Abstract
Among the genes regulated by estrogen receptor (ER) are miRNAs that play a role in breast cancer signaling pathways. To determine whether miRNAs are involved in ER-positive breast cancer progression to hormone independence, we profiled the expression of 800 miRNAs in the estrogen-dependent human breast cancer cell line MCF7 and its estrogen-independent derivative MCF7:2A (MCF7:2A) using NanoString. We found 78 miRNAs differentially expressed between the two cell lines, including a cluster comprising let-7c, miR-99a, and miR-125b, which is encoded in an intron of the long non-coding RNA LINC00478. These miRNAs are ER targets in MCF7 cells, and nearby ER binding and their expression is significantly decreased in MCF7:2A cells.
The expression of these miRNAs was interrogated in patient samples profiled in The Cancer Genome Atlas (TCGA). Among luminal tumors, these miRNAs are expressed at higher levels in luminal A vs. B tumors. While their expression is uniformly low in luminal B tumors, they are lost only in a subset of luminal A patients. Interestingly, this subset with low expression of these miRNAs had worse overall survival compared with luminal A patients with high expression. We confirmed that miR-125b directly targets HER2 and that let-7c also regulates HER2 protein expression. In addition, HER2 protein expression and activity is negatively correlated with let-7c expression in TCGA. In summary, we identified an ER-regulated miRNA cluster that regulates HER2, is lost with progression to estrogen independence, and may serve as a biomarker of poor outcome in ER+ luminal A breast cancer patients.
Keywords: estrogen receptor, miRNA, luminal breast cancer, nuclear receptor, cistrome
INTRODUCTION
The estrogen receptor (ER) is an estrogen-regulated transcription factor that controls the transcription of numerous coding and non-coding RNAs and is a key target for therapy in ER+ breast cancers (1, 2). In breast cancer, ER acts predominantly by binding to distal enhancer sites to mediate transcription (3). Downstream effectors of ER activity in breast cancer include genes with pro-oncogenic functions including survival and growth. It has been known for more than 40 years that a primary determinant of the response of breast cancers to endocrine therapy is the expression of ER, leading to the first stratification of breast cancer into ER+ and ER− subsets. More recently, refined subsets have been identified by gene expression profiles characteristic of clinical subtypes in which ER may play different roles (4–6).
microRNAs (miRNAs) are small non-coding RNAs ~22 bp in length that regulate the expression of genes by targeting the 3′ UTRs of mRNAs. These molecules have been demonstrated to play important roles in normal development and physiology as well as regulating a number of disease processes including breast cancer (7–9). miRNAs have been reported to be generally downregulated in cancers, and their loss leads to the increased expression of targeted genes, notably including oncogenes that lead to cancer progression. In breast cancer, a number of miRNAs have been reported to be abnormally regulated (10–13). ER has also been reported to regulate the expression of a number of miRNAs in response to its ligand estradiol (E2) (14–17).
Here, we report the identification of miRNAs directly regulated by ER and differentially expressed in the estrogen-dependent ER+ breast cancer cell line MCF7 and its hormone-independent derivative MCF7:2A. The let-7c/miR-99a/miR-125b cluster is expressed in MCF7 cells where it is directly targeted by ER and both expression and ER binding are lost in MC7:2A cells. Expression of this miRNA cluster is uniformly low in luminal B breast cancers, which have a worse outcome than luminal A. Within the luminal A subtype, low expression of the cluster predicts for poor patient outcome. We find that two members of the cluster, let-7c and mirR-125b, inhibit HER2 protein expression and increased expression of the HER2 protein in luminal A tumors lacking expression of these miRNA may mediate their poor outcome.
MATERIALS AND METHODS
Cell culture and reagents
MCF7 cells were grown in high-glucose DMEM (Invitrogen) supplemented with 2 mM L-glutamine, 10% (vol/vol) heat-inactivated FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin (Invitrogen) in a humidified incubator at 37°C and 5% CO2. The MCF7:2A, MCF7:5C, and MCF7:LTLT cell lines were grown in phenol red-free high-glucose DMEM (Invitrogen) supplemented with 2 mM L-glutamine, 5% (vol/vol) heat-inactivated FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). The MCF7:LTLT cells were also supplemented with 1 μM letrazole. The MCF7:2A and MCF7:5C cell lines were obtained from V. Craig Jordan and the MCF7:LTLT cell line was obtained from Angela Brodie. The Dharmacon anti-miRs and miRNA mimics were obtained from ThermoFisher (Pittsburgh, PA).
NanoString
A total of 2×106 MCF7 and MCF7:2A cells growing in the exponential phase were seeded in 6-well plates and cultured for 2 days. The cells were then harvested for total RNA using the miRNeasy Kit (Qiagen). A total of 100 ng of total RNA was assayed using the Human nCounter miRNA Assay 2.0 Kit following the manufacturer’s instructions (NanoString). Differences in miRNA expression were analyzed using the NanoSTRIDE software program (18) with default settings. Clustering of the differentially expressed genes and heatmap generation was performed using the GenePattern Server (genepattern.broadinstitute.org). The volcano plot displaying the significance of the miRNA differences was produced using R version 3.0.2.
RT-PCR
For RT-PCR, total RNA was isolated using a combination of TRIzol (Sigma) and the RNeasy Mini Kit (Qiagen). First-strand cDNA, which was created using the Quantitect Reverse Transcription Kit (Qiagen) following the manufacturer’s protocol, was assayed using Taqman miRNA assays (Life Technologies, Inc.), and the level of U6 RNA was used as a control. The expression of LINC00478 was measured using the Power SYBR Green PCR Master Mix (Life Technologies, Inc.) with the following primers: 5′-GATCTGAGAACGCTGTCTGG-3′ (forward) and 5′-AGAGTCTCCCTCCTGCTTCC-3′ (reverse). For the Ago1 experiments, the following primers were used: HER2: 5′-CTGGTGGATGCTGAGGAGTA-3′ (forward) and 5′-TCCAGCCCTAGTGTCAGGTC-3′ (reverse), Myc: 5′-CTGGTGCTCCATGAGGAGA-3′ (forward) and 5′-CTCTGACCTTTTGCCAGGAG-3′ (reverse), p21: 5′-GGAAGACCATGTGGACCTGT-3′ (forward) and 5′-GGCGTTTGGAGTGGTAGAAA-3′ (reverse).
