Abstract
While urine has been an easily accessible and feasible matrix for human biomonitoring, analytical measurements in internal tissues and organs can provide more accurate exposure assessments to understand disease etiology. This is especially important for the endocrine active compound, bisphenol A (BPA), where studies investigating internal doses at sensitive periods of human development are currently lacking. Herein, BPA concentrations, BPA-specific metabolizing enzyme gene expression, and global DNA methylation were characterized across three matched tissues from elective pregnancy terminations of 2nd trimester human fetuses: the placenta, liver, and kidney (N=12 each; N=36 total). Compared to liver (free: 0.54-50.5 ng/g), BPA concentrations were lower in matched placenta (<0.05-25.4 ng/g) and kidney (0.08-11.1 ng/g) specimens. BPA-specific metabolism gene expression of GUSB, UGT2B15, STS, and SULT1A1 differed across each tissue type; however, conjugation and deconjugation expression patterns were similar across the fetus. Average LINE1 and CCGG global methylation were 58.3 and 59.2% in placenta, 79.5 and 66.4% in fetal liver, and 77.9 and 77.0% in fetal kidney, with significant tissue-specific DNA methylation differences in both LINE1 (p-value <0.001) and CCGG content (p-value <0.001). Total BPA concentrations were positively associated with global methylation for the placenta only using the LINE1 assay (p-value: 0.002), suggesting organ-specific biological effects after fetal exposure. Utilizing sensitive human clinical specimens, results are informative for BPA toxicokinetics and toxicodynamics assessment in the developing human fetus.
Keywords: bisphenol A, xenobiotic metabolism, DNA methylation, human fetus
1. Introduction
Characterizing internal dose, defined as the absorbed concentration, is essential for understanding a compound’s toxicokinetic and toxicodynamic properties. Therefore, biomonitoring focus has recently shifted from urinary measurements to quantification of xenobiotics in circulating blood and tissue, especially for human bisphenol A (BPA) studies (Vandenberg, Gerona et al. 2014). BPA is a synthetic estrogen, ubiquitously present in a variety of consumer products including food and beverage containers, thermal paper, electronics, and medical supplies (Vandenberg, Chahoud et al. 2010). Urinary measurements are convenient and commonly analyzed to understand BPA exposure in human populations, (Calafat, Ye et al. 2008; Zhang, Alomirah et al. 2011) but BPA measurements in tissue provide evidence for contact at target sites with implications for organ-specific biological effects. Animal and in vitro studies have established links between BPA exposure across various doses and adverse health outcomes including altered body weight, impaired brain development, altered reproductive function, changes in immune function and metabolism, and increased cancer susceptibility (Maffini, Rubin et al. 2006; Chapin, Adams et al. 2008; Vandenberg, Ehrlich et al. 2013). While epidemiological studies are beginning to reveal BPA’s risk in humans (Rochester 2013), efforts to properly characterize human exposures, especially in vulnerable populations, are ongoing. A growing number of studies are quantifying BPA in biological fluids like blood, saliva, and breast milk, (Padmanabhan, Siefert et al. 2008; Vandenberg, Chahoud et al. 2012; Zhang, Sun et al. 2013) but only a handful of studies have reported BPA concentrations in tissue (Fernandez, Arrebola et al. 2007; Zhang, Cooke et al. 2011; Nahar, Liao et al. 2013).
The rate of BPA absorption, distribution, metabolism, and excretion is well studied in adult animal models. The few studies that have attempted to determine BPA toxicokinetics in humans, however, are limited based on dose frequency, analytical techniques, sample size, and selection of sample population (Volkel, Colnot et al. 2002; Volkel, Bittner et al. 2005; Teeguarden, Calafat et al. 2011). Traditional methods of assessing BPA toxicokinetics are especially difficult in human pregnant adults and the developing fetus, but when tissues are available, biomonitoring with sensitive analytical techniques are a first step to addressing the BPA knowledge gap. In our previous work, gene expression of metabolism enzymes important for BPA detoxification including UDP-glucuronyltransferase (UGT2B15) and sulfotransferase (SULT1A1), and enzymes important for BPA activation including β-glucuronidase (GUSB) and steroid sulfatase (STS), differed significantly in human fetal livers compared to adult livers (Nahar, Liao et al. 2013). Findings suggest the importance of characterizing BPA toxicokinetic properties across the developing human organism.
The exposure-disease relationships identified through epidemiological studies currently lack important mechanistic data that inform direct alterations in structure and function. These subtle exposure-dependent changes can gradually manifest as diseases much later in life, most likely occurring through epigenetic mechanisms (Gluckman, Hanson et al. 2011). DNA methylation at cytosine-guanine dinucleotides (CpG sites) is one commonly studied epigenetic marker that undergoes extensive reprogramming during pre-implantation and gametogenesis in early fetal development followed by tissue- dependent epigenetic differentiation (Faulk and Dolinoy 2011). Several animal studies have already identified methylation as an important intermediate in BPA related adverse health outcomes (Ho, Tang et al. 2006; Bromer, Zhou et al. 2010; Anderson, Nahar et al. 2012). Research addressing human BPA-methylation associations is necessary, especially throughout early development.
