick J, Robbins J, Woodhead J, Lehman W, Craig R. Electron microscopy and 3D reconstruction of F-actin decorated with cardiac myosin-binding protein C (cMyBP-C) J Mol Biol. 2011;410:214–25. doi: 10.1016/j.jmb.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Offer G, Moos C, Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extractions, purification and characterization. J Mol Biol. 1973;74:653–76. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 3.Flashman E, Redwood C, Moolman-Smook J, Watkins H. Cardiac myosin binding protein C: its role in physiology and disease. Circ Res. 2004;94:1279–89. doi: 10.1161/01.RES.0000127175.21818.C2. [DOI] [PubMed] [Google Scholar]
- 4.Barefield D, Sadayappan S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J Mol Cell Cardiol. 2010;48:866–75. doi: 10.1016/j.yjmcc.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig R, Lee KH, Mun JY, Torre I, Luther PK. Structure, sarcomeric organization, and thin filament binding of cardiac myosin-binding protein-C. Pflugers Arch. 2014;466:425–31. doi: 10.1007/s00424-013-1426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett PM, Furst DO, Gautel M. The C-protein (myosin binding protein C) family: regulators of contraction and sarcomere formation? Rev Physiol Biochem Pharmacol. 1999;138:203–34. doi: 10.1007/BFb0119628. [DOI] [PubMed] [Google Scholar]
- 7.Govada L, Carpenter L, da Fonseca PC, Helliwell JR, Rizkallah P, Flashman E, Chayen NE, Redwood C, Squire JM. Crystal structure of the C1 domain of cardiac myosin binding protein-C: implications for hypertrophic cardiomyopathy. J Mol Biol. 2008;378:387–97. doi: 10.1016/j.jmb.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 8.Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? EMBO J. 1995;14:1952–60. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta MK, Robbins J. Post-translational control of cardiac hemodynamics through myosin binding protein C. Pflugers Arch. 2014;466:231–6. doi: 10.1007/s00424-013-1377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW, 2nd, Klevitsky R, Seidman CE, Seidman JG, Robbins J. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circ Res. 2005;97:1156–63. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong CW, Stelzer JE, Greaser ML, Powers PA, Moss RL. Acceleration of crossbridge kinetics by protein kinase A phosphorylation of cardiac myosin binding protein C modulates cardiac function. Circ Res. 2008;103:974–82. doi: 10.1161/CIRCRESAHA.108.177683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stelzer JE, Patel JR, Walker JW, Moss RL. Differential roles of cardiac myosin-binding protein C and cardiac troponin I in the myofibrillar force responses to protein kinase A phosphorylation. Circ Res. 2007;101:503–11. doi: 10.1161/CIRCRESAHA.107.153650. [DOI] [PubMed] [Google Scholar]
- 13.Sequeira V, Witjas-Paalberends ER, Kuster DW, van der Velden J. Cardiac myosin-binding protein C: hypertrophic cardiomyopathy mutations and structure-function relationships. Pflugers Arch. 2014;466:201–6. doi: 10.1007/s00424-013-1400-3. [DOI] [PubMed] [Google Scholar]
- 14.Kuster DW, Bawazeer AC, Zaremba R, Goebel M, Boontje NM, van der Velden J. Cardiac myosin binding protein C phosphorylation in cardiac disease. J Muscle Res Cell Motil. 2012;33:43–52. doi: 10.1007/s10974-011-9280-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pepe FA, Drucker B. The myosin filament. III. C-protein. J Mol Biol. 1975;99:609–17. doi: 10.1016/s0022-2836(75)80175-6. [DOI] [PubMed] [Google Scholar]
- 16.Craig R, Offer G. The location of C-protein in rabbit skeletal muscle. Proc R Soc Lond B Biol Sci. 1976;192:451–61. doi: 10.1098/rspb.1976.0023. [DOI] [PubMed] [Google Scholar]
- 17.Craig R. Structure of A-segments from frog and rabbit skeletal muscle. J Mol Biol. 1977;109:69–81. doi: 10.1016/s0022-2836(77)80046-6. [DOI] [PubMed] [Google Scholar]
- 18.Bennett P, Craig R, Starr R, Offer G. The ultrastructural location of C- protein, X-protein and H-protein in rabbit muscle. J Muscle Res Cell Motil. 1986;7:550–67. doi: 10.1007/BF01753571. [DOI] [PubMed] [Google Scholar]