Cell growth assays
To determine the rate of growth in the presence of miRNA mimics, 2.3 × 105 MCF7:2A cells/ml were seeded into 6-well plates. The following day, the cells were transfected with 20 pmol of let-7c, miR-99a, or miR-125b miRIDIAN microRNA Mimics (ThermoFisher) or a negative control using the Lipofectamine RNAiMAX transfection reagent (Life Technologies, Inc.) following the manufacturer’s protocol. The cells were incubated at 37°C under 5% CO2, passaged into 96-well plates the following day (day 0), and allowed to proliferate. Triplicate wells were counted on days 1, 3, and 5 to determine the rate of growth.
Luciferase assays
A total of 3 × 104 HEK 293 cells were seeded into 96-well plates. Twenty-four hours after plating, the cells were transfected with a psiCHECK2 vector encoding the entire 3′ UTR of HER2 fused downstream of the renilla luciferase gene and the firefly luciferase gene as a reporter with Lipofectamine 2000 following the manufacturer’s instructions. After incubation for 48 h, the cells were lysed in 1X Passive Lysis Buffer and assayed with the Dual-Luciferase® Reporter Assay System (Promega) to measure the renilla luciferase activity and that of firefly luciferase, which served as a transfection control.
Ago1 RNA immunoprecipitation
The Ago1 complex was immunoprecipitated as described in (19). Briefly, A total of 2 × 106 MCF7 and MCF7:2A cells in the growth phase were seeded in 10 cm plates. After 24 h, the cells were harvested in 400 μl lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES, pH 7.0, 0.5% Nonidet P-40) supplemented with 100 U/ml RNase Out (Invitrogen Cat# 10777-019) and Complete Protease Inhibitor Cocktail (Roche). The lysates were centrifuged, and 50 μl was set aside for input.
A total of 2 μg anti-Ago1 antibody (Abcam #ab5070) was prebound to protein A Dynabeads (Life Technologies). The antibody and lysate mixture was incubated overnight at 4°C. The next morning, the beads were collected by magnetic separation, and they were treated with DNaseI in NT2 buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM MgCl2, and 0.05% Nonidet P-40) for 10 min at 37°C. The beads were then washed twice with NT2 buffer, treated with proteinase K to digest protein, and resuspended in 300 μl acid-phenol:chloroform (Ambion). The solution was centrifuged for 1 min at 14,000 rpm at RT, the upper layer was collected, and the RNA was ethanol precipitated in the presence of GlycoBlue (Life Technologies, Inc.). The obtained RNA was resuspended in 30 ml water and used to generate cDNA and subsequent RT-PCR analysis.
Transfection and Immunoblotting
MCF7 and MCF7:2A cells were transfected with 20 pmol of miRIDIAN microRNA anti-miRs or miRNA mimics as described above. Cells were incubated for five days, and whole-cell extracts were then harvested in RIPA buffer (Tris-Buffered Saline, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.004% sodium azide). Protein lysates were quantified using the BCA Protein Assay Kit (Pierce), and they were then separated in 4–12% NuPAGE Bis-Tris SDS/PAGE Protein Gels (Life Technologies) followed by transfer onto a PVDF membrane. The membrane was blotted with anti-HER2 (2165; Cell Signaling Technologies) and β-actin (4967; Cell Signaling Technologies) antibodies followed by incubation with a secondary donkey anti-rabbit antibody (Pierce). The blots were developed using the Western Blotting Luminol Reagent (Santa Cruz).
Patient sample analysis
For patient sample analysis, data were extracted from the Breast Invasive Carcinoma provisional dataset in The Cancer Genome Atlas (TCGA) using the cBioPortal for Cancer Genomics CGDS-R version 1.1.19 package in R version 3.0.2. Kaplan-Meir analysis was performed using the Survival package version 2.37-7, and significance was determined using the log-rank test.
RESULTS
miRNAs are differentially expressed in MCF7:2A vs. MCF7 cells
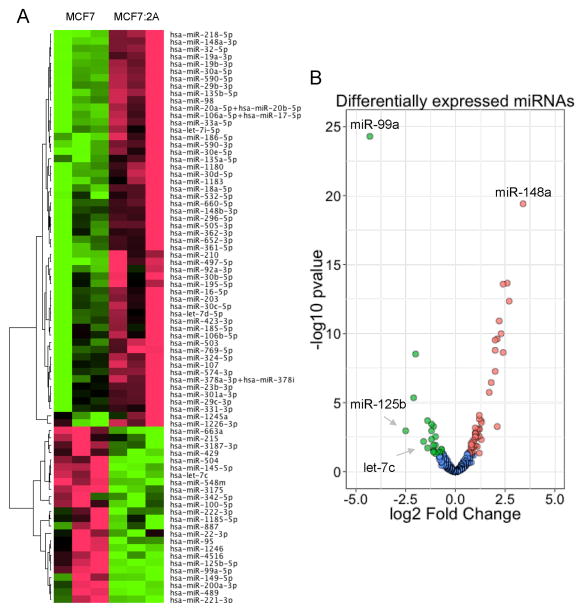
To identify candidate miRNAs that may play a role in endocrine resistance, we compared miRNA expression between estrogen-dependent MCF7 cells and the estrogen-independent derivative cell line MCF7:2A using the nCounter NanoString platform. Using RNA derived from MCF7 and MCF7:2A cells under standard culturing conditions, we found that a number of miRNAs are differentially expressed (Figure 1A). Of the 800 miRNAs assayed by this method, 78 (9.8%) had significant differential expression (p < 0.05, 1.5 fold) in the two cell lines including 54 that were downregulated and 24 that were upregulated in MCF7:2A cells as compared with MCF7 cells (Table 1). Of these miRNAs, 57 are located within annotated sequences including coding and noncoding RNAs, and 21 are intergenic (Table 1). The top upregulated miRNA was miR-148a (fold change: 10.6, p-value: 3.9 × 10−20), and the top downregulated miRNA was miR-99a (fold change: −19.7, p-value: 5.1 × 10−25; Figure 1B). We found that the miR-17-92a cluster, previously been shown to be regulated by ER was upregulated, and that miR-221/-222, which was previously shown to regulate ER expression, was downregulated (16, 20) in MCF7:2A vs. MCF7 cells. In addition, the clusters miR-497/miR-195, miR-590-3p/miR-590-5p, and miR-30e/miR-30e were significantly upregulated in MCF7:2A cells, while the let-7c/miR-99a/miR-125b cluster was downregulated (Supplementary Figure 1).