Using matched placenta, liver, and kidney specimens from 12 different 2nd trimester human fetuses, this study examines tissue-specific BPA concentrations, metabolism gene expression, and global DNA methylation. Characterizing BPA and biological changes at these specific organs is important because there may be major consequences in the organism’s ability to respond to subsequent endogenous and exogenous agents throughout development. Studies have also found significant associations between exposure and outcomes related to these organs; for example, in animal models, perinatal BPA exposure has been linked to adult-onset liver tumors (Weinhouse, Anderson et al. 2014) and altered expression of calcium transport genes in the kidney (Kim, An et al. 2013). Also, increased BPA levels in human placental samples have been associated with preeclampsia (Leclerc, Dubois et al. 2014), low birth weight and small for gestation age (Troisi, Mikelson et al. 2014). Little is known, however, about BPA’s levels and effects in 2nd trimester human placenta, fetal kidney, and fetal liver, which are known to be metabolically active during pregnancy.
2. Methods and Materials
2.1 Clinical Sample Selection
Pre-existing human fetal tissue samples were obtained from the NIH-funded University of Washington Laboratory for the Study of Human Embryology (LSHE) fetal tissue bank (2R24 HD000836-47). These human clinical samples were procured from voluntary 2nd trimester pregnancy terminations after surgery and proper consent from donors. Other than gestational age, and occasionally sex and race, no identifiable or traceable information regarding the subjects was provided. Thus, samples met the criteria for IRB exemption for human subject research (UM IRB exemption: HUM00024929). After procurement, samples were immediately flash frozen and subsequently stored in polypropylene tubing at −80 °C, prior to shipment to the University of Michigan (UM) on dry ice. In a previous study, 50 fetal liver specimens were utilized for chemical and biological assessment (Nahar, Liao et al. 2013). For this project, we employed only a subset of the fetal liver specimens that had matching tissue. In particular, we identified N=12 fetal kidney and N=12 placental tissues matching BPA-characterized fetal liver samples (subjects A-L), for which sufficient volume was available for BPA quantification and DNA/RNA extraction. The final sample gestational age ranged from 91 to 115 days.
2.2 Tissue BPA Analysis and Quality Control
Tissue BPA concentrations in fetal liver were previously measured at the Wadsworth Center (New York State Department of Health), indicating broad exposure ranging from below limit of quantification (LOQ) up to 96.8 ng/g of total tissue BPA (Nahar, Liao et al. 2013). All flash-frozen matched fetal specimens were homogenized before processing. Like liver specimens, both free BPA and BPA conjugates were quantified in matched kidney and placental samples using high-performance liquid chromatography coupled with API 2000 electrospray triple-quadrupole mass spectrometer (HPLC ESI-MS/MS). Free BPA and conjugated BPA were homogenized from tissue weighing 550-710 mg and extracted following a similar protocol.
Several quality assurance and quality control measurements were implemented for method validation and to check for laboratory contamination, including procedural blanks and spiked BPA standards (Nahar, Liao et al. 2013). Results indicated an average recovery of 108% (±15%) for spiked BPA and 86% (±27%) for spiked 13C12-BPA in all samples. When an external calibration curve was created using 10 μL of standards ranging from 0.05-100 ng/mL, the calibration coefficient was >0.99. Sample concentrations falling below the LOQ (0.05 ng/g) for HPLC ESI-MS/MS were assigned a value of 0.035 ng/g, estimated from dividing LOQ by the square root of 2. The analytical laboratory participated in several proficiency testing programs for BPA analysis and validated methods were used in the analysis (Vandenberg, Gerona et al. 2014).
2.3 RNA and DNA Extraction
Total RNA and DNA were isolated from matched frozen liver, kidney, and placental tissue using the AllPrep DNA/RNA/Protein kit following manufacturer’s instructions (Qiagen, Valencia, CA). Approximately 10-20 mg of tissue was homogenized in 600 μL of Buffer RLT solution (containing 1% β-mercaptoethanol) for 2 min at 20 Hz (2x) in the TissueLyser II (Qiagen). DNA quantity and purity were measured using the Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE), while RNA concentration and purity were assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). High quality mRNA was extracted from most specimens, with RNA integrity numbers (RIN) averaging 9.5 for kidney, 7.1 for placenta, and 7.6 for liver.
2.4 cDNA Synthesis and Real-time Quantitative PCR
To produce complementary DNA, 1 μg of total RNA template was processed using the iScript cDNA synthesis Kit (Bio-Rad, Hercules, CA) as per manufacturer’s instructions. The thermocycler settings for cDNA synthesis included incubation at 25 °C for 5 min, 42 °C for 60 min, and 90 °C for 5 min. Four BPA-related metabolism genes (UGT2B15, GUSB, SULT1A1, and STS) along with three reference genes (GAPDH, UBC, and B2M) were previously assessed in fetal liver tissue using optimized assays (Nahar, Liao et al. 2013). Utilizing the same primer pairs, we also amplified each gene in matched kidney and placenta samples using the same cycle parameters, running each unique reaction out in triplicate.
The CFX Manager Version 1.6 software (Bio-Rad) was used to calculate the average threshold cycle (CT) across the triplicate runs. Results are reported as normalized expression with standard deviation. The normalized expression of a particular target gene for each sample is calculated by normalizing the relative quantity (RQ=Efficiency primers [CT(minimum)-CT(target sample)]) of the gene of interest to that of the reference genes.
Normalized Expression = RQ gene of interest/ [(RQGAPDH * RQUBC * RQB2M)^(1/3)]
2.5 Bisulfite Conversion and Methylation Analysis
For the production of bisulfite converted DNA (BSC), 1 μg of genomic DNA was treated with sodium bisulfite solution using the Qiagen Epitect kit automated on the Qiacube (Qiagen, Valencia, CA) following manufacturer’s protocol. The converted DNA was diluted in 20 μL of EB Buffer, yielding a final concentration of approximately 50 ng/μL. Epitect methylated and unmethylated human BSC standards (Qiagen) were purchased and used as positive controls.