- 19.Luther PK, Bennett PM, Knupp C, Craig R, Padron R, Harris SP, Patel J, Moss RL. Understanding the organisation and role of myosin binding protein C in normal striated muscle by comparison with MyBP-C knockout cardiac muscle. J Mol Biol. 2008;384:60–72. doi: 10.1016/j.jmb.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okagaki T, Weber FE, Fischman DA, Vaughan KT, Mikawa T, Reinach FC. The major myosin-binding domain of skeletal muscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. J Cell Biol. 1993;123:619–26. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freiburg A, Gautel M. A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur J Biochem. 1996;235:317–23. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert R, Cohen JA, Pardo S, Basu A, Fischman DA. Identification of the A-band localization domain of myosin binding proteins C and H (MyBP-C, MyBP-H) in skeletal muscle. J Cell Sci. 1999;112:69–79. doi: 10.1242/jcs.112.1.69. [DOI] [PubMed] [Google Scholar]
- 23.Ratti J, Rostkova E, Gautel M, Pfuhl M. Structure and interactions of myosin-binding protein C domain C0: cardiac-specific regulation of myosin at its neck? J Biol Chem. 2011;286:12650–8. doi: 10.1074/jbc.M110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhuiyan MS, Gulick J, Osinska H, Gupta M, Robbins J. Determination of the critical residues responsible for cardiac myosin binding protein C's interactions. J Mol Cell Cardiol. 2012;53:838–47. doi: 10.1016/j.yjmcc.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruen M, Gautel M. Mutations in beta-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J Mol Biol. 1999;286:933–49. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 26.Starr R, Offer G. The interaction of C-protein with heavy meromyosin and subfragment-2. Biochem J. 1978;171:813–6. doi: 10.1042/bj1710813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ababou A, Gautel M, Pfuhl M. Dissecting the N-terminal Myosin Binding Site of Human Cardiac Myosin-binding Protein C. Structure and myosin binding of domain C2. Journal of Biological Chemistry. 2007;282:9204–9215. doi: 10.1074/jbc.M610899200. [DOI] [PubMed] [Google Scholar]
- 28.Ababou A, Rostkova E, Mistry S, Le Masurier C, Gautel M, Pfuhl M. Myosin binding protein C positioned to play a key role in regulation of muscle contraction: structure and interactions of domain C1. J Mol Biol. 2008;384:615–30. doi: 10.1016/j.jmb.2008.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruen M, Prinz H, Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Lett. 1999;453:254–9. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- 30.Kunst G, Kress KR, Gruen M, Uttenweiler D, Gautel M, Fink RH. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circ Res. 2000;86:51–8. doi: 10.1161/01.res.86.1.51. [DOI] [PubMed] [Google Scholar]
- 31.Moos C, Mason CM, Besterman JM, Feng IN, Dubin JH. The binding of skeletal muscle C-protein to F-actin, and its relation to the interaction of actin with myosin subfragment-1. J Mol Biol. 1978;124:571–86. doi: 10.1016/0022-2836(78)90172-9. [DOI] [PubMed] [Google Scholar]
- 32.Moos C. Fluorescence microscope study of the binding of added C protein to skeletal muscle myofibrils. J Cell Biol. 1981;90:25–31. doi: 10.1083/jcb.90.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto K. The binding of skeletal muscle C-protein to regulated actin. FEBS Lett. 1986;208:123–7. doi: 10.1016/0014-5793(86)81545-9. [DOI] [PubMed] [Google Scholar]
- 34.Kulikovskaya I, McClellan G, Flavigny J, Carrier L, Winegrad S. Effect of MyBP-C binding to actin on contractility in heart muscle. J Gen Physiol. 2003;122:761–74. doi: 10.1085/jgp.200308941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaffer JF, Kensler RW, Harris SP. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J Biol Chem. 2009;284:12318–27. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weith A, Sadayappan S, Gulick J, Previs MJ, Vanburen P, Robbins J, Warshaw DM. Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. J Mol Cell Cardiol. 2012;52:219–27. doi: 10.1016/j.yjmcc.