Figure 1. Differentially expressed miRNAs in MCF7:2A vs. MCF7 cells.
MCF7 and MCF7:2A cells were grown under standard culturing conditions, and small RNAs were extracted from each cell line. Each sample was then assayed for the expression of miRNA using nCounter NanoString assays. A) Heatmap demonstrating the differentially expressed miRNAs found in the MCF7:2A and MCF7 cells including 54 upregulated and 24 downregulated miRNAs. B) Volcano plot demonstrating the profile of the differentially expressed miRNAs in MCF7:2A vs. MCF7 cells. This plot demonstrates the fold change (x-axis) and significance level expressed as the −log10 p-value (y-axis). The green circles represent the miRNAs downregulated in the MCF7:2A compared with MCF7 cells, and the red circles represent the miRNAs upregulated in the MCF7:2A compared with MCF7 cells. The blue circles indicate miRNAs that were not significantly expressed. Significance was determined with a p-value cutoff of 0.05 and a 1.5 fold change.
Table 1.
miRNAs differentially regulated in MCF7:2A vs. MCF7 cells.
| miRNA | Fold Change | pvalue | Location | ER binding site |
|---|---|---|---|---|
| hsa-miR-148a-3p | 10.6 | 3.9E-20 | Intergenic | No |
| hsa-miR-20a-5p/hsa-miR-20b-5p | 6.5 | 4.4E-13 | MIR17HG | No |
| hsa-miR-218-5p | 6.1 | 2.2E-14 | SLIT3 | Yes |
| hsa-miR-19a-3p | 5.3 | 2.3E-09 | MIR17HG | No |
| hsa-miR-19b-3p | 5.3 | 2.6E-14 | MIR17HG | No |
| hsa-miR-106a-5p/hsa-miR-17-5p | 4.9 | 1.0E-10 | Intergenic | No |
| hsa-miR-32-5p | 4.6 | 1.2E-11 | TMEM245 | Yes |
| hsa-miR-590-5p | 4.3 | 2.4E-10 | EIF4H | Yes |
| hsa-miR-92a-3p | 4.3 | 5.4E-04 | MIR17HG | Yes |
| hsa-miR-30a-5p | 4.0 | 1.6E-09 | Intergenic | No |
| hsa-miR-135b-5p | 4.0 | 5.4E-08 | Intergenic | Yes |
| hsa-miR-29b-3p | 4.0 | 2.9E-10 | Intergenic | Yes |
| hsa-miR-210 | 3.5 | 3.5E-07 | MIR210HG | Yes |
| hsa-miR-18a-5p | 3.2 | 1.8E-06 | MIR17HG | No |
| hsa-miR-30b-5p | 2.5 | 4.7E-03 | Intergenic | Yes |
| hsa-miR-660-5p | 2.5 | 2.1E-04 | CLCN5 | No |
| hsa-miR-33a-5p | 2.5 | 2.8E-04 | SREBF2 | No |
| hsa-miR-590-3p | 2.3 | 7.7E-04 | EIF4H | Yes |
| hsa-miR-1180 | 2.3 | 1.5E-04 | B9D1 | Yes |
| hsa-miR-98 | 2.3 | 8.2E-05 | HUWE1 | Yes |
| hsa-miR-296-5p | 2.3 | 2.7E-03 | Intergenic | No |
| hsa-miR-1245a | 2.3 | 4.5E-02 | COL3A1 | No |
| hsa-miR-186-5p | 2.3 | 8.6E-04 | ZRANB2 | No |
| hsa-miR-497-5p | 2.1 | 3.1E-03 | MIR497HG | No |
| hsa-miR-16-5p | 2.1 | 1.4E-03 | DLEU2 | Yes |
| hsa-miR-135a-5p | 2.1 | 1.5E-02 | GLYCTK | Yes |
| hsa-miR-203 | 2.1 | 8.8E-04 | Intergenic | Yes |
| hsa-let-7i-5p | 2.0 | 6.7E-04 | Intergenic | Yes |
| hsa-miR-30d-5p | 2.0 | 1.7E-03 | Intergenic | Yes |
| hsa-miR-30e-5p | 2.0 | 1.5E-03 | NFYC | Yes |
| hsa-miR-652-3p | 2.0 | 4.2E-03 | TMEM164 | Yes |
| hsa-let-7d-5p | 2.0 | 1.8E-03 | MIRLET7DHG | No |
| hsa-miR-324-5p | 2.0 | 2.9E-03 | ACADVL | Yes |
| hsa-miR-30c-5p | 2.0 | 3.0E-03 | NFYC | Yes |
| hsa-miR-505-3p | 2.0 | 1.3E-02 | Intergenic | No |
| hsa-miR-23b-3p | 1.9 | 3.9E-02 | C9ORF3 | Yes |
| hsa-miR-423-3p | 1.9 | 5.3E-03 | NSRP1 | No |
| hsa-miR-503 | 1.9 | 7.2E-03 | MGC16121 | Yes |
| hsa-miR-362-3p | 1.9 | 1.7E-02 | CLCN5 | No |
| hsa-miR-107 | 1.9 | 1.3E-02 | PANK1 | No |
| hsa-miR-195-5p | 1.7 | 1.7E-02 | MIR497HG | No |
| hsa-miR-1183 | 1.7 | 1.3E-02 | SP4 | No |
| hsa-miR-361-5p | 1.7 | 8.4E-03 | CHM | No |
| hsa-miR-574-3p | 1.7 | 1.9E-02 | FAM114A1 | Yes |
| hsa-miR-148b-3p | 1.7 | 1.1E-02 | COPZ1 | Yes |
| hsa-miR-1226-3p | 1.7 | 5.0E-02 | DHX30 | Yes |
| hsa-miR-301a-3p | 1.7 | 4.0E-02 | SKA2 | No |
| hsa-miR-362-3p | 1.6 | 4.5E-02 | CLCN5 | No |
| hsa-miR-769-5p | 1.6 | 3.9E-02 | Intergenic | No |
| hsa-miR-378a-3p/hsa-miR-378i | 1.6 | 3.9E-02 | PPARGC1B | Yes |
| hsa-miR-185-5p | 1.6 | 4.3E-02 | TANGO2 | Yes |
| hsa-miR-106b-5p | 1.6 | 4.7E-02 | MCM7 | Yes |
| hsa-miR-331-3p | 1.6 | 3.7E-02 | Intergenic | No |