Two independent DNA methylation assays were employed using the Pyrosequencing platform (Pyrosequencing, Westborough, MA) for global DNA methylation analysis: 1) the long interspersed transposable element-1 (LINE1) assay, which interrogates promoter DNA methylation of repetitive elements, specifically long interspersed (LI) retrotransposons, throughout the genome, and 2) the Luminometric Methylation Assay (LUMA), detects methylation at 5’-CCGG-3’ sequences throughout the genome, irrespective of methylation at repetitive elements. These commonly used assays help evaluate DNA instability; while they explore different regions, simultaneous examination of both assays help better capture methylation across the entire genome.
For amplification of LINE1, 2 μL of BSC DNA was added to a 30 μL PCR reaction containing 15 μL of HotStarTaq master mix (Qiagen), 375 nM forward primer, and 187.5 nM of biotin-labeled reverse primer. LINE1 product amplification was set up using the following PCR cycling parameters: 95 °C for 14.5 minutes followed by 45 cycles of 95 °C for 30 sec, 58 °C for 30 sec, and 72 °C for 30 sec, then by 5 min at 72 °C. After amplicon verification via gel electrophoresis, 10 μL of PCR product was added to LINE1 sequencing primers and analyzed using a pre-determined sequence to analyze run. Each amplicon was run in duplicate in the PyroMark™Q96 MD Pyrosequencing System.
The PyroMark software computes % methylation at 4 CpG sites across the assay for each sample and control. Cycling parameters and primer sequences used for the assessment were obtained from a previously published study (Virani, Dolinoy et al. 2012).
The restriction digest based assay for LUMA requires unconverted genomic DNA and MspI, HpaII, and EcoRI restriction enzymes, as described previously (Karimi, Johansson et al. 2006; Bjornsson, Sigurdsson et al. 2008). For each sample digest, two parallel reactions were set up containing 300 ng of genomic DNA, an internal standard enzyme (5 units of EcoRI), and 10x Buffer Tango™ with BSA (Fermentes). We incubated these mixtures at 37 °C for 4 hours after adding 5 units of a methylation insensitive (MspI) enzyme to one reaction, and 5 units of methylation sensitive enzyme (HpaII) to the other. Digested products were assessed in duplicates using the PyroMark MD SNP software using the nucleotide dispensation sequence order: GTGTCACATGTGTG. Percent global methylation results were calculated based on the ratio of digested versus undigested CCGG sites.
2.6 Statistical Analysis
Univariate analyses were conducted across all exposure, gene expression, and global DNA methylation variables across the different tissue types. Mean differences across matched tissue were evaluated using ANOVA with Tukey post-hoc analysis. While parametric tests were conducted, nonparametric results were also conducted and reported given the skewed distribution of BPA in tissue. To test the association between BPA and mRNA expression, we identified the Spearman’s correlation coefficient and p-value between BPA species and individual assays for each tissue type.
Spearman’s correlation analysis was also used to test the association between total BPA concentration and average LUMA methylation for each tissue type. The LINE1 assay evaluates % methylation across 4 adjacent CpG sites per reaction. To account for correlations between sites for each reaction, a linear mixed effects model was used. While total BPA concentration was held as the fixed effects and percent methylation was used as the dependent variable, CpG site number was used as the random effect. For each model tested using kidney, liver, and placental samples, the inter-subject variability was higher than the variability across CpG sites. All analyses were also tested with free BPA. All statistical analyses for this project were conducted using the stats, epicalc, and lme4 packages in R (version 2.14.2).
3. Results
3.1 BPA Concentrations Across Tissues
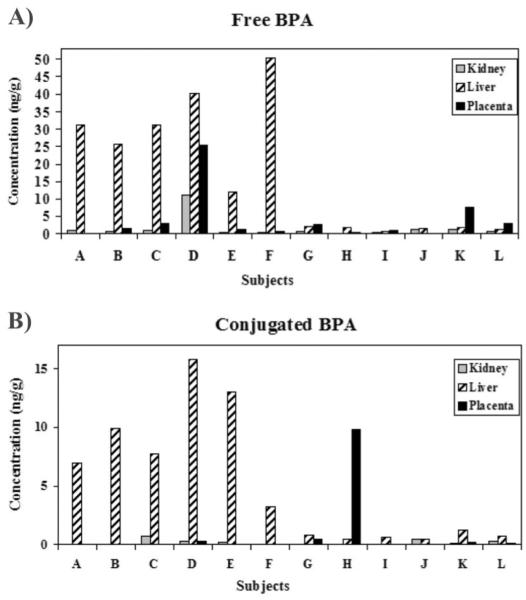
From a pre-existing set of clinical samples previously characterized for fetal liver BPA concentrations (Nahar, Liao et al. 2013), free and conjugated BPA was characterized in matching placenta and fetal kidneys within N=12 individuals (labeled A- L). This subset of fetal livers exhibited free BPA concentrations at 0.54-50.5 ng/g (median: 7.09 ng/g). Free BPA in matched kidneys ranged from 0.08-11.1 ng/g (median: 0.54 ng/g), and in matched placenta from <LOQ-25.4 ng/g (median: 1.36 ng/g; Figure 1A). Conjugated BPA concentrations in the same 12 individuals (Figure 1B) were lower than free BPA, ranging from 0.45-15.9 ng/g (median: 2.26 ng/g) in fetal liver, <LOQ-0.75 ng/g (median: 0.09 ng/g) in fetal kidney, and <LOQ-9.81 ng/g (median: 0.07 ng/g) in placenta. In general, concentrations significantly differed across matched fetal kidney, fetal liver, and placenta with p-values at 0.005 for free BPA and 0.004 for conjugated BPA. The majority of subjects exhibited the highest BPA concentrations in liver compared to matched kidney and placenta.