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Dijk SJ, Bezold KL, Harris SP. Earning stripes: myosin binding protein-C interactions with actin. Pflugers Arch. 2014;466:445–50. doi: 10.1007/s00424-013-1432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mun JY, Previs MJ, Yu HY, Gulick J, Tobacman LS, Beck Previs S, Robbins J, Warshaw DM, Craig R. Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc Natl Acad Sci U S A. 2014;111:2170–5. doi: 10.1073/pnas.1316001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Previs MJ, Beck Previs S, Gulick J, Robbins J, Warshaw DM. Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science. 2012;337:1215–8. doi: 10.1126/science.1223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfuhl M, Gautel M. Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): who does what, with what, and to whom? J Muscle Res Cell Motil. 2012;33:83–94. doi: 10.1007/s10974-012-9291-z. [DOI] [PubMed] [Google Scholar]
- 41.Moolman-Smook J, Flashman E, de Lange W, Li ZL, Corfield V, Redwood C, Watkins H. Identification of novel interactions between domains of myosin binding protein-C that are modulated by hypertrophic cardiomyopathy missense mutations. Circulation Research. 2002;91:704–711. doi: 10.1161/01.res.0000036750.81083.83. [DOI] [PubMed] [Google Scholar]
- 42.Squire JM, Luther PK, Knupp C. Structural evidence for the interaction of C-protein (MyBP-C) with actin and sequence identification of a possible actin-binding domain. J Mol Biol. 2003;331:713–24. doi: 10.1016/s0022-2836(03)00781-2. [DOI] [PubMed] [Google Scholar]
- 43.Witt CC, Gerull B, Davies MJ, Centner T, Linke WA, Thierfelder L. Hypercontractile properties of cardiac muscle fibers in a knock-in mouse model of cardiac myosin-binding protein-C. J Biol Chem. 2001;276:5353–9. doi: 10.1074/jbc.M008691200. [DOI] [PubMed] [Google Scholar]
- 44.Squire JM, Sjostrom M, Luther P. Fine structure of the A-band in cryo-sections II: evidence for a length-determining protein in the thick filament of vertebrate skeletal muscle. Proc. Sixth Eur. Cong. Electron Microsc.; 1976. pp. 91–95. Tal International. [Google Scholar]
- 45.Squire JM, Harford JJ, Edman AC, Sjostrom M. Fine structure of the A- band in cryo-sections. III. Crossbridge distribution and the axial structure of the human C-zone. J Mol Biol. 1982;155:467–94. doi: 10.1016/0022-2836(82)90482-x. [DOI] [PubMed] [Google Scholar]
- 46.Huxley HE, Brown W. The low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J Mol Biol. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- 47.Flashman E, Korkie L, Watkins H, Redwood C, Moolman-Smook JC. Support for a trimeric collar of myosin binding protein C in cardiac and fast skeletal muscle, but not in slow skeletal muscle. FEBS Lett. 2008;582:434–8. doi: 10.1016/j.febslet.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 48.Razumova MV, Shaffer JF, Tu AY, Flint GV, Regnier M, Harris SP. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay: Evidence for long-lived cross-bridges. J Biol Chem. 2006;281:35846–54. doi: 10.1074/jbc.M606949200. [DOI] [PubMed] [Google Scholar]
- 49.Huxley HE. Recent x-ray diffraction and electron microscope studies of striated muscle. J Gen Physiol. 1967;50:71–83. doi: 10.1085/jgp.50.6.71. Suppl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Squire JM, Roessle M, Knupp C. New X-ray diffraction observations on vertebrate muscle: organisation of C-protein (MyBP-C) and troponin and evidence for unknown structures in the vertebrate A-band. J Mol Biol. 2004;343:1345–63. doi: 10.1016/j.jmb.2004.08.084. [DOI] [PubMed] [Google Scholar]