| hsa-miR-29c-3p | 1.6 | 4.7E-02 | Intergenic | Yes |
| hsa-miR-22-3p | −1.6 | 2.4E-02 | MIR22HG | Yes |
| hsa-miR-222-3p | −1.6 | 3.8E-02 | Intergenic | No |
| hsa-miR-95 | −1.6 | 2.4E-02 | ABLIM2 | Yes |
| hsa-miR-145-5p | −1.7 | 4.2E-02 | MIR143HG | Yes |
| hsa-miR-663a | −1.7 | 3.8E-02 | LOC284801 | No |
| hsa-miR-215 | −1.9 | 3.7E-02 | IARS2 | No |
| hsa-miR-4516 | −2.0 | 1.2E-02 | PKD1 | Yes |
| hsa-miR-504 | −2.0 | 2.9E-03 | FGF13 | No |
| hsa-miR-887 | −2.0 | 4.9E-02 | FBXL7 | No |
| hsa-miR-3187-3p | −2.1 | 2.8E-02 | LPPR3 | Yes |
| hsa-miR-1185-5p | −2.1 | 3.4E-02 | Intergenic | No |
| hsa-miR-342-5p | −2.1 | 3.8E-02 | EVL | Yes |
| hsa-let-7c | −2.1 | 5.2E-04 | LINC00478 | Yes |
| hsa-miR-548m | −2.1 | 1.2E-02 | Intergenic | No |
| hsa-miR-3175 | −2.3 | 4.5E-03 | CHD2 | Yes |
| hsa-miR-200a-3p | −2.3 | 1.1E-03 | Intergenic | Yes |
| hsa-miR-100-5p | −2.3 | 3.6E-04 | MIG100HG | No |
| hsa-miR-149-5p | −2.6 | 1.9E-02 | GPC1 | Yes |
| hsa-miR-429 | −2.6 | 2.0E-04 | Intergenic | Yes |
| hsa-miR-221-3p | −3.0 | 6.5E-03 | Intergenic | No |
| hsa-miR-125b-5p | −4.0 | 3.0E-09 | LINC00478 | Yes |
| hsa-miR-1246 | −4.3 | 4.4E-06 | Intergenic | No |
| hsa-miR-489 | −5.7 | 1.1E-03 | CALCR | Yes |
| hsa-miR-99a-5p | −19.7 | 5.1E-25 | LINC00478 | Yes |
Because the ER is responsible for the transcriptional regulation of genomic targets in MCF7 and MCF7:2A cells (3, 21), we next sought to determine which of the differentially expressed miRNAs are direct ER targets. ER binding sites are located within 30 kb for 965 of the 1,595 miRNAs annotated in miRBase (version 19), including 631 miRNAs contained within the introns of coding or noncoding RNAs and 334 in intergenic regions. Of the miRNAs with an ER binding site within 30 kb of their start sites, 47 were differentially expressed in MCF7 vs. MCF7:2A. When we examined the ER binding sites located near miRNAs with decreased expression in MCF7:2A, we found that binding at these sites is also lost despite significant ER binding at other sites within these cells (Supplementary Figure 2).
The miR-7c locus is downregulated in MCF7:2A cells
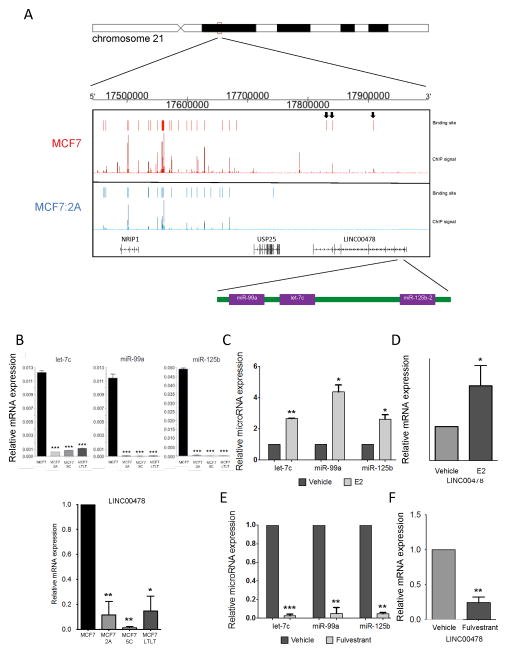
The most significantly underexpressed miRNA in MCF7:2A cells compared with parental MCF7 cells is miR-99a. This miRNA is encoded in the intronic sequence of the long non-coding RNA (lncRNA) LINC00478 together with let-7c and miR-125b (Figure 2A), which are also downregulated in MCF7:2A cells (Figure 1). Examination of ER binding near this miRNA cluster demonstrates that there is a loss of ER binding activity at this locus in MCF7:2A vs. MCF7 cells (Figure 2A). Interestingly, ER binding at the nearby NRIP1 gene is not lost.
Figure 2. The let-7c/miR-99a/miR-125b cluster is regulated by the ER.
A) The top panel represents a schematic of the genomic location of the let-7c/miR-99a/miR-125b cluster within chromosome 21. The ER ChIP-Seq signal derived from both MCF7 (shown in red) and MCF7:2A (shown in blue) cells is shown demonstrating a loss of ER signal at the loci near the let-7c/miR-99a/miR-125b cluster. The ER binding sites within LINC00478 lost in MCF7:2A cells are indicated with arrows. B) The relative expression level of let-7c, miR-99a, and miR-125b (top) and LINC00478 (bottom) is shown in the MCF7, MCF7:2A, MCF7:5C, and MCF7:LTLT cell lines. C) and D) E2 regulates the expression of the cluster miRNAs and primary transcript. MCF7 cells were treated with E2 for 3 h, and the level of let-7c, miR-99a, miR-125b, and LINC00478 expression was determined by RT-PCR. E) and F) Fulvestrant treatment leads to loss of the cluster miRNAs and LINC00478. MCF7 cells were treated with fulvestrant for 48 h, and the level of let-7c, miR-99a, miR-125b, and LINC00478 expression was determined by RT-PCR. *, p < 0.01; **, p < 0.01, ***; p < 0.0001.