Figure 1. BPA concentrations differ across tissue and individual fetuses.
Across N=12 individuals (labeled: A-L), three tissue types were analyzed for BPA concentrations: placenta, fetal liver, and fetal kidney. Two separate graphs for all subjects and tissues display (A) free BPA concentrations and (B) conjugated BPA concentrations. (A) Across N=12 subjects, free BPA was detectable across all kidney samples, ranging from 0.08 to 11.1 ng/g (median: 0.54 ng/g). Free BPA was also detectable across all liver specimens, with concentrations ranging from 0.54 to 50.5 ng/g (median 7.09 ng/g). Placental free BPA, however, was detectable in 10 out of 12 samples with concentrations ranging from <LOQ to 25.4 (median: 1.36 ng/g). In general, free tissue BPA concentrations across the 12 individuals significantly differed across the three tissue types (p-value: 0.005). (B) Conjugated BPA concentrations were detectable in 7 out of 12 kidney specimens, with a median of 0.09 ng/g and maximum of 0.75 ng/g. Concentrations were detectable across all fetal liver samples, however, and ranged from 0.45 to 15.9 ng/g (median: 2.26 ng/g). In placental specimens, conjugated BPA was detected in only 5 out of 12 samples, ranging from <LOQ-9.81 ng/g (median: 0.07 ng/g). Across the 12 subjects, conjugated BPA concentrations also significantly differed from one tissue type to another (p-value: 0.004).
3.2 BPA-specific Metabolism Gene Expression Across Tissue
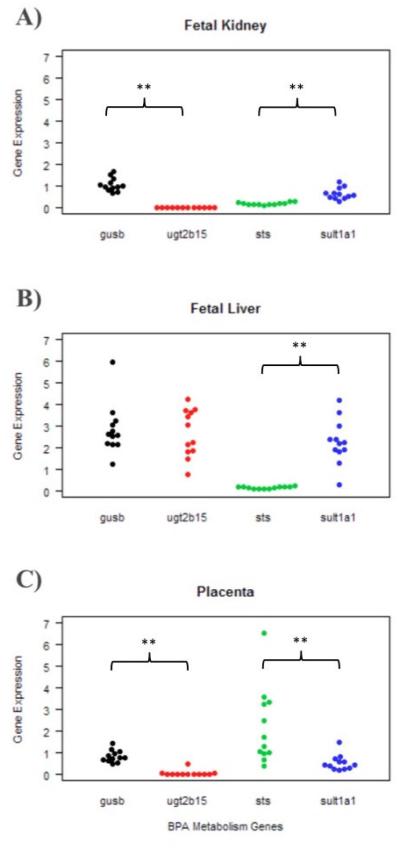
The placenta, liver, and kidney are organs that have some metabolic capacity in the 2nd trimester developing fetus. Each tissue was analyzed for mRNA expression of xenobiotic metabolizing enzymes important in the conjugation (UGT2B15, SULT1A1) of BPA or deconjugation (GUSB, STS) of its metabolites. Figure 2 illustrates the normalized expression patterns of these four genes separated by tissue type. In general, the three organs have distinct metabolic profiles with varying mRNA levels. For the glucuronidation metabolic pathway, although the two genes did not significantly differ from one another in liver (p-value: 0.727), kidney and placenta GUSB expression was significantly higher than UGT2B15 expression (p-values <0.001). For the sulfation metabolic pathway, SULT1A1 expression was significantly higher than STS expression in kidney and liver specimens (p-values <0.001). The placenta mRNA profile was different with STS expression significantly higher than SULT1A1 (p-value <0.001).
Figure 2. BPA-related metabolism gene expression profiles are generally similar across tissue.
The dot plot displays the normalized gene expression values for all 12 subjects for four genes: GUSB, UGT2B15, STS, and SULT1A1. Each panel shows the expression profiles by tissue type: (A) fetal kidney (B) fetal liver and (C) placenta. GUSB expression was significantly higher than UGT2B15 expression within fetal kidney and placenta, while SULT1A1 expression was significantly higher than STS expression in fetal kidney and fetal liver samples (p-values <0.01 as represented by **). When an ANOVA test was conducted across the three tissues for GUSB, UGT2B15, STS, and SULT1A1 separately, there were significant tissue-dependent differences in expression (p-values <0.001).
3.3 BPA Concentration and Metabolism Gene Expression Associations Across Tissues
The SULT1A1 and UGT2B15 enzymes contribute to higher concentrations of BPA-glucuronide and BPA-sulfate conjugates, while the STS and GUSB deconjugation enzymes contribute to higher concentrations of the parent free compound. Since a composite conjugated BPA concentration was measured instead of BPA-sulfate and BPA-glucuronide separately, we only assessed the correlation between free BPA and the four metabolism genes for each tissue. Neither Pearson nor Spearman correlations indicate significant associations between free BPA and metabolism gene expression for each tissue type (p-values >0.200).