- 51.Luther PK, Craig R. Modulation of striated muscle contraction by binding of myosin binding protein C to actin. Bioarchitecture. 2011;1:277–283. doi: 10.4161/bioa.1.6.19341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luther PK, Winkler H, Taylor K, Zoghbi ME, Craig R, Padron R, Squire JM, Liu J. Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proc Natl Acad Sci U S A. 2011;108:11423–8. doi: 10.1073/pnas.1103216108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Obermann WM, Gautel M, Steiner F, van der Ven PF, Weber K, Fürst DO. The structure of the sarcomeric M band: localization of defined domains of myomesin, M-protein, and the 250-kD carboxy-terminal region of titin by immunoelectron microscopy. J Cell Biol. 1996;134:1441–1453. doi: 10.1083/jcb.134.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ackermann MA, Hu LY, Bowman AL, Bloch RJ, Kontrogianni-Konstantopoulos A. Obscurin interacts with a novel isoform of MyBP-C slow at the periphery of the sarcomeric M-band and regulates thick filament assembly. Mol Biol Cell. 2009;20:2963–78. doi: 10.1091/mbc.E08-12-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin B, Govindan S, Lee K, Zhao P, Han R, Runte KE, Craig R, Palmer BM, Sadayappan S. Cardiac Myosin Binding Protein-C Plays No Regulatory Role in Skeletal Muscle Structure and Function. PLoS ONE. 2013;8:e69671. doi: 10.1371/journal.pone.0069671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, Turnbull DH, Georgakopoulos D, Kass D, Bond M, Niimura H, Schoen FJ, Conner D, Fischman DA, Seidman CE, Seidman JG. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J Clin Invest. 1999;104:1235–44. doi: 10.1172/JCI7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanson J, O'Brien EJ, Bennett PM. Structure of the myosin-containing filament assembly (A-segment) separated from frog skeletal muscle. J Mol Biol. 1971;58:865–71. doi: 10.1016/0022-2836(71)90045-3. [DOI] [PubMed] [Google Scholar]
- 58.Rome E, Offer G, Pepe FA. X-ray diffraction of muscle labelled with antibody to C-protein. Nat New Biol. 1973;244:152–4. doi: 10.1038/newbio244152a0. [DOI] [PubMed] [Google Scholar]
- 59.Martin-Fernandez ML, Bordas J, Diakun G, Harries J, Lowy J, Mant GR, Svensson A, Towns-Andrews E. Time-resolved X-ray diffraction studies of myosin head movements in live frog sartorius muscle during isometric and isotonic contractions. J Muscle Res Cell Motil. 1994;15:319–48. doi: 10.1007/BF00123484. [DOI] [PubMed] [Google Scholar]
- 60.Reconditi M, Brunello E, Fusi L, Linari M, Martinez MF, Lombardi V, Irving M, Piazzesi G. Sarcomere-length dependence of myosin filament structure in skeletal muscle fibres of the frog. J Physiol. 2014;592:1119–1137. doi: 10.1113/jphysiol.2013.267849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trinick J. Titin as a scaffold and spring. Cytoskeleton. Curr Biol. 1996;6:258–60. doi: 10.1016/s0960-9822(02)00472-4. [DOI] [PubMed] [Google Scholar]
- 62.Whiting A, Wardale J, Trinick J. Does titin regulate the length of muscle thick filaments? J Mol Biol. 1989;205:263–268. doi: 10.1016/0022-2836(89)90381-1. [DOI] [PubMed] [Google Scholar]
- 63.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci U S A. 2008;105:2386–90. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Al-Khayat HA, Kensler RW, Squire JM, Marston SB, Morris EP. Atomic model of the human cardiac muscle myosin filament. Proc Natl Acad Sci U S A. 2013;110:318–23. doi: 10.1073/pnas.1212708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ackermann MA, Patel PD, Valenti J, Takagi Y, Homsher E, Sellers JR, Kontrogianni-Konstantopoulos A. Loss of actomyosin regulation in distal arthrogryposis myopathy due to mutant myosin binding protein-C slow. FASEB J. 2013;27:3217–28. doi: 10.1096/fj.13-228882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ackermann MA, Kontrogianni-Konstantopoulos A. Myosin binding protein-C slow: a multifaceted family of proteins with a complex expression profile in fast and slow twitch skeletal muscles. Front Physiol. 2013;4:391. doi: 10.3389/fphys.2013.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Govindan S, McElligott A, Muthusamy S, Nair N, Barefield D, Martin JL, Gongora E, Greis KD, Luther PK, Winegrad S, Henderson KK, Sadayappan S. Cardiac myosin binding protein-C is a potential diagnostic biomarker for myocardial infarction. J Mol Cell Cardiol. 2012;52:154–64. doi: 10.1016/j.yjmcc.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, Powers PA, Moss RL. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 69.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.