All three miRNAs in this cluster are also downregulated in two additional estrogen-independent derivatives of MCF7 cells, MCF7:5C and MCF7:LTLT (Figure 2B) (22, 23). The downregulation of these miRNAs parallels the expression of their primary transcript LINC00478 in MCF7 vs. MCF7:2A, MCF7:5C, and MCF7:LTLT cells (Figure 2B, bottom panel). To determine whether these miRNAs and primary transcript are estrogen regulated, we measured their expression in response to E2. Treatment of MCF7 cells with E2 for 3 h demonstrated an increased in let-7c, miR-99a, miR-125b, and LINC00478 (Figure 2C, D). Conversely, treatment with the ER antagonist fulvestrant led to a decrease in the level of LINC00478 (Figure 2F) and the cluster miRNAs (Figure 2E), suggesting that ER regulates this lncRNA together with the miRNA cluster.
The let-7c/miR-99a/miR-125b cluster is underexpressed in luminal B breast cancers and subset of luminal A tumors that demonstrate poor outcome
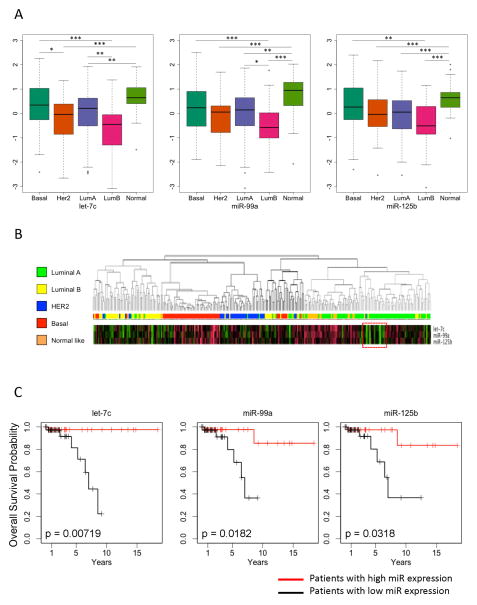
We next sought to determine whether the let-7c/miR-99a/miR-125b cluster is clinically relevant. We first examined the expression of these miRNAs in patient samples derived from The Cancer Genome Atlas (TCGA) for which mRNA and miRNA expression profiling was performed (285 cases). The let-7c, miR-99a, and miR-125b expression levels were highly correlated in the patient samples (r = 0.84 for let-7c/miR-99a; r = 0.73 for let-7c/miR-125b; r = 0.71 for miR-99a/miR-125b; Supplementary Figure 3). We next segregated the patient samples into clinical subgroups based on PAM50 classification (24) and then examined the expression level of let-7c, miR-99a, and miR-125b in the different clinical subgroups. The expression of all three miRNAs was highest in normal-like tumors and lowest in luminal B cancers (Figure 3A). In the luminal A and luminal B subsets, which comprise most of the ER+ breast cancers, we found a significant decrease in the let-7c and miR-99a expression level in luminal B compared with luminal A tumors (p < 0.001 and p < 0.01, respectively) and a trend toward reduced miR-125b expression in these same subsets. Interestingly, within the luminal A subset, we observed a significant fraction with low levels of the expression of these miRNAs (Figure 3B).
Figure 3. The expression of miR-99a, miR-125b, and let-7c is lowest in patients with luminal B breast cancer and predicts outcome in luminal A breast cancer.
A) Patients with breast cancer from TCGA who were profiled for their mRNA and miRNA expression were analyzed for the expression of let-7c, miR-99a, miR-125b in the different PAM50 clinical subsets. All three miRNAs are expressed at the lowest levels in patients with luminal B breast cancer. *, p < 0.01; **, p < 0.001; p < 0.0001. B) The TCGA patients from A were clustered via hierarchical clustering, and the expression of let-7c, miR-99a, miR-125b is shown for each of the patient subsets. The dotted red box demonstrates the subset of luminal A patients with lower expression of the let-7c/miR-99a/miR-125b cluster C) Kaplan-Meier plot demonstrating the overall survival probability for patients with luminal A breast cancer based on the expression of let-7c, miR-99a, and miR-125b.
We next sought to determine whether the let-7c/miR-99a/miR-125b cluster was correlated with the clinical outcome of each of the different subsets. While no correlation was found between the expression of these miRNAs and outcome in the basal, Her2, luminal B, and normal-like subsets (Supplementary Figure 4), there was significant correlation between the expression of the let-7c/miR-99a/miR-125b cluster and overall survival in the luminal A subset (Figure 3C). Patients in the luminal A subset who express higher levels of these miRNAs have significantly better survival than those expressing lower levels of miR-99a, let-7c, and miR-125b (Figure 3C). Furthermore, the low-expressing luminal A subset has a similar outcome as luminal B patients (Supplementary Figure 5). Because low expression of this cluster in patients with luminal A breast cancer indicates poor outcome and the luminal B subset is characterized by the low expression of this cluster and poor outcome (24), these data suggest that low let-7c/miR-99a/miR-125b expression is predictive of poor outcome for ER+ patients.
let-7c, miR-99a, and miR-125b inhibit MCF7:2A cell growth and target HER2
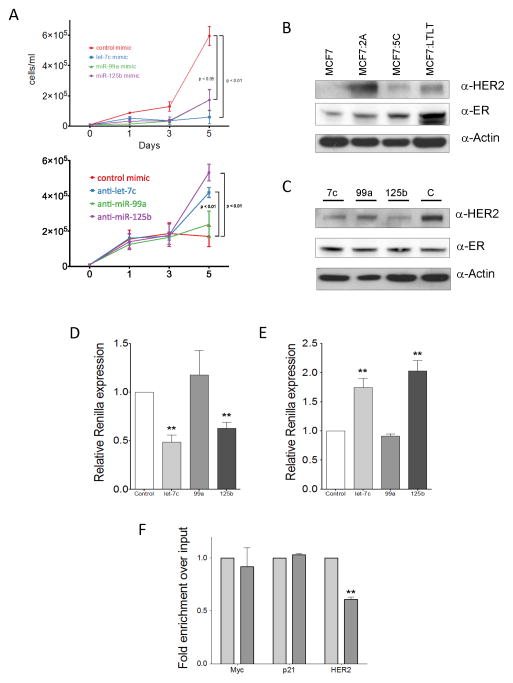
We next sought to determine whether the let-7c/miR-99a/miR-125b cluster has an effect on cell growth. MCF7:2A cells were transfected with each of the individual miRNAs, and the number of cells was counted every other day for five days. While there was little to no difference in the growth rate of MCF7:2A cells transfected with a miRNA mimic control compared with untransfected cells, there was a significant decrease in the growth rate of cells transfected with miRNA mimics for let-7c, miR-99a and miR-125b (Figure 4A, top panel). In addition, when we transfected MCF7 cells with anti-miRs targeting each of the miRNAs we found that anti-miRs directed against let-7c and miR-125b significantly increased the growth of MCF7 cells, while the growth effects of anti-miR-99a were not insignificant. Together, these data suggest that loss of the let-7c/miR-99a/miR-125b cluster in MCF7:2A cells provides a growth advantage by permitting the expression of downstream miRNA targets.