3.4 Global Methylation Across Tissues
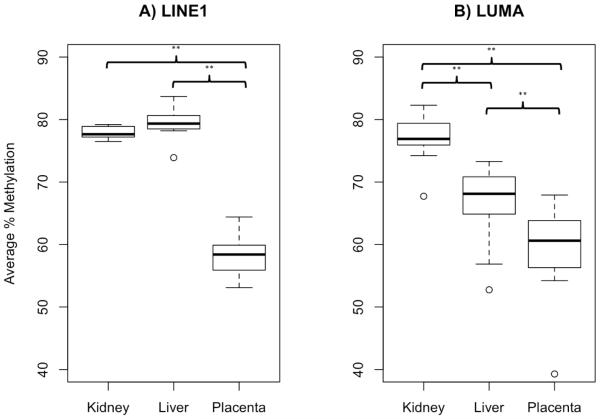
Methylation at LINE1 and CCGG was measured across three matched tissues to assess global DNA methylation. Average LINE1 methylation across N=12 subjects was 77.9% in kidney, 79.5% in liver, and 58.3% in placenta, while the average LUMA methylation was 77.0% in kidney, 66.4% in liver, and 59.2% in placenta. Although LUMA methylation was highly variable compared to LINE1, average methylation values were comparable especially across the kidney and placenta samples. Both LINE1 and LUMA assays exhibited significant hypomethylation in placenta compared to matched liver and kidneys (p-values <0.001, Figure 3).
Figure 3. Global methylation levels differ across tissue.
Box plots represent the 1st quartile, median, and 3rd quartile global % methylation values for placenta, fetal liver, and fetal kidney using the assay for (A) LINE1 on the left panel and (B) LUMA on the right panel. Values greater than 1.5 times the interquartile range were demarcated as outliers. Average LINE1 methylation was 77.9% in kidney, 79.5% in liver, and 58.3% in placenta, while average LUMA methylation was 77% in kidney, 66.4% in liver, and 59.2% in placenta. Percent global methylation measured by LINE1 and LUMA were significantly different across tissues with significant hypomethylation in placenta compared to either fetal liver or fetal kidney (p-values <0.001 as represented by **).
3.5 BPA Concentration and Global Methylation Associations Across Tissues
While free BPA is the only species that can actively bind to hormone receptors, it is uncertain whether both free and conjugated forms influence epigenetic processes. Therefore, we reported the relationship between global methylation and total BPA concentrations instead of the separate BPA species. As listed in Table 1, the Spearman’s correlation between average % methylation using LUMA and total BPA was not significant for kidney, liver, and placenta (p-values: 0.921, 0.276, and 0.534, respectively). Similarly, we did not identify any significant associations between average LUMA methylation and free BPA (p-values for kidney: 0.852, liver: 0.266, placenta: 0.336).
Table 1.
Associations Between BPA (Free and Total) Concentration and Global Methylation
| BPA | LUMA | LINE1† | |||
|---|---|---|---|---|---|
|
|
|||||
| Spearman’s correlation |
p-value | β1 Estimate | p-value | ||
| Free | Kidney | 0.063 | 0.852 | 0.073 | 0.388 |
| Liver | 0.350 | 0.266 | 0.020 | 0.544 | |
| Placenta | 0.305 | 0.336 | 0.269 | <0.001* | |
|
| |||||
| Total | Kidney | −0.035 | 0.921 | 0.066 | 0.424 |
| Liver | 0.347 | 0.276 | 0.021 | 0.425 | |
| Placenta | −0.199 | 0.534 | 0.225 | 0.002* | |
Mixed effects model controlling for random effect by CpG site
p-value <0.05 show significant association
To consider site-specific differences across 4 adjacent LINE1 CpG sites, a mixed effects model was used to test the relationship between total BPA and LINE1 methylation at each site, separately for each tissue. The β1 estimates in Table 1 for the BPA- methylation relationship were not significant for kidney and liver, but significant for placenta. Accounting for CpG site-specific variation, with every 1 ng/g increase in total BPA, there was a 0.23% increase in LINE1 methylation in placental samples (p-value <0.002). The correlation specifically between free BPA and LINE1 methylation appears stronger, with a β1 value of 0.269 and p-value of <0.001.
4. Discussion
The internal dose for nutrients, pharmaceuticals, and toxicants may differ across tissues within an individual, depending on the fat content, capacity for uptake and elimination, and metabolic potential of an organ. The selection of biological specimen for exposure assessment depends on tissue availability, cost and ease of extraction, optimization of environmental analysis, and expected adverse effects. Research addressing internal dose of BPA across various human tissues is currently limited. BPA concentrations have been reported across several postpartum organs in adults, detecting 1-10 ng/g of free BPA across liver, brain, and adipose tissues in 11 human subjects (Geens, Neels et al. 2012). In our study, we quantified both free and conjugated BPA concentrations across three organs from individuals representing a sensitive population. In general, BPA concentrations were highest in the 2nd trimester fetal liver compared to placenta and fetal kidney. Given concentration differences across matched tissues, accessible tissues like placenta that are used as a proxy may be inappropriate for understanding internal dose at other target sites.
Our tissue BPA results add to a growing body of literature suggesting detectable concentrations across the maternal-fetal compartments throughout human development (Schonfelder, Wittfoht et al. 2002; Jimenez-Diaz, Zafra-Gomez et al. 2010; Zhang, Cooke et al. 2011). Across various biomonitoring studies, urinary and serum BPA levels have been reported in the ng/mL or parts per billion range (Vandenberg, Chahoud et al. 2012), similar to the fetal tissue levels observed in this study. The exact range of tissue concentration and detection frequency from one population to another may differ, but is likely explained by the various sensitivities and analytical techniques employed. The HPLC-MS/MS method utilized is an important strength of this project, given its high sensitivity (LOQ= 0.05 ng/g) and use as the “gold standard” for BPA analysis (Vandenberg, Chahoud et al. 2010; Vandenberg, Gerona et al. 2014). Even with stringent quality controls throughout tissue processing and analysis in the laboratory, BPA contamination from tissue acquisition and storage is a major concern for all human BPA biomonitoring studies (Vandenberg, Gerona et al. 2014). The findings from this study suggest that BPA contamination during tissue acquisition is negligible in this sample set, supporting a recent review reporting that contamination is well controlled in most labs and should not be used to discount evidence of true exposure (Vom Saal and Welshons 2014). Assuming similar instrument use4 and consistent background contamination at the collection site, any BPA contamination occurring throughout surgical processing would be expected to have distributed similarly to the various matched tissues within a single donor. In this study, several subjects with measurable levels of free BPA in liver showed undetectable concentrations in matching placenta and low concentrations in kidney, suggesting that free BPA was attributed to in utero instead of ex vivo contamination. Furthermore, tissue storage in polypropylene tubing at −80 °C with minimal freeze-thaw events should negate contamination during storage.