Figure 4. The let-7c/miR-99a/miR-125b cluster regulates the growth of breast cancer cells and downregulates HER2.
A) MCF7:2A cells were transfected with the indicated miRNA mimics (top) or anti-miRs (bottom, split into a 96-well plate and allowed to grow for five days. Cells were counted on days one, three, and five to determine the growth rate. B) HER2 is expressed at a higher level in estrogen-independent cell lines. Western blot demonstrating HER2 expression in the MCF7, MCF7:2A, MCF7:5C, and MCF7:LTLT cell lines. The expression level of ER is also shown together with that of β-actin, which served as a loading control. C) HER2 is downregulated by let-7c and miR-125b overexpression. MCF7:2A cells were transfected with miRNA mimics for let-7c, miR-99a, and miR-125b. Cells were harvested after five days, and the level of HER2 expression was measured. The middle panel shows the expression of ER, which was unchanged with miRNA treatment. D) let-7c and miR-125b target HER2. A vector encoding the 3′-UTR of HER2 was transfected in HEK293 in the presence of miRNA mimics (D) and anti-miRs (E). The level of renilla luciferase expression was measured after 48 days and normalized to that of firefly luciferase. F) HER2 mRNA association with the Ago1 complex is lost in MCF7:2A cells. The Ago1 complex was immunoprecipiated from MCF7 and MCF7:2A cells, and the associated level of HER2 in each cell line as normalized to input total RNA was quantified by RT-PCR. The levels of associated Myc, p21, and HER2 mRNA are shown.
We next sought to identify targets that may be responsible for the growth of these cells. A previous study reported that miR-125b targets HER2 in an in vitro system (25). HER2 has also been shown to be responsible for the growth and activity of MCF7 cells that have been selected for estrogen-independent growth (22, 26) and is expressed at a higher level in MCF7:2A, MCF7:5C, and MCF7:LTLT cells compared with MCF7 cells. This expression pattern is in contrast with the level of ER protein expression, which is similar in the MCF7, MCF7:2A, and MCF7:5C cells and elevated in the MCF7:LTLT cells (Figure 4B). To determine whether HER2 protein expression is under miRNA control, we transfected MCF7:2A cells with miRNA mimics and measured the HER2 protein expression level in these cells after a period of five days. As expected, the miR-125 mimic led to a decrease in HER2 protein expression as measured by western blot (Figure 4C), whereas the miR-99a mimic had little to no effect; however, let-7c also led to a decrease in HER2 protein expression (Figure 4C). In addition, we found a significant decrease in the level of HER2 mRNA expression with let-7c overexpression (Supplementary Figure 6). In contrast, no difference in HER2 mRNA level was found for miR-125b overexpression as changes in mRNA level need not correlate with miRNA-mediated changes in protein expression. To further confirm that the HER2 protein is targeted by these miRNAs, we cloned the 3′-UTR of HER2, the gene that encodes the HER2 protein, downstream of renilla luciferase and determined changes in the level of luciferase activity in the presence of the mimics and anti-miRs of this miRNA cluster. Co-transfection of the HER2-UTR luciferase plasmid with let-7c led to a decrease in reporter expression that was similar to that for miR-125b. In contrast, transfection with the mimic for miR-99a had no effect (Figure 4D). In addition, co-transfection of the HER2 3′-UTR luciferase reporter with anti-miRs confirmed that let-7c and miR-125b act through the HER2 3′-UTR (Figure 4E). These data suggest that let-7c and miR-125b regulate HER2 at the protein level. In contrast to miR-125b, which has been previously demonstrated to directly target the HER2 3′-UTR, let-7c is not predicted to target the HER2 3′-UTR. Thus, we attempted to determine the sequences targeted by let-7c in the HER2 3′-UTR by examining sites predicted by the Probability of Interaction by Target Accessibility (PITA) algorithm, which takes into account the free energy of base pair binding for potential sites (27)(Supplementary Figure 7A). However, mutation of these sites could not block the let-7c mediated reduction in luciferase activity, suggesting that the effects on the HER2 3′-UTR mediated by let-7c may be indirect (Supplementary Figure 7B). In examining targets previously reported to be regulated by let-7c that could mediate the effects of let-7c on HER2 expression, we found that there is strong downregulation of Dicer mediated by let-7c overexpression (Supplementary Figure 7C). This observation suggests that the mechanism involved in upregulated HER2 protein expression in patients in response to let-7c overexpression includes a reduction in Dicer protein.
To further confirm that the HER2 gene is regulated by miRNAs in MCF7 cells, we examined its association with the Ago1 complex, which plays a role in translational silencing mediated by miRNA. We performed immunoprecipitation of the Ago1 complex in MCF7 and MCF7:2A cells and measured the level of associated HER2 mRNA (Figure 4F). In contrast to the levels of the Myc or p21 mRNA in the Ago1 complex which are equivalent in MCF7 and MCF7:2A cells, the level of HER2 mRNA associated with the Ago1 complex is significantly reduced in MCF7:2A cells compared with MCF7 cells. These data support the conclusion that there is less miRNA-mediated regulation of HER2 expression in MCF7:2A cells compared with MCF7 cells, leading to greater HER2 protein expression in these cells.