Identifying toxicokinetic characteristics, such as prevailing enzymatic detoxification reactions, may explain tissue differences in BPA concentrations observed within subjects. Therefore, we examined mRNA expression of genes specific for BPA metabolism across placenta, fetal liver, and fetal kidney. In this study, we noted distinct tissue-specific expression profiles, with the highest SULT1A1, GUSB, and UGT2B15 mRNA levels in fetal liver compared to other tissues. As previously reported, GUSB expression was higher than UGT2B15 expression and SULT1A1 expression was higher than STS expression across fetal organs (Nahar, Liao et al. 2013; O'Shaughnessy, Monteiro et al. 2013). While sulfation reactions are generally higher in the developing fetus for xenobiotic defense, high STS activity is necessary in the placenta for the regulation of hormones, including estrone and DHEA (dehydroepiandrosterone) (Stanley, Hume et al. 2005). These mRNA expression patterns may indicate the production of higher free BPA and lower conjugated BPA species across fetal compartments, but functional assays are better at directly assessing metabolism and must be considered for future studies. Also, future biomonitoring studies will require separate measurements of BPA-glucuronide and BPA-sulfate, as new evidence reveals that BPA-sulfate may be the predominant conjugate in the human fetus (Gerona, Woodruff et al. 2013).
Simply correlating BPA concentrations with mRNA expression did not reveal significant associations for any particular organ. The null results suggest that other factors may influence BPA toxicokinetics such as 3'-phosphoadenosine-5'-phosphosulfate and UDP-glucuronic acid cofactor concentrations, protein modification of enzymes, and xenobiotic transporters for absorption and elimination across tissue. Variability in the transporters specific for free BPA or its conjugates, although currently unknown, may help explain the different concentrations observed in our matched tissues. For future studies, proper assessment of BPA toxicokinetics will require an all-encompassing investigation that includes enzymatic activity and transporter function in addition to metabolism gene expression and tissue biomonitoring.
Despite bioaccumulation, individual organs may exhibit differential sensitivity to BPA, eliciting distinct biological responses. Epigenetic regulatory markers such as DNA methylation may be a suitable biomarker for assessing short-term biological effects from exposure, especially if longitudinal data is unavailable for characterizing phenotypic changes. As previously observed, baseline global methylation levels were higher in embryonic compared to extra embryonic tissues like the placenta in this clinical sample set (Ehrlich, Gama-Sosa et al. 1982; Fuke, Shimabukuro et al. 2004; Novakovic and Saffery 2010; Price, Cotton et al. 2012; Sant, Dolinoy et al. 2013). Using the less variable LINE1 assay, we identified a small but significant positive association between BPA concentration and methylation only in the placenta. Given its role in nutrient and waste exchange across maternal-fetal compartments, minute changes in the placental epigenome may have large implications for fetal development. In a recent study, prenatal exposure to xenoestrogen mixtures has been associated with decreased methylation at repetitive elements in term placenta from male, but not female, offspring (Vilahur, Bustamante et al. 2014). In this study, however, BPA is associated with LINE1 hypermethylation in 2nd trimester placental tissue, suggesting potential differences in DNA methylation patterning by tissue sample age. Global methylation assays are crude, efficient methods to assess exposure-dependent changes, but BPA may influence methylation in a locus or gene-specific manner. Furthermore, epigenome-wide methylation patterns are not only tissue-specific but also cell-specific (Michels, Binder et al. 2013). Thus, once cell-specific methylation techniques are optimized and cost- effective, BPA’s impact on cell-specific methylation changes should be assessed.
5. Conclusions
This is the first study to report environmentally relevant BPA concentrations and altered capacity for metabolism across several matched tissue types utilizing sensitive human specimens. Characterizing tissue BPA, expression of BPA-specific metabolism genes, and subsequent exposure-dependent regulatory changes across tissue is especially important in vulnerable populations and will be informative for BPA toxicokinetic and toxicodynamic assessment.
Highlights.
BPA is found in matched human fetal liver, kidney, and placenta
BPA-specific metabolic profiles are similar across tissue
BPA-associated alterations to global methylation occur in placenta
ACKNOWLEDGEMENTS
The authors would like to thank the University of Washington Laboratory for the Study of Human Embryology (2R24 HD000836-47) for human tissue samples. This research was supported by NIH grant ES017524 and the University of Michigan National Institutes of Environmental Health Sciences (NIEHS) Core Center P30 ES017885. Also, support for MSN was provided by Institutional Training Grant T32 ES007062. The funding sources had no involvement in the data collection, interpretation of data, and the writing of the report.