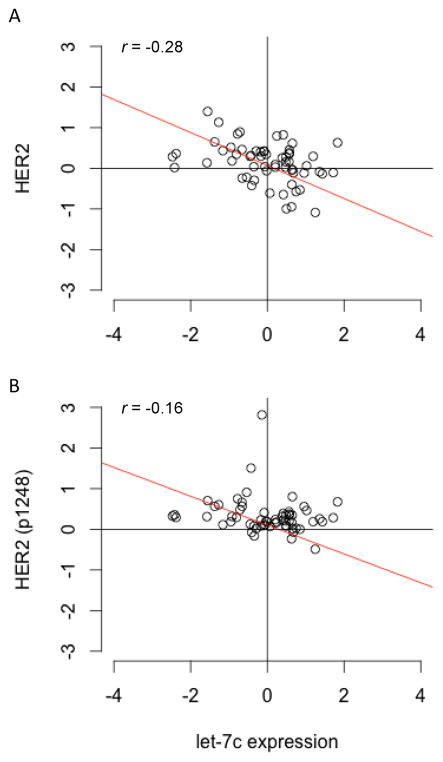
HER2 protein expression and activity is negatively correlated with let-7c expression
In order to validate our cell model findings in actual patient samples, we examined whether there is a correlation between HER2 protein expression and activity and the expression of let-7c and miR-125b miRNAs in patient samples using HER2 protein expression and phosphorylation data obtained from the TCGA cohort (Figure 5 and Supplementary Figure 7). We found that let-7c levels are significantly negatively correlated with HER2 protein expression (Figure 5A; r = −0.28) in the luminal A subset of patients. In addition, there was a similar negative correlation with the expression of the Tyr1248 phosphorylated form of HER2 (Figure 5B; r= −0.16), suggesting that HER2 expression and activity are negatively regulated by the miRNA let-7c. In contrast, no significant correlation was found between miR-125b and HER2 protein expression or activity (Supplementary Figure 7). These data suggest that let-7c may be an important determinant of HER2 protein expression and pathway activation in ER+ breast cells.
Figure 5. HER2 protein expression and activity is negatively correlated with let-7c expression.
Luminal A breast cancer patient samples from TCGA for which protein expression data were generated were examined for their HER2 (A) and phosphorylated HER2 (B) expression levels. A negative correlation was found for both HER2 (A) and phosphorylated HER2 (B) protein expression, suggesting that HER2 expression and activity is negatively associated with let-7c miRNA expression in patients with breast cancer.
DISCUSSION
Understanding the factors underlying the acquisition of endocrine resistance in ER+ breast cancers not only allows for the prediction of outcome but more importantly may identify novel therapeutic strategies to overcome resistance. Expression profiling of mRNA genes has provided important insights into both breast cancer subtypes and increased precision in predicting which patients may benefit from endocrine therapy (4, 28). More recently, miRNA expression levels have been explored both for predictive biomarker development and therapeutic target identification. Expression of miRNAs has been reported to be generally decreased during cancer progression (9). By examining the miRNA expression profile of cell lines modeling estrogen-dependent and estrogen-independent ER+ cancers, we found that expression of the let-7c/miR-99a/miR-125b cluster is decreased during the progression to endocrine resistance. In data derived from a large cohort of primary breast cancers, this miRNA cluster was found to be uniformly reduced in luminal B tumors, a subset characterized by its aggressiveness, lower ER expression and poorer survival in comparison with luminal A cancers (6, 29, 30). More significantly, luminal A tumors, which generally have more favorable outcome and a better response to endocrine therapy (31, 32), could be subdivided based on the expression of this miRNA cluster. High cluster expression led to characteristically favorable outcome, whereas low cluster expression reflected patients with poor outcome.
Patient outcome could be directly related to the proteins targeted by the differentially expressed miRNAs; thus, we examined the expression of HER2, which was previously described as a miR-125b target. Surprisingly, we found that let-7c also regulates HER2 expression. We found a negative correlation between let-7c miRNA expression and the expression of HER2 protein and phosphorylated HER2 in TCGA patient samples, but no correlation was found for miR-125b. These data suggest that let-7c may be the most clinically relevant miRNA within the let-7c/miR-99a/miR-125b cluster. HER2 expression has been correlated with the expression of lin28 and its homolog lin28b (33). These proteins bind the stem loop of let-7 family member precursors to directly inhibit the Drosha- and Dicer-mediated processing of their primary-miRNA precursors into mature let-7 miRNAs (34–38). Moreover, Lin28 expression determines the expression of the let-7 family in tumors and cell lines (33, 39).
Previous studies have shown that the let-7 family controls the cell cycle, is associated with increased proliferation, and blocks tumorigenicity (40–42). Moreover, Lin28 is transcriptionally regulated by Myc, which is an ER-regulated gene that is upregulated with progression to hormone independence (43, 44). This protein is also targeted by let-7, suggesting a regulatory loop involving Lin28, let-7, and Myc (45–47). As we found that let-7c could also target HER2, our data suggest that let-7 family members may be directly involved in the regulation of HER2 in Lin28-negative breast tumors.
Because many mRNAs are predicted to be targeted by the let-7c/miR-99a/miR-125b cluster, other targets of these miRNAs may also be significantly regulated in breast cancer. The mTOR protein, which is a downstream effector of the PI3K pathway (48), has been reported to be regulated by miR-99a (49); thus, it would be interesting to determine whether this miR-99a targets the expression of mTOR, which has also been reported to play a role in endocrine resistance (50–52). In addition, all three miRNAs are predicted to target insulin-like growth factor 1 receptor (IGF1R), which is a growth factor receptor that, like HER2, has been reported to be upregulated in estrogen-deprived breast cancer cells and is thought to be responsible for breast cancer cell signaling pathways. Thus, loss of expression of this miRNA cluster may play a role in the acquisition of endocrine resistance through the upregulation of multiple growth factor signaling pathways.
In summary, we have identified a number of miRNAs differentially expressed in estrogen-dependent vs. estrogen-independent cells and have demonstrated that the let-7c/miR-99a/miR-125b cluster is group of miRNAs that regulate HER2 protein expression and when lost may lead to worse outcome for patients with luminal A tumors.
Supplementary Material
Acknowledgments
Financial support: This study was supported by grants from Susan G. Komen for the Cure (to MB), the NCI (P01 CA080111 to MB) and NIDDK (R01 DK074967 to MB).
The authors would like thank Dr. Dipanjan Chowdhury and his laboratory for helpful discussions for the miRNA experiments. We would also like to thank Drs. V. Craig Jordan and Angela Brodie for providing the estrogen-independent MCF7 cell lines. This study was supported by grants from Susan G. Komen for the Cure (to MB), the NCI (P01 CA080111 to MB) and NIDDK (R01 DK074967 to MB).
Footnotes
Potential conflicts of interest: The authors declare no potential conflicts of interest.