Abbreviations
- BPA
Bisphenol A
- BSC
Bisulfite Converted
- CpG
Cytosine guanine dinucleotides
- CT
Threshold cycle
- GUSB
β-glucuronidase
- HPLC ESI-MS/MS
High-performance liquid chromatography coupled with API 2000 electrospray triple-quadrupole mass spectrometer
- LINE1
Long interspersed transposable element-1
- LSHE
Laboratory for the Study of Human Embryology
- LOQ
Limit of Quantification
- LUMA
Luminometric Methylation Assay
- RIN
RNA integrity numbers
- STS
Steroid sulfatase
- SULT1A1
Sulfotransferase isoform 1A1
- UGT2B15
UDP-glucuronosyltransferase isoform 2B15
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson OS, Nahar MS, et al. Epigenetic responses following maternal dietary exposure to physiologically relevant levels of bisphenol A. Environ Mol Mutagen. 2012;53(5):334–342. doi: 10.1002/em.21692. doi: 310.1002/em.21692. Epub 22012 Mar 21629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson HT, Sigurdsson MI, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299(24):2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromer JG, Zhou Y, et al. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J. 2010;24(7):2273–2280. doi: 10.1096/fj.09-140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafat AM, Ye X, et al. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003-2004. Environ Health Perspect. 2008;116(1):39–44. doi: 10.1289/ehp.10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin RE, Adams J, et al. NTP-CERHR expert panel report on the reproductive and developmental toxicity of bisphenol A. Birth Defects Res B Dev Reprod Toxicol. 2008;83(3):157–395. doi: 10.1002/bdrb.20147. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, et al. Amount and distribution of 5- methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Dolinoy DC. Timing is everything: the when and how of environmentally induced changes in the epigenome of animals. Epigenetics. 2011;6(7):791–797. doi: 10.4161/epi.6.7.16209. Epub 2011 Jul 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez MF, Arrebola JP, et al. Bisphenol-A and chlorinated derivatives in adipose tissue of women. Reprod Toxicol. 2007;24(2):259–264. doi: 10.1016/j.reprotox.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Fuke C, Shimabukuro M, et al. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann Hum Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. Pt 3. [DOI] [PubMed] [Google Scholar]
- Geens T, Neels H, et al. Distribution of bisphenol-A, triclosan and n- nonylphenol in human adipose tissue, liver and brain. Chemosphere. 2012;87(7):796–802. doi: 10.1016/j.chemosphere.2012.01.002. doi: 710.1016/j.chemosphere.2012.1001.1002. Epub 2012 Jan 1024. [DOI] [PubMed] [Google Scholar]
- Gerona RR, Woodruff TJ, et al. Bisphenol-A (BPA), BPA Glucuronide, and BPA Sulfate in Midgestation Umbilical Cord Serum in a Northern and Central California Population. Environ Sci Technol. 2013;47(21):12477–12485. doi: 10.1021/es402764d. doi: 12410.11021/es402764d. Epub 402013 Oct 402767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, et al. The role of developmental plasticity and epigenetics in human health. Birth Defects Res C Embryo Today. 2011;93(1):12–18. doi: 10.1002/bdrc.20198. doi: 10.1002/bdrc.20198. [DOI] [PubMed] [Google Scholar]
- Ho SM, Tang WY, et al. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer research. 2006;66(11):5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Diaz I, Zafra-Gomez A, et al. Determination of Bisphenol A and its chlorinated derivatives in placental tissue samples by liquid chromatographytandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878(32):3363–3369. doi: 10.1016/j.jchromb.2010.10.021. [DOI] [PubMed] [Google Scholar]
- Karimi M, Johansson S, et al. LUMA (LUminometric Methylation Assay)--a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res. 2006;312(11):1989–1995. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Kim S, An BS, et al. Effects of octylphenol and bisphenol A on the expression of calcium transport genes in the mouse duodenum and kidney during pregnancy. Toxicology. 2013;303:99–106. doi: 10.1016/j.tox.2012.10.023. (doi): 10.1016/j.tox.2012.1010.1023. Epub 2012 Nov 1018. [DOI] [PubMed] [Google Scholar]
- Leclerc F, Dubois MF, et al. Maternal, placental and fetal exposure to bisphenol A in women with and without preeclampsia. Hypertens Pregnancy. 2014;33(3):341–348. doi: 10.3109/10641955.2014.892607. doi: 310.3109/10641955.10642014.10892607. Epub 10642014 Apr 10641911. [DOI] [PubMed] [Google Scholar]
- Maffini MV, Rubin BS, et al. Endocrine disruptors and reproductive health: the case of bisphenol-A. Mol Cell Endocrinol. 2006:254–255. doi: 10.1016/j.mce.2006.04.033. 179-186. [DOI] [PubMed] [Google Scholar]
- Michels KB, Binder AM, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods. 2013;10(10):949–955. doi: 10.1038/nmeth.2632. doi: 910.1038/nmeth.2632. [DOI] [PubMed] [Google Scholar]
- Nahar MS, Liao C, et al. Fetal liver bisphenol A concentrations and biotransformation gene expression reveal variable exposure and altered capacity for metabolism in humans. J Biochem Mol Toxicol. 2013;27(2):116–123. doi: 10.1002/jbt.21459. doi: 110.1002/jbt.21459. Epub 22012 Dec 21453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B, Saffery R. DNA methylation profiling highlights the unique nature of the human placental epigenome. Epigenomics. 2010;2(5):627–638. doi: 10.2217/epi.10.45. doi: 610.2217/epi.2210.2245. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Monteiro A, et al. Steroidogenic enzyme expression in the human fetal liver and potential role in the endocrinology of pregnancy. Mol Hum Reprod. 2013;19(3):177–187. doi: 10.1093/molehr/gas059. [DOI] [PubMed] [Google Scholar]
- Padmanabhan V, Siefert K, et al. Maternal bisphenol-A levels at delivery: a looming problem? J Perinatol. 2008;28(4):258–263. doi: 10.1038/sj.jp.7211913. doi: 210.1038/sj.jp.7211913. Epub 7212008 Feb 7211914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price EM, Cotton AM, et al. Different measures of "genome-wide" DNA methylation exhibit unique properties in placental and somatic tissues. Epigenetics. 2012;7(6):652–663. doi: 10.4161/epi.20221. doi: 610.4161/epi.20221. Epub 22012 Jun 20221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochester JR. Bisphenol A and human health: a review of the literature. Reprod Toxicol. 2013;42:132–55. doi: 10.1016/j.reprotox.2013.08.008. (doi): 10.1016/j.reprotox.2013.1008.1008. Epub 2013 Aug 1030. [DOI] [PubMed] [Google Scholar]
- Sant KE, Dolinoy DC, et al. Inhibition of proteolysis in histiotrophic nutrition pathways alters DNA methylation and one-carbon metabolism in the organogenesis-stage rat conceptus. J Nutr Biochem. 2013;24(8):1479–1487. doi: 10.1016/j.jnutbio.2012.12.007. doi: 1410.1016/j.jnutbio.2012.1412.1007. Epub 2013 Feb 1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonfelder G, Wittfoht W, et al. Parent bisphenol A accumulation in the human maternal-fetal-placental unit. Environ Health Perspect. 2002;110(11):A703–707. doi: 10.1289/ehp.110-1241091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley EL, Hume R, et al. Expression profiling of human fetal cytosolic sulfotransferases involved in steroid and thyroid hormone metabolism and in detoxification. Mol Cell Endocrinol. 2005;240(1-2):32–42. doi: 10.1016/j.mce.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Teeguarden JG, Calafat AM, et al. Twenty-four hour human urine and serum profiles of bisphenol a during high-dietary exposure. Toxicol Sci. 2011;123(1):48–57. doi: 10.1093/toxsci/kfr160. doi: 10.1093/toxsci/kfr1160. Epub 2011 Jun 1024. [DOI] [PubMed] [Google Scholar]
- Troisi J, Mikelson C, et al. Placental concentrations of bisphenol A and birth weight from births in the Southeastern U.S. Placenta. 2014;35(11):947–952. doi: 10.1016/j.placenta.2014.08.091. doi: 910.1016/j.placenta.2014.1008.1091. Epub 2014 Sep 1016. [DOI] [PubMed] [Google Scholar]
- Vandenberg LN, Chahoud I, et al. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ Health Perspect. 2010;118(8):1055–1070. doi: 10.1289/ehp.0901716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg LN, Chahoud I, et al. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Cien Saude Colet. 2012;17(2):407–434. doi: 10.1590/s1413-81232012000200015. [DOI] [PubMed] [Google Scholar]
- Vandenberg LN, Ehrlich S, et al. Low dose effects of bisphenol A: An integrated review of in vitro, laboratory animal, and epidemiology studies. Endocrine Disruptors. 2013;1(1):0–1. [Google Scholar]
- Vandenberg LN, Gerona RR, et al. A round robin approach to the analysis of bisphenol a (BPA) in human blood samples. Environ Health. 2014;13(1):25. doi: 10.1186/1476-069X-13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilahur N, Bustamante M, et al. Prenatal exposure to mixtures of xenoestrogens and repetitive element DNA methylation changes in human placenta. Environ Int. 2014;71:81–7. doi: 10.1016/j.envint.2014.06.006. (doi): 10.1016/j.envint.2014.1006.1006. Epub 2014 Jun 1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virani S, Dolinoy DC, et al. Delivery type not associated with global methylation at birth. Clin Epigenetics. 2012;4(1):8. doi: 10.1186/1868-7083-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkel W, Bittner N, et al. Quantitation of bisphenol A and bisphenol A glucuronide in biological samples by high performance liquid chromatography- tandem mass spectrometry. Drug Metab Dispos. 2005;33(11):1748–1757. doi: 10.1124/dmd.105.005454. [DOI] [PubMed] [Google Scholar]
- Volkel W, Colnot T, et al. Metabolism and kinetics of bisphenol a in humans at low doses following oral administration. Chem Res Toxicol. 2002;15(10):1281–1287. doi: 10.1021/tx025548t. [DOI] [PubMed] [Google Scholar]
- Vom Saal FS, Welshons WV. Evidence that bisphenol A (BPA) can be accurately measured without contamination in human serum and urine and that BPA causes numerous hazards from multiple routes of exposure. Mol Cell Endocrinol. 2014;7(14):00308–00306. doi: 10.1016/j.mce.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhouse C, Anderson OS, et al. Dose-dependent incidence of hepatic tumors in adult mice following perinatal exposure to bisphenol A. Environ Health Perspect. 2014;122(5):485–491. doi: 10.1289/ehp.1307449. doi: 410.1289/ehp.1307449. Epub 1302014 Jan 1307417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cooke GM, et al. GC-MS analysis of bisphenol A in human placental and fetal liver samples. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879(2):209–214. doi: 10.1016/j.jchromb.2010.11.031. [DOI] [PubMed] [Google Scholar]
- Zhang T, Sun H, et al. Blood and urinary bisphenol A concentrations in children, adults, and pregnant women from china: partitioning between blood and urine and maternal and fetal cord blood. Environ Sci Technol. 2013;47(9):4686–4694. doi: 10.1021/es303808b. doi: 4610.1021/es303808b. Epub 302013 Apr 303804. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Alomirah H, et al. Urinary bisphenol A concentrations and their implications for human exposure in several Asian countries. Environ Sci Technol. 2011;45(16):7044–7050. doi: 10.1021/es200976k. [DOI] [PubMed] [Google Scholar]