References
- 1.Charpentier AH, Bednarek AK, Daniel RL, Hawkins KA, Laflin KJ, Gaddis S, et al. Effects of estrogen on global gene expression: identification of novel targets of estrogen action. Cancer research. 2000;60:5977–83. [PubMed] [Google Scholar]
- 2.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–74. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 3.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, et al. Genome-wide analysis of estrogen receptor binding sites. Nature genetics. 2006;38:1289–97. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 4.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell cycle. 2008;7:2485–92. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- 8.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. The Journal of pathology. 2010;220:126–39. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Mulrane L, McGee SF, Gallagher WM, O’Connor DP. miRNA dysregulation in breast cancer. Cancer research. 2013;73:6554–62. doi: 10.1158/0008-5472.CAN-13-1841. [DOI] [PubMed] [Google Scholar]
- 11.Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–82. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 12.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer research. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 13.O’Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast cancer research: BCR. 2010;12:201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS, et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic acids research. 2009;37:4850–61. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic acids research. 2009;37:2584–95. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15732–7. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klinge CM. Estrogen Regulation of MicroRNA Expression. Current genomics. 2009;10:169–83. doi: 10.2174/138920209788185289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brumbaugh CD, Kim HJ, Giovacchini M, Pourmand N. NanoStriDE: normalization and differential expression analysis of NanoString nCounter data. BMC bioinformatics. 2011;12:479. doi: 10.1186/1471-2105-12-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Molecular cell. 2011;41:210–20. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. The Journal of biological chemistry. 2008;283:31079–86. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Magnani L, Stoeck A, Zhang X, Lanczky A, Mirabella AC, Wang TL, et al. Genome-wide reprogramming of the chromatin landscape underlies endocrine therapy resistance in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E1490–9. doi: 10.1073/pnas.1219992110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabnis G, Schayowitz A, Goloubeva O, Macedo L, Brodie A. Trastuzumab reverses letrozole resistance and amplifies the sensitivity of breast cancer cells to estrogen. Cancer research. 2009;69:1416–28. doi: 10.1158/0008-5472.CAN-08-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pink JJ, Jiang SY, Fritsch M, Jordan VC. An estrogen-independent MCF-7 breast cancer cell line which contains a novel 80-kilodalton estrogen receptor-related protein. Cancer research. 1995;55:2583–90. [PubMed] [Google Scholar]
- 24.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. The Journal of biological chemistry. 2007;282:1479–86. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 26.Knowlden JM, Hutcheson IR, Jones HE, Madden T, Gee JM, Harper ME, et al. Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology. 2003;144:1032–44. doi: 10.1210/en.2002-220620. [DOI] [PubMed] [Google Scholar]
- 27.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nature genetics. 2007;39:1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 28.Sorlie T. Molecular portraits of breast cancer: tumour subtypes as distinct disease entities. European journal of cancer. 2004;40:2667–75. doi: 10.1016/j.ejca.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 29.Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25:1239–46. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 30.Creighton CJ. The molecular profile of luminal B breast cancer. Biologics: targets & therapy. 2012;6:289–97. doi: 10.2147/BTT.S29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:5678–85. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 32.Ignatiadis M, Sotiriou C. Luminal breast cancer: from biology to treatment. Nature reviews Clinical oncology. 2013;10:494–506. doi: 10.1038/nrclinonc.2013.124. [DOI] [PubMed] [Google Scholar]
- 33.Sakurai M, Miki Y, Masuda M, Hata S, Shibahara Y, Hirakawa H, et al. LIN28: a regulator of tumor-suppressing activity of let-7 microRNA in human breast cancer. The Journal of steroid biochemistry and molecular biology. 2012;131:101–6. doi: 10.1016/j.jsbmb.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. Rna. 2008;14:1539–49. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. The Journal of biological chemistry. 2008;283:21310–4. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 36.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature cell biology. 2008;10:987–93. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 37.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Molecular cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 39.Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3384–9. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 41.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends in molecular medicine. 2008;14:400–9. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer research. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 43.Miller TW, Balko JM, Ghazoui Z, Dunbier A, Anderson H, Dowsett M, et al. A gene expression signature from human breast cancer cells with acquired hormone independence identifies MYC as a mediator of antiestrogen resistance. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:2024–34. doi: 10.1158/1078-0432.CCR-10-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeng MH, Shupnik MA, Bender TP, Westin EH, Bandyopadhyay D, Kumar R, et al. Estrogen receptor expression and function in long-term estrogen-deprived human breast cancer cells. Endocrinology. 1998;139:4164–74. doi: 10.1210/endo.139.10.6229. [DOI] [PubMed] [Google Scholar]
- 45.Lan FF, Wang H, Chen YC, Chan CY, Ng SS, Li K, et al. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16(INK4A) International journal of cancer Journal international du cancer. 2011;128:319–31. doi: 10.1002/ijc.25336. [DOI] [PubMed] [Google Scholar]
- 46.Koscianska E, Baev V, Skreka K, Oikonomaki K, Rusinov V, Tabler M, et al. Prediction and preliminary validation of oncogene regulation by miRNAs. BMC molecular biology. 2007;8:79. doi: 10.1186/1471-2199-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer research. 2007;67:9762–70. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 48.Cidado J, Park BH. Targeting the PI3K/Akt/mTOR pathway for breast cancer therapy. Journal of mammary gland biology and neoplasia. 2012;17:205–16. doi: 10.1007/s10911-012-9264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oneyama C, Ikeda J, Okuzaki D, Suzuki K, Kanou T, Shintani Y, et al. MicroRNA-mediated downregulation of mTOR/FGFR3 controls tumor growth induced by Src-related oncogenic pathways. Oncogene. 2011;30:3489–501. doi: 10.1038/onc.2011.63. [DOI] [PubMed] [Google Scholar]
- 50.Boulay A, Rudloff J, Ye J, Zumstein-Mecker S, O’Reilly T, Evans DB, et al. Dual inhibition of mTOR and estrogen receptor signaling in vitro induces cell death in models of breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:5319–28. doi: 10.1158/1078-0432.CCR-04-2402. [DOI] [PubMed] [Google Scholar]
- 51.Miller TW, Forbes JT, Shah C, Wyatt SK, Manning HC, Olivares MG, et al. Inhibition of mammalian target of rapamycin is required for optimal antitumor effect of HER2 inhibitors against HER2-overexpressing cancer cells. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:7266–76. doi: 10.1158/1078-0432.CCR-09-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baselga J, Campone M, Piccart M, Burris HA, 3rd, Rugo HS, Sahmoud T, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. The New England journal of medicine. 2012;366:520–9